Abstract
Background
The clock-controlled RNA-binding protein AtGRP7 influences circadian oscillations of its own transcript at the post-transcriptional level. To identify additional targets that are regulated by AtGRP7, transcript profiles of transgenic plants constitutively overexpressing AtGRP7 (AtGRP7-ox) and wild type plants were compared.
Results
Approximately 1.4% of the transcripts represented on the Affymetrix ATH1 microarray showed changes in steady-state abundance upon AtGRP7 overexpression. One third of the differentially expressed genes are controlled by the circadian clock, and they show a distinct bias of their phase: The up-regulated genes preferentially peak around dawn, roughly opposite to the AtGRP7 peak abundance whereas the down-regulated genes preferentially peak at the end of the day. Further, transcripts responsive to abiotic and biotic stimuli were enriched among AtGRP7 targets. Transcripts encoding the pathogenesis-related PR1 and PR2 proteins were elevated in AtGRP7-ox plants but not in plants overexpressing AtGRP7 with a point mutation in the RNA-binding domain, indicating that the regulation involves RNA binding activity of AtGRP7. Gene set enrichment analysis uncovered components involved in ribosome function and RNA metabolism among groups of genes upregulated in AtGRP7-ox plants, consistent with its role in post-transcriptional regulation.
Conclusion
Apart from regulating a suite of circadian transcripts in a time-of-day dependent manner AtGRP7, both directly and indirectly, affects other transcripts including transcripts responsive to abiotic and biotic stimuli. This suggests a regulatory role of AtGRP7 in the output of the endogenous clock and a complex network of transcripts responsive to external stimuli downstream of the AtGRP7 autoregulatory circuit.
Background
AtGRP7 (Arabidopsis thaliana glycine-rich RNA-binding protein 7) is an RNA-binding protein with an N-terminal RNA recognition motif and a C-terminal glycine-rich domain. It is under control of the circadian clock and has been implicated in stress responses and floral transition [1-6].
The circadian clock is an endogenous timekeeping device that provides the organism with an approximately 24-hour time. The core clockwork comprises transcriptional feedback loops with positively and negatively acting proteins that directly or indirectly regulate their own expression and thus generate their own 24-h rhythm [7-9]. The core oscillator is composed of interconnected morning and evening loops. In the morning loop, the two MYB-type transcription factors CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL) activate two pseudo response regulators, PRR7 and PRR9 that feed back to repress CCA1 and LHY [10-12]. In the evening loop, TOC1 (TIMING of CAB EXPRESSION 1) represses GI (GIGANTEA) which in turn contributes to TOC1 activation [13]. In shoots, these two loops are interlocked through reciprocal regulation of CCA1/LHY and TOC1 [14-18].
The core oscillator imparts rhythmicity on downstream transcripts to generate output rhythms. Among those transcripts is AtGRP7 which oscillates with a peak in the evening and is directly controlled by the CCA1 and LHY clock proteins [2,14,19,20]. Notably, AtGRP7 itself influences the oscillations of its own transcript at the post-transcriptional level [20,21]. AtGRP7 binds to its own pre-mRNA and promotes the formation of an alternatively spliced transcript that retains part of the intron including a premature termination codon [22,23]. This unproductively spliced transcript form is short-lived and is degraded via the Nonsense-mediated decay (NMD) pathway [24]. Apart from this AtGRP7 also influences the AtGRP8 transcript encoding a related RNA-binding protein. This negative feedback loop is thought to represent a slave oscillator as part of clock output signalling [25,26].
RNA-binding proteins are involved in almost all aspects of RNA metabolism. Upon transcription and throughout their life, mRNAs are bound by a suite of proteins that define pre-mRNA processing, lifetime, export from the nucleus and translation [27,28]. In higher plants, RNA-binding proteins perform a crucial role in key developmental processes such as floral transition and flower development or stress tolerance [29-31]. The targets of these RNA-binding proteins and their mode of action are known in only a few cases, however [32,33].
In order to obtain insights into cellular processes AtGRP7 may be involved in we set out to globally identify potential AtGRP7 target transcripts. About 300 transcripts were found to be differentially expressed in plants constitutively overexpressing AtGRP7 (AtGRP7-ox). About one third of these are controlled by the circadian clock and they show a certain bias towards specific circadian phases. Furthermore, transcripts associated with responses to stress and to abiotic or biotic stimuli were prevalent. Monitoring for enrichment of gene sets revealed that components associated with various aspects of RNA metabolism predominate among transcripts with higher abundance in AtGRP7-ox plants.
Results
Identification of genes differentially expressed in plants constitutively over-expressing AtGRP7
Transcripts regulated by the RNA-binding protein AtGRP7 are expected to have altered expression levels in transgenic plants constitutively overexpressing AtGRP7, as observed for endogenous AtGRP7 whose abundance is depressed by the elevated AtGRP7 protein level [20]. Therefore, differences in the mRNA complement of wt and AtGRP7-ox plants were analyzed on the Affymetrix ATH1 microarray. To filter out any line- or accession-specific effects, independent transgenic lines in the C24 background, line RS13 [20], and in the Col background, line G [6], were assayed. Transgenic lines and the wild types were grown in parallel in long days (16 h light, 8 h darkness) and harvested at the circadian maximum of AtGRP7 expression (zt12, zeitgeber time 12, 12 hrs after lights on).
The RankProduct algorithm was employed to compare transcript profiles of the AtGRP7-ox lines to the respective wild types [34]. Transcripts corresponding to 153 probe sets were present at an elevated level in AtGRP7-ox plants (Additional file 1) and 161 were present at a reduced level (Additional file 2) with a signal log ratio > 0.6 in all three experiments (p < 0.05). Among the transcripts most strongly reduced in AtGRP7-ox plants was AtGRP8 previously shown to be under negative control by AtGRP7 [20,24], validating the strategy.
