Abstract
AIMS
Static and dynamic models (incorporating the time course of the inhibitor) were assessed for their ability to predict drug–drug interactions (DDIs) using a population-based ADME simulator (Simcyp®V8). The impact of active metabolites, dosing time and the ability to predict inter-individual variability in DDI magnitude were investigated using the dynamic model.
METHODS
Thirty-five in vivo DDIs involving azole inhibitors and benzodiazepines were predicted using the static and dynamic model; both models were employed within Simcyp for consistency in parameters. Simulations comprised of 10 trials with matching population demographics and dosage regimen to the in vivo studies. Predictive utility of the static and dynamic model was assessed relative to the inhibitor or victim drug investigated.
RESULTS
Use of the dynamic and static models resulted in comparable prediction success, with 71 and 77% of DDIs predicted within two-fold, respectively. Over 40% of strong DDIs (>five-fold AUC increase) were under-predicted by both models. Incorporation of the itraconazole metabolite into the dynamic model resulted in increased prediction accuracy of strong DDIs (80% within two-fold). Bias and imprecision in prediction of triazolam DDIs were higher in comparison with midazolam and alprazolam; >50% of triazolam DDIs were under-predicted regardless of the model used. Predicted inter-individual variability in the AUC ratio (coefficient of variation of 45%) was consistent with the observed variability (50%).
CONCLUSIONS
High prediction accuracy was observed using both the Simcyp dynamic and static models. The differences observed with the dose staggering and the incorporation of active metabolite highlight the importance of these variables in DDI prediction.
Keywords: CYP3A4, drug-drug interactions, Simcyp
WHAT THIS PAPER ADDS
Prediction of DDIs for 35 clinical studies incorporating a representative range of drug–drug interactions, with multiple studies across different inhibitors and victim drugs.
Assessment of whether the inclusion of the time course of inhibition in the dynamic model improves prediction in comparison with the static model.
Investigation of the impact of different inhibitor and victim drug parameters on DDI prediction accuracy including dosing time and the inclusion of active metabolites. Assessment of ability of the dynamic model to predict inter-individual variability in the DDI magnitude.
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
The prediction of drug–drug interactions (DDIs) from in vitro data usually utilizes an average dosing interval estimate of inhibitor concentration in an equation-based static model.
Simcyp®, a population-based ADME simulator, is becoming widely used for the prediction of DDIs and has the ability to incorporate the time-course of inhibitor concentration and hence generate a temporal profile of the inhibition process within a dynamic model.
Introduction
Metabolic drug–drug interactions (DDIs) continue to be a major concern in drug development emphasizing the need to optimize predictions of the interaction potential from in vitro data. Current quantitative prediction of DDIs can be achieved from the ratio of the area under the plasma concentration–time curve (AUC) following multiple dosing of inhibitor in comparison with the control state. This can be retrieved with a ‘static’ model equation incorporating a number of in vitro and in vivo parameters (equation 1). Alternatively, a more comprehensive physiologically-based pharmacokinetic approach such as the Simcyp® population-based absorption, distribution metabolism and excretion (ADME) simulator can be used [1–9]).
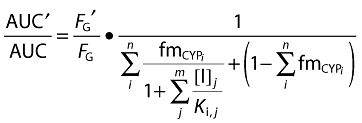 |
equation 1 |
where [I]j is the inhibitor concentration, Ki,j is the inhibition constant, fmCYPi is the fraction of substrate drug metabolized by the inhibited pathway via a cytochrome P450 (CYP) enzyme, (1–ΣfmCYPi) represents clearance via other CYP enzymes and/or renal clearance, FG is the fraction escaping metabolism in the intestine in the presence (denoted by ′) or absence of inhibitor. The terms i and j indicate the existence of multiple enzymes (n) and inhibitors (m), respectively.
The inhibitor concentration at the enzyme active site cannot be experimentally measured and previous DDI predictions have been attempted using various [I] values as a surrogate. This includes the use of average plasma total or unbound concentration or hepatic input concentration of the inhibitor [4, 6, 8, 10, 11]. Despite much debate [12–15], there is currently no consensus on which surrogate concentration should be utilized, although the use of unbound drug is accepted as more theoretically relevant [4, 16, 17]. The situation is confounded by the impact of other parameters in the equation influencing prediction success, for example the absorption rate constant (ka), fmCYP or FG values. Thus, predictions may actually be influenced to a greater extent by differences in the choice of the values for these inhibitor and victim drug related parameters, rather than just Ki or the [I] itself [15, 18, 19].
