Abstract
We present a simple approach in electrophoretic DNA separation and fluorescent monitoring that allows to identify the insertion or deletion of base-pairs in DNA probe molecules from genetic samples, and to perform intrinsic calibration/referencing for highly accurate DNA analysis. The principle is based on dual-point, dual-wavelength laser-induced fluorescence excitation using one or two excitation windows at the intersection of integrated waveguides and microfluidic channels in an optofluidic chip and a single, color-blind photodetector, resulting in a limit of detection of ~200 pM for single-end-labeled DNA molecules. The approach using a single excitation window is demonstrated experimentally, while the option exploiting two excitation windows is proposed theoretically.
OCIS codes: (260.2510) Fluorescence, (280.1415); Optical sensing and sensors, (130.2755) Glass waveguides
1. Introduction
Lab-on-a-chip (LOC) [1] systems have become increasingly popular for DNA analysis. In microchip capillary electrophoresis (MCE) [2] labeled DNA molecules are separated in a microfluidic (MF) channel. Typically, the molecules are monitored by laser-induced fluorescence (LIF) [3] either in a confocal setting using bulky bench-top optics [4] or in an integrated optofluidic approach [5]. To enhance analysis capabilities, multi-wavelength fluorescence sensing has been implemented, mostly in bulk capillaries [6], e.g. by use of a broadband light source such as a Xe lamp and extracting the multiple wavelengths with monochromators and complex optical schemes to achieve an unambiguous separation of different wavelengths. Other reports include the use of multiple photodiodes [7], external wavelength selective gratings [8], color CCD arrays [9], or external spectrum analyzers [10].
In several fundamentally important diagnostic applications only two independent samples, each consisting of a number of differently sized DNA molecules, have to be monitored simultaneously, hence only two excitation beams and detection of fluorescence signals at merely two wavelengths are required. These applications include, firstly, internal calibration of the set-up during a MCE experiment by adding to the sample under investigation a reference sample with several DNA molecules of known base-pair sizes, thereby making the experiment insensitive to environmental influences such as operating temperature, condition of the sieving gel matrix and inner wall coating of the MF channels, or changes of the applied electric field. Secondly, an unknown, potentially malign sample exhibiting single base-pair insertions or deletions can be separated together with its healthy counterpart, thereby providing unprecedented sizing accuracy to the experiment.
In this paper we propose a simple method for spatial and wavelength duplexing, which applies dual-point, dual-wavelength detection from either one or two detection windows (DWs). It does not require any external apparatus to separate the wavelengths, thereby allowing for fluorescence detection with a single, ultrasensitive photomultiplier tube (PMT), which enables an ultra-low limit of detection. After introducing the set-up used for on-chip DNA separation and fluorescence detection in Section 2, we describe two applications, namely detection of single base-pair insertion/deletion in genetic samples through a single DW, which we demonstrate experimentally in Section 3, and internal calibration of DNA separation through two DWs, which we propose theoretically in Section 4.
2. Experimental approach to highly accurate dual-wavelength fluorescent DNA analysis
Optofluidic chips were fabricated in a two-step procedure. Firstly, the MF channel network and MF reservoirs were patterned photolithographically and wet-etched into fused silica glass and then sealed off by bonding another piece of fused silica glass on top. This commercially mass-produced chip (LioniX BV) has dimensions of 55 mm width × 5.5 mm depth × 1 mm height and the MF channels have a cross section of ~110 µm depth and ~50 µm height. In a second step, we applied femtosecond-laser micromachining to post-process optical waveguides (WGs) into the bulk of such LOCs on a flexible, chip-by-chip basis [11] for integrated fluorescence excitation of DNA molecules [12]. An elliptical WG cross section was obtained, with a height of ~50 µm, in order to excite the maximum possible volume of the MF channel, while its width is ~12 µm in order to retain a high spatial resolution along the direction of DNA flow and separation. Single-mode optical fibers carrying the excitation laser light were butt-coupled to the end-facets of WGs in the optofluidic chip with coupling losses < 2 dB. The WG propagation losses were in the range of 0.5–0.9 dB/cm at the wavelength of 543 nm [11].
