Abstract
Background
Unigene sequences constitute a rich source of functionally relevant microsatellites. The present study was undertaken to mine the microsatellites in the available unigene sequences of sugarcane for understanding their constitution in the expressed genic component of its complex polyploid/aneuploid genome, assessing their functional significance in silico, determining the extent of allelic diversity at the microsatellite loci and for evaluating their utility in large-scale genotyping applications in sugarcane.
Results
The average frequency of perfect microsatellite was 1/10.9 kb, while it was 1/44.3 kb for the long and hypervariable class I repeats. GC-rich trinucleotides coding for alanine and the GA-rich dinucleotides were the most abundant microsatellite classes. Out of 15,594 unigenes mined in the study, 767 contained microsatellite repeats and for 672 of these putative functions were determined in silico. The microsatellite repeats were found in the functional domains of proteins encoded by 364 unigenes. Its significance was assessed by establishing the structure-function relationship for the beta-amylase and protein kinase encoding unigenes having repeats in the catalytic domains. A total of 726 allelic variants (7.42 alleles per locus) with different repeat lengths were captured precisely for a set of 47 fluorescent dye labeled primers in 36 sugarcane genotypes and five cereal species using the automated fragment analysis system, which suggested the utility of designed primers for rapid, large-scale and high-throughput genotyping applications in sugarcane. Pair-wise similarity ranging from 0.33 to 0.84 with an average of 0.40 revealed a broad genetic base of the Indian varieties in respect of functionally relevant regions of the large and complex sugarcane genome.
Conclusion
Microsatellite repeats were present in 4.92% of sugarcane unigenes, for most (87.6%) of which functions were determined in silico. High level of allelic diversity in repeats including those present in the functional domains of proteins encoded by the unigenes demonstrated their use in assay of useful variation in the genic component of complex polyploid sugarcane genome.
Background
Sugarcane (Saccharum sp.) is a complex polyploid belonging to the family Poaceae of the tribe Andropogoneae. It is an important commercial sugar producing crop and a source of approximately 50% of the world's sugar and alcohol. The polyploid/aneuploid nature with variation in chromosome number has been largely responsible for its genetic and taxonomic complexity [1]. Characterization of such large genomes is greatly facilitated by the use of molecular markers. Microsatellite or simple sequence repeat (SSR) markers are being preferred because of their co-dominant inheritance, reproducibility, multi-allelic nature, chromosome-specific location and wide genomic distribution. These markers are amenable to high throughput genotyping due to multiplexing and efficient resolution of amplicons by automated fragment analysis [2,3].
In sugarcane, Cordeiro et al. (2000) [2] and Parida et al. (2009a) [4] developed a large number of microsatellite markers from the genomic sequences. Pinto et al. (2004; 2006) [5,6] and more recently Oliveira et al. (2009) [7] also designed such markers from the sugarcane ESTs, which were used for constructing high resolution functional genetic linkage map of Saccharum spp. [8]. However, information on a limited number of these genic microsatellite markers is available in the public domain. Recently, the EST sequences have been assembled into unigenes [9], which is expected to provide non-redundant, locus specific and novel gene-based functional markers for sugarcane having a large genome not amenable to complete sequencing. The unigene sequences of sugarcane have not yet been analyzed for their microsatellite constitution and compared with the other small genome members of the grass family.
In India, systematic breeding of sugarcane has resulted in the development of a number of varieties with high productivity and stress tolerance by inter-specific hybridization [10]. However, the genetic base of modern Indian sugarcane cultivars is considered narrow due to use of a limited number of parental species clones in cross hybridization and repeated intercrossing of hybrids [11]. Understanding the extent of natural variation at molecular level is essential to develop new strategies for sugarcane improvement. Earlier, molecular markers such as RAPD (Randomly Amplified Polymorphic DNA), AFLP (Amplified Fragment Length Polymorphism), and maize and sugarcane genomic microsatellites have been used for this purpose [4,12-15]. No effort has yet been made to understand the genetic diversity of Indian sugarcane cultivars based on functionally relevant genic regions of its complex genome.
The present study was undertaken to mine the available unigene sequences of sugarcane (Saccharum sp.) to understand the microsatellite structure and distribution in the expressed genic component of the genome, assess their functional significance in silico, design primers from the flanking regions of the identified microsatellites, assess the efficiency of a set of fluorescent dye labeled primers in genotyping using automated fragment analysis system and determine functional diversity among different species, related genera and Indian varieties of sugarcane.
Results
Frequency, distribution and organization of microsatellites in sugarcane unigenes
The type, frequency and relative distribution of the microsatellites in the unigene sequences of sugarcane are given in Table 1. The perfect microsatellite (excluding the mononucleotides) frequency in the unigenes of sugarcane was one in every 10.9 kb and the proportion of microsatellite carrying unigenes was 3.7% (584 out of 15,582). When 1,871 (12%) mononucleotide microsatellites were included, the proportion of unigenes carrying microsatellites increased to 17.4%. The mononucleotides in sugarcane showed a strong bias (84.6%) towards A/T repeat, with the majority (89%) being 9 to 30 bases long and the remaining (11%) extending up to 69 bases (T69). A total of 841 perfect microsatellites were identified in 584 unigene sequences of sugarcane. One hundred sixty-seven (28.6%) of these unigenes contained multiple microsatellites (that accounted for 424 microsatellites) which were interrupted by more than 100 nucleotides and the remaining 417 (71.4%) unigenes had a single microsatellite each. In addition to the perfect microsatellites, we identified 183 compound microsatellites, of which 74.9% were interrupting types and the rest being non-interrupting types (Table 1). The trinucleotide repeat motifs were the most prevalent (73.1%) class of microsatellites followed by dinucleotide (23.8%), tetranucleotide (1.8%), pentanucleotide (0.83%) and hexanucleotide (0.47%) repeat motifs (Table 1). The GC rich repeat motifs GCA/GCC/GCG/GCT (23%) coding for alanine were most abundant followed by arginine (AGA/AGG/CGA/CGC/CGG/CGT, 22%) and glycine (GGA/GGC/GGT, 11%) (see Additional file 1). Among the dinucleotide repeats, the most and least frequent motifs were GA and CG, which accounted for 21% and 1.4% of all the microsatellites, respectively (see Additional file 2).
Table 1.
Distribution of microsatellites in the unigene sequences of sugarcane
| Characters under study | Unigenes* |
|---|---|
| Number of sequences examined | 15,594 |
| Size (bp) of examined sequences | 9,17,43,95 |
| Number of identified perfect microsatellites | 2,712 (17.4) |
| Number of perfect microsatellite containing sequences | 2,230 (14.3) |
| Number of perfect microsatellite (excluding mononucleotides) containing sequences | 584 (3.7) |
| Number of sequences containing more than one perfect microsatellites | 167 (28.6) |
| Number of sequences containing single and unique perfect microsatellites | 417 (71.4) |
| Number of mononucleotides | 1,871 (12) |
| Number of dinucleotides | 200 (23.8) |
| Number of trinucleotides | 615 (73.1) |
| Number of tetranucleotides | 15 (1.8) |
| Number of pentanucleotides | 7 (0.83) |
| Number of hexanucleotides | 4 (0.47) |
| Number of perfect microsatellites excluding mononucleotides | 841 (5.4) |
| Size (kb) of sequences containing one perfect microsatellite | 10.9 |
| Number of perfect class I microsatellites | 207 (24.6) |
| Size (kb) of sequences containing one perfect class I microsatellite | 44.3 |
| Number of primer pairs designed for perfecta microsatellites | 810 (96.3) |
| Number of compound class I microsatellite containing sequences | 183 |
| Size (kb) of sequences containing one compound class I microsatellite | 50.1 |
| Number of compound class I microsatellites | 183 (1.2) |
| Number of compound interrupting class I microsatellites | 137 (74.9) |
| Number of compound non-interrupting class I microsatellites | 46 (25.1) |
| Number of primer pairs designed for compound class I microsatellites | 151 (82.5) |
*The number in the bracket is the proportion expressed in percentage
aMononucleotides to hexanucleotides repeated up to 100 times without any interruption at a locus
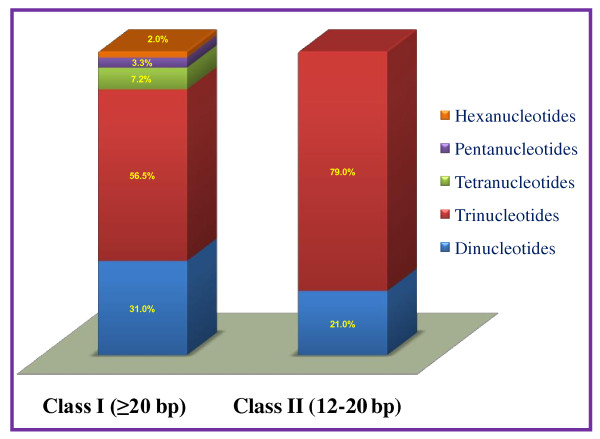
Out of 841 perfect microsatellites identified in sugarcane, 587 (69.8%) were found in the ORFs and the remaining were present either in the 3'UTRs (102, 12.1%) or in the 5'UTRs (152, 18.1%). The trinucleotide repeat motifs were significantly more frequent (about 86%) in the ORFs. In contrast, the GA-rich dinucleotide repeat motifs were more in the 5' (49%) and 3' (32%) UTRs. The density of longer motif containing perfect class I microsatellites was one in every 44.3 kb sequences, which accounted for 24.6% (207) of the total 841 microsatellites identified (Table 1, Figure 1).
Figure 1.
Frequency and relative distribution of class I and class II microsatellite repeats in sugarcane unigenes. Comparative distribution of long hypervariable class I and potentially variable class II microsatellite repeats in the unigenes of sugarcane. Trinucleotide was the most abundant repeat-motif in both class I (56.5%) and class II (79%) category, which was followed by dinucleotide motifs.