Active metabolites are an important consideration for DDI prediction models, particularly if the metabolite circulates at a sufficiently high concentration [15, 20] or has a potent effect on different pathways or shows a different interaction mechanism to the parent drug [20, 21]. Isoherranen et al. [22], report that the majority of clinically important inhibitors possess circulating metabolites. However, in vitro evaluation of the inhibition potential of the circulating metabolites was only reported for approximately 30% of these. In addition, multiple inhibitors are not commonly included in DDI assessment leading to a potential under-estimation of the magnitude of DDI [4, 12, 14–16, 20, 23, 24]. One example of a clinically important metabolite is hydroxy-itraconazole. This metabolite has lower clearance compared with the parent and equal circulatory concentration in plasma and has been suggested to be an important contributor in overall CYP3A inhibition, despite lower potency compared with the parent drug (Ki of 14.4 nm compared with 1.3 nm for the parent drug) [25–28].
The Simcyp simulator provides a comprehensive framework for DDI prediction by allowing simulations to incorporate a range of individual pharmacokinetic parameters in a virtual patient population. The Monte Carlo method of generating virtual entities with randomly assigned characteristics is used [9, 15]. It includes the impact of demographic factors (e.g. age and gender), physiological factors (e.g. blood flow), genetic factors (e.g. CYP polymorphisms) and pathological factors [4, 7, 9]. As a result, patient populations that are specifically at risk of DDIs can be identified that are unlikely to be represented in a small clinical trial population [15, 29, 30]. Simulations can be performed using either a static or dynamic approach. The static model is based on the use of single time-averaged inhibitor and substrate concentrations, analogous to the principles underlying the static model shown in equation 1. In addition, the dynamic model incorporates the time-course of the concentrations and hence generates a temporal profile of the inhibition process [31–33]. The additional ability to incorporate multiple inhibitors and active metabolites in the dynamic model allows a more comprehensive and meaningful prediction [7, 9].
The equation-based static (equation 1) and Simcyp dynamic model were previously compared in a dataset of 100 DDIs including a range of inhibitors and victim drugs metabolized by a variety of the CYP enzymes [4]. Use of the Simcyp dynamic model resulted in the most accurate predictions (64% within 2-fold). However, parameter values (e.g. fup, Ki, FG, fmCYP) differed between the two models, as well as the choice of [I] value used in the equation-based static model (equation 1). Differences in the values of the input parameters between the models are likely to result in different AUC ratio predictions, considering the sensitivity of the model to certain parameter estimates (e.g. fmCYP[1, 16, 34]). Therefore, although the results are valid for a basic comparison of the models, assessment of the impact of specific factors, for example, the inclusion of the time course, was confounded by the issues outlined above.
The objectives of the current study were to compare directly and evaluate the static and dynamic model approaches to DDI prediction and to assess the impact of including the time-course for the inhibition process. The ability to assess both models within Simcyp allowed a valid comparison, as other factors were kept consistent. For this study, a set of 35 DDIs between three azole inhibitors (fluconazole, ketoconazole and itraconazole) and three benzodiazepine substrates (alprazolam, midazolam, triazolam) encompassing different properties was used. The impact of different inhibitor and victim drug properties on DDI prediction was also assessed, namely the incorporation of active metabolite (itraconazole) and inhibitor dosing time relative to the victim drugs. A number of additional factors were assessed for itraconazole-triazolam interactions considering the poor prediction success observed in preliminary analysis, namely variability in itraconazole ka, use of differential triazolam in vitro clearance and permeability data. Finally, the ability of Simcyp to predict inter-individual variability in the magnitude of DDI was assessed and compared with the reported data in the individuals in the case of a ketoconazole/itraconazole interaction with triazolam.
