Abstract
We report development of a continuous scanning procedure and the use of a time delay integration (TDI) line scan camera for a light-sheet based microscope called a thin-sheet laser imaging microscope (TSLIM). TSLIM is an optimized version of a light-sheet fluorescent microscope that previously used a start/stop scanning procedure to move the specimen through the thinnest portion of a light-sheet and stitched the image columns together to produce a well-focused composite image. In this paper, hardware and software enhancements to TSLIM are described that allow for dual sided, dual illumination lasers, and continuous scanning of the specimen using either a full-frame CCD camera and a TDI line scan camera. These enhancements provided a ~70% reduction in the time required for composite image generation and a ~63% reduction in photobleaching of the specimen compared to the start/stop procedure.
OCIS codes: (180.0180) Microscopy, (220.0220) Optical design and fabrication, (170.3880) Medical and biological imaging, (180.2520) Fluorescence microscopy, (040.1490) Cameras, (180.6900) Three-dimensional microscopy
1. Introduction
A thin-sheet laser imaging microscope (TSLIM) is a device for nondestructive optical sectioning of organisms and thick tissues such as the mouse cochlea, zebrafish brain/inner ear and rat brain [1]. TSLIM is a modular device that can image transparent tissues such as zebrafish embryos or fixed and cleared transparent tissues that have been labeled with a fluorochrome. It is similar to other light sheet based microscopes in which specimens are illuminated by a light-sheet, and fluorescent tissue planes are recorded by a camera that is mounted orthogonal to the light-sheet. By moving in the z-axis, serial optical sections are obtained and three-dimensional (3D) reconstructions of structures can be generated for visualization and morphometry. However, since the light sheet is thinnest only in a narrow region (i.e., the beam waist) an x-axis scanning procedure was used to produce a well-focused composite image across the full width of the specimen. The original description of TSLIM [1] used a single green laser and a full-frame CCD camera in which the center 40 lines of the CCD array were collected using a start/stop procedure and an x-axis composite image was produced by stitching these image columns together. The present report describes hardware and software solutions that increase image quality and speed of optical sectioning, which also reduces photobleaching of the specimen. The use of a TDI line scan camera and our software and hardware features can also be used for other light-sheet based microscopes to reduce scanning times and specimen photobleaching.
Light-sheet microscopy was introduced into the current literature by Voie and colleagues [2,3] who used it to image ear tissues but were unaware of a much earlier publication in 1903 [4] describing light-sheet microscopy. It is technologically a relatively simple procedure whereby a thin sheet of light illuminates a plane in a transparent volume and that plane, or optical section, is observed orthogonal to the light-sheet. It has been used to image organisms in a plane of water [5] but its most common application is to optically section thick tissues that have been labeled with a fluorochrome and made chemically transparent. Light-sheet microscopes have been developed by a number of different investigators [1–8] and this technology is rapidly evolving. TSLIM [1] was developed by optimizing components of previous light-sheet microscopes and this report describes software and hardware enhancements to TSLIM that significantly improve its capabilities. Specifically, TSLIM now features a line scan camera in addition to the full-frame CCD camera, hardware triggering of the cameras, and software that allows for continuous scanning of the image in the x-axis.
2. Methods and results
The preferred way to produce a sheet of light is by using a cylindrical lens. In the present version of TSLIM a green solid-state laser with a wavelength of 532-nm (Optotronics, VAI50532) is used to produce a light-sheet and their light path is shown in Fig. 1 . The laser beam is expanded and collimated using a 10X Galilean beam expander (Edmund Optics, NT55-579) and passed through, an f = 50.8 mm cylindrical lens (Newport CKX525-C). The light-sheet is rotated 90 degrees by passing through the image side of an NA = 0.14 EO infinity-corrected long working distance 5X Plan Apochromat microscopic objective (Edmund Optics, NT59-876). The light-sheet is projected through the side of an open top, glass or quartz specimen chamber (Starna, 3G20; DLC300Q20). The specimen is attached to a Delrin® (DuPont) rod that is suspended in the center of the chamber which contains either a clearing solution for fixed and cleared specimens or an aqueous solution for live specimen imaging. The illuminated plane within the tissue specimen is observed perpendicular to the light-sheet using either a 1X or 2X objective lens (NA = 0.25, NA = 0.50) on a horizontally mounted, Olympus micro MVX10 microscope. Images are captured using either a full-frame CCD digital camera (Retiga 2000R) or a TDI line scan camera (Dalsa, Piranha HS-40-04K40).