Development of unigene derived microsatellite (UGMS) markers and evaluation of their polymorphic potential
The primer pairs could be designed for 810 perfect microsatellites that was 96.3% of the total microsatellites (841) identified in the present investigation. The primer sequences flanking all the perfect UGMS including 207 class I microsatellites with their Tm values and product sizes are given in the Additional file 3. Besides, the primer sequences for 151 compound class I UGMS were designed and provided in the Additional file 4. To validate the UGMS markers, 176 primer-pairs designed from different microsatellite containing unigenes were used in PCR amplification (see Additional file 3). One hundred sixty seven (94.9%) of these produced clear and reproducible amplicons, whereas remaining nine (5.1%) did not give amplification in the S. officinarum from which sequence the primers were designed. To verify that the primers did amplify the expected microsatellite repeat-motifs, the amplified products obtained with 19 of the primers in all the Saccharum species and related genera as well as cereals were sequenced and the presence of the target microsatellite motifs as well the flanking sequences was observed in all the cases (see Additional file 5).
In silico polymorphism analysis was confined to Saccharum officinarum and five cereal species namely, rice, wheat, maize, Sorghum and barley for which unigene sequences were available in the database. The polymorphism based on variation in microsatellite repeat length was observed for a maximum of 163 primers (46.6%) between sugarcane and barley followed by 161 (42.5%) between sugarcane and wheat and least (92, 17.8%) between sugarcane and Sorghum (see Additional file 6). The actual level of polymorphism detected by automated fragment analysis using 47 fluorescent dye labeled primers was much higher than that based on in silico analysis although the trend was maintained. S. officinarum had maximum polymorphism with barley (92.7%) followed by wheat (90.6%), rice (85.8%), maize (67.7%) and Sorghum (61.4%). Forty-three (91.5%) of the 47 primers detected polymorphism (mean PIC of 0.85) among the 41 genotypes belonging to Saccharum species, related genera, tropical and sub-tropical Indian sugarcane varieties and five cereal species (Table 2). This included 21 (95.5%, mean PIC of 0.81) amplifying dinucleotide repeats, 16 (84.2%, PIC of 0.73) trinucleotide repeats, two (100%, PIC of 0.87) tetranucleotide repeats, two (100%, PIC of 0.86) pentanucleotide repeats and two (100%, PIC of 0.83) amplifying hexanucleotide repeats. All the 38 (100%, mean PIC of 0.82) primers that targeted amplification of class I repeats and five (55.6%, PIC of 0.55) of the nine that targeted class II repeats showed polymorphism among Saccharum species, genera, varieties and cereal species. The microsatellites in the 5' and 3'UTR sequences showed higher potential for polymorphism (15 out of 16, 94%, mean PIC of 0.83) as compared to that from the coding regions (28 out of 31, 90%, mean PIC of 0.77).
Table 2.
Evaluation of the amplification efficiency and polymorphic potential of 47 fluorescent dye labeled primers
| Polymorphic potential | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sl. No. | Unigene Accession IDs' | Class I UGMS primersA |
Repeat-motifs | Location | Forward primer sequences (5'-3') | Reverse primer sequences (5'-3') | Putative functions | Actual annealing temperature (°C) | No. of locus | Total no. of alleles amplified | Size (bp) of allele (s) amplified | Type of allele size distribution | No. of heterozygous loci | Among sugarcane species and genera | Among sugarcane varieties | |
| P/MB | PIC | |||||||||||||||
| 1 | CA297715 | UGSuM2a | (AT)43 | CDS | CTGTGTATATGTTCGTAGTTTG | CACTTAGTCACACTCTCACACAC | Sucrose phosphate synthase | 55 | 2 | 20 | 206-302 | Step-wise | 1 | P | P | 0.86 |
| UGSuM2b | 4 | 481-512 | Mixed | |||||||||||||
| 2 | CA278792 | UGSuM5 | (TA)28 | CDS | TCACATCCATCATCCACAGC | TCCAATGCAAGCAAACTCAC | Maize-Cyclin III | 55 | 1 | 23 | 80-170 | Step-wise | 0 | P | P | 0.84 |
| 3 | AY596609 | UGSuM11 | (TA)21 | 3'UTRs | TGGTAACCCTAGGCAGGTGA | GTGCACCAGATTTGGATGGT | Fructose-bisphosphate aldolase | 56 | 1 | 23 | 92-190 | Step-wise | 0 | P | P | 0.83 |
| 4 | CA279221 | UGSuM15a | (CCTCGC)6 | CDS | GTTTAAGACAAGATGGTGTAGATG | TACATATTTACATTGTTACTCCGC | Hypothetical protein | 56 | 2 | 17 | 170-247 | Mixed | 1 | P | P | 0.80 |
| UGSuM15b | 3 | 360-378 | Step-wise | |||||||||||||
| 5 | CA278282 | UGSuM16 | (AT)18 | CDS | GCGTCTTCATCATCTGCAAC | GCGTCTTCATCATCTGCAAC | Pathogenesis-related PRMS protein | 55 | 1 | 22 | 150-244 | Step-wise | 0 | P | P | 0.82 |
| 6 | CA253277 | UGSuM17 | (AG)18 | CDS | TTTCCATTCTTCCATTCAACTG | GGCAGGCTGAGAGACTGTTC | Abcisic acid-protein kinase | 55 | 1 | 10 | 90-188 | Step-wise | 1 | P | P | 0.78 |
| 7 | CA227482 | UGSuM18a | (GA)18 | CDS | GGCGAGAGAGAGAGAGAGAGAG | AGGTGGAGATCTTGAGGTAGGC | Glycine decarboxylase | 56 | 2 | 20 | 96-193 | Mixed | 1 | P | P | 0.81 |
| UGSuM18b | 2 | 326-348 | Step-wise | |||||||||||||
| 8 | CA126180 | UGSuM20a | (TCA)12 | CDS | ATCCCTTATGCTACAGAAATGT | TTAGCCTAGAGGTTTGATTGAT | Acetyl-CoA synthetase | 54 | 2 | 16 | 190-259 | Step-wise | 1 | P | P | 0.83 |
| UGSuM20b | 4 | 392-428 | Step-wise | |||||||||||||
| 9 | CA177414 | UGSuM21 | (AGGA)9 | CDS | CGCTCCCTCACCGTCATT | CTCCGCATCCTCGTCACC | Transcription regulator protein | 62 | 1 | 20 | 160-252 | Step-wise | 0 | P | P | 0.81 |
| 10 | CA223153 | UGSuM22a | (GCG)12 | 3'UTRs | CTCCCTCCTCCTCCCGTTG | CTCTTGGGTGTGAACCAG | Polyadenylate-binding protein | 64 | 2 | 19 | 96-187 | Mixed | 1 | P | P | 0.85 |
| UGSuM22b | 3 | 322-358 | Step-wise | |||||||||||||
| 11 | CA122659A | UGSuM24a | (TTTTC)7 | 5'UTRs | CTGTACAACAGCAATTATGAATCT | CTCGACTACGAGAGGATATGAT | Hypothetical protein | 55 | 3 | 12 | 240-294 | Mixed | 1 | P | P | 0.86 |
| UGSuM24b | 4 | 398-428 | Step-wise | |||||||||||||
| UGSuM24c | 5 | 531-571 | Step-wise | |||||||||||||
| 12 | BU103692A | UGSuM26 | (CT)17 | 5'UTRs | CTCGATCCCAGAGAGCTCCACAG | AGTACCGAATTCATTAAACTCCT | Beta-amylase | 55 | 1 | 21 | 250-346 | Step-wise | 0 | P | P | 0.85 |
| 13 | CA073284 | UGSuM27a | (GGC)11 | CDS | CTGCAGTACGGTCCGGAATC | GTACCACCATGGCTCTAGCTTC | 30 S ribosomal protein S16 | 60 | 2 | 2 | 50-54 | Mixed | 1 | P | P | 0.84 |
| UGSuM27b | 22 | 154-288 | Mixed | |||||||||||||
| 14 | AY596606 | UGSuM33a | (AGC)10 | CDS | CGAGGCACTGAACCCATATC | TGTTTGAACTGGATGGCGTA | Hypothetical protein | 58 | 3 | 17 | 92-189 | Mixed | 1 | P | P | 0.84 |
| UGSuM33b | 2 | 293-305 | Step-wise | |||||||||||||
| UGSuM33c | 2 | 431-491 | Step-wise | |||||||||||||
| 15 | CA268640 | UGSuM34a | (AAG)10 | CDS | TTACAAATGTAGCCTTGCCTTG | ATCTTTCCTTGCTTGCCTCTC | Soluble acid invertase | 61 | 2 | 19 | 99-178 | Mixed | 1 | P | P | 0.83 |
| UGSuM34b | 4 | 333-351 | Step-wise | |||||||||||||
| 16 | CA131350 | UGSuM41 | (CCG)10 | CDS | ATCATTCTCCATCATTTCTCA | AGGCTCTTCAACCGTGCT | Unknown protein | 54 | 1 | 20 | 120-210 | Step-wise | 0 | P | P | 0.80 |
| 17 | CA133924 | UGSuM42a | (CTCTCC)5 | 5'UTRs | TTCATACAGAAGAACCTCCAC | TCCATCAGAGACAAGCAGA | Auxin-independent growth promoter | 54 | 3 | 12 | 130-212 | Mixed | 1 | P | P | 0.83 |
| UGSuM42b | 8 | 325-367 | Step-wise | |||||||||||||
| UGSuM42c | 2 | 475-487 | Step-wise | |||||||||||||
| 18 | CA136599 | UGSuM43a | (CCG)10 | 3'UTRs | CAAAGTGCTGTAGGGCTG | TTCAATGGGTGATAAGTGTGT | Ribose-phosphate pyrophosphokinase 1 | 55 | 2 | 17 | 90-187 | Mixed | 1 | P | P | 0.85 |