Overrepresentation of circadian transcripts among AtGRP7 targets
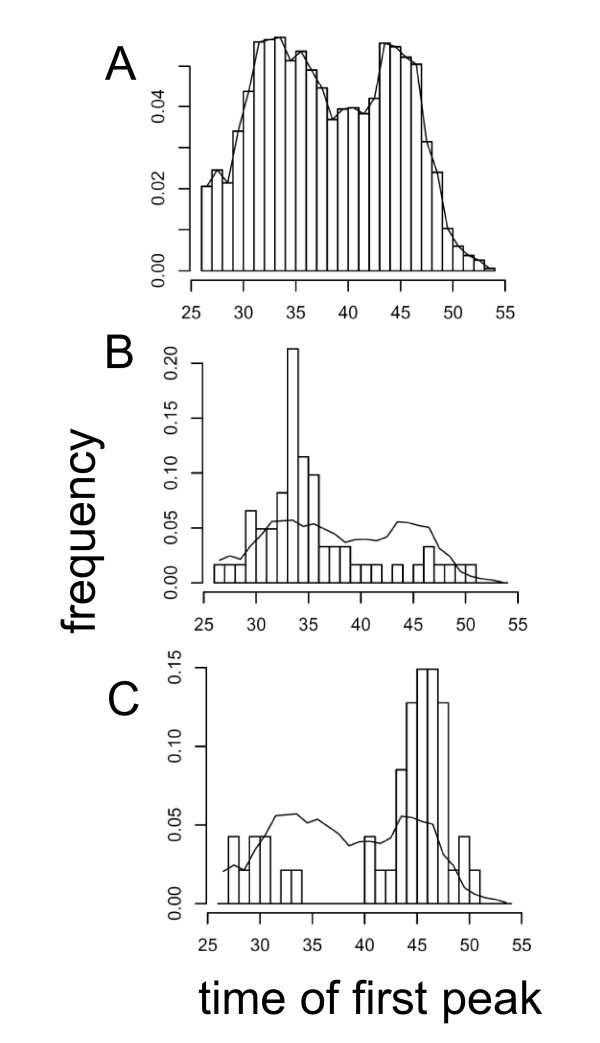
To determine the proportion of rhythmic transcripts among the AtGRP7 targets, we compared the differentially expressed genes to a published dataset scoring 15.4% of the Arabidopsis genes as circadian-regulated [35]. In this experiment, eight-day-old seedlings were entrained in 12-hr-light/12-hr-dark cycles before transfer to constant light and harvested at 4-hr-intervals, starting 26 h after the last dark-light transition. Thus, subjective dawn corresponds to zt24 and zt48, respectively, and subjective dusk corresponds to zt36. AtGRP7, also named CCR2 (COLD AND CIRCADIAN REGULATED 2), peaked around zt36 in this experiment. Among the transcripts we found to be differentially expressed in AtGRP7-ox plants rhythmic transcripts are significantly enriched. 47 of the 153 probe sets with elevated levels in AtGRP7-ox, corresponding to 30.7% (p = 5.27E+7) (Additional file 1) and 61 of the 161 probe sets with reduced levels corresponding to 37.9% (p = 7.91E+13) (Additional file 2) are among those classified as rhythmic in the Edwards dataset. Edwards and coworkers have shown a nearly uniform distribution of the peaks across all time points (Figure 1A) implicating a complex network downstream of the core oscillator in conveying different phases upon clock-controlled output genes. Notably, the genes differentially expressed in AtGRP7-ox are biased toward specific circadian phases. A large fraction of transcripts with reduced level in AtGRP7-ox mainly peaks between zt30 and zt35, in the second half of the subjective day (Figure 1B). This suggests that AtGRP7 has a mostly negative effect on transcripts oscillating with a similar phase. Conversely, transcripts upregulated in AtGRP7-ox mainly peak between zt44 and zt50 in the Edwards dataset (Figure 1C), i.e. towards the end of the subjective night and thus in antiphase to AtGRP7.
Figure 1.
Phase of transcripts differentially expressed in AtGRP7-ox. A) Time of first peak after release from light-dark cycles into continuous light of all transcripts scored rhythmic by COSOPT (pMMC-β < 0.05) according to Edwards et al. (2006). B) Corresponding histogram of genes by Edwards et al. (2006) with reduced levels in AtGRP7-ox plants. C) Corresponding histogram of genes by Edwards et al. with elevated levels in AtGRP7-ox plants.
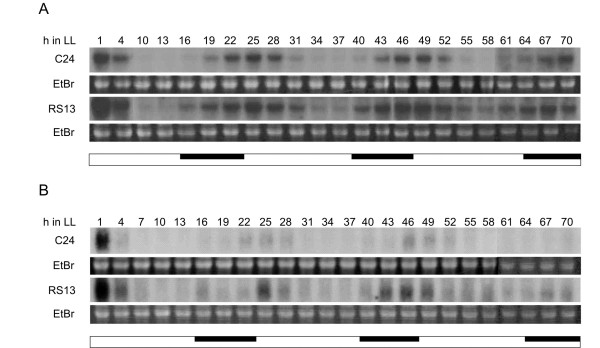
To determine how AtGRP7 affects rhythmic downstream genes, oscillations of selected candidate target transcripts were compared between wt and AtGRP7-ox plants under free-running conditions. Plants were grown in long days for two weeks and subsequently transferred to continuous light and harvested at 3-h intervals for three days. SALT TOLERANCE HOMOLOGUE (STH) encoding a B-box zinc finger protein [36] which shows a mean SLR of 1.5 in all three AtGRP7-ox vs. wt comparisons at zt12 oscillates with a maximum around subjective dawn both in wt and AtGRP7-ox plants (Figure 2A). In the AtGRP7-ox plants, the STH peak is higher and broader while the phase is maintained. HY5-HOMOLOG (HYH) encoding a bZip transcription factor involved in phyB signalling [36] which shows a mean SLR of 1.4 in all three AtGRP7-ox vs. wt comparisons at zt12 (Figure 2B) oscillates at a higher level in AtGRP7-ox plants.
Figure 2.
Influence of AtGRP7 overexpression on rhythmic target transcripts. C24 wt and the AtGRP7-ox plants RS13 were grown in long days for two weeks, transferred to continuous light and harvested at 3-h intervals for three days. The RNA gel blots were hybridized with an STH probe (A) and an HYH probe (B). A representative northern blot of two independent time courses is shown. The ethidium bromide-stained agarose gel is shown to confirm equal loading. The inserted dark bar indicates subjective night.
Characterization of non-circadian transcripts among AtGRP7 targets
The observation that one third of AtGRP7 targets are clock-controlled is in line with a function of the AtGRP7 feedback loop in clock output but the fact that two thirds are not classified as rhythmic [35] points to additional processes AtGRP7 may be involved in. The distribution of genes into ontology categories corresponding to plant GOSLIM revealed that the categories "response to stress" and "response to abiotic or biotic stimulus" were significantly enriched among transcripts with reduced levels in AtGRP7-ox plants (p ≤ 1.1E-2 and p ≤ 1.1E-3, respectively) and were also prevalent among transcripts with elevated levels (Table 1, table 2). Further, significant enrichment among transcripts with reduced abundance in AtGRP7-ox was found for transcripts upregulated by the phytohormones methyl jasmonate (p ≤ 3.2E-8) and abscisic acid (p ≤ 1.8E-6).
Table 1.