The lay-out of our optofluidic chip is presented in Fig. 1 . Each of the two DWs consists of a region in which two WGs carrying two different excitation wavelengths intersect the MCE separation channel in plane at two nearby locations. From the MF crossing junction, at which the separation commences, the four WGs are distanced by 2.0 cm and 2.1 cm (WG 1 and WG 2 within DW1, respectively) as well as 3.5 cm and 3.6 cm (WG 3 and WG 4 within DW2, respectively). Depending on the application (see Sections 3 and 4) only a single DW or both DWs can be used. In the latter case, the order of the two excitation wavelengths is swapped between the two DWs to ensure unambiguous analysis.
Fig. 1.
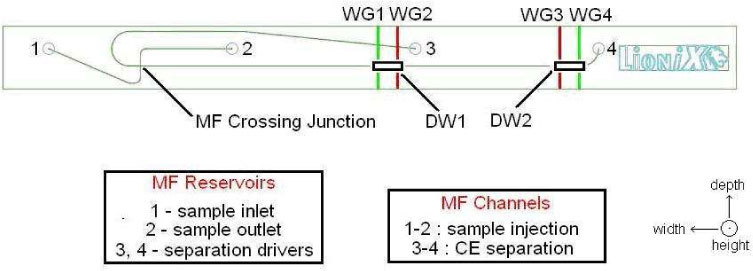
Layout of the optofluidic chip indicating the MF reservoirs, MF channels, and the two DWs, each comprising two WGs crossing the MCE separation channel perpendicularly in plane
Prior to the experimental runs the inner walls of the MF channel network were coated with a polymer to suppress the electro-osmotic flow and minimize adsorption of DNA molecules on the MF channel wall [13]. Subsequently, the channels were filled with a sieving gel matrix consisting of hydroxypropyl-cellulose [14] to maximize the resolution of the DNA CE separation. The reagents were sterilized, filtered, and stored at 269 K until their use. The CE sample loading and separation protocol was based on actuation voltages of up to 1.5 kV, provided by a MF control system (CapiliX BV) and delivered via Pt electrodes integrated into the MF reservoirs. Application of a high voltage forced the negatively charged DNA molecules to migrate into the CE injection channel from sample inlet reservoir 1 to sample outlet reservoir 2. By switching the voltages at all four reservoirs simultaneously a well-confined plug of DNA molecules – with a volume of ~605 picoliters at the crossing junction of the two MF channels – was injected into the CE separation channel toward separation driver reservoir 4. The electric field applied to the CE separation channel was ~180 V/cm and the corresponding speed of the DNA molecules was ~320 μm/s. During their migration along the CE separation channel the DNA molecules contained in the plug volume were separated according to their size.
A cooled PMT (H7421-40, Hamamatsu Photonics K.K.) was built onto the output port of an inverted microscope (DMI5000M, Leica Microsystems GmbH) with the objective lens (10 × , NA = 0.25) aligned to collect light from either of the two DWs. In the experiments presented in Section 3 we used DW2. Through a butt-coupled fiber-array unit, 0.9 mW of power from the 543-nm and 633-nm lines of a green and a red He-Ne laser were coupled into WG3 and WG4, respectively. We estimate ~0.4 mW of power from each laser to be incident on the MF channel. Upon laser excitation of fluorescently labeled, migrating DNA molecules in the CE separation channel through such a WG, a sharp fluorescent segment 12 µm in width is observed along the intersection of WG and MF channel [12]. An appropriate multi-band filter (XF57, Omega Optical, Inc.) ensured that only the desired fluorescence signals reached the PMT. With this setup, separation of a set of DNA molecules in the diagnostically relevant size range (150-1000 bp) and investigation of the migration times of the individual molecules as a function of their a priori known sizes resulted in a mean square deviation from the expected behavior of < 1% – corresponding to a relative sizing accuracy of > 99% [15]. A limit of detection of 2.1 pM was estimated with intercalating dyes as fluorescent labels [15].