Methods
Virtual trials
The static and dynamic approaches were compared using the Simcyp population-based ADME simulator (Version 8.10, SP1). The underlying concepts and principles of Simcyp have been previously described [7, 32]. This allowed assessment of the ability of each model to predict the magnitude of DDI (through change in the AUC) of 35 published interactions. The selected DDIs included fluconazole, ketoconazole and itraconazole as inhibitors and alprazolam, midazolam and triazolam as victim drugs; all DDIs were reversible inhibition interactions involving CYP3A. The criterion for the inclusion of the study was the oral administration of both substrate and inhibitor. The DDIs in the current analysis were classified according to the fold change in the AUC of the victim drug either as a weak (AUC ratio <2), moderate (2 to 5-fold increase in AUC) or strong DDI (AUC ratio >5), analogous to the FDA guidelines for the assessment of potential inhibitors of CYP3A4 [5]. Five studies (representing 12 data points due to different dose or dosing schedule) in the database [35–39] reported DDIs after single dose of the inhibitors and were used for additional analysis and comparison with those studies reporting DDIs at steady-state inhibitor concentrations. The AUC ratio used for comparison from each study was calculated from AUC(0,∞). The only exception was the study between ketoconazole and itraconazole with triazolam [40], where the AUC ratio was calculated from the last time point in the study (AUC(0,t)). Whenever available the mean AUC(0,∞) ratio for each study was calculated from values reported for the individual subjects.
In order to keep as many variables as consistent as possible, the simulations for both the static and dynamic models were performed in Simcyp, allowing direct assessment of the impact of inclusion of the time course. For assessment of DDIs, each simulation trial had matching population demographics (number of subjects, age and male : female ratio) and dosage regimen (dose and timing of all inhibitor and victim drug doses) to the reported clinical study. Ten of these trials were performed for each study in the dataset in order to assess variability across groups of subjects. Simulations were based on the systemic plasma concentration of the inhibitor, the use of a one compartmental model [41] and a population of healthy, fasted subjects assuming a 250 ml fluid intake with dosing. In cases when the age range was given as a mean ± SD, the distribution was estimated or if the age was not reported the default range was used (18–65 years with the proportion of females at 0.34). If the dose timing details were not reported, simulations were undertaken with the victim drug dosed 1 h after the final inhibitor dose, as this was the most common dosing schedule (Table 1). The simulations were terminated at least two inhibitor half-lives after the final inhibitor dose in order to allow assessment of the AUC where it is approaching infinity. Model principles are based on those reported by Ito et al. [56], but also include intestinal wall metabolism as described previously [32, 57]. The gut and the liver are represented as separate compartments and the other organs are included in a single systemic compartment [32]. The dynamic model is based on analogous principles to the static model, but it allows the incorporation of the time-variant intestinal and hepatic metabolism. Drug metabolism is described using the Michaelis-Menten relationship and incorporates changes of active enzymes with time in both liver and intestine. The differential equations and assumptions of these methods are described fully in Rowland-Yeo et al. [32].
Table 1.
Summary details including the dosing details and observed AUC ratio of the 35 in vivo DDI studies between three azoles and three benzodiazepines used for assessment of the static and dynamic models