| UGSuM43b | 2 | 332-368 | Step-wise | |||||||||||||
| 19 | CA139800 | UGSuM44a | (CT)15 | 5'UTRs | TCCATCAAGCCGTTCCTC | GCCAAGCAGATAAAGAAGTG | Rudimentary enhancer | 55 | 2 | 16 | 220-308 | Step-wise | 1 | P | P | 0.84 |
| UGSuM44b | 1 | 419 | - | |||||||||||||
| 20 | CA171090 | UGSuM45a | (AAAAG)6 | CDS | ATCTCCTCTTATTCGTTCTGG | AGCAGCGTCTTATCTGGG | PAP fibrillin | 56 | 3 | 5 | 91-141 | Step-wise | 1 | P | P | 0.79 |
| UGSuM45b | 2 | 267-287 | Step-wise | |||||||||||||
| UGSuM45c | 2 | 368-393 | Step-wise | |||||||||||||
| 21 | CA196477 | UGSuM46a | (GAC)10 | CDS | ACTCCTCCCGCCTCCACTAC | CTCACCGAAGCAATCAAG | Hypothetical protein | 60 | 2 | 15 | 101-188 | Mixed | 1 | P | P | 0.81 |
| UGSuM46b | 3 | 341-377 | Step-wise | |||||||||||||
| 22 | CA228772 | UGSuM47a | (GCC)10 | CDS | ATTTATGGAGGAAGAAACGG | ATTACAAACAAGAAGAGCGG | Transport protein particle component | 55 | 2 | 3 | 82-97 | Step-wise | 1 | P | P | 0.80 |
| UGSuM47b | 14 | 215-270 | Mixed | |||||||||||||
| 23 | CA161416 | UGSuM50 | (TC)14 | CDS | CTACTGCCGAGGAAAGATCG | GGAAAAGTTTGTGGCAAGGA | Hypothetical protein | 58 | 1 | 16 | 94-188 | Step-wise | 0 | P | P | 0.80 |
| 24 | CA228375 | UGSuM73a | (CGC)8 | 5'UTRs | CTTTCAACCTCTACACCTCCAC | ACTAGAAGACTGAGAAGAACCAGT | 40 S ribosomal protein S11 | 55 | 2 | 13 | 101-186 | Mixed | 1 | P | P | 0.82 |
| UGSuM73b | 5 | 354-375 | Step-wise | |||||||||||||
| 25 | CA261182 | UGSuM74 | (ACA)8 | CDS | TCAGCAGCTGTGAAGTTTCATT | CGTCTCTTTTGGGTTTCATCTC | Transcription regulator protein | 55 | 1 | 18 | 190-271 | Step-wise | 0 | P | P | 0.81 |
| 26 | CA219230 | UGSuM75a | (TA)12 | CDS | TTGTGCTGATGTTTCCTGCT | CAAGAGAAGATGCCATTAGCC | Patatin-like protein | 55 | 2 | 13 | 94-176 | Step-wise | 1 | P | P | 0.83 |
| UGSuM75b | 4 | 333-351 | Step-wise | |||||||||||||
| 27 | CA093071 | UGSuM96a | (AT)11 | CDS | TCAAACCAGGATCTAAGCTCAC | GGTAGTGCCATTGAGGTTGC | Putative apyrase | 57 | 2 | 11 | 205-290 | Mixed | 1 | P | P | 0.81 |
| UGSuM96b | 5 | 408-458 | Step-wise | |||||||||||||
| 28 | CA229840 | UGSuM97a | (GA)11 | 5'UTRs | GCGAGAGAGATAGAGGGAGAGA | AGGTGCCGTTCATGAGGTAGT | Glycine decarboxylase | 56 | 2 | 19 | 240-334 | Step-wise | 1 | P | P | 0.85 |
| UGSuM97b | 2 | 453-469 | Step-wise | |||||||||||||
| 29 | CA112979 | UGSuM149a | (AGC)7 | CDS | GTTCAATCAAATCCCTCTCCTC | AGCTTGGTCAGCTCCTCATCGTT | Ubiquitin-specific protease 4 (UBP4) | 60 | 2 | 4 | 104-141 | Mixed | 0 | P | P | 0.78 |
| UGSuM149b | 3 | 276-297 | Step-wise | |||||||||||||
| 30 | CA116368 | UGSuM150a | (GGC)7 | CDS | ACACTGACCGATGGATCCTCTT | ATCAACGTGGACCAGATCTTCTT | hAT family dimerisation domain protein | 60 | 2 | 12 | 90-167 | Mixed | 1 | P | P | 0.76 |
| UGSuM150b | 2 | 279-291 | Step-wise | |||||||||||||
| 31 | CA280782 | UGSuM177a | (TGC)7 | CDS | GGTGCTGTCCCTATCACTAC | GCCCTTGTTTCTTTGTCTACT | Hypothetical protein | 55 | 2 | 3 | 150-167 | Mixed | 0 | P | P | 0.79 |
| UGSuM177b | 3 | 287-314 | Step-wise | |||||||||||||
| 32 | CA291445 | UGSuM178 | (GAC)7 | 5'UTRs | GGACTACTACGACTACTGCGA | ACCTTGCTTACATCTTCCTCT | O-diphenol-O-methyl transferase | 54 | 1 | 15 | 100-196 | Step-wise | 0 | P | P | 0.80 |
| 33 | CA244023 | UGSuM186 | (AG)10 | CDS | AACATTTCGGCATTTGAAGC | GGTCTTTCTTGGGGATCTCTC | Ubiquitin C-terminal hydrolase | 56 | 1 | 4 | 190-232 | Step-wise | 0 | P | M | 0.73 |
| 34 | CA231668 | UGSuM187a | (CT)10 | CDS | CAACAATTGTCGAAGCCTCTC | TTTGCTTACCCCCTGTTGAC | ATP synthase | 58 | 2 | 10 | 200-264 | Step-wise | 0 | P | M | 0.83 |
| UGSuM187b | 6 | 376-418 | Step-wise | |||||||||||||
| 35 | AY596560 | UGSuM188 | (GA)10 | 3'UTRs | CCCAAGCGAGCTAGAGAGAG | TCTTCTTTCCTTCGCACAGC | Hypothetical protein | 55 | 1 | 19 | 95-186 | Mixed | 0 | P | P | 0.81 |
| 36 | CA241232 | UGSuM189a | (CT)10 | CDS | CCGCGACTCTCCTCTCTCT | GTTCTTCTCGGCGTTCCTC | Auxin-regulated protein | 55 | 2 | 15 | 106-200 | Step-wise | 1 | P | P | 0.80 |
| UGSuM189b | 3 | 345-377 | Step-wise | |||||||||||||
| 37 | CA133642 | UGSuM196a | (AAAG)5 | 5'UTRs | GCTACTATGGACAACAGGG | ATGAAGAGACGAGACGAAGA | Cinnamoyl CoA reductase | 54 | 2 | 18 | 90-181 | Mixed | 1 | P | P | 0.81 |
| UGSuM196b | 2 | 373-393 | Step-wise | |||||||||||||
| 38 | CA134472 | UGSuM197 | (GA)10 | 3'UTRs | GAAGGAGCAGCAGCGCCAGT | GATTTGCCGTCCTAGGGTTT | Epsin N-terminal homology domain | 56 | 1 | 22 | 200-290 | Step-wise | 0 | P | P | 0.84 |
| 39 | CA297648 | UGSuM343a | (CAG)6 | 5'UTRs | ACTCCTCCTCCTCGCCGT | TCTTGTTGTAGTAGCCCTTGT | SOUL heme-binding family protein | 62 | 2 | 17 | 250-320 | Mixed | 1 | P | M | 0.87 |
| UGSuM343b | 3 | 438-450 | Step-wise | |||||||||||||
| 40 | CA300679 | UGSuM344 | (CTC)6 | CDS | CTATCCTCTTGTTGGGTCCT | TCCGCACCTCCGTTCACC | Nucleoside diphosphate kinase protein | 55 | 1 | 1 | 260 | Step-wise | 0 | M | M | 0.0 |
| 41 | CA084691 | UGSuM345 | (TC)8 | CDS | TATACAAGAATGAAAGGTGAGAGA | AAGCATACTCCCTCTATCTCTATG | DC1 domain-containing protein | 55 | 1 | 2 | 210-220 | Step-wise | 0 | M | P | 0.70 |
| 42 | CA093455 | UGSuM346 | (AG)8 | CDS | TATACGTAGTAGTGATGATGACCG | CTCCTTCGTCCAGTACCAGTAG | DNA-binding protein DF1 | 60 | 1 | 4 | 150-180 | Step-wise | 1 | P | M | 0.71 |
| 43 | CA110745 | UGSuM347a | (CT)8 | 5'UTRs | TCTGGCTTTATCGTAACTTGTAT | GAGCCTCGTTTGGGTGGCTTTC | Expressed protein | 55 | 2 | 5 | 230-254 | Step-wise | 1 | P | M | 0.74 |
| UGSuM347b | 1 | 365 | - | |||||||||||||
| 44 | CA112979 | UGSuM348 | (CG)8 | CDS | CTACCTCCTCGTCTCCTCCCTCTT | AACAAGGAATATGGTCCCTGAG | Unknown protein | 61 | 1 | 2 | 240-261 | Mixed | 0 | M | M | 0.0 |
| 45 | CA116458 | UGSuM349a | (TC)8 | CDS | CAAGATGTACCCGGACATGGCT | TGCTATACTAGCTATCTCCTTCCT | Unknown protein | 55 | 2 | 3 | 220-236 | Step-wise | 0 | P | M | 0.73 |
| UGSuM349b | 2 | 385-395 | Step-wise | |||||||||||||
| 46 | CA123971 | UGSuM464 | (ACA)5 | 5'UTRs | GGCTACTTCAGACACGCA | TCTACGCATCAACCTCTCA | SNF2-domain-containing protein | 55 | 1 | 1 | 220-232 | Step-wise | 0 | M | M | 0.0 |
| 47 | CA125310 | UGSuM465 | (GCA)5 | CDS | GCTAACCAACATCAGCAGT | AGGAGATTGACGAAGAAGAAG | Transducin family protein | 53 | 1 | 2 | 260-279 | Mixed | 0 | M | M | 0.0 |
AUGSuM stands for unigene derived sugarcane microsatellite primers as indicated in the Additional file 3
BPolymorphic (P) and Monomorphic (M)
Due to high polyploidy and heterozygosity of the Saccharum genome, most (28, 65.1%) of the primers amplified two to three different loci (each locus being designated as an UGMS marker) yielding a total of 60 loci with the number of alleles per locus ranging from 2 to 22 and the allele size varying from 82 to 408 bp across sugarcane species, genera and varieties (Figure 2) as well as five cereal species. The remaining 15 (34.9%) primers gave single locus amplification (Table 2) with 1 to 23 alleles each. Overall, the 43 polymorphic primers amplified an average of 7.42 alleles per UGMS marker locus with a total of 722 alleles across 74 loci. Forty-two (73 UGMS marker loci with 720 alleles, 97.7% polymorphic, mean PIC of 0.81) of the 43 informative primers revealed polymorphism between sugarcane species and related genera, whereas 37 (66 UGMS marker loci with 678 alleles, 86% polymorphic, PIC of 0.74) primers detected polymorphism among the Indian commercial sugarcane varieties (Figure 3). Twenty-six (34.7%) of the 75 UGMS marker loci showed heterozygosity in different genotypes used. Maximum heterozygous loci was detected in five sugarcane species (14, 18.7%) followed by commercial varieties (9, 12%) and minimum (4, 5.3%) in the three related genera and five cereal species.