3. Monitoring of single base-pair insertion / deletion through a single DW
Dual-point, dual-wavelength fluorescence monitoring is valuable in diagnostic applications to compare an unknown sample, e.g. from a gene region under investigation for base-pair insertion / deletion, with a reference sample, e.g. from the same gene region corresponding to a healthy person. The base-pair difference between the corresponding analyte DNA molecules would be small (1-2 bp), if any, thus challenging the resolution capabilities of standard MCE systems. Here we propose to determine these differences by exclusively labeling the two samples, flowing them against each other, and monitoring their separation with dual-point, dual-wavelength fluorescence excitation / detection through a single DW. If no insertion / deletion is present, the two species migrate with the same speed (with an error of < 1%), but will nevertheless be optically separated due to the spatial separation between the two excitation WGs in the DW.
While diagnostically relevant genetic probes are usually double-stranded DNA molecules sized >100 bp, clinically produced by a polymerase chain reaction, for this proof of principle we used commercially produced (Molecular Probes, Inc.) single-stranded DNA primer molecules sized 19 and 20 nucleotides (nt), end-labeled with either Alexa Fluor 647 (AF647) or Cyanine 3 (Cy3). These two fluorophores can be selectively excited in the red (633 nm, coupled through WG3) and in the green (543 nm, coupled through WG4), respectively.
Initially, only a single species (19-nt-AF647) was injected in the separation channel, with either the red or green He-Ne laser turned on. In the former case a strong signal peak was observed when the fluorescently labeled DNA plug crossed the excitation WG, while in the latter case the measured signal equaled the PMT baseline signal. This procedure was repeated for the same molecule labeled with the other dye (19-nt-Cy3), leading to the complementary result (i.e., strong signal peak with the green He-Ne laser and no signal with the red He-Ne laser). This confirmed that cross-excitation between the two fluorophores was, for all practical purposes, absent. Based on the signal-to-noise ratio of the electropherograms and the known concentrations of single-end-labeled DNA molecules in the plugs, we estimate for this experiment a limit of detection of ~200 pM. The limit of detection achieved earlier (2.1 pM) is valid for DNA molecules with intercalating dyes that attach at numerous positions on the same molecule, leading to a much larger effective fluorescence intensity.
The two differently labeled, but equally sized DNA molecules were then mixed and the resulting polychromatic mixture was resolved into the individual monochromatic DNA components by dual-point, dual wavelength excitation in the MCE separation channel. Figure 2(a) depicts the corresponding electropherogram. Since cross-excitation is absent, the two peaks appearing after a migration time of ~108-110 s and separated by ~1.8 s correspond to the two species, each excited by the corresponding laser wavelength of 633 nm or 543 nm coupled into the adjacent WGs. This is further confirmed by repeating the MCE separation of the two molecules twice, alternately turning off one of the lasers. The corresponding electropherograms displayed in Fig. 2(b) show a good match with the corresponding peak heights and positions in Fig. 2(a). This result demonstrates the impact of the described dual-point, dual-wavelength technique on the DNA separation resolution. Even equally sized, but differently labeled molecules that naturally flow at the same speed and could electrophoretically never be separated, have nevertheless been optically separated by the inherent spatial separation of the two WGs carrying the corresponding unique excitation wavelengths. By changing the distance between the adjacent WGs, the effective separation can be adjusted.
Fig. 2.