GOSLIM categorization in biological processes of genes expressed at reduced levels in AtGRP7-ox plants.
| n genes selection |
expected frequencies |
log enrichments |
p values enriched |
p values depleted |
|
|---|---|---|---|---|---|
| other physiological processes | 22 | 22.4 | -0.025 | 1 | 1 |
| response to stress | 15 | 5.2 | 1.517 | 1.14E-03 | 1 |
| response to abiotic or biotic stimulus | 16 | 7.1 | 1.186 | 1.05E-02 | 1 |
| signal transduction | 3 | 6.2 | -1.045 | 1 | 1 |
| other cellular processes | 2 | 7.4 | -1.884 | 1 | 0.297 |
| other biological processes | 12 | 13.8 | -0.202 | 1 | 1 |
| biological process unknown | 63 | 68.8 | -0.128 | 1 | 1 |
| other metabolic processes | 59 | 53.1 | 0.153 | 1 | 1 |
| developmental processes | 4 | 5.1 | -0.349 | 1 | 1 |
| cell organization and biogenesis | 4 | 4.0 | -0.007 | 1 | 1 |
| transcription | 10 | 12.2 | -0.282 | 1 | 1 |
| protein metabolism | 21 | 23.8 | -0.183 | 1 | 1 |
| DNA or RNA metabolism | 3 | 4.3 | -0.524 | 1 | 1 |
| transport | 16 | 12.4 | 0.369 | 1 | 1 |
| electron transport or energy pathways | 7 | 8.3 | -0.248 | 1 | 1 |
Table 2.
GOSLIM categorization in biological processes of genes expressed at elevated levels in AtGRP7-ox plants.
| n genes selection |
expected frequencies |
log enrichments |
p values enriched |
p values depleted |
|
|---|---|---|---|---|---|
| other physiological processes | 21 | 21.3 | -0.018 | 1 | 1 |
| response to stress | 9 | 4.9 | 0.854 | 0.425 | 1 |
| response to abiotic or biotic stimulus | 12 | 6.7 | 0.845 | 0.252 | 1 |
| signal transduction | 7 | 5.9 | 0.251 | 1 | 1 |
| other cellular processes | 8 | 7.0 | 0.189 | 1 | 1 |
| other biological processes | 13 | 13.1 | -0.013 | 1 | 1 |
| biological process unknown | 59 | 65.4 | -0.149 | 1 | 1 |
| other metabolic processes | 56 | 50.4 | 0.151 | 1 | 1 |
| developmental processes | 4 | 4.8 | -0.276 | 1 | 1 |
| cell organization and biogenesis | 3 | 3.8 | -0.349 | 1 | 1 |
| transcription | 11 | 11.6 | -0.071 | 1 | 1 |
| protein metabolism | 23 | 22.7 | 0.021 | 1 | 1 |
| DNA or RNA metabolism | 5 | 4.1 | 0.287 | 1 | 1 |
| transport | 15 | 11.8 | 0.349 | 1 | 1 |
| electron transport or energy pathways | 8 | 7.9 | 0.018 | 1 | 1 |
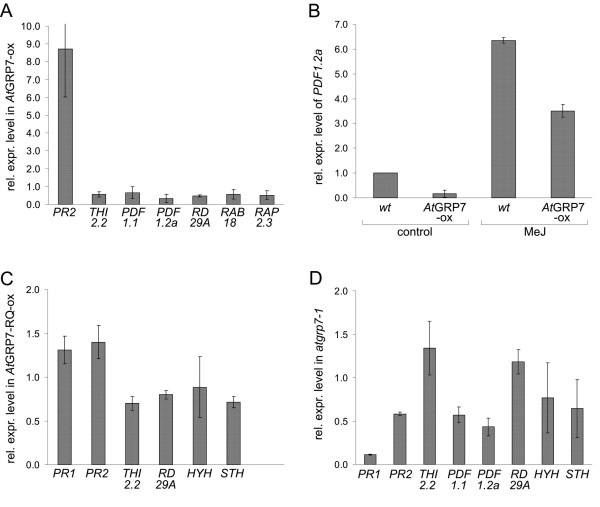
To validate a differential expression of these candidate targets, a suite of them was monitored in several independent AtGRP7-ox plants and the corresponding wt plants using RT-PCR and Realtime PCR on zt12 RNA. The transcripts encoding the pathogenesis-related proteins PR1, PR2 encoding a β-glucanase and PR5 encoding an antimicrobial thaumatin-like protein are present at elevated levels in AtGRP7-ox plants (Figure 3A, Additional file 3). Because two cold- and ABA-regulated transcripts, RD29A (COR78) and RAB18, were confirmed to have reduced levels in AtGRP7-ox plants (Figure 3A), we tested another cold-induced transcript, COR15A, which was also reduced in AtGRP7-ox plants (Additional file 3). Transcripts encoding the plant defensins PDF1.1 and PDF1.2a, RAP2.3/ERF72/AtEBP encoding a member of the ethylene response factor subfamily B2 of AP2-domain transcription factors implicated in PDF1.2a activation [37,38] and the antimicrobial thionin THI2.2 are present at reduced levels in AtGRP7-ox plants (Figure 3A, Additional file 3).
Figure 3.
Analysis of selected AtGRP7 candidate target transcripts in transgenic lines with elevated or reduced AtGRP7 levels. A) AtGRP7-ox and wt plants were grown in LDs and harvested around the circadian maximum. Expression levels were determined by qRT-PCR and normalized to PTB expression. Shown is the expression in AtGRP7-ox plants relative to wt. B) Transcript levels of the JA-responsive PDF1.2a in MeJ-treated plants. Independent AtGRP7-ox lines and the corresponding wt plants were grown in liquid culture in LDs and harvested 24 h after MeJ treatment. Expression levels were determined by qRT-PCR and normalized to PTB. The level in untreated wt plants is set to 1. C) Mutation of a conserved arginine in the AtGRP7 RNA recognition motif interferes with regulation of target transcripts. qRT-PCR was performed on RNA from AtGRP7-RQ-ox and wt plants grown in LDs and harvested at zt12 for the analysis of PR1, PR2, THI2.2 and RD29A, and at zt3 for the analysis of HYH and STH, respectively. Data were normalized to PTB and the levels in AtGRP7-RQ-ox plants are expressed relative to wt. D) qRT-PCR was performed on RNA from the T-DNA line atgrp7-1 and Col-0 wt plants grown in LDs and harvested at zt12 for the analysis of PR1, PR2, THI2.2, PDF1.1, PDF1.2a and RD29A, and at zt3 for the analysis of HYH and STH, respectively. Data were normalized to PTB and the levels in atgrp7-1 are expressed relative to wt.
Because methyl jasmonate (MeJ)-induced transcripts were overrepresented among genes with reduced levels in AtGRP7-ox plants, we investigated a potential role of AtGRP7 in the response of the AtGRP7 target gene PDF1.2a to MeJ treatment. PDF1.2a accumulated 24 hrs after addition of MeJ both in wt and independent AtGRP7-ox plants (Figure 3B). PDF1.2a levels in MeJ-treated AtGRP7-ox plants still remained lower than in MeJ treated wt plants. Thus, overexpression of AtGRP7 does not prevent the response to MeJ but MeJ does not overcome the negative regulation by AtGRP7.