| Inhibitor | Inhibitor dose | Dosing start day and time | Dosing interval (h) | Number of doses | Victim drug | Victim drug dose | Victim drug dosing time | Observed AUC ratio | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Fluconazole | 400 mg | Day 1, 13.00 h | SD | 1 | Midazolam | 7.5 mg | Day 1, 14.00 h | 3.73 | [35] |
| Fluconazole | 100 mg | Day 1 | SD | 1 | Midazolam | 3 mg | Day 1, +2 h | 2.16 | [37] |
| Fluconazole | 200 mg | Day 1 | SD | 1 | Midazolam | 3 mg | Day 1, +2 h | 3.32 | [37] |
| Fluconazole | 400 mg | Day 1 | SD | 1 | Midazolam | 3 mg | Day 1, +2 h | 4.93 | [37] |
| Fluconazole | 400 mg | Day 1, 13.00 h | SD | 1 | Midazolam | 7.5 mg | Day 1, 15.00 h | 3.51 | [39] |
| Fluconazole | 400 mg/200 mg | 400 mg Day 1, 13.00 h, 200 mg Day 2–6, 13.00 h | 24 | 6 | Midazolam | 7.5 mg | Day 6, 15.00 h | 3.60 | [39] |
| Fluconazole | 100 mg | Day 1–4, 14.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 15.00 h | 2.46 | [42] |
| Fluconazole | 50 mg | Day 1–4, 13.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 14.00 h | 1.63 | [43] |
| Fluconazole | 100 mg | Day 1–4, 13.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 14.00 h | 2.05 | [43] |
| Fluconazole | 400 mg/200 mg | 400 mg Day 1, 13.00 h, 200 mg Day 2–4, 13.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 14.00 h | 4.42 | [43] |
| Ketoconazole | 200 mg | Day 1–3, 08.00 h | 12 | 5 | Alprazolam | 1 mg | Day 3, 09.00 h | 3.98 | [44] |
| Ketoconazole | 200 mg | Day 1–3 | 12 | 5 | Alprazolam | 1 mg | Day 3 | 1.76 | [45] |
| Ketoconazole | 200 mg | Day 1–3 | 12 | 5 | Midazolam | 0.075 mg | Day 3 | 6.47 | [46] |
| Ketoconazole | 200 mg | Day 1–12 | 24 | 12 | Midazolam | 10 mg | Day 12, +1 h | 6.56 | [47] |
| Ketoconazole | 400 mg | Day 1–4, 14.00 h | 24 | 4 | Midazolam | 7.5 mg | Day 4, 15.00 h | 15.9 | [48] |
| Ketoconazole | 200 mg | Day 1–2 | 12 | 3 | Midazolam | 6 mg | Day 1, +12 h* | 16.0 | [49] |
| Ketoconazole | 200 mg | Day 1–2 | 12 | 3 | Midazolam | 5 mg | Day 1, +13 h* | 8.10 | [50] |
| Ketoconazole | 400 mg | Day 1–10 | 24 | 10 | Midazolam | 5.5 mg | Day 9 | 9.51 | [51] |
| Ketoconazole | 200 mg | Day 1, 07.00 h | SD | 1 | Midazolam | 2 mg | Day 1, 09.00 h | 4.95 | [36] |
| Ketoconazole | 200 mg | Day 1, 09.00 h | SD | 1 | Midazolam | 2 mg | Day 1, 09.00 h | 6.45 | [36] |
| Ketoconazole | 200 mg | Day 1–3, 08.00 h | 12 | 5 | Triazolam | 0.25 mg | Day 3, 09.00 h | 13.7 | [44] |
| Ketoconazole | 400 mg | Day 1–4, 14.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 15.00 h | 9.17 | [40] |
| Ketoconazole | 200 mg | Day 1–2 | 16 | 2 | Triazolam | 0.125 mg | Day 2, +1 h | 9.16 | [52] |
| Itraconazole | 200 mg | Day 1–6, 08.00 h | 24 | 6 | Alprazolam | 0.8 mg | Day 4, 09.00 h | 2.66 | [53] |
| Itraconazole | 100 mg | Day 1–4, 13.00 h | 24 | 4 | Midazolam | 7.5 mg | Day 4, 15.00 h | 5.75 | [54] |
| Itraconazole | 200 mg | Day 1–4, 11.00 h | 24 | 4 | Midazolam | 7.5 mg | Day 4, 13.00 h | 6.16 | [55] |
| Itraconazole | 200 mg | Day 1–4, 11.00 h | 24 | 4 | Midazolam | 7.5 mg | Day 8, 13.00 h | 2.51 | [55] |
| Itraconazole | 200 mg | Day 1–4, 14.00 h | 24 | 4 | Midazolam | 7.5 mg | Day 4, 15.00 h | 10.8 | [48] |
| Itraconazole | 200 mg | Day 1, 13.00 h | SD | 1 | Midazolam | 7.5 mg | Day 1, 15.00 h | 3.42 | [39] |
| Itraconazole | 200 mg | Day 1–6, 13.00 h | 24 | 6 | Midazolam | 7.5 mg | Day 6, 15.00 h | 6.64 | [39] |
| Itraconazole | 200 mg | Day 1, 15.00 h | SD | 1 | Triazolam | 0.25 mg | Day 2, 15.00 h | 3.57 | [38] |
| Itraconazole | 200 mg | Day 2, 03.00 h | SD | 1 | Triazolam | 0.25 mg | Day 2, 15.00 h | 4.33 | [38] |
| Itraconazole | 200 mg | Day 2, 12.00 h | SD | 1 | Triazolam | 0.25 mg | Day 2, 15.00 h | 4.48 | [38] |
| Itraconazole | 200 mg | Day 2, 15.00 h | SD | 1 | Triazolam | 0.25 mg | Day 2, 15.00 h | 3.11 | [38] |
| Itraconazole | 200 mg | Day 1–4, 14.00 h | 24 | 4 | Triazolam | 0.25 mg | Day 4, 15.00 h | 10.5 | [40] |
If exact timings were not reported, the substrate dosing time is given as plus a number of hours after the final inhibitor dose.