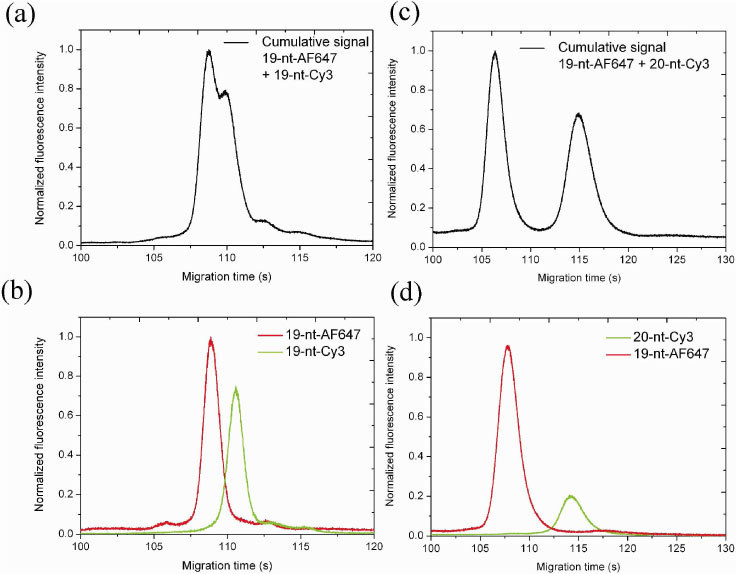
Electropherograms depicting the MCE separation of two fluorescently labeled DNA molecules: (a) cumulative signal during simultaneous dual-wavelength excitation of migrating 19-nt-AF647 and 19-nt-Cy3 molecules; (b) individual signals detected during different flow experiments applying single-wavelength excitation with only one of the two lasers switched on, temporally superimposed on each other; (c) and (d) the same for 19-nt-AF647 and 20-nt-Cy3 molecules.
Furthermore, considering the potential application, i.e. the detection of a genetic abnormality, to visualize the case of single base-pair insertion / deletion, we repeated the experiment with a new sample mixture (19-nt-AF647 + 20-nt-Cy3), where the size difference of a single base-pair between the two molecules was chosen to represent the potential application scenario. The corresponding electropherograms are shown in Figs. 2(c) and 2(d), with an evidently larger peak separation of ~6.2 s. The lack of perfect match in the temporal positions of the peaks and their intensities between Fig. 2(a) and 2(b) and Fig. 2(c) and 2(d) is attributed to slight, unintentional differences in the experimental flow parameters. Such effects justify the need for an internal calibration / referencing as described in Section 4.
4. Intrinsic referencing / calibration of DNA analysis by monitoring through two DWs
MCE separation and, consequently, sizing accuracy depend critically on multiple flow parameters, such as fluid and chip temperature, condition of the sieving gel matrix and MF wall coating, and applied electric field. Hence, for achieving single-base-pair accuracy of DNA analysis – for molecules in the diagnostically relevant range of 150-1000 bp – in an optofluidic chip, reliable intrinsic calibration of the CE separation setup is required. In our approach, a set of sample molecules under investigation and a set of well-known reference molecules (for calibration) can be exclusively labeled with two different fluorophores and separated simultaneously in a single MCE experiment. The reference molecules provide an intrinsic ‘ruler’ allowing one to accurately sort the sample molecules according to their size independently of the fluctuating experimental conditions. Building upon the experimental proof of principle presented in Section 3, we discuss our approach for intrinsic calibration by means of a numerical simulation based on data obtained from earlier experiments [15].
Monitoring through two well separated DWs, each comprising a WG pair carrying two different excitation wavelengths whose spatial order is swapped in DW2 (Fig. 1), provides the following advantages. (i) An unfortunate size difference between two differently labeled DNA species may occur, such that the difference in migration speed exactly compensates for the distance between the two excitation WGs within one DW. As a result, the two fluorescence peaks are simultaneously detected by the color-blind PMT in this DW. However, the peaks do not coincide in the other DW, because the swapping of the excitation wavelength would indeed enhance the effect of different migration speeds instead of creating a degeneracy. (ii) From the double detection, additional information can be deduced about flow speed, plug broadening, etc. (iii) Low-concentrated species that disappear under the background noise owing to plug broadening while migrating to DW2, will still give rise to weak fluorescence peaks at DW1, thus providing extra information that would otherwise be lost.