Figure 2.
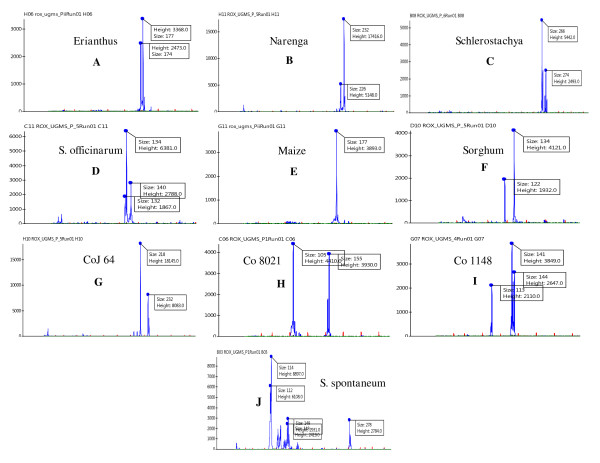
Different UGMS allele types detected by automated fragment analysis. Distribution of various allele types detected in Saccharum species, genera, commercial varieties and five cereal species using the automated fragment analysis. A, C, D, F and H: Multiple alleles in a single locus showing step-wise distribution, B, G and I: Multiple alleles in a single locus showing mixed distribution, E: Single allele in a unique locus, and J: Multiple loci showing both stepwise and mixed distribution in different loci.
Figure 3.
Fragment length polymorphism detected among Indian sugarcane varieties using fluorescent dye labeled primers. Allelic variation in a representative set of 14 commercial Indian tropical and sub-tropical sugarcane varieties. Fourteen alleles were amplified by a class I UGMS marker (UGSuM17) designed from the unigene encoding Abcisic acid inducible protein kinase that contained (AG)18 repeat-motif. The amplicons generated by the fluorescent dye labeled primers were resolved in MegaBACE automated DNA sequencer and analyzed in Fragment Profiler software. Fragment size (bp) and average peak height for all the amplified alleles are indicated.
Molecular basis of UGMS polymorphism and its functional significance
Comparison of fragment size (bp) of variant alleles amplified at 75 polymorphic UGMS loci with changes in number of repeats among Saccharum complex, varieties and five cereal species revealed purely step-wise allelic distribution for 51 (68%) loci, while remaining 24 (32%) loci showed mixed type of allele size distribution (Figure 2). To elucidate the distribution pattern of UGMS alleles showing fragment length polymorphism, 10 amplified size variant alleles each of two microsatellite primers namely, UGSuM2 and UGSuM27 showing both step-wise and mixed type of allele distribution were sequenced. High quality sequence alignment revealed that the size variant alleles showing step-wise distribution contained the expected microsatellite repeat sequences with conserved primer binding sites, but corresponded exactly to the step-wise multiples of the number of repeat units. The size variation of sequenced alleles was explained by differences in the number of repeat units. Mixed type of allele distribution resulted from both variation in the number of repeat-units and insertions/deletions in the sequences flanking the UGMS repeats. For instance, UGSuM2 (Figure 4A) and UGSuM27 (Figure 4B) showing both step-wise and mixed type allele size distribution had the expected (AT)n and (GGC)n UGMS motifs, respectively with varying number of repeat-units in different sugarcane species, genera, varieties and cereals along with a small stretch of nucleotide insertion/deletion in the flanking regions of target microsatellites.
Figure 4.
DNA sequence alignment depicting the molecular basis of UGMS fragment length polymorphism among Saccharum complex, varieties and five cereals. Multiple sequence alignment of 10 size variant alleles showing both stepwise and mixed distributions amplified from sugarcane species, genera, varieties and five cereals for the two UGMS marker loci namely, UGSuM2 (A) and UGSuM27 (B). Variation in the number of repeats of (AT)n and (GGC)n microsatellite motifs at UGSuM2 and UGSuM27 loci, respectively and additional nucleotide insertions/deletions in the flanking sequences of the repeats are highlighted.
In order to assess the functional significance of the UGMS, gene ontology classification of 767 unigenes carrying perfect and compound microsatellites was carried out. Six hundred seventy-two (87.6%) of these (see Additional files 3 and 4) could be functionally annotated and were shown to encode enzymes for sugar metabolism (39%), structural proteins (26%), transcription and translation factors (18%), signal transduction pathway proteins (8%), cell growth and development factors (5%) and disease resistance proteins (4%) (see Additional file 7). Three hundred sixty-four (54.2%) of 672 unigenes contained microsatellite repeats in 89 different functional domains of proteins encoded by these unigenes (see Additional file 8) which included cytochrome b/c oxidase, chlorophyll A/B binding, FAD and ubiquitin binding, sucrose synthase, alpha-amylase, acetyl COA, aldehyde dehydrogenase, glyceraldehyde-3-phosphate, lipase, S-adenosylmethionine, leucine rich repeat (LRR), ferritin, protein kinase, chitinase, basic leucine zipper (bZIP), zinc finger, TATA box, Myb and WRKY DNA binding domains (Figure 5). Fifteen primers designed from 15 different unigenes targeting such functional domains (see Additional file 9) that gave single locus amplification and step-wise allele distribution (Table 2) were selected for validation. All of these primers gave fragment length polymorphism among Saccharum complex, varieties and five cereals based on variation in the number of UGMS repeat-units within the functional domains.
Figure 5.
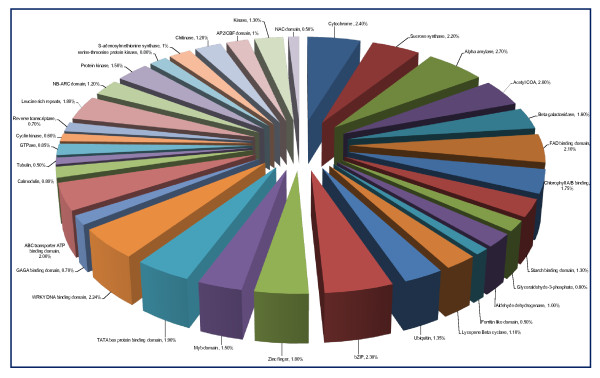
Functional annotation of unigenes carrying microsatellites in their functional domains. Functional annotation of 364 unigenes carrying microsatellites in the functional domains of encoded proteins. These unigenes corresponded maximum (47.7%) to the domains responsible for photosynthesis (cytochrome b/c and chlorophyll A/B binding domains) and carbohydrate metabolism (sucrose synthase and alpha amylase domains) followed by transcription factor associated basic leucine zipper, zinc finger, TATA box, Myb and WRKY DNA binding domains (22%), and minimum to the abiotic and biotic stress related leucine rich repeat, protein kinase and chitinase domains (5%).
One of these primers (UGSuM26) showing polymorphism targeted the amylase catalytic domain of β-amylase (Figure 6). To understand the possible biological significance of the variable UGMS repeat in the amylase domain, the structure predicted from the aminoacid sequences of the functional domain was analyzed in silico. It revealed variation in the active binding site involved in formation of the Ca2+ ligand complexes due to the presence of variable number of repeats encoding Leucine-Serine aminoacid residues (Figure 6). This would most likely alter the function of amylase gene in respect of carbohydrate metabolism in sugarcane. Similarly, the expansion/contraction of microsatellite motifs (AG)n encoding Arginine-Glutamine aminoacid residues was observed in the protein kinase functional domain of another sugarcane unigene which was amplified by the primer UGSuM17. In silico analysis suggested a novel secondary protein structure and catalytic domain binding site in the variant form that was different from the native form which contained (AG)18 microsatellite repeats (see Additional file 10).
Figure 6.
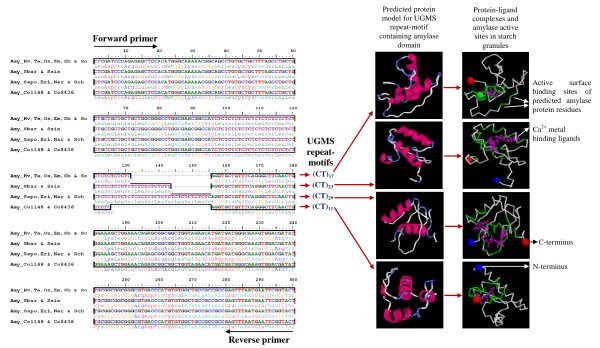
Alignment, and predicted protein structure and catalytic domain binding sites depicting the functional relevance of microsatellite carrying unigenes. Multiple sequence alignment of the four size variant alleles showing step-wise distribution in the amylase catalytic domain amplified from sugarcane species, genera, varieties and five cereals using primer (UGSuM26) for β-amylase unigene. Variation in the number of repeat-units of CT and the encoding repeated tracts of Leucine-Serine aminoacid residues at a microsatellite locus predicted different three dimensional protein structures and protein-ligand complex binding sites which are highlighted.