Fig. 1.
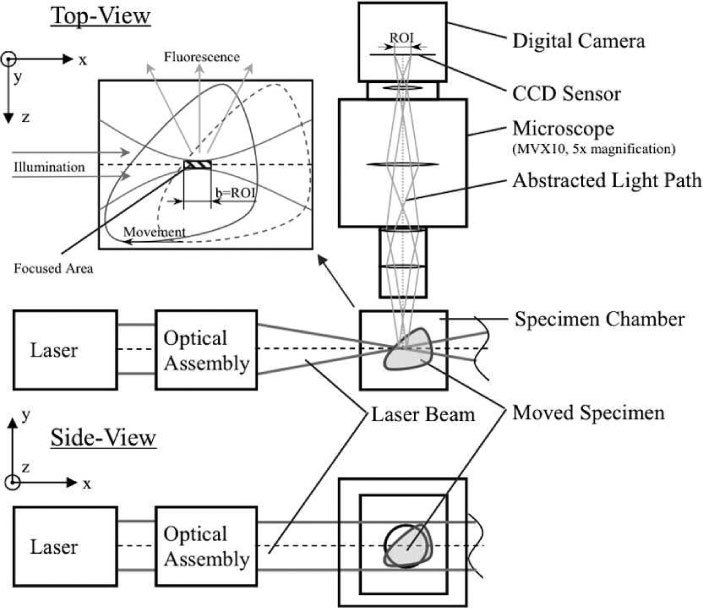
Top-view and side-view of TSLIM’s light-sheet pathway. A solid state laser beam is converted in an optical assembly (beam expander, cylindrical lens and microscope objective) and passes through the specimen chamber containing the tissue. The thinnest portion of the light-sheet is at focal point of the microscope objective. The illuminated plane in the tissue is collected by a microscope objective and recorded using a digital camera. Upper left panel: the specimen is moved through the beam waist to produce a well-focused imaged across the width of the specimen.
As can be seen in Fig. 1, the light-sheet is thinnest in only a small x-dimension at the focal point of the microscope objective. The thinnest region of a focused light-sheet is called the beam waist w0. The width of the area in which w0 increases to √2·w0 is called the Rayleigh range xR. Twice the Rayleigh range is called the confocal parameter b, an area of the beam where its thickness remains approximately constant. However, calculations pertaining to the parameters of the light-sheet were provided in a previous publication describing TSLIM [1].
A thicker light-sheet has a longer confocal parameter but a thicker light sheet decreases the ability to resolve fine structures due to light scattering. Thus, producing thinner light-sheets is desirable, but comes at the cost of a shorter confocal parameter and focal point [1]. If the width of the tissue fits within the confocal parameter, then a well-focused image is observed. However, if the width of the tissue is greater than the confocal parameter, then it does not appear to be well focused across its width. The region of the confocal parameter can also be referred to as the region of interest (ROI) within the tissue.
2.1 Start stop procedure (SSP)
Since the thinnest portion of the light-sheet covers only a small region of a large tissue to be imaged, either the specimen or the beam needs to be moved to produce a well-focused image across the width of the specimen. For simplicity we fixed the light-sheet and its beam waist at the focal plane of the Olympus microscope and moved the specimen in the x,y,z-axes through the light-sheet using Newport 423 linear stages and three Newport motors. This was similar to a procedure developed by Buytaert and Dirckx [8]. Figure 1 shows (in the upper left panel) TSLIM’s original start/stop procedure that was used to move the specimen in the x-direction and to collect columns of image data whose width are less than or equal to the confocal parameter of the light-sheet.