In principle, such a set-up can be operated with a single PMT, e.g., by collecting the fluorescence from the two DWs by two fibers glued to the top of the chip [11] or two 3D-integrated WGs [16], and transporting the signals to the same PMT, thereby simplifying the whole setup and reducing its size. In this case, to achieve an unambiguous detection, it is a critical prerequisite to choose a sufficiently large distance between the two DWs, such that the first peak in DW2 appears only after the last peak in DW1 has disappeared. Since our experimental setup requires detection by a PMT through a microscope, only one of the two DWs can be monitored at a time. Hence, it is currently not possible in our setup to monitor both DWs simultaneously.
Therefore, to demonstrate the potential of dual-point, dual-wavelength monitoring for internal size calibration, we present a simulation of a MCE experiment based on the logarithmic relationship between DNA size and migration speed, as well as the square-root dependence of plug broadening on the DNA size, as established in an earlier experimental analysis [15]. The Poissonian dependence of the electropherogram peak width on the DNA size is represented in Fig. 3 as the evidently broader peaks in DW2 in comparison with DW1, with the broadening being related to the DNA size by a square-root dependence, while the migration times of the DNA molecules have been simulated based on their estimated migraton speeds, which depend on their sizes. In this simulation, we consider the separation of a (usually unknown) DNA sample under investigation, consisting of green-labeled molecules S1 (250 bp), S2 (300 bp), S3 (450 bp), and S4 (700 bp), flown against a (known) DNA reference consisting of red-labeled molecules R1 (150 bp), R2 (355 bp), and R3 (1000 bp).
Fig. 3.
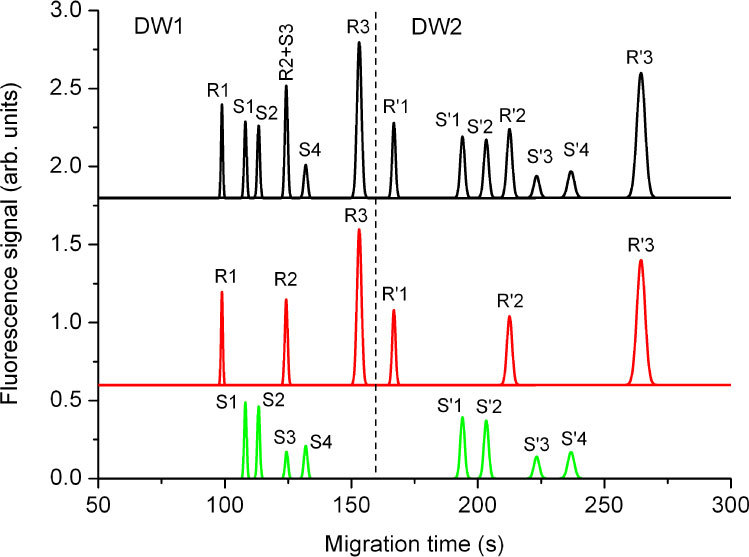
Simulated electropherograms as would be detected from the two DWs with swapped excitation wavelengths during an experiment with internal calibration using a green-labeled DNA sample (“S”) consisting of four different molecule sizes (250 bp, 300 bp, 450 bp, and 700 bp) and a red-labeled DNA reference (“R”) consisting of three different molecule sizes (150 bp, 355 bp, and 1000 bp). Fluorescence signals S were excited through WG1, R through WG2, R' through WG3 and S' through WG4.