Assessment of functional genetic diversity
The pair-wise similarity among 36 genotypes belonging to Saccharum species, related genera and 28 tropical and sub-tropical Indian varieties of sugarcane based on 43 fluorescent dye labeled polymorphic primers revealed a broad range from 0.12 to 0.91 with an average of 0.32 similarity index. The similarity among the sugarcane species varied from 0.24 (IJ-76-3-1-9 and 1151) to 0.88 (IJ-76-3-1-9 and IM-76-256) with an average of 0.36. Among the species, S. officinarum showed maximum similarity (0.88) with S. robustum followed by S. barberi with S. sinense (similarity of 0.82). S. spontaneum was most divergent from the rest of the Saccharum species. The similarity of the three genera of sugarcane, namely, Erianthus, Narenga and Sclerostachya with the sugarcane species varied from 0.22 to 0.90. Erianthus had the least similarity with the Saccharum species. Similarity among the 28 Indian tropical and sub-tropical sugarcane varieties varied from 0.33 (Bo 91 and Co 8021) to 0.84 (Co 8371 and Co 8021) with an average of 0.40. The pair-wise similarity between the sugarcane species and varieties varied from 0.20 to 0.78 with average of 0.35, while between the three related genera and varieties, it ranged from 0.13 to 0.80 with an average of 0.24. The average similarity between the sub-tropical and tropical varieties (0.40) was slightly lower than the average pair-wise similarity measures within the sub-tropical (0.47) and tropical varietal groups (0.43).
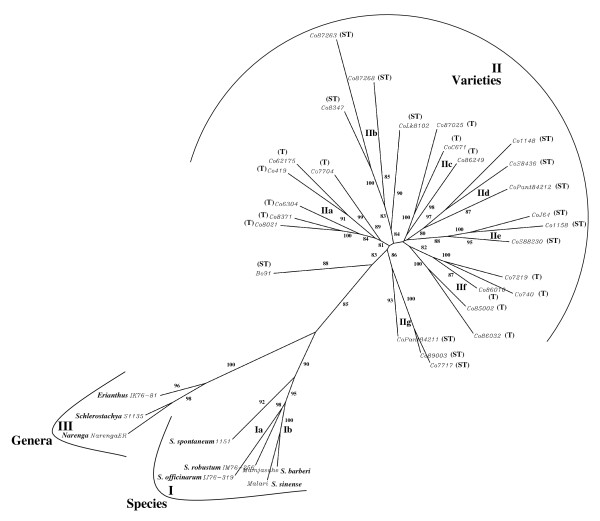
The genetic relationship among the Saccharum species, related genera, and tropical and sub-tropical varieties is depicted in Figure 7. The UGMS markers clearly discriminated all the 36 genotypes from each other and resulted in a definitive grouping among different genera and species of Saccharum with high bootstrap values (81 to 100) that corresponded well with their known pedigree relationships. All the clones of five Saccharum species were included in the major cluster I in which S. officinarum and S. robustum clones were sub-clustered (Ia) with 98% occurrence in boot strap analysis; S. barberi and S. sinense clones grouped together in a separate sub-cluster (Ib) with 100% occurrence and S. spontaneum grouped distinctly from these sub-clusters with 92% occurrence. The three related genera of Saccharum being highly divergent from the Saccharum species and varieties, grouped in a separate cluster (III) with 98% occurrence.
Figure 7.
Phylogenetic tree depicting genetic relationships among Saccharum complex and Indian varieties. Unrooted phylogenetic tree depicting the genetic relationships among the Saccharum species, related genera and 28 commercial Indian tropical and sub-tropical sugarcane varieties based on Nei and Li's similarity coefficient using 47 fluorescent dye labeled primers. Bootstrap values are indicated at the corresponding node for each cluster. Molecular classification corresponded to the known evolutionary and pedigree relationship as well as the adaptive environment. T and ST denote the tropical and sub-tropical region of adaptation, respectively.
All the tropical and sub-tropical varieties were included in a distinct and separate cluster (II) with seven distinct sub-clusters (Figure 7) with high (88) bootstrap value. The tropical variety Co 62175 grouped in a major sub-cluster IIa with its tropical male parent Co 419 (similarity 0.71) with a bootstrap value of 99%. In this sub-cluster (IIa), the other tropical varieties namely, Co 8021, Co 8371, Co 7704 and Co 6304 were also grouped together with average similarity of 0.77 (supported by 93% occurrences in bootstrap) possibly because of their common ancestry involving Co 419. The two sub-tropical varieties, Co 8347 and Co 87263, and three other sub-tropical varieties, CoPant 84211, Co 89003 and Co 7717, in spite of having a common ancestor Co 775, were included in two distantly placed major sub-clusters; IIb and IIg, respectively. It could be due to inclusion of two diverse sub-tropical varieties, Co 87268 and CoLk 8102 having common parent Black Cheribon within the sub-cluster IIb. Similarly, the clustering of two tropical varieties namely, CoC 671 and Co 87025 in the sub-cluster IIc with average similarity of 0.69 (supported by 100% occurrences) and five tropical varieties, Co 7219, Co 740, Co 86010, Co 86032 and Co 85002 together in another separate large sub-cluster (IIf) with average similarity of 0.64 (supported by 92% occurrences) was observed even though they had one common distant progenitor parent Black Cheribon. The grouping of sub-tropical varieties namely, Co 1148, CoS 8436 and CoPant 84212 in cluster IId with an average similarity of 0.70 (88% occurrences in bootstrap) was clearly influenced by a common ancestry involving Co 1148. The sub-tropical varieties, CoJ 64, Co 1158 and CoS 88230 included in the cluster IIe had common progenitor parent PoJ 2878 that could have influenced their clustering with a high bootstrap value (94.3%).
Discussion
Unigene resources representing the transcriptome of an organism provide opportunities to understand the sequence organization in the genic regions of complex polylploids and allow design of sequence based robust markers for various genotyping applications. Considering the availability of a large unigene database of sugarcane in public domain, this resource was studied for the presence and functional relevance of different microsatellite repeats. The frequency of microsatellites in sugarcane unigenes (1/10.9 kb) was lower than that obtained in rice, Sorghum, barley and maize (1/3.6 kb, 1/5.9 kb, 1/8.9 kb and 1/9 kb, respectively) but similar to wheat (1/10.6 kb). In polyploids, extensive loss of duplicated genes and chromosomal rearrangement after polyploidization possibly have resulted in shortening and loss of microsatellite repeat-motifs from the genic coding sequences leading to dosage compensation, developmental stability and functional plasticity [16,17]. It would be of interest to explore the mechanism that restricts repeat expansion in genic regions of wheat and sugarcane leading to lower microsatellite frequency in these species.
The observed frequency of the mononucleotides (12%) in sugarcane was much less than that reported earlier [18] in the unigenes of maize (75.8%), wheat (71%), barley (42.4%) and rice (41.6%), but similar to Sorghum (13%). This suggested a lack of correspondence with genome size and ploidy. The presence of longer (69 bases or more) A/T mononucleotide repeat-motifs in sugarcane is comparable to earlier observations on the nature and the frequency of such repeats in the genomic and EST sequences of cereals [19]. The abundance of trinucleotide microsatellite motifs in the sugarcane unigenes is consistent with the earlier observations based on EST sequences of sugarcane [3,5] and unigene sequences of cereals [18]. The limited expansion of non-triplet microsatellites in the unigenes of sugarcane could be due to selection against frameshift mutations in the coding regions resulting from length changes in non-triplet repeats. We observed that more than 50% of the identified trinucleotide repeat motifs in sugarcane were GC-rich, possibly due to their high GC content [3,5] and consequent usage bias in the coding sequences [20]. The abundance of GC-rich trinucleotide repeat motifs coding for small and hydrophilic aminoacid alanine and GA-rich dinucleotide motifs in sugarcane coding sequences parallels their abundance in cereal exons [18]. The GA-rich dinucleotide UGMS motifs were observed particularly in the 5' and 3' (32%) UTRs with balanced (46 to 52%) GC content and therefore would support better amplification as polymorphic markers [21].
The unigenes being longer in higher quality sequences offer advantages over the EST sequence resources for the development of microsatellite markers. In the present study, 961 (810 perfect and 151 compound) primer-pairs were designed from the 767 different unigene sequences carrying microsatellite repeats in the expressed component of the sugarcane genome. The primers designed from the unigene sequences flanking the microsatellite motifs were highly efficient with amplification success rate of 94.9% that suggested the utility of the unigene database in designing sequence based robust genic markers. The paucity of usable and robust sequence based markers in sugarcane has been a major limitation in genetic analysis in this important sugar crop. The UGMS markers developed by us have been placed in the public domain and thus would be immediately useful in various genotyping applications in sugarcane.