Since great accuracy and speed is not necessary in the y-axis, this direction is manually controlled by a Newport NSC200, which drives an open-loop Newport CMA-25PP motor with a bi-directional repeatability of 4 µm and velocity of 0.4 mm/s. However, in the x- and z-axes, Newport’s computer guided ESP301 motion controller in combination with Newport’s LTA-HS closed loop servomotors (with a typical bi-directional repeatability of 0.6 µm and velocity of 5 mm/s) are used to position the specimen. A LabVIEW program coordinates image acquisition and motor positioning simultaneously. The image acquisition routine consists of grabbing a column of image data from the camera and moving the specimen the width of that column and then collecting another column. These columns are stitched together to form a composite image across the full width of the specimen.
Figure 2(a) shows the ROI (i.e., the confocal paramater) in an image of the mouse cochlea. The Rayleigh range xR was calculated [1] with the 5X Plan Apo microscopic objective to approximately 34 µm and thus the confocal parameter b is 68 µm. To the left and the right of the ROI, the image appears out-of-focus due to greater beam thickness and light scattering. In addition, a blur is present in the lower left of the image, which is due to the presence of a high density structure (i.e. bone) in the cochlea that lies between the optical section and collecting lens of the Olympus microscope. However, Fig. 2(b) shows that stitching of multiple ROIs across the width of the specimen, results in improved focus of specimen across its width. Higher magnification of these stitched image columns [Fig. 2(c)] reveals some stitching artifacts due to image column misalignments.
Fig. 2.
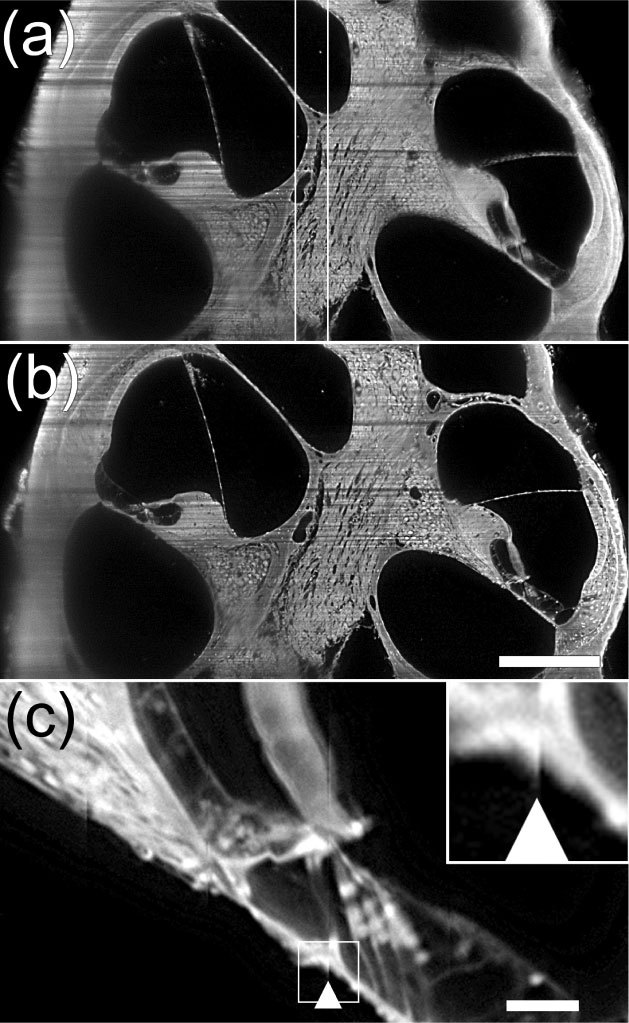
Optical section of a mouse cochlea before and after stitching. (a). Double white lines indicate the confocal region of the light-sheet where the specimen is in focus. Note that to the left and right of this region the tissue appears to be out-of-focus due to a thicker beam. (b). After stitching of image columns focus appears improved across the width of the specimen. Bar = 250 µm for panels a,b. (c). A higher magnification of 2b (above scale bar) showing a stitching artifact (arrowhead) and two less noticeable artifacts to the left of the arrowhead. Insert in upper left is a 3X magnification of the region outlined in main Fig. and shows the stitching artifact in detail. Bar = 50 µm.