Figure 3 (top, black curve) shows a simulated electropherogram that would be generated if the fluorescence is detected by a color-blind PMT simultaneously from both DWs as a result of laser excitation (with a mutually swapped order of excitation wavelengths) through the four WGs. The distance between the two DWs and/or the minimum and maximum sizes of DNA molecules are chosen such that all DNA molecules migrate first through DW1 and then through DW2, i.e., the luminescence signals occurring from the two DWs are separated in time, as indicated by the dashed line.
The deconvolution of the detected signal into the individual signals obtained from the sample and reference DNA molecules is presented in Fig. 3 (center, red curve, and bottom, green curve). The sizes of the reference molecules are chosen such that the smallest and largest base-pair sizes belong to reference molecules, hence they can be easily identified. Based on the a priori known sizes of the reference molecules, one can make use of the known dependence of the relative migration time on DNA size [15] to identify the third, medium-sized reference molecule in both windows, because its peak principally shifts from DW1 to DW2 in the same manner with respect to the peaks of the unknown molecules as the first and last reference molecule, and calibrate the remaining peaks of the electropherogram for varying environmental conditions, i.e., to accurately determine the sizes of the corresponding sample molecules based on their migration times relative to the migration times of the known reference molecules. The third, medium-sized reference molecule provides an indication of the experimental deviation from the expected migration due to parameter drifts. While the R2 and S3 peaks incidentally overlap in DW1 owing to their unfortunate size difference, they are well resolved as R'2 and S'3 in DW2 as a result of swapping the excitation wavelengths. In such a setting, it is straight forward to identify each peak in both parts of the diagram, DW1 and DW2, thus enabling unambiguous analysis of two sets of exclusively color-labeled DNA molecules of different origin by detection with a single color-blind PMT.
5. Conclusions
We have analyzed the electrophoretic separation of fluorescently labeled DNA molecules along a microfluidic channel with integrated waveguides in a dual-point, dual-wavelength setting. Using a single detection window, we demonstrate highly accurate detection of single-nucleotide insertion/deletion, as is relevant in genetic diagnostics – detection of anomalies in genetic samples obtained from a patient with respect to their healthy genetic counterparts. Furthermore, based on earlier experimental DNA separation data, we present a simulation of a separation experiment in a setup that uses two detection windows, with the two excitation wavelengths swapped in the second one, for internal calibration/referencing of DNA sizes. This can be a highly effective approach for intrinsically eliminating the undesirable effects of several flow parameters on calibration of a MCE separation system by flowing a well-known reference sample simultaneously with the unknown sample. The results presented in this paper bear the potential of leading to a new generation of compact optofluidic devices for use in point-of-care diagnostics.
Acknowledgements
We acknowledge financial support by the EU Project HIBISCUS (IST-2005-034562).
References and links
- 1.Manz A., Graber N., Widmer H. M., “Miniaturized total chemical analysis systems: A novel concept for chemical sensing,” Sens. Actuators B Chem. 1, 244–248 (1990). [Google Scholar]
- 2.Landers J. P., “Molecular diagnostics on electrophoretic microchips,” Anal. Chem. 75(12), 2919–2927 (2003). 10.1021/ac0301705 [DOI] [PubMed] [Google Scholar]
- 3.Kuswandi B., Nuriman J., Huskens J., Verboom W., “Optical sensing systems for microfluidic devices: a review,” Anal. Chim. Acta 601(2), 141–155 (2007). 10.1016/j.aca.2007.08.046 [DOI] [PubMed] [Google Scholar]
- 4.Johnson M. E., Landers J. P., “Fundamentals and practice for ultrasensitive laser-induced fluorescence detection in microanalytical systems,” Electrophoresis 25(21-22), 3513–3527 (2004). 10.1002/elps.200406086 [DOI] [PubMed] [Google Scholar]
- 5.Psaltis D., Quake S. R., Yang C., “Developing optofluidic technology through the fusion of microfluidics and optics,” Nature 442(7101), 381–386 (2006). 10.1038/nature05060 [DOI] [PubMed] [Google Scholar]
- 6.R. Iten, “Apparatus for emitting and detecting light in a nucleic acid amplification reaction,” Eur. Patent 1962084 (2007).