For most (87.6%) of the unigene sequences from which the primers were designed, putative functions have been predicted. For instance, about 39% and 4% of the primers were from sequences related to sugar metabolic enzymes and disease resistance, respectively. Interestingly, 54% of the microsatellite repeats were present in various functional domains of proteins encoded by the unigenes. Correlation between fragment length polymorphism due to variable number of UGMS repeat-units in the functional domains and alteration of the predicted protein structure and active ligand binding sites suggested functional relevance of the genic microsatellites. The evolutionary and adaptive advantages of such variable microsatellite repeats affecting the structure and function of the encoded proteins to generate favorable alleles for relaxation of environmental stress impact under the action of high natural selection pressure through modulation of mutation/recombination in these loci have been reported in many eukaryotes [22]. For instance, in Saccharomyces cerevisiae, the expansion and contraction of microsatellite repeats in the coding regions of protein kinase genes have provided greater adaptability to various abiotic and biotic stresses [22]. Sugarcane is a tropical crop. However, varieties adapted to subtropical condition have been developed in India. Variation in microsatellites in the coding regions of these two groups was evident. For example, the tropical sugarcane varieties Co7219 and Co740 contained (AG)18 microsatellite repeats whereas varieties Co89003 and Co7717 adapted to sub-tropical condition showed contraction of AG repeats to (AG)10 leading to alteration of protein structure and possibly function. Understanding the adaptive significance of such variation is of relevance that needs further experimentation. Designing of genic microsatellite markers targeting functional domains in solanaceous crops like tomato and pepper [23] was reported. Association of such markers with many traits including diseases like neuronal disorders [22] and cancers [24] in humans based on expansion/contraction of repeated tracts of microsatellites encoding aminoacid residues in the functional domains of proteins that changes their secondary structure and function has been demonstrated. The information generated in this study thus would allow selection of candidate gene-based markers for rapidly establishing marker-trait linkages in sugarcane.
The efficiency of sugarcane UGMS markers to detect polymorphism within Saccharum complex and among five cereals was evaluated in silico that was further validated experimentally. In silico polymorphism based on variation in length of the UGMS repeat among S. officinarum and each of five cereal species could be due to divergent microsatellite evolution in these lineages [25]. The actual level of polymorphism detected by automated fragment analysis using fluorescent dye labeled primers was much higher than that based on in silico analysis. The expected relationship in evolution was reflected by actual polymorphism observed between sugarcane and cereals. With the use of automated fragment analysis system, all the allelic variants could be captured efficiently in a set of 36 sugarcane genotypes and five cereal species, thereby providing a platform for rapid, automated and large-scale genotyping of sugarcane. Further, the allele size information generated would enable multiplexing of the UGMS markers, thus making them useful in various high-throughput genotyping applications. The observed polymorphism (97.7%) among the members of the Saccharum species and related genera is higher than that estimated earlier using the fluorescent dye labeled sugarcane genomic (90%, [2]) and EST derived microsatellite markers (81%, [3]). The higher potential of UGMS markers particularly the longer class I di- and tetra-nucleotide repeat-motifs to detect polymorphism as compared to the class I trinucleotide and class II motifs reflected correspondence between the type and length of repeats, and the level of polymorphism as observed earlier in sugarcane [4] and rice [21,26]. Higher polymorphic potential of UGMS markers derived from the UTRs than that from the conserved coding sequences, which are constrained by purifying selection [25,27] suggested the utility of unigenes having such repeat-motifs as a source of polymorphic microsatellite markers.
The level of inter-varietal polymorphism (86%, mean PIC of 0.74) detected by the fluorescent dye labeled primers was higher than the level reported previously with the labeled sugarcane EST derived (38%, PIC of 0.23, [3]) microsatellite markers. The discrepancies could be due to use of different sets of markers and genotypes. However, genotyping using the automated fragment analysis system in this study revealed comparable level of polymorphism with the genic and genomic microsatellite markers in sugarcane. It suggested that the genic microsatellite markers developed in this study would be highly informative and useful in sugarcane genetics, genomics and breeding.
Unigene sequences usually have advantages of unique identity and position in the transcribed regions of the genome. If this is the case then primers designed from the unigene sequences flanking the microsatellite repeats should amplify unique single locus. In contrast, in the present study, multiple loci and thus amplification of multiple sequences of the same gene was observed for 28 (65.1%) of the 43 primers designed from different microsatellite carrying unigenes. This is possibly due to poor representation of unigene sequences in the database that was scanned for microsatellite repeats. Alternatively, all the copies of a microsatellite carrying gene that are PCR amplified might not be transcribed due to dosage compensation leading to silencing of all but one copy in the large polyploid sugarcane genome [16,28]. In spite of high polyploidy and heterozygosity of the sugarcane genome, 15 (34.9%) of the 43 polymorphic UGMS primers amplified a single discrete locus with step-wise allele distribution and showed fragment length polymorphism across the members of Saccharum complex and varieties in an automated fragment analysis system. This could be due to polyploidization followed by selective gene loss [29,16,28] resulting in retention of a single copy during evolution of Saccharum. Gene ontology analysis of these 14 unigenes showed correspondence with the genes coding for basic metabolic process of energy generation, degradation of cellular building blocks, DNA recombination and repair proteins which played a vital role in plant biology and thus remained as single copy without alteration.
Distribution pattern of size variant alleles amplified at UGMS loci showed higher proportionate (68%) distribution of alleles showing step-wise mutation than that of mixed allele distribution (32%). High-quality sequence alignment of the size variant alleles showing both step-wise and mixed distributions confirmed the presence of variable number of repeat-units in different amplified alleles and additional insertions/deletions in the flanking regions of microsatellite repeat-motifs, which contributed to the UGMS fragment length polymorphism observed in the Saccharum complex, varieties and cereal species. Such complex pattern of allele distribution and fragment length polymorphism at microsatellite loci due to variation in the copy number of microsatellite repeats and insertions/deletions in the flanking sequences of microsatellite motifs have been observed earlier in species like maize with a large genome size [30].
It is important to evaluate molecular diversity existing among the members of the Saccharum species and related genera, since major varietal improvement in sugarcane was through inter-specific hybridization. Use of S. spontaneum and Erianthus is vital since they carry many traits of agricultural importance including tolerance to biotic and abiotic stresses [14]. Identification of true inter-specific and inter-generic hybrids is crucial for successful transfer of the target traits. Besides, monitoring of introgression is required to verify the transfer of the target genomic regions from the wild relatives to the cultivated genetic backgrounds. 97.7% of markers revealed inter-specific and inter-generic polymorphism and thus would enable precise identification of inter-specific and inter-generic hybrids, and also provide opportunities to assess transfer of genic regions to desirable genetic backgrounds, thereby offering advantages over the random markers such as RAPD and AFLP.
Evaluation of molecular diversity in a set of 28 commercial Indian tropical and sub-tropical sugarcane varieties using unigene based genic microsatellite markers revealed a wider range (0.33 to 0.84 with an average of 0.40) of genetic similarity than the level detected previously with RAPD (0.59 to 0.81 with average of 0.71, [12]), maize microsatellite (0.40 to 0.73 with average of 0.64, [13]), sugarcane genomic microsatellite (0.39 to 0.82 with average of 0.53, [4]) and AFLP (0.52 to 0.83 with average of 0.62, [14]) markers. Hence, high efficiency of UGMS markers in assaying functional diversity in the transcribed component of the sugarcane genome makes them valuable for understanding diversity pattern, and variety identification. The commercial Indian sugarcane varieties used in this study were bred for either tropical or subtropical agro-climatic conditions with differential contribution of S. spontaneum, the most variable Saccharum species [14]. The random markers assaying genetic variation largely in different non-genic genomic regions that contribute to large genome size [31] would be of little relevance to phenotypic selection exercised during variety development. In contrast, the genic microsatellite markers assaying variation in the transcribed non-repetitive regions of the genome might be directly related to the phenotypic variation [25]. Our results thus suggested that the genetic base of the Indian sugarcane varieties is not very narrow particularly in the genic regions of the genome, which is most likely due to selection for wider adaptability of these varieties. The adaptive environment as well as parentage of these varieties corresponded well with the clustering pattern obtained using a small set of these markers, that further suggested the usefulness of the designed markers in realistic assessment of genetic diversity.
Conclusion
The present study identified microsatellites in sugarcane unigenes and assessed their functional relevance. A total of 961 primer-pairs were designed targeting 767 different unigenes carrying microsatellite repeats in the expressed component of the sugarcane genome, which would extend the accessibility of such microsatellite markers to researchers for many genetic studies in sugarcane. Precise allele sizing in automated fragment analysis system has encouraging implications to various high-throughput genotyping applications in sugarcane. Assessment of functional genetic diversity revealed that the genetic base of the Indian sugarcane varieties is not narrow.
Methods
Mining of microsatellites in the unigenes of sugarcane and assessment of their functional relevance
Fifteen thousand five hundred ninty-four sugarcane (Saccharum sp.) unigenes (Build 13.0, 19th Feb' 2008) comprising of 9.17 million base sequences were acquired in FASTA format from the recent NCBI FTP UniGene repository database of S. officinarum [32] in batches and searched for microsatellites using a perl scripting language based program MISA (MIcroSAtellite) [33] considering complementary sequence of repeat-motifs (like AG, GA, TC and CT) in the same class [18]. The identified microsatellites were characterized as perfect (monomers to hexamers repeated up to 100 times without any interruption), compound (non-interrupting) with at least two different repeat-motifs without any interruption, compound (interrupting) with a maximum of 100 nucleotides interposing two microsatellite repeat-motifs, class I (≥20 nucleotides) and class II (12 to 20 nucleotides) types. The microsatellite (excluding monomers) containing unigenes were used in a batch module for designing forward and reverse primers employing the microsatellite primer discovery tool of BatchPrimer3 [34]. All default options except for the optimum and maximum primer sizes of 22 and 24 nucleotides, respectively, were used. The putative functions for these unigenes were determined using NCBI BLASTX search tool [35] as against the nr-protein database. The BLASTX output was annotated into four functional categories viz, exact, putative, unknown and hypothetical, and then extracted to Excel sheets. Different structural components namely, coding and untranslated regions (5'UTRs and 3'UTRs) in the unigene sequences was determined using the software tools FGENESH [36] and UTRScan [37], respectively. The aminoacid sequences encoded by the predicted coding nucleotide regions of the microsatellite carrying unigenes were analyzed using the software Pfam [38] to determine the presence of functional domain/protein family within the UGMS. The aminoacid sequences of the functional domain carrying microsatellites was analyzed further using the I-TASSER automated web server [39,40] for prediction of three dimensional protein structure and active binding sites with ligands. Five different protein models and active binding sites were predicted at significant cut-off confidence (C ≥ -1.5) and binding site (BS ≥ 0.5) scores. The high quality protein model of correct topology and protein-ligand complex active binding sites was identified based on high C- and BS- scores.