In order to produce a stitched image without columns artifacts like shown in Fig. 2(c), it is crucial to find a relationship between the moved distance m (μm) and the strip width wROI (pixel). Since the magnification M of the microscope is the factor between its image and the original, it is also the major parameter in the relationship between m and wROI. But because m is defined in μm and wROI in pixels, the correction factor p, defined as the pixel size of the camera, is also included in Eq. (1).
| (1) |
To ensure that the stitched photos consist of completely focused strips, m is chosen smaller than the confocal parameter of 68 μm. Thus, a typical wROI is 40 pixels or 59.2 μm with a 5X zoom and the Retiga 2000R camera (pixel size p2000R = 7.4 μm). To produce a picture of 1200 pixels with 40 pixels per column, it takes ~60 seconds and 30 cycles of starting/stopping the servomotor to produce a composite image using a 15mW green laser. To optically section a whole mouse cochlea with approximate dimensions of 2 x 1.5 mm it took up to 5 hrs and 350 images with a 5 μm z-step size (distance between the pictures in z-direction). A consequence of this prolonged light exposure is a loss of fluorescence intensity in the specimen, i.e, photobleaching. This effect is increased by subsequent sections of the specimen that are exposed to the beam before being collected since the unfocused area of the beam is significantly larger than the z-step size. Our previous measurements indicated a loss of approximately 8.85% in fluorescence intensity to complete one composite image [1]. Although the effect is much less than wide-field or confocal microscopy it is detrimental and decreases the signal to noise ratio, contrast, and picture quality.
In addition, artifacts occur in the image due to misalignment of the pixel array of the camera to the direction of movement by the servomotors as well as position deviations. The latter is due to a difference between the acquired slice width of 40 pixels (59.2 μm) and the distance moved by the motor, which exceeds the size of one pixel. Hence the ROI needs to include overlapping pixels to minimize the stitching artifacts. However, that procedure requires a significant image processing and the overlapping pixels are still visible in the final image. That is why the strip width wROI has to be reduced in order to reduce the influence of that distance and improve the image acquisition. This strategy will also reduce the effect of the misalignment of the pixel array of the camera chip to the moving direction, which makes it even more crucial to consider this requirement for an improvement of the image acquisition. A similar optimization strategy for improving the image quality by reducing the strip width wROI was previously described by Buytaert and Dirckx [8].
2.2 Hardware mastered scan procedure (HMSP)
Since the specimen was already being moved, we decreased the image collection time by implementing a continuous x-axis scanning procedure. A slice could be acquired “on the fly” without starting and stopping the mechanics, resulting in a significant time savings and reducing photobleaching of the specimen. However, such a procedure required accurate timing between the motor position and the image acquisition. A TTL trigger pulse was generated for the camera by converting and dividing the encoder signals from the Newport LTA-HS servomotor. A bypass card was used to break the appropriate signal lines of the encoder out of the servo-controller connection. The RS 422 signal was then converted to a TTL signal using a Microbotics Inc SLC10422-TTL converter. TTL signal data were then acquired by a National Instruments PCI-6221 DAQmx board with an SCB-68 connector block. The board is equipped with two counters, which can be armed with different counting, timing or generating tasks. One counter is used to count the incoming encoder signals and output a TTL impulse after a certain amount of encoder ticks.
The relationship i between the input and output of ticks depends on the zoom M, the resolution of the encoder si, the pixel size of the camera p and the strip width wROI by Eq. (2).
| (2) |
The resolution of the encoder si was determined by moving the motor 1 mm and counting the encoder signals. One encoder impulse for the Newport LTA-HS is equivalent to si = 0.14 μm which is important for the calculation of the relationship i. For example, the Retiga 2000R at 5x magnification p/M = 1.48 µm/pixel and when wROI = 9 lines, an output signal has to be generated after every 94 input ticks of the encoder. This signal goes directly over the digital output via the SCB-68 connector block into the optic-coupled input of the camera where it initiates an exposure of the chip. The pictures are picked up on demand by a loop in the controlling LabVIEW program. The whole concept for a hardware triggered system is summarized in Fig. 3 .
Fig. 3.