The exception was where the substrate was given with or 1 h after the second inhibitor dose.
SD, Single dose.
All of the inhibition input parameters for the azole inhibitors are shown in Table 2; Simcyp default values were used for all other input parameters and the coefficients of variation for these parameters were kept as set by the software during the simulations (30% in most cases), as performed previously [41]. The values of Ki and fup utilized for fluconazole and ketoconazole were taken from Brown et al. [1], and those for itraconazole (and hydroxy-itraconazole when included in simulation) from Isoherranen et al. [25] (Table 2). Some of the input parameters vary from Simcyp default values; for example, default ketoconazole values are 0.015 µm for CYP3A4 Ki[58] and fup is 0.029. All the Ki values used were obtained in human liver microsomes from Caucasian donors. Hence the assumption was that the Ki reflected the inhibition of CYP3A4. In the case of itraconazole [25] it was reported that the microsomes had no significant amounts of CYP3A5 protein. Inhibition of CYP3A5 as a potentially contributing enzyme to the clearance of these three victim drugs [59] was included whenever the information on CYP3A5 inhibition by azole inhibitors was available. For example, CYP3A5 Ki of 0.109 and 84.6 µm[58] were incorporated in the assessment of ketoconazole and fluconazole, respectively; no such information was available for itraconazole. Recent studies have reported a minor contribution (approximately 6%) of N-glucuronidation to midazolam clearance in vitro[60, 61]. It is important to note that the glucuronidation metabolic pathway was not available as a pathway for midazolam in Version 8.10, SP1 of the software used for the current analysis. Klieber et al. [61] reported no significant inhibition of midazolam N-glucuronidation in the presence of ketoconazole.
Table 2.
Input parameters for the azole inhibitors used in Simcyp simulations. Values for ka are Simcyp defaults, where CYP3A4 Ki and fup data for fluconazole and ketoconazole were from [1] and itraconazole and hydroxy-itraconazole from [25]
| Inhibitor | Ki (CYP3A4) (µm) | fup | ka (h−1) |
|---|---|---|---|
| Fluconazole | 11.9 | 0.890 | 1.28 |
| Ketoconazole | 0.0420 | 0.017 | 1.90 |
| Itraconazole | 0.0013 | 0.036 | 0.62 |
| Hydroxy-itraconazole | 0.0144 | 0.0045 | – |
The fraction absorbed for all drugs was assumed to be 1. In the case of ketoconazole, a default hepatic uptake value of 2.07 was used, consistent with previous reports [52, 62]; for other inhibitors no uptake was assumed. Default clearances were used for all inhibitors; oral clearance values of 13.3 and 1.5 l h−1 were used for ketoconazole and fluconazole, respectively [45, 46]. In the case of itraconazole and hydroxy-itraconazole, in vitro Km and Vmax data obtained in recombinant baculovirus CYP3A4 [25] and corrected for the corresponding intersystem extrapolation factors [15] were used. The impact of the inclusion of the most abundant and potent metabolite of itraconazole, hydroxy-itraconazole, in dynamic DDI prediction was also assessed. The impact of different ka values for itraconazole was explored, based on the 0.58–0.64 h−1 range calculated from the clinical 90% confidence intervals of time to reach maximal plasma drug concentration (tmax) and half-life (t1/2) [63]; impact of a wider range of this parameter (0.3–1 h−1) was additionally investigated.
Impact of victim drug properties and dose timing
The data showing the accuracy of the DDI predictions were also compared for each victim drug in order to identify any trends. Values of FG and fmCYP are calculated intrinsically within Simcyp; output values for FG estimated using the Qgut model were 0.99, 0.59 and 0.91 for alprazolam, midazolam and triazolam, respectively (in all the cases fraction unbound in the intestine was assumed to be 1) [32, 57, 64]. The contribution of CYP3A4 and CYP3A5 was estimated from in vitro clearance data after correcting the simulated plasma clearance for renal excretion. Contribution of renal clearance was 18% for alprazolam, whereas it was negligible for midazolam and triazolam (1–1.5% of total clearance). Proportional contribution of CYP3A4 to hepatic clearance of midazolam and triazolam was 85 and 86%, respectively, and the remaining fraction metabolized was associated with CYP3A5.