Evaluation of polymorphic potential
To assess the amplification success rate of microsatellite markers designed from sugarcane unigenes, 176 primers were used to amplify one genotype of S. officinarum. Forty-seven, including thirty-eight class I and nine class II markers of these, labeled with 6-carboxyflourescein (6-FAM) dye phosphoramidites, were used to genotype one accession each belonging to five Saccharum species and three related genera, and twenty-eight commercial Indian tropical and sub-tropical sugarcane varieties (Table 3) for evaluating their polymorphic potential and assessment of genetic diversity. Five cereal species namely, rice, wheat, maize, Sorghum and barley were included in the experiment for comparison. Standard PCR constituents and cycling conditions except for annealing temperature, which varied from 55°C to 64°C depending on the primers, were used for PCR amplification. The amplified products were purified, mixed with 3.75 μl of MegaBACE formamide loading buffer and 0.25 μl of internal-lane MegaBACE™ET 550-R ROX size standard, denatured, cooled and resolved in automated MegaBACE 1000 DNA sequencer (Amersham Biosciences, Piscataway NJ, USA). The electropherogram containing trace files were analyzed and automated allele calling was carried out using the Binning Peak Post Processor tool of MegaBACE™Fragment Profiler Software Version 1.2 (Amersham Biosciences, Piscataway NJ, USA). The actual allele size (bp) was determined and fragment length polymorphism among the sugarcane genotypes and cereal species was identified. The average peak height and peak quality of alleles generated for all the UGMS loci were graphed and alleles exhibiting average peak height and quality of ≥ 1500 and ≥ 6.0 fluorescence units [41,42] respectively were considered for sizing and base-pair estimation. Allele binning was performed for individual UGMS loci for precise and accurate allele size determination and discrimination of homozygous and heterozygous allele types [42,43] for each genotype.
Table 3.
List of genotypes belonging to five cereal species and Saccharum complex used in the study
| Sl. No. | Genus and species | Clones/varieties | Parentage/origin | Region of adaptation |
|---|---|---|---|---|
| 1 | Hordeum vulgare | AK559 | Unknown | - |
| 2 | Triticum aestivum | Kalyansona | PJ"S" × GB 55 | - |
| 3 | Oryza sativa | IR64 | IR5857-33-2-1 × IR-2061-465-1-5-5 | - |
| 4 | Zea mays | KA509 | India | - |
| 5 | Sorghum bicolor | Pusa chari6 | India | - |
| 6 | Saccharum officinarum | IJ-76-3-1-9 | Indonesia | - |
| 7 | S. barberi | Mamjasahe | India | - |
| 8 | S. sinense | Malari | Unknown | - |
| 9 | S. robustum | IM-76-256 | Indonesia | - |
| 10 | S. spontaneum | 1151 | India | - |
| 11 | Narenga | NarengaER | Unknown | - |
| 12 | Sclerostachya | S1135 | Unknown | - |
| 13 | Erianthus | IK76-81 | Indonesia | - |
| 14 | Saccharum spp. | Co 1158 | Co 421 × Co 419 | Sub-tropical |
| 15 | Saccharum spp. | CoJ 64 | Co 976 × Co 617 | Sub-tropical |
| 16 | Saccharum spp. | CoS 88230 | Co 1148 × Co 775 | Sub-tropical |
| 17 | Saccharum spp. | Bo 91 | Bo 55 × Bo 43 | Sub-tropical |
| 18 | Saccharum spp. | CoS 8436 | MS 68/47 × Co 1148 | Sub-tropical |
| 19 | Saccharum spp. | Co 1148 | P 4383 × Co 301 | Sub-tropical |
| 20 | Saccharum spp. | CoPant 84212 | Co 1148 × Co 775 | Sub-tropical |
| 21 | Saccharum spp. | Co 87268 | Bo 91 × Co 62399 | Sub-tropical |
| 22 | Saccharum spp. | CoLk 8102 | Co 1158 GC | Sub-tropical |
| 23 | Saccharum spp. | Co 7717 | Co 419 × Co 775 | Sub-tropical |
| 24 | Saccharum spp. | Co 87263 | Co 312 × Co 6806 | Sub-tropical |
| 25 | Saccharum spp. | Co 89003 | Co 7314 × Co 775 | Sub-tropical |
| 26 | Saccharum spp. | CoPant 84211 | Co 6806 × Co 6912 | Sub-tropical |
| 27 | Saccharum spp. | Co 8347 | Co 419 × CoC 671 | Sub-tropical |
| 28 | Saccharum spp. | Co 7219 | Co 449 × Co 658 | Tropical |
| 29 | Saccharum spp. | Co 740 | P 3247 × P 4775 | Tropical |
| 30 | Saccharum spp. | Co 86010 | Co 740 × Co 7409 | Tropical |
| 31 | Saccharum spp. | Co 86032 | Co 62198 × CoC 671 | Tropical |
| 32 | Saccharum spp. | Co 85002 | Co 62198 × (-) | Tropical |
| 33 | Saccharum spp. | Co 8021 | Co 740 × Co 6806 | Tropical |
| 34 | Saccharum spp. | Co 8371 | Co 740 × Co 6806 | Tropical |
| 35 | Saccharum spp. | Co 6304 | Co 419 × Co 453 | Tropical |
| 36 | Saccharum spp. | Co 419 | PoJ 2878 × Co 290 | Tropical |
| 37 | Saccharum spp. | Co 62175 | Co 951 × Co 419 | Tropical |
| 38 | Saccharum spp. | Co 7704 | Co 740 × Co 6806 | Tropical |
| 39 | Saccharum spp. | Co 86249 | CoJ 64 × CoA 7601 | Tropical |
| 40 | Saccharum spp. | CoC 671 | Q 63 × Co 775 | Tropical |
| 41 | Saccharum spp. | Co 87025 | Co 7704 × Co 62198 | Tropical |
Variation in the fragment size (bp) of amplified alleles at each polymorphic UGMS marker locus was compared with changes in the number of microsatellite repeat-units at that target locus, and the occurrence of "stepwise" and "mixed" type of allele size distribution was inferred. When the allele size differences strictly corresponded to the variation in the number of repeat-units, it was considered as stepwise distribution. A mixed allele distribution was assumed when the allele size differences could partly be explained by the stepwise model. Multiple amplicons obtained by a primer-pair with peaks ≥ 1500 fluorescence units showing ≥ 100 bp allele size differences were binned into different loci. To confirm that the primers amplified the target microsatellite repeat motifs in different species and genera, the amplified products were purified using Micropon PCR purification kits (Millipore, Bedford, MA, USA) and sequenced two times in both forward and reverse directions using a capillary-based Automated DNA Sequencer (MegaBACE 1000, Amersham Biosciences, Piscataway NJ, USA). The trace files were base called, checked for quality and assembled into contigs [44]. The high quality sequences thus obtained were used for interspecies comparison using CLUSTALW multiple sequence alignment tool employing BIOEDIT software [45]. Ten size variant amplicons each of three microsatellite markers (UGSuM2, UGSuM26 and UGSuM27) showing both step-wise and mixed type of allele size distribution in sugarcane species, genera, varieties and cereals were eluted, purified, cloned in pGEM-T Easy Vector (Promega, USA) and sequenced as described above.
Assessment of functional genetic diversity
The polymorphic information content (PIC) was calculated using the formula, PIC = 1- ∑Pij2 [46], where Pij is the frequency of the jth allele for ith locus summed across all alleles for the locus. Cluster analysis among the 36 genotypes of Saccharum species, varieties and related genera was based on Nei and Li similarity coefficient [47] using the un-weighted pair group method (UPGMA) in PowerMarker Version 3.0 [48,49] software. The confidence limits of UPGMA based dendrogram was determined by 1000 bootstrap replicates and bootstrap of 50% majority rule consensus unrooted phylogenetic tree was constructed.
List of abbreviations
Sugarcane, Unigenes, Microsatellites, Functional genetic diversity
Authors' contributions
SKP conducted mining of UGMS, marker design, large-scale genotyping, polymorphism survey, functional diversity estimation and drafted the manuscript. AP was involved in genotyping and sequencing. KG and TRS participated in microsatellite mining and data analysis. PSS and NKS helped in data analyses, interpretation and drafting of the manuscript. TM designed the study, guided data analysis and interpretation, participated in drafting and correcting the manuscript and gave the final approval of the version to be published. All authors have read and approved the final manuscript.
Supplementary Material
Distribution of codon repeats along with the corresponding aminoacids in the unigenes of sugarcane.
Frequency and abundance of microsatellite repeat motifs and distribution of class I and class II motifs in the unigenes of sugarcane.
Primers designed from sugarcane unigenes carrying perfect microsatellite repeat-motifs along with the expected amplicon size and putative unigene function.
Primers designed from sugarcane unigenes carrying compound class I microsatellite repeat-motifs along with the expected amplicon size and putative unigene function.
Alignment showing the presence of class I UGMS repeat-motifs in sugarcane species, related genera and five cereal species.
In silico polymorphism of sugarcane UGMS loci at sequence level in five cereal species.
Functional annotation of 672 unigenes carrying microsatellites of sugarcane.
Primers targeting the microsatellite repeats present in the functional domains of proteins encoded by the unigenes.
Primers targeting the microsatellite repeats located within the functional domains of proteins encoded by sugarcane unigene sequences which gave single locus and step-wise allele amplification.
Alignment depicting the functional significance of sugarcane UGMS markers targeting the microsatellite repeat-motif (AG)n located within the protein kinase domain of unigene.
Contributor Information
Swarup K Parida, Email: swarupdbt@gmail.com.