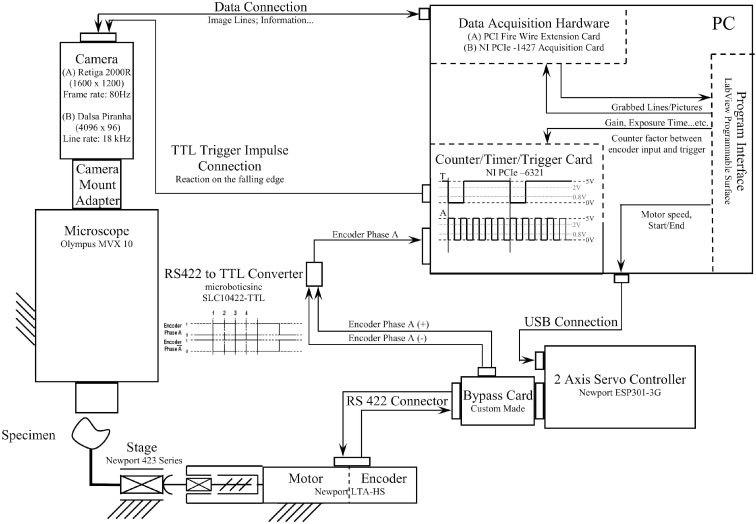
Hardware triggering and scanning with both, the Retiga 2000R and the Dalsa HS-40-04K40 camera. The encoder signal of Newport’s LTA-HS servomotor in x-direction of the ESP 301 motion controller is bypassed over a custom made card and an RS422-to-TTL converter and goes to a National Instrument NI PCIe – 6321 Counter/Timer/Trigger Card. This card is programmed to divide the rising edge of the received signal by a constant factor and the result is an output signal. The signal triggers the Retiga 2000R and the Dalsa HS-40-04K40 by a connection to the camera’s trigger input channel.
With this system, acquisition is triggered by a hardware signal generated by the motor position. That means that there is a constant time gap between the arrival of the specimen at the desired position and the exposure of the camera, which makes the system real-time capable. This prevents artifacts from being produced by asynchronies between the software acquisition loop and the hardware motor position during the scan. One significant improvement of this system was that the produced composite images were almost completely free from the previous stitching artifacts using the start/stop procedure. Only very rarely did an artifact line appear due to a slight movement of the specimen in-between two strips. These artifacts were rare and minimal as long as the slice width was small (roughly under 10 lines). However, since Eq. (2) usually does not return an integer result for the relationship i between the input and output of ticks, any result has to be rounded, which can lead to significant deviations with respect to the entire image since this deviation is compounded over the whole size of the image. Since a 3D reconstruction is used to measure volumes, areas and lengths of certain parts of the specimen, we did not want to exceed a ± 1% limit regarding the accuracy of the images.
To compensate for the integer problem of the parameter i, the abilities of the NI PCIe - 6321 data acquisition card were used. The resolution was raised by increasing the number of steps per slice i, to minimize the rounding error over the whole image. Since the counters on the card can use either the rising or the falling edge, the signal contains more information than only the rising edge that is used for dividing the signal.
This information can be used to increase the resolution of i by leading the incoming signal from the encoders to a digital input, which created an event on both the falling and the rising edges. A counter generates on each event a signal of two periods with a frequency that is four times higher than the one that comes from the encoder (because it is triggered on the falling and rising edge of the encoder). This frequency can be calculated over the speed that is given on the motor. The signal is counted on the rising edge by the second counter to create an output signal that is used to trigger the camera. The resolution was then four times higher than before and this increased the accuracy and created a pattern that does not exceed the ± 1% limit over the whole magnification range.
The scanning speed is now only limited by the frame rate of the camera and the motion blur, which is caused by an exposure time that exceeds the time of the specimen’s movement of one pixel. The Retiga 2000R is capable of 100 frames per second (10 ms per picture for 5 ms exposure time) but for the calculation of the speed, only 80 fps are used (20 fps conservatively). It can be converted into a speed limit by using the magnification M and the number of lines wROI, according to Eq. (3).
| (3) |
Furthermore the motion blur time limit depends on exposure time tE itself, the magnification M, and the pixel size p, according to Eq. (4).
| (4) |
Since pixel size p is a constant parameter given by the camera, Eq. (3) and (4) show, that the maximum speed is either limited by the number of lines per slice or the exposure time for certain magnification configurations. That is why both have to be considered to realize an optimized scan speed. The acquisition of one strip, with a width of 5 pixels and an exposure time of 5 ms, was accomplished in 30 to 40 ms. This procedure allowed us to scan 1200 pixels in 7.2 sec according to Eq. (5).
| (5) |
To allow time for building the image, the acquisition was only feasible in one direction. That is why the total scan time with stitching was about 15 sec per image, which is faster than the scan time reported by Buytaert and Dirckx [8] using 200 pixel lines. This procedure reduced total stack collection time from 3.5 to 5 hr for the start/stop method to 1.5 hr for the continuous scanning method, which was a significant improvement.