We have shown previously that site-specific mutation of a single arginine to glutamine within the RNA recognition motif impairs both in vitro binding of recombinant AtGRP7 to its pre-mRNA and in vivo function [22]. Therefore we investigated the steady-state abundance of selected putative AtGRP7 targets in transgenic plants constitutively overexpressing the mutant protein (AtGRP7-RQ-ox). Real time PCR showed that levels of PR1, PR2, THI2.2 and RD29A remained similar to wt levels in plants harvested at zt12 (Figure 3C). For HYH and STH with a morning peak, plants were harvested at zt3. Again, HYH and STH levels remained similar to wt levels (Figure 3C). The fact that the candidate targets were affected by high levels of the authentic AtGRP7 protein but not of the mutant protein indicates that the regulation is based on the AtGRP7 RNA-binding activity. Nevertheless, this does not unambiguously imply direct binding of AtGRP7 to these transcripts, as overexpression of regulatory RNA-binding proteins, similar to the overexpression of transcription factors leads to direct and indirect effects on the transcriptome. To begin to understand how AtGRP7 may influence the differentially expressed transcripts we assessed their steady-state abundance in the atgrp7-1 T-DNA insertion line that lacks AtGRP7 [4]. PR1 and PR2 transcript levels were reduced in atgrp7-1, suggesting that their expression closely correlates with the AtGRP7 level (Figure 3D).
Also HYH and STH levels were weakly reduced at the time of their circadian maximum. The levels of THI2.2 and RD29A remained mostly unchanged and levels of PDF1.1 and PDF1.2a were reduced in atgrp7-1, suggesting either that elevated AtGRP7 levels have a slightly negative effect but reduced AtGRP7 levels are not sufficient to cause their upregulation or that they are influenced indirectly.
Analysis of gene set enrichment
Overall, changes in expression of most candidate target genes were moderate (Additional file 1, Additional file 2). Therefore we subjected the expression data to PAGE (parametric analysis of gene set enrichment) [39], an improved tool to analyze overrepresentation of groups of genes that employs predefined gene sets. It relies on the assumption that differential expression manifests itself more clearly at the level of coregulated genes than at the level of individual genes. Thus, PAGE is a complementary approach to uncover genes differentially expressed with a small fold change (below the cut-off level). Its significance comes from the possibility to detect entire gene sets, with narrower definition than GO categories, that are co-ordinately up- or down-regulated to a small degree.
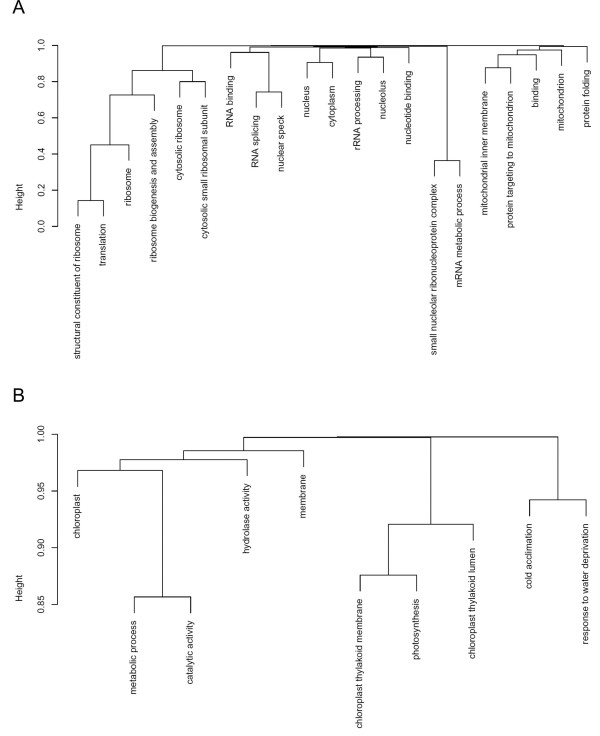
Most prominent among gene sets upregulated in AtGRP7-ox plants were structural constituents of the ribosome and functions associated with ribosome biogenesis and assembly (Figure 4A, Table 3). The next categories were RNA binding and small nucleolar ribonucleoprotein complexes, followed by rRNA processing, nucleolus and RNA splicing including the SR (serine arginine rich) proteins RSZ22 and RSZ32. This may point to an involvement of the RNA-binding protein AtGRP7 in the modulation of RNA processing and translational activity. Two transcripts encoding proteins participating in pre-mRNA splicing, the snRNP core protein D1 and the U5 snRNP helicase (Additional file 1), were confirmed to be expressed at higher levels in independent AtGRP7-ox lines (data not shown). Transcripts with reduced abundance in AtGRP7-ox comprise functions associated with chloroplasts (Figure 4B, Table 4). This may relate to the observation of a slightly reduced chlorophyll content in AtGRP7-ox plants (Streitner, unpublished).
Figure 4.
Gene sets enriched among transcripts differentially expressed in AtGRP7-ox. PAGE [39] was used to calculate the Z score for predefined gene sets from the signal log ratio between wt and AtGRP7-ox plants and to infer statistical significance against standard normal distribution. A) Agglomerative clustering of groups of genes expressed at a higher level in AtGRP7-ox plants. B) Agglomerative clustering of groups of genes expressed at a reduced level in AtGRP7-ox plants.
Table 3.
Gene sets enriched among transcripts expressed at elevated levels in AtGRP7-ox plants.
| GO term | Z | p | p adjusted |
|---|---|---|---|
| structural constituent of ribosome | -16.62418 | 4.66E-62 | 2.19E-59 |
| translation | -15.1101 | 1.39E-51 | 3.26E-49 |
| ribosome | -15.00005 | 7.34E-51 | 1.15E-48 |
| ribosome biogenesis and assembly | -12.17341 | 4.31E-34 | 5.06E-32 |
| cytosolic ribosome | -12.06825 | 1.55E-33 | 1.46E-31 |
| RNA binding | -9.410485 | 4.94E-21 | 3.87E-19 |
| small nucleolar ribonucleoprotein complex | -8.51926 | 1.61E-17 | 1.08E-15 |
| cytosolic small ribosomal subunit | -7.678024 | 1.62E-14 | 8.44E-13 |
| nucleus | -7.240035 | 4.49E-13 | 2.11E-11 |
| mitochondrial inner membrane | -7.047071 | 1.83E-12 | 7.81E-11 |
| mRNA metabolic process | -6.383831 | 1.73E-10 | 6.76E-09 |
| binding | -5.912648 | 3.37E-09 | 1.22E-07 |
| nucleotide binding | -5.874369 | 4.24E-09 | 1.42E-07 |
| rRNA processing | -5.858142 | 4.68E-09 | 1.42E-07 |
| nucleolus | -5.853125 | 4.82E-09 | 1.42E-07 |
| RNA splicing | -5.83756 | 5.30E-09 | 1.46E-07 |
| protein targeting to mitochondrion | -5.507412 | 3.64E-08 | 9.24E-07 |
| cytoplasm | -5.502979 | 3.73E-08 | 9.24E-07 |
| mitochondrion | -5.485297 | 4.13E-08 | 9.70E-07 |
| protein folding | -5.425586 | 5.78E-08 | 1.29E-06 |
| nuclear speckles | -5.229014 | 1.70E-07 | 3.64E-06 |
Table 4.