- 7.Zhu L., Stryjewski W. J., Soper S. A., “Multiplexed fluorescence detection in microfabricated devices with both time-resolved and spectral-discrimination capabilities using near-infrared fluorescence,” Anal. Biochem. 330(2), 206–218 (2004). 10.1016/j.ab.2004.03.047 [DOI] [PubMed] [Google Scholar]
- 8.Götz S., Karst U., “Wavelength-resolved fluorescence detector for microchip capillary electrophoresis separations,” Sens. Actuators B Chem. 123(1), 622–627 (2007). 10.1016/j.snb.2006.08.027 [DOI] [Google Scholar]
- 9.Karger A. E., Harris J. M., Gesteland R. F., “Multiwavelength fluorescence detection for DNA sequencing using capillary electrophoresis,” Nucleic Acids Res. 19(18), 4955–4962 (1991). 10.1093/nar/19.18.4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugino H., Ozaki K., Shirasaki Y., Arakawa T., Shoji S., Funatsu T., “On-chip microfluidic sorting with fluorescence spectrum detection and multiway separation,” Lab Chip 9(9), 1254–1260 (2009). 10.1039/b815765k [DOI] [PubMed] [Google Scholar]
- 11.Vazquez R. M., Osellame R., Nolli D., Dongre C., van den Vlekkert H. H., Ramponi R., Pollnau M., Cerullo G., “Integration of femtosecond laser written optical waveguides in a lab-on-chip,” Lab Chip 9(1), 91–96 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Dongre C., Dekker R., Hoekstra H. J. W. M., Pollnau M., Martinez-Vazquez R., Osellame R., Cerullo G., Ramponi R., van Weeghel R., Besselink G. A. J., van den Vlekkert H. H., “Fluorescence monitoring of microchip capillary electrophoresis separation with monolithically integrated waveguides,” Opt. Lett. 33(21), 2503–2505 (2008). 10.1364/OL.33.002503 [DOI] [PubMed] [Google Scholar]
- 13.Martinez Vazquez R., Osellame R., Cretich M., Chiari M., Dongre C., Hoekstra H. J. W. M., Pollnau M., Vlekkert H., Ramponi R., Cerullo G., “Optical sensing in microfluidic lab-on-a-chip by femtosecond-laser-written waveguides,” Anal. Bioanal. Chem. 393(4), 1209–1216 (2009). 10.1007/s00216-008-2399-8 [DOI] [PubMed] [Google Scholar]
- 14.Sanders J. C., Breadmore M. C., Kwok Y. C., Horsman K. M., Landers J. P., “Hydroxypropyl cellulose as an adsorptive coating sieving matrix for DNA separations: artificial neural network optimization for microchip analysis,” Anal. Chem. 75(4), 986–994 (2003). 10.1021/ac020425z [DOI] [PubMed] [Google Scholar]
- 15.Dongre C., van Weerd J., Besselink G. A. J., van Weeghel R., Vazquez R. M., Osellame R., Cerullo G., Cretich M., Chiari M., Hoekstra H. J. W. M., Pollnau M., “High-resolution electrophoretic separation and integrated-waveguide excitation of fluorescent DNA molecules in a lab on a chip,” Electrophoresis 31(15), 2584–2588 (2010). 10.1002/elps.201000126 [DOI] [PubMed] [Google Scholar]
- 16.Crespi A., Gu Y., Ngamsom B., Hoekstra H. J. W. M., Dongre C., Pollnau M., Ramponi R., van den Vlekkert H. H., Watts P., Cerullo G., Osellame R., “Three-dimensional Mach-Zehnder interferometer in a microfluidic chip for spatially-resolved label-free detection,” Lab Chip 10(9), 1167–1173 (2010). 10.1039/b920062b [DOI] [PubMed] [Google Scholar]