The speed and resolution is only limited by the camera frame rate and exposure time and the system has the ability to trigger the camera at an appropriate speed without requiring properties that cannot be fulfilled by a line scan camera. Thus, that system is now suitable to be combined with a line scan camera with a comparable pixel size to the one of the Retiga 2000R.
2.3 Implementation of a Dalsa TDI line scan camera
With HMSP and its hardware solution, the system had the ability to supply a line scan camera with an appropriate trigger signal to achieve a fast and reliable scan. There were only two questions that had to be addressed with regard to using a line scan camera over a full-frame CCD camera; 1) could the confocal parameter be localized within the optical section and adjusted without a full-frame image, and 2), was the line scan camera sensitive enough for low light, fluorescent imaging?
To solve the first problem, that was already identified as crucial in the Buytaert and Dirckx [8] paper, we selected a line scan camera with a high resolution in one dimension and more than one line in the other. The best solution was Dalsa with its High Sensitivity TDI Line Scan Cameras because of its Area and TDI Modes. Area Mode (reading out the entire chip in one exposure) was important for stack alignment and localization of the confocal parameter within the optical section because it allows the user to view a 96 x 4096 pixel picture. TDI Mode solved the second problem of low-light imaging.
Instead of exposing the chip only once to the object as in a regular line scan camera, the TDI Mode allows for the integration of multiple exposures of the same, moving object. That means the entire chip of 16 to 96 lines is exposed to the object but only the last line of the camera is read out. The other ones are waiting to be accumulated with their following lines after the next exposure. The effect is that the integration time is spread out over several (depending on the available number of lines) short exposures, which increases the speed significantly for low light applications.
A TDI line scan camera has several requirements. First, high accuracy is required for the trigger signal, since the object has to be exactly one pixel size shifted for the next exposure compared to the last one. An altering trigger signal would cause an exposure at the wrong position of the specimen, which would lead to a blurry image. Second, the trigger signal has to be provided with a higher frequency because the integration time is spread out over 16 to 96 exposures. But with the HMSP we are able to provide both, an accurate trigger signal and a high frequency, which made this camera very suitable for TSLIM.
A third consideration is that the confocal parameter cannot be reduced to the width of one pixel as with a regular line scan camera, since at least 16 lines are involved in the picture acquisition. However, the combination of the wide field of 4096 pixels and a 10X magnification of the Olympus microscope results in a wider field of the specimen that is available in a single frame from the line scan camera [Fig. 4(c) ] compared to the Retiga 2000R. But the advantage of more pixels can also be used to increase the magnification by a factor of ~2.5 X compared to the Retiga 2000R image. The resolution with the Dalsa camera increases while maintaining the same field of view as the Retiga 2000R. This is demonstrated in Fig. 4ab using a raw, unprocessed image of Claudius cells/basilar membrane taken from a low power image of the entire cochlea at a 5X magnification with the Retiga 2000R in Fig. 4(a) compared with a 10X magnification and the Dalsa in Fig. 4(b) (magnification increased by the factor of 2 leads to a comparable picture size). According to Eq. (1) we achieved a higher resolution and a lower confocal parameter limit of 11.2 µm with that system, compared to 59.2 µm for the Retiga, including a pixel size p = 7 µm for the Dalsa camera. Thus the confocal parameter limit was reduced to a significantly smaller confocal parameter than with a regular CCD camera while not only retaining but increasing the scanning speed.
Fig. 4.