Gene sets enriched among transcripts expressed at reduced levels in AtGRP7-ox plants.
| GO term | Z | p | p adjusted |
|---|---|---|---|
| chloroplast | -11.5152 | 1.11E-30 | 5.20E-28 |
| chloroplast thylakoid membrane | -9.640974 | 5.37E-22 | 1.26E-19 |
| cold acclimation | -7.550362 | 4.34E-14 | 6.80E-12 |
| chloroplast thylakoid lumen | -6.959285 | 3.42E-12 | 3.44E-10 |
| membrane | -6.949736 | 3.66E-12 | 3.44E-10 |
| hydrolase activity | -6.410412 | 1.45E-10 | 1.14E-08 |
| response to water deprivation | -5.926906 | 3.09E-09 | 2.07E-07 |
| metabolic process | -5.873516 | 4.27E-09 | 2.51E-07 |
| photosynthesis | -5.681972 | 1.33E-08 | 6.26E-07 |
| catalytic activity | -5.316148 | 1.06E-07 | 4.53E-06 |
Discussion
Transcript profiling has identified about 300 transcripts with altered expression in transgenic lines overexpressing the clock regulated RNA-binding protein AtGRP7. Gratifyingly, among the transcripts most strongly reduced in AtGRP7-ox plants is AtGRP8 previously shown to be under negative control by AtGRP7 [20], validating the strategy used to identify candidate targets.
One third of the differentially expressed genes are controlled by the circadian clock, in line with the proposed function of the AtGRP7 feedback loop as a slave oscillator in clock output [40,41]. Binding of the morning-phased LHY and CCA1 clock proteins to the promoters of some morning-specific genes shows that rhythmic transcripts can be directly controlled by the core oscillator proteins [14]. Other oscillating transcripts presumably are regulated via signalling intermediates that receive timing cues from the circadian clock and in turn convey rhythmicity upon downstream transcripts [42]. For example, circadian oscillations of the MYB factor EARLY PHYTOCHROME RESPONSIVE 1 (EPR1) are controlled by CCA1 and LHY [43]. EPR1 negatively autoregulates, presumably at the transcriptional level and moreover activates the morning-specific LHC (LIGHT HARVESTING CHLOROPYLL BINDING PROTEIN) genes. Thus, EPR1 may represent a slave oscillator downstream of the core oscillator that contributes to a phase-specific transcriptional program [43]. AtGRP7 is the first example of a molecular slave oscillator that autoregulates at the posttranscriptional level. Notably, the rhythmic transcripts that are affected by AtGRP7 overexpression show a distinct phase bias: The up-regulated transcripts preferentially peak around dawn, roughly opposite to the AtGRP7 peak abundance whereas the down-regulated transcripts preferentially peak at the end of the day (Figure 1). In accordance with this, the morning-phased STH and HYH transcripts show higher peak levels in an extended time course over three days in LL (Figure 2). Previously, we have found that the AtGRP8 transcript, which cycles in phase with AtGRP7, is strongly downregulated in AtGRP7-ox plants but also retains rhythmicity over three days in LL [22]. To begin to understand the relation between the core oscillator and the AtGRP7 slave oscillator and their respective downstream transcripts, we monitored recently published datasets for the phase distribution of transcripts controlled by TOC1 and LHY with reference to the Edwards dataset [35,44,45]. Transcripts that are elevated in TOC1-ox plants at zt16 in 16 h light-8 h dark cycles [45] peak in the second half of the night, i.e. at time points opposite to TOC1 itself (peak at zt36), and transcripts with reduced abundance have a more uniform phase distribution throughout the light phase with a bias towards the evening, similar to TOC1 (Additional file 4). From the datasets comparing lhy plants expressing elevated LHY levels grown in 8 h light-16 h dark cycles to wt we chose zt0 when LHY peaks [44]. Of 1503 transcripts elevated in lhy, 348 are rhythmic with peaks roughly opposite to LHY itself (Additional file 4). Of 1748 transcripts reduced in lhy, 255 are rhythmic with a broad distribution during the night and around dawn. Thus, both AtGRP7 and TOC1 with preferential expression in the evening as well as LHY with a dawn peak have a bias towards negatively affecting the abundance of similarly phased transcripts and positively affecting the abundance of oppositely phased transcripts. To obtain a detailed picture of the RNA networks controlled by core and slave oscillators, respectively, further transcript profiling of plants mis-expressing the components harvested under identical photoperiods around the clock will be required.
PR1, PR2 and PR5 which are expressed at elevated levels in independent transgenic AtGRP7-ox lines are associated with salicylic acid (SA)-mediated defence pathways [46]. Conversely, PDF1.2a encoding a plant defensin, a target of jasmonic acid and ethylene signalling, and RAP2.3 encoding an ethylene response factor implicated in PDF1.2a activation are expressed at reduced levels in AtGRP7-ox. Thus, high AtGRP7 levels correlate with increased expression of SA-responsive PR transcripts and decreased expression of the JA-responsive transcripts. Antagonisms between the SA and JA pathways were observed during defence responses [47]. Notably, the atgrp7-1 mutant lacking AtGRP7 shows increased susceptibility to Pseudomonas syringae DC3000 [4]. AtGRP7 is ADP-ribosylated by the Pseudomonas syringae type III effector protein HopU1. This modification depends on the conserved Arginine residue that is crucial for RNA binding activity and in vivo function and is suggested to interfere with a defence-related function of AtGRP7 [4,22]. Whether the elevated levels of PR1, PR2 and PR5 in AtGRP7-ox plants may point to a role of AtGRP7 in processing of defence-related transcripts remains to be tested. Alternatively, the hormonal balance could be altered in these plants and cause a general stress response. In line with this, overexpression of AtGRP7 entails reduced levels of the JA-responsive PDF1.2a but does not prevent its induction by MeJ (Figure 3B). Our data also indicate that transcripts regulated by the phytohormone ABA are prevalent among AtGRP7 targets. Previously a considerable overlap of the circadian transcriptome with ABA-related genes has been noted [48,49].
The observation that several target transcripts are affected by elevated levels of the authentic AtGRP7 but not of the AtGRP7-RQ mutant protein shows that the effect depends on the RNA-binding activity. Negative regulation in AtGRP7-ox plants but not AtGRP7-RQ-ox plants has been observed for the endogenous AtGRP7 and AtGRP8 transcripts [22]. Both are regulated post-transcriptionally via binding of AtGRP7 to the pre-mRNAs that entails alternative splicing and degradation through NMD [24]. It seems conceivable that AtGRP7 may interact with similar binding sites in some of the candidate target transcripts, thus controlling their stability or splicing.