Awadhesh Pandit, Email: awadheshpandit@gmail.com.
Kishor Gaikwad, Email: kish2012@yahoo.com.
Tilak R Sharma, Email: trsharma@nrcpb.org.
Prem Shankar Srivastava, Email: pss410@rediffmail.com.
Nagendra K Singh, Email: nksingh@nrcpb.org.
Trilochan Mohapatra, Email: tm@nrcpb.org.
Acknowledgements
The work presented in the manuscript was funded by the Department of Biotechnology (DBT), Government of India. We are thankful to the NCBI for making available their databases, Institute of Plant Genetics and Crop Research (IPK) for the availability of microsatellite search tool MISA and Dr. Athiappan Selvi, Sugarcane Breeding Institute, Coimbatore for providing the sugarcane genotypes.
References
- Grivet L, Arruda P. Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol. 2002;5:122–127. doi: 10.1016/S1369-5266(02)00234-0. [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Taylor GO, Henry RJ. Characterization of microsatellite markers from sugarcane (Saccharum spp.), a highly polyploid species. Plant Sci. 2000;155:161–168. doi: 10.1016/S0168-9452(00)00208-9. [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ. Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross-transferable to Erianthus and Sorghum. Plant Sci. 2001;160:1115–1123. doi: 10.1016/S0168-9452(01)00365-X. [DOI] [PubMed] [Google Scholar]
- Parida SK, Kalia SK, Kaul S, Dalal V, Hemaprabha G, Selvi A, Pandit A, Singh A, Gaikwad K, Sharma TR, Srivastava PS, Singh NK, Mohapatra T. Informative genomic microsatellite markers for efficient genotyping applications in sugarcane. Theor Appl Genet. 2009;118:327–338. doi: 10.1007/s00122-008-0902-4. [DOI] [PubMed] [Google Scholar]
- Pinto LR, Oliveira KM, Ulian EC, Garcia AA, De Souza AP. Survey in the sugarcane expressed sequence tag database (SUCEST) for simple sequence repeats. Genome. 2004;47:795–804. doi: 10.1139/g04-055. [DOI] [PubMed] [Google Scholar]
- Pinto LR, Oliveira KM, Marconi T, Garcia AAF, Ulian EC, deSouza AP. Characterization of novel sugarcane expressed sequence tag microsatellites and their comparison with genomic SSRs. Plant Breed. 2006;125:378–384. doi: 10.1111/j.1439-0523.2006.01227.x. [DOI] [Google Scholar]
- Oliveira KM, Pinto LR, Marconi TG, Mollinari M, Ulian EC, Chabregas SM, Falco MC, Burnquist W, Garcia AAF, Souza AP. Characterization of new polymorphic functional markers for sugarcane. Genome. 2009;52:191–209. doi: 10.1139/G08-105. [DOI] [PubMed] [Google Scholar]
- Oliveira KM, Pinto LR, Marconi TG, Margarido GRA, Pastina MM, Teixeira LHM, Figueira AV, Ulian EC, Garcia AAF, Souza AP. Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed. 2007;20:189–208. doi: 10.1007/s11032-007-9082-1. [DOI] [Google Scholar]
- Pontius JU, Wagner L, Schuler GD. UniGene: a unified view of the transcriptome. The NCBI Handbook. National Center for Biotechnology Information, Bethesda, Md; 2003. [Google Scholar]
- Thuljaram Rao J. In: Sugarcane varietal improvement. Naidu M, et al, editor. Sugarcane Breeding Institute, Coimbatore, India; 1987. Sugarcane origin, taxonomy, breeding and varieties. [Google Scholar]
- Daniels J, Roach BT. In: Sugarcane improvement through breeding. Heintz DJ, editor. Elsevier Press, Amsterdam; 1987. Taxonomy and evolution; pp. 7–84. [Google Scholar]
- Nair NV, Selvi A, Srinivasan TV, Pushpalatha KN. Molecular diversity in Indian sugarcane cultivars as revealed by randomly amplified DNA polymorphisms. Euphytica. 2002;127:219–225. doi: 10.1023/A:1020234428681. [DOI] [Google Scholar]
- Selvi A, Nair NV, Balasundaram N, Mohapatra T. Evaluation of maize microsatellite markers for genetic diversity analysis and fingerprinting in sugarcane. Genome. 2003;46:394–403. doi: 10.1139/g03-018. [DOI] [PubMed] [Google Scholar]
- Selvi A, Nair NV, Noyer JL, Singh NK, Balasundaram N, Bansal KC, Koundal KR, Mohapatra T. Genomic constitution and genetic relationship among the tropical and subtropical Indian sugarcane cultivars revealed by AFLP. Crop Sci. 2005;45:1750–1757. doi: 10.2135/cropsci2004.0528. [DOI] [Google Scholar]
- Selvi A, Nair NV, Noyer JL, Singh NK, Balasundaram N, Bansal KC, Koundal KR, Mohapatra T. AFLP analysis of the phenetic organization and genetic diversity in the sugarcane complex, Saccharum and Erianthus. Genet Resour Crop Evol. 2006;53:831–842. doi: 10.1007/s10722-004-6376-6. [DOI] [Google Scholar]
- Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2009;13:1–7. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang M, Tuskan GA. Characterization of microsatellites in the coding regions of Populus genome. Mol Breed. 2010.
- Parida SK, Dalal V, Singh NK, Mohapatra T. Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet. 2006;112:808–817. doi: 10.1007/s00122-005-0182-1. [DOI] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nature Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Temnykh S, Declerk G, Lukashover A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length-variation, transposon associations and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: Structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Yu J-K, Paik H, Choi JP, Han JH, Choe J-K, Hur C-G. Functional domain marker (FDM): an in silico demonstration in solanaceae using simple sequence repeats (SSRs) Plant Mol Biol. 2009;28:352–356. [Google Scholar]
- Rao AA, Sridhar GR, Mudunuri SB, Vamsidhar E, Kishore G. Computational analysis of mutations in colon cancer genes reveals a possible role of microsatellite in mutagenesis. J Proteomics Bioinform. 2008;S1:S041–S045. doi: 10.4172/jpb.s1000007. [DOI] [Google Scholar]
- Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends Biotech. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Parida SK, Dalal V, Singh AK, Singh NK, Mohapatra T. Genic non-coding microsatellites in the rice genome: characterization, marker design and use in assessing genetic and evolutionary relationships among domesticated groups. BMC Genomics. 2009;10:140. doi: 10.1186/1471-2164-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida SK, Yadava DK, Mohapatra T. Microsatellites in Brassica unigenes: Relative abundance, marker design and use in comparative physical mapping and genome analysis. Genome. 2010;53:55–67. doi: 10.1139/G09-084. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim E-D, Chen ZJ. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci. 2009;106:2295–2300. doi: 10.1073/pnas.0807350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. doi: 10.1023/A:1006392424384. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Mitchell SE, Kresovich S, Goodman M, Doebley J. Microsatellites in Zea - variability, patterns of mutations and use for evolutionary studies. Theor Appl Genet. 2002;104:436–450. doi: 10.1007/s001220100694. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidizaion predating divergence of the cereals and its consequences for comparative genomics. Proc Natl Acad Sci. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI FTP UniGene repository database of S. officinarum. ftp://ftp.ncbi.nih.gov/repository/UniGene/Saccharum_officinarum
- MIcroSAtellite identification tool. http://pgrc.ipk-gatersleben.de/misa
- BatchPrimer3 microsatellite primer designing tool. http://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi
- NCBI BLASTX search tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi
- FGENESH gene structure prediction program. http://linux1.softberry.com/berry.phtml
- UTRScan for prediction of 5' and 3' UTR functional elements. http://utrdb.ba.itb.cnr.it/tool/utrscan
- Pfam 24.0 protein families and functional domains database. http://pfam.sanger.ac.uk
- I-TASSER automated web server for protein structure and function predictions. http://zhanglab.ccmb.med.umich.edu/I-TASSER
- Zhang Y. I-TASSER server for protein 3 D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn JR, Temnykh SV, Paul EM, McCouch SR. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L.) Crop Sci. 2002;42:2092–2099. doi: 10.2135/cropsci2002.2092. [DOI] [Google Scholar]
- Pessoa-Filho M, Belo A, Alcochete AAN, Rangel PHN, Ferreira ME. A set of multiplex panels of microsatellite markers for rapid molecular characterization of rice accessions. BMC Plant Biol. 2007;21:7–23. doi: 10.1186/1471-2229-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair MW, Hedetale V, McCouch SR. Fluorescent-labeled microsatellite panels useful for detecting allelic diversity in cultivated rice (Oryza sativa L.) Theor Appl Genet. 2002;105:449–457. doi: 10.1007/s00122-002-0921-5. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base calling sequencer traces using Phred II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- BIOEDIT software. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PowerMarker version 3.0. http://www.powermarker.net
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of codon repeats along with the corresponding aminoacids in the unigenes of sugarcane.
Frequency and abundance of microsatellite repeat motifs and distribution of class I and class II motifs in the unigenes of sugarcane.
Primers designed from sugarcane unigenes carrying perfect microsatellite repeat-motifs along with the expected amplicon size and putative unigene function.
Primers designed from sugarcane unigenes carrying compound class I microsatellite repeat-motifs along with the expected amplicon size and putative unigene function.
Alignment showing the presence of class I UGMS repeat-motifs in sugarcane species, related genera and five cereal species.
In silico polymorphism of sugarcane UGMS loci at sequence level in five cereal species.
Functional annotation of 672 unigenes carrying microsatellites of sugarcane.
Primers targeting the microsatellite repeats present in the functional domains of proteins encoded by the unigenes.
Primers targeting the microsatellite repeats located within the functional domains of proteins encoded by sugarcane unigene sequences which gave single locus and step-wise allele amplification.
Alignment depicting the functional significance of sugarcane UGMS markers targeting the microsatellite repeat-motif (AG)n located within the protein kinase domain of unigene.