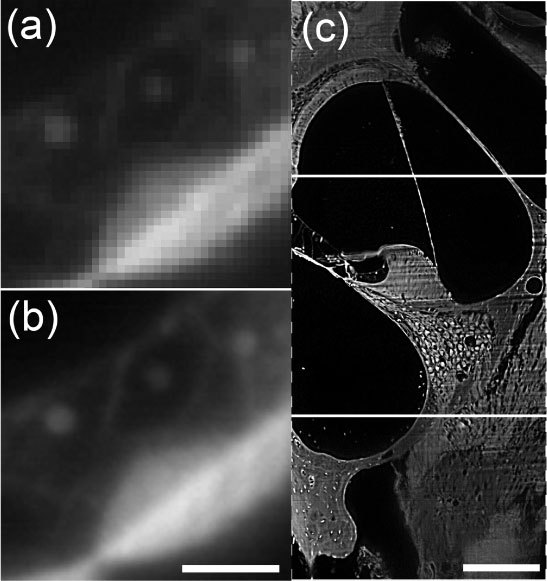
Comparison between full frame (a) and line scan camera (b) with a higher magnification. Due to increased resolution by the line scan camera the image can be enlarged more without showing pixelation. Bar = 20µm. (c) Comparison between size of a Retiga CCD image in the center panel with the image using the Dalsa camera (full image) at the same magnification. The Dalsa camera allows for collection of a larger view of the specimen. The dashed lines indicate that the width of the image is not limited because it can be scanned in that dimension. Bar = 150µm.
We chose the HS-40-04K40 model for TSLIM because it balances a high vertical resolution of 4096 pixels with a sensitive 7 μm pixel size in a chip size that is still compatible with the MVX10 Microscope. Since the pixel size is different from the Retiga 2000R camera, the accuracy had to be recalculated, but remains close to the ± 1% limit. The Dalsa camera is a medium configuration Camera Link camera with 36 kHz readout speed connected to a NI PCIe-1427 Camera Link card, as shown in Fig. 3. This card only allowed us to run the camera in base configuration with 18 kHz readout speed, which was adequate given our low light conditions.
Since a line scan camera exposes the frame over the entire time between two trigger signals, rather than only exposing it over the exposure time, no time is lost between acquisitions. Combined with the technology of the TDI mode, it takes approximately 5 sec to scan a full-frame, high resolution image (4096x3000 pixels) of the entire cochlea with a 10x zoom and 32 integrations without any blur adjustments. That is not only significantly faster than the HROPFOS [8] scan but it also allows for greater enlargement of the image without pixelation.
Due to a decrease in the time that it takes to collect a composite image by continuous scanning, photobleaching of the fluorochrome within a specimen should also be reduced. To test that hypothesis, three 100 pixel square, similar homogenous regions (i.e., compact bone) within the mouse cochlea (prepared as described in previous publication [1]) at three different z-stack locations were imaged repeatedly for 50 times and the intensity measured after each composite was produced by the three methods described in this paper (start/stop, continuous CCD scan, and continuous line scan). Figure 5 shows that generation of 50 composite images that took 1550, 750, and 475 sec for each method respectively and fluorescence intensity was retained by 65, 80, 87% respectively. Thus, the line scan camera provided a ~70% reduction in the time required for composite image generation and a ~63% reduction (35%-13%/35%). in photobleaching of the specimen.
Fig. 5.
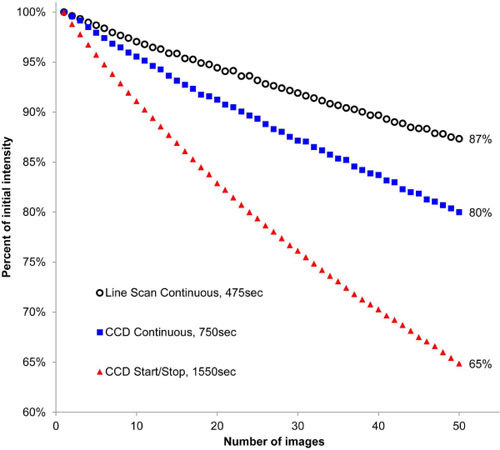
Photobleaching of a specimen following 50 repeated imaging of the same plane of an optical section. The start/stop procedure using the CCD camera takes 1550 sec to image 50 sections and retains 65% of the fluorescence. A continuous scan using the CCD takes only 750 sec and retains 80% of the fluorescence. A continuous scan with the line scan camera takes only 475 sec and retains 87% of the fluorescence making it the best choice for TSLIM imaging.