So far, a minimal AtGRP7 binding site identified in its 3'UTR [23] was not found to be prevalent in the 5'UTRs or 3'UTRs of either the upregulated or downregulated transcripts (Lewinski and Staiger, unpublished). However, computational identification of RNA substrates based on conserved binding motifs is not straightforward because in addition to the sequence context structural features of the RNA are relevant. Thus, programs for RNA sequence alignment have to be informed by structure [50]. To unequivocally demonstrate direct regulation, in vivo binding of the targets by AtGRP7 will have to be demonstrated by precipitating AtGRP7-containing mRNP particles from transgenic plants expressing epitope-tagged AtGRP7 and identification of co-precipitated transcripts.
Transcripts that are controlled directly by AtGRP7 may be affected in the opposite way by reduced levels of AtGRP7. Several transcripts such as PR1 and PR2 that show higher levels in AtGRP7-ox indeed are present at reduced levels in the atgrp7-1 T-DNA insertion line [4]. Several other transcripts that are altered in AtGRP7-ox plants remain at wt levels in atgrp7-1 or are even changed in the same direction. This may indicate that they respond to elevated levels of AtGRP7 but that AtGRP7 is not limiting. Alternatively, altered steady-state abundance could also be a secondary consequence of AtGRP7 overexpression, but nevertheless may be biologically meaningful. For example, it could result from changes in transcription rate as a consequence of modulation of activators or repressors. This could be assessed by measuring the effect of a high AtGRP7 concentration upon promoter-reporter gene constructs.
Constitutive overexpression of AtGRP7 promotes the transition to flowering [6]. In the present comparison between AtGRP7-ox and wt plants transcripts related to flowering time control are not prevalent. Presumably target transcripts associated with the role of AtGRP7 in floral transition have not been identified in the long-day grown plants because the floral promotive effect of AtGRP7 manifests itself mostly under short-day conditions [6].
Overall, in the AtGRP7-ox plants only 1.4% of the transcripts present on the ATH1 Chip are altered, and the changes are moderate (Additional file 1, Additional file 2). In addition to posttranscriptional regulation AtGRP7 may exert control on downstream targets also at the translational level which is not revealed by transcript profiling. In line with this, gene sets comprising structural components of ribosomes and functions associated with ribosome biogenesis and assembly are enriched among transcripts elevated in AtGRP7-ox plants. Notably, in Chlamydomonas reinhardtii the clock-regulated RNA-binding protein CHLAMY1 has been shown to repress translation of enzymes involved in CO2- and N-metabolism [51,52]. That RNA-binding proteins affect multiple facets of post-transcriptional control is not without precedent: Polypyrimidine tract-binding protein, also known as hnRNP I, binds pyrimidine-rich regions in introns to regulate alternative splicing but also is responsible for time-of-day dependent degradation of the mRNA encoding the mammalian clock gene Period2 [53].
Conclusion
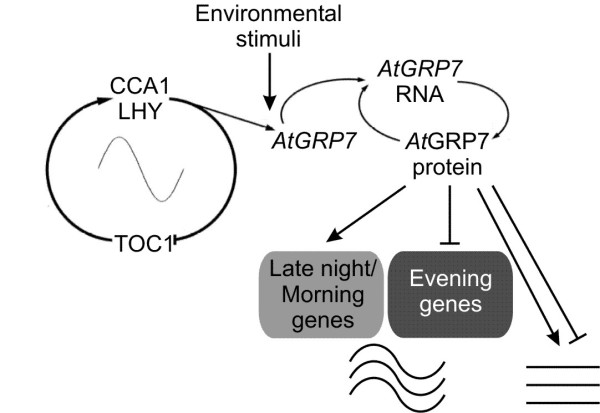
Expression of the RNA-binding protein AtGRP7 is trigged by the circadian clock. In turn, it affects accumulation of rhythmic transcripts in a time-of-day dependent manner: Transcripts peaking in the evening like AtGRP7 itself mainly are expressed at reduced levels in AtGRP7-ox plants whereas transcripts peaking in the morning mainly are expressed at elevated levels (Figure 5). Further, AtGRP7 directly and indirectly affects a suite of other transcripts including hormone responsive and pathogenesis-related and cold-regulated transcripts. Based on these findings, AtGRP7 is placed in clock output signalling, transducing timing information from the circadian clock upon downstream targets. Additionally, AtGRP7 that itself is influence by external stimuli including cold appears to be embedded in an environmental response network (Figure 5).
Figure 5.
Conceptual model of the AtGRP7 feedback loop. AtGRP7 transcript oscillations are under control of the core oscillator [14,60]. Furthermore, AtGRP7 expression is influenced by environmental stimuli including cold. AtGRP7 itself negatively autoregulates at the post-transcriptional level. Downstream targets of AtGRP7 include circadian transcripts that are affected in a time-of-day dependent manner: AtGRP7 negatively affects steady-state abundance of evening-phased transcripts and positively affects steady-state abundance of late night/morning genes. Further, transcripts responsive to environmental signals are prevalent among AtGRP7 targets. Arrows indicate activation, blunt lines indicate negative interaction.
Methods
Plant growth and treatment
The genotypes used were C24, RS13 (AtGRP7-ox in C24 background) [20], Col, D and G (AtGRP7-ox in Col background) [22], AtGRP7RQ-ox [22] and the T-DNA insertion line atgrp7-1 [4,6]. Seeds were surface-sterilised, stratified at 4°C for two days, germinated and grown on half-strength MS medium [54] supplemented with 0.5% sucrose and 0.5 g MES/l in long days (16-hr light/8-hr dark cycles) at 20°C. After about ten days seedlings of comparable size were transferred to MS plates without sucrose.
For JA treatment, seeds were germinated in liquid half-strength MS medium supplemented with 0.5% sucrose and 0.5 g MES/l and incubated on a rotary shaker in long days. After 12 days 50 μM MeJ was added and control samples were treated with 0.2% ethanol. Plants were harvested after 24 h.
RNA isolation for transcript profiling on microarrays
The aerial part of plants with about eight true leaves were harvested at zt12, the time of the circadian maximum of AtGRP7 transcript abundance. RNA was isolated using Trizol. Total RNA was treated with DNaseI and further purified using the RNeasy kit (Qiagen, Hilden, Germany).