4. Conclusions
As far as we know this is the first description of the implementation of a line scan camera with the TDI technology for light-sheet microscopy. The Dalsa camera is well suited for imaging fluorescent specimens using scanned, thin-sheet laser microscopy. The TDI mode of the camera offers high sensitivity and sophisticated imaging technology to not only provide high resolution and low noise images, but also a fast scanning speed. This leads to a more efficient collection of the fluorescence and less photobleaching of the specimen. In the area mode, it also allows for alignment of the specimen and localization of the confocal region within the optical section. TSLIM has been enhanced with hardware and software additions that provide for high quality, rapid scans of large tissues such as the mouse cochlea. Due to the increased number of pixels in the line scan camera it is possible to image larger specimens within a single frame and to discriminate fine details in smaller specimens thanks to higher pixel density. This is important when one wants to contain the full height and width of a tissue or specimen within one picture of an image stack. In addition, TSLIM also allows us to decrease the beam waist and confocal parameter (above a limit of 11.2µm) and thus reveal finer subcellular structures. These enhancements to TSLIM makes it well suited for producing high resolution, optical sections of large tissues.
Acknowledgements
This work was supported by the National Institutes of Deafness and Communication Disorders (NIDCD) grant (RO1DC007588-04) and American Recovery and Reinvestment funds (RO1DC007588-03S1) and by the Capita Foundation (La Mesa, CA). This research was conducted while Peter Schacht was a visiting fellow at the University of Minnesota. He performed the engineering aspects of the project and wrote the manuscript. He is currently an engineering graduate student at the Technische Universität Ilmenau and received a grant to study at the University of Minnesota. Shane B. Johnson is a junior scientist in the Santi laboratory and performed the histological aspects of this study and wrote the manuscript. Peter A. Santi is a professor of Otolaryngology and Neuroscience at the University of Minnesota. He wrote grants that funded this research, proposed and supervised the work, and wrote the manuscript. The authors would like to thank Matthias Hillenbrand for critically reading of the manuscript and for his useful comments. The animal care and use for this study was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Minnesota.
References and links
- 1.Santi P. A., Johnson S. B., Hillenbrand M., GrandPre P. Z., Glass T. J., Leger J. R., “Thin-sheet laser imaging microscopy for optical sectioning of thick tissues,” Biotechniques 46(4), 287–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voie A. H., Burns D. H., Spelman F. A., “Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens,” J. Microsc. 170(Pt 3), 229–236 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Voie A. H., “Imaging the intact guinea pig tympanic bulla by orthogonal-plane fluorescence optical sectioning microscopy,” Hear. Res. 171(1-2), 119–128 (2002). 10.1016/S0378-5955(02)00493-8 [DOI] [PubMed] [Google Scholar]
- 4.Siedentopf H., Zsigmondy R., “Über Sichtbarmachung und Groessenbestimmung ultramikroskopischer Teilchen, mit besonderer Anwendung auf Goldrubinglaesern,” Annalen der Physik 10, 1–39 (1903). [Google Scholar]
- 5.Fuchs E., Jaffe J. S., Long R. A., Azam F., “Thin laser light sheet microscope for microbial oceanography,” Opt. Express 10(2), 145–154 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Huisken J., Swoger J., Del Bene F., Wittbrodt J., Stelzer E. H., “Optical sectioning deep inside live embryos by selective plane illumination microscopy,” Science 305(5686), 1007–1009 (2004). 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- 7.Dodt H.-U., Leischner U., Schierloh A., Jährling N., Mauch C. P., Deininger K., Deussing J. M., Eder M., Zieglgänsberger W., Becker K., “Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain,” Nat. Methods 4(4), 331–336 (2007). 10.1038/nmeth1036 [DOI] [PubMed] [Google Scholar]
- 8.Buytaert J. A. N., Dirckx J. J. J., “Design and quantitative resolution measurements of an optical virtual sectioning three-dimensional imaging technique for biomedical specimens, featuring two-micrometer slicing resolution,” J. Biomed. Opt. 12(1), 014039 (2007). 10.1117/1.2671712 [DOI] [PubMed] [Google Scholar]