Array hybridization
Synthesis of cDNA and biotinylated cRNA were performed as recommended by Affymetrix (Santa Clara, USA). Total RNA (20 μg) was used to prepare cDNA with SuperscriptII Reverse transcriptase (Invitrogen) according to the manufacturer's instructions with an oligo(dT)24-T7 oligonucleotide (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG(dT)24). The cDNA was subjected to in vitro transcription in the presence of 2 mM of each biotin-11-CTP and biotin-16-UTP (ENZO Life Sciences, Farmingdale, NY) with the MegaScript High Yield Transcription Kit (Ambion, Austin, TX). After purification of the cRNA on RNeasy columns (Qiagen, Hilden, Germany), 15 μg of cRNA was fragmented in a volume of 40 μl, denatured for 5 min at 99°C and hybridized to the arrays for 16 h. Washing and detection of labelled cRNA with streptavidin-phycoerythrin were performed according to the manufacturer's instructions. The arrays were scanned using Affymetrix 3000 7G confocal scanner. Affymetrix Arabidopsis ATH1 GeneChips(r) were used throughout the experiment (Affymetrix, Santa Clara, CA). The exact list of probes present on the arrays can be obtained from the manufacturer's website http://www.affymetrix.com. Analysis was based upon annotations compiled by TAIR (http://www.arabidopsis.org, version 2007-5-2).
Data analysis
Raw data were processed with MAS (Microarray suite) 5.0 from Affymetrix. Signal values were derived from Affymetrix *.cel files using GCRMA [55]. All data processing was performed using the statistic language R (version 2.6.2) that is freely available at http://www.r-project.org/[56].
Coefficients of variation (cv) between replicates as a quantitative measure of data quality and consistency between replicates were calculated as described previously [57]. Differentially expressed genes were identified using the RankProd package in R [34] that inherently corrects for multiple testing. Probe sets were called significantly differentially expressed when q < 0.05. To enrich for biologically relevant changes, only probe sets with a minimal fold change of 1.5 were selected. Differentially expressed genes were grouped into collapsed functional gene ontology categories (GOSLIM, obtained from http://www.arabidopsis.org). Furthermore, enrichment of detailed GO categories (obtained from TAIR) was tested. In this case, multiple-testing correction was according to [58] with a critical p-value of 1E-2. Grouping according to preferred phase of circadian expression was based on data from Edwards et al. [35]. Grouping according to phytohormone regulation was based on data from Nemhauser et al. [59]. The significance of enrichment was estimated based on the hypergeometric test and multiple-testing correction according to Bonferroni. PAGE (parametric analysis of gene set enrichment) was performed as described [39] using a critical p-value of 1E-6 after multiple testing correction according to Benjamini and Hochberg [58].
RNA analysis
Isolation of total RNA and hybridization of RNA gel blots were performed as described [60]. For semiquantitative RT PCR, retrotranscribed RNA was amplified with Taq Polymerase. To determine the linear range of amplification for each primer pair, samples were withdrawn after 24, 26, 28, 30, 32 and 34 cycles. PCR products were separated on agarose gels and either visualized by Ethidium-bromide staining or transferred to a nylon membrane and hybridized with radiolabeled cDNA probes.
For Real time PCR, duplicate samples were analysed in a MJ research Opticon DNA Engine. Total RNA was treated with DNaseI and reverse-transcribed using Superscript II (Invitrogen). 20 ng of retrotranscribed RNA was amplified with the Eppendorf Real MasterMix kit using an initial denaturation step of 2 min, followed by 45 cycles of 20 sec at 94°C, 30 sec at 60°C and 40 sec at 68°C. CT values were determined and relative expression levels for the analyzed transcripts were calculated based on non-equal efficiencies for each primer pair [61,62]. Data were normalized to transcripts encoding the translation initiation factor eIF-4A-1 (At3g13920), PTB (At3g01150) and PPR (At5g55840) [63]. Shown are the mean expression levels +/- s.d.. The absence of amplification products from genomic DNA was confirmed in non-retrotranscribed controls. Primers are listed in Additional file 5.
Authors' contributions
C.S. and C.K. performed experiments, L. H. analyzed the Affymetrix data set and performed data mining, D.S. designed the experiments and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Transcripts present at an elevated level in AtGRP7-ox plants. Additional file 1 contains a list of the transcripts present at an elevated level in AtGRP7-ox plants with a signal log ratio > 0.6. Transcripts classified as rhythmic according to Edwards et al. (2006) are marked by an asterisk. The median cv (coefficient of variation) values between the C24 replicates and the RS13 replicates were 2.4% and 2.7%, respectively, demonstrating the high quality of the data.
Transcripts present at a reduced level in AtGRP7-ox plants. Additional file 2 contains a list of the transcripts present at a reduced level in AtGRP7-ox plants with a signal log ratio < -0.6. Transcripts classified as rhythmic according to Edwards et al. (2006) are marked by an asterisk.
RT-PCR analysis of selected AtGRP7 candidate target transcripts in AtGRP7-ox plants. Additional file 3 shows a figure with expression data of targets in AtGRP7-ox and wt plants grown in LDs and harvested around the circadian maximum. RT-PCR products were separated on agarose gels and visualized by Ethidium bromide-staining. The THI2.2 transcript was detected by hybridization with a 32P labelled probe. The gels show results representative for several independent transgenic lines in the Col or C24 background.
Phase distribution of transcripts controlled by LHY and TOC1. Additional file 4 shows diagrams of the phases of LHY and TOC1 target transcripts. A) Phase of transcripts differentially expressed in transgenic plants overexpressing the oscillator component TOC1 (data from [45]). 245 transcripts present at an elevated level in TOC1-ox plants and 160 transcripts present at a reduced level in TOC1-ox plants harvested at zt16 in LDs were interrogated for their first peak after release from light-dark cycles into continuous light in the Edwards dataset of transcript scored rhythmic by COSOPT [35]. B) Phase of transcripts differentially expressed in lhy mutants expressing elevated levels of the oscillator component LHY (data from [44]). GC-RMA normalized data for lhy and Col plants harvested at zt0 in SDs were downloaded. Transcripts expressed with a signal-log ratio < 1 were interrogated for their first peak after release from light-dark cycles into continuous light in the Edwards dataset of transcripts scored rhythmic by COSOPT [35].
PCR primers. Additional file 5 contains a list of the PCR primers used for transcript analysis by RT-PCR, eal-Time RT-PCR and amplification of hybridization probes.
Contributor Information
Corinna Streitner, Email: corinna.streitner@uni-bielefeld.de.
Lars Hennig, Email: lars.hennig@vbsg.slu.se.
Christin Korneli, Email: christin.korneli@uni-bielefeld.de.
Dorothee Staiger, Email: dorothee.staiger@uni-bielefeld.de.
Acknowledgements
We thank Fabian Rudolf, Elisabeth Detring and Kristina Neudorf for expert technical assistance, Bianca Löhr for cDNAs of MeJ-treated plants and Martin Lewinski for sequence alignments. We acknowledge Prof. Wilhelm Gruissem for his support and for the opportunity to perform microarray hybridizations at the Functional Genomics Center Zurich, and Tino Köster for critical reading of the manuscript. This work was supported by the DFG (STA 653/2 and SFB 613).
References
- Schmidt F, Marnef A, Cheung M-K, Wilson I, Hancock J, Staiger D, Ladomery M. A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol Biol Rep. 2010;37(2):839–845. doi: 10.1007/s11033-009-9636-x. [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104(3):1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Jiang L, Song S, Jing R, Xu G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell Mol Biol Lett. 2006;11:526–535. doi: 10.2478/s11658-006-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008;55:455–466. doi: 10.1111/j.1365-313X.2008.03518.x. [DOI] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D. The small glycine-rich RNA-binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 2008;56:239–250. doi: 10.1111/j.1365-313X.2008.03591.x. [DOI] [PubMed] [Google Scholar]
- Mas P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008;18(6):273–281. doi: 10.1016/j.tcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- McClung CR. Comes a time. Current opinion in plant biology. 2008;11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Staiger D. Clockwork Green - the circadian oscillator in Arabidopsis. Biological Rhythm Research. 2006;37:335–352. doi: 10.1080/09291010600804577. [DOI] [Google Scholar]
- Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17(3):791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. Pseudo-response regulators, PRR9, PRR7, and PRR5, play together essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiology. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15(1):47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Molecular systems biology. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/S0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/S0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41(9):1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322(5909):1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994;5(6):799–813. doi: 10.1046/j.1365-313X.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33(2):361–371. doi: 10.1046/j.1365-313X.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M. Changes of conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J American Chemical Society. 2008;130:9507–9513. doi: 10.1021/ja801994z. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic acids research. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Heintzen C. The circadian system of Arabidopsis thaliana: forward and reverse genetic approaches. Chronobiol Int. 1999;16(1):1–16. doi: 10.3109/07420529908998708. [DOI] [PubMed] [Google Scholar]
- Staiger D. RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philo Trans R Soc Lond B Biol Sci. 2001;356(1415):1755–1759. doi: 10.1098/rstb.2001.0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1548. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell. 2003;4:53–66. doi: 10.1016/S1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(17):11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13(6):1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Boss PK, Hadfield J, Dean C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. Journal of experimental botany. 2006;57:3379–3386. doi: 10.1093/jxb/erl073. [DOI] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG. The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA. Dev Cell. 2010;18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1-3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2008;72(6):1539–1549. doi: 10.1271/bbb.80041. [DOI] [PubMed] [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(11):5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Pan L, Kawai-Yamada M, Yu LH, Yamamura S, Koyama T, Kitajima S, Ohme-Takagi M, Sato F, Uchimiya H. Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 2005;138(3):1436–1445. doi: 10.1104/pp.105.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf F, Wehrle F, Staiger D. Slave to the rhythm. The Biochemist. 2004;26:11–13. [Google Scholar]
- Staiger D, Streitner C, Rudolf F, Huang X. In: Endogenous plant rhythms. Hall A, McWatters H, editor. Blackwell Publishers; 2006. Multiple and slave oscillators; pp. 57–83. full_text. [Google Scholar]
- Brown SA, Schibler U. The ins and outs of circadian timekeeping. Curr Opin Genet Dev. 1999;9(5):588–594. doi: 10.1016/S0959-437X(99)00009-X. [DOI] [PubMed] [Google Scholar]
- Kuno N, Moller SG, Shinomura T, Xu X, Chua N-H, Furuya M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell. 2003;15:2476–2488. doi: 10.1105/tpc.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. A Morning-Specific Phytohormone Gene Expression Program underlying Rhythmic Plant Growth. PLoS biology. 2008;6(9):e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. The EMBO journal. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4(6):645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis. Current opinion in plant biology. 1999;2(4):280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome biology. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49(3):481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J. et al. The RNA structure alignment ontology. RNA. 2009;15(9):1623–1631. doi: 10.1261/rna.1601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltenberger H, Schneid C, Grosch JO, Bareiss A, Mittag M. Identification of target mRNAs for the clock-controlled RNA-binding protein Chlamy 1 from Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;265(1):180–188. doi: 10.1007/s004380000406. [DOI] [PubMed] [Google Scholar]
- Kiaulehn S, Voytsekh O, Fuhrmann M, Mittag M. The presence of UG-repeat sequences in the 3'-UTRs of reporter luciferase mRNAs mediates circadian expression and can determine acrophase in Chlamydomonas reinhardtii. Journal of biological rhythms. 2007;22(3):275–277. doi: 10.1177/0748730407301053. [DOI] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic acids research. 2009;37(1):26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia plantarum. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model based background adjustement for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria; 2009. [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003;17(12):1540–1553. doi: 10.1101/gad.257403 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126(3):467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Staiger D, Apel K. Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol Gen Genet. 1999;261(4-5):811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38(2):366–379. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcripts present at an elevated level in AtGRP7-ox plants. Additional file 1 contains a list of the transcripts present at an elevated level in AtGRP7-ox plants with a signal log ratio > 0.6. Transcripts classified as rhythmic according to Edwards et al. (2006) are marked by an asterisk. The median cv (coefficient of variation) values between the C24 replicates and the RS13 replicates were 2.4% and 2.7%, respectively, demonstrating the high quality of the data.
Transcripts present at a reduced level in AtGRP7-ox plants. Additional file 2 contains a list of the transcripts present at a reduced level in AtGRP7-ox plants with a signal log ratio < -0.6. Transcripts classified as rhythmic according to Edwards et al. (2006) are marked by an asterisk.
RT-PCR analysis of selected AtGRP7 candidate target transcripts in AtGRP7-ox plants. Additional file 3 shows a figure with expression data of targets in AtGRP7-ox and wt plants grown in LDs and harvested around the circadian maximum. RT-PCR products were separated on agarose gels and visualized by Ethidium bromide-staining. The THI2.2 transcript was detected by hybridization with a 32P labelled probe. The gels show results representative for several independent transgenic lines in the Col or C24 background.
Phase distribution of transcripts controlled by LHY and TOC1. Additional file 4 shows diagrams of the phases of LHY and TOC1 target transcripts. A) Phase of transcripts differentially expressed in transgenic plants overexpressing the oscillator component TOC1 (data from [45]). 245 transcripts present at an elevated level in TOC1-ox plants and 160 transcripts present at a reduced level in TOC1-ox plants harvested at zt16 in LDs were interrogated for their first peak after release from light-dark cycles into continuous light in the Edwards dataset of transcript scored rhythmic by COSOPT [35]. B) Phase of transcripts differentially expressed in lhy mutants expressing elevated levels of the oscillator component LHY (data from [44]). GC-RMA normalized data for lhy and Col plants harvested at zt0 in SDs were downloaded. Transcripts expressed with a signal-log ratio < 1 were interrogated for their first peak after release from light-dark cycles into continuous light in the Edwards dataset of transcripts scored rhythmic by COSOPT [35].
PCR primers. Additional file 5 contains a list of the PCR primers used for transcript analysis by RT-PCR, eal-Time RT-PCR and amplification of hybridization probes.