In the case of triazolam, virtual trials were also performed using mean triazolam CLint from several sources of human liver microsomal data (30.3 ± 11.1 and 22.3 ± 11.6 µl min−1 mg−1 protein for 1′-hydroxy and 4-hydroxytriazolam, respectively [52, 64–67]). Impact of variability in triazolam in vitro clearance data on the DDI prediction was assessed due to availability of data from individual donors of human liver microsomes [52]. To investigate the impact of the timing of dose on the magnitude of itraconazole DDIs, dynamic simulations were performed with all victim drugs using the most common inhibitor dosing schedule (i.e. 200 mg itraconazole day−1 for 4 days, with the inclusion of hydroxy-itraconazole in simulations). The administration of the substrate dose (0.5 mg, 5 mg and 0.25 mg of alprazolam, midazolam and triazolam, respectively) was varied from −10 to +24 h from the last itraconazole dose. Where available, observed AUC ratios from the studies were compared with those obtained by virtual trials to assess the ability of the dynamic model to predict the actual differences seen as a result of changes in the timing of the dose.
Inter-individual variability in the AUC ratios
One clinical study in the subset reported the AUC data from each individual subject for the DDIs between either ketoconazole or itraconazole with triazolam as a victim drug [40]. These individual AUC ratios were therefore used to assess the ability of the dynamic model to predict inter-individual variability in the AUC ratio. This was assessed using 10 virtual trials with the same number of subjects as in the study (n = 9); median, minimum and maximum predicted AUC ratios of each individual trial and all 10 trials in total were compared with the observed data. The simulation of the DDI involving itraconazole was performed with the inclusion of hydroxy-itraconazole and triazolam human liver microsomal clearance data, as outlined above. To investigate further variation in the virtual trial results, the data on CYP3A5 genotype and CYP3A4 abundance in the liver and gut were studied for the individuals in the simulated trials. No such data were available for the clinical studies included in the current dataset and information on the CYP3A5 status of the subjects involved in the DDI clinical studies was generally limited [68, 69].
Data analysis
Predictions within 2-fold of the observed AUC ratio were considered successful, although the number within 1.5-fold of the in vivo values was also assessed. The average fold error (afe) was used to assess bias in the DDI prediction, and the mean and root mean squared prediction error (mse and rmse, respectively) provided a measure of precision of the DDI prediction (equations 2–4) [70, 71]. In addition, the weighted afe of the predictions performed in the dynamic model was calculated by correction for the number of subjects in the corresponding clinical trial.
| equation 2 |
 |
equation 3 |
| equation 4 |
where n represents number of studies.
Results
A database of 35 studies reporting DDIs between the azole inhibitors (fluconazole, ketoconazole and itraconazole) and CYP3A benzodiazepine substrates (alprazolam, midazolam and triazolam) was collated in order to assess the static and dynamic prediction of DDIs assessed within Simcyp. Table 1 provides the dosing details of the inhibitors and victim drugs and the observed AUC ratios for the studies investigated. In the current dataset, 51% of the interactions investigated were moderate (2 to 5-fold increase in AUC of the victim drug), 43% were strong (AUC ratio >5) and the remainder (6%) were weak (AUC ratio < 2).
Comparison between static and dynamic DDI predictions
Figure 1 shows the comparison of the predicted AUC ratios from virtual trials obtained by the dynamic and static model within Simcyp, classified according to the inhibitors or victim drugs in the dataset. Breakdown of studies where the predicted magnitude of DDIs was greater by the dynamic model (dynamic : static ratio >1) and vice versa is illustrated in Table 3. The dynamic : static model ratio of predicted DDI magnitude ranged from 0.4 for an itraconazole DDI with midazolam, up to 1.73 in the case of an itraconazole DDI with triazolam. For 14/35 DDIs in the dataset, the difference between the models was minimal and predicted AUC ratios by the static and dynamic model were within 20% of each other (Figure 1). Nearly half of the interactions in the dataset (49%) had a dynamic : static ratio of <0.8, indicating higher AUC ratio predictions obtained using the static model. This was particularly apparent for DDIs involving fluconazole and ketoconazole with 70% and 54% of study ratios in this range, respectively. The only inhibitor with a tendency for increased predictions via the dynamic model was itraconazole. This trend was particularly evident for strong DDIs where the average ratio between the models was 1.28 (Table 3). DDIs with triazolam as the victim drug had the most comparable mean ratio between the models (0.93) despite the wide range observed (0.51–1.73). No trend was observed between the potency of DDI and the dynamic : static ratio when assessed per victim drug (Figure 1).
Figure 1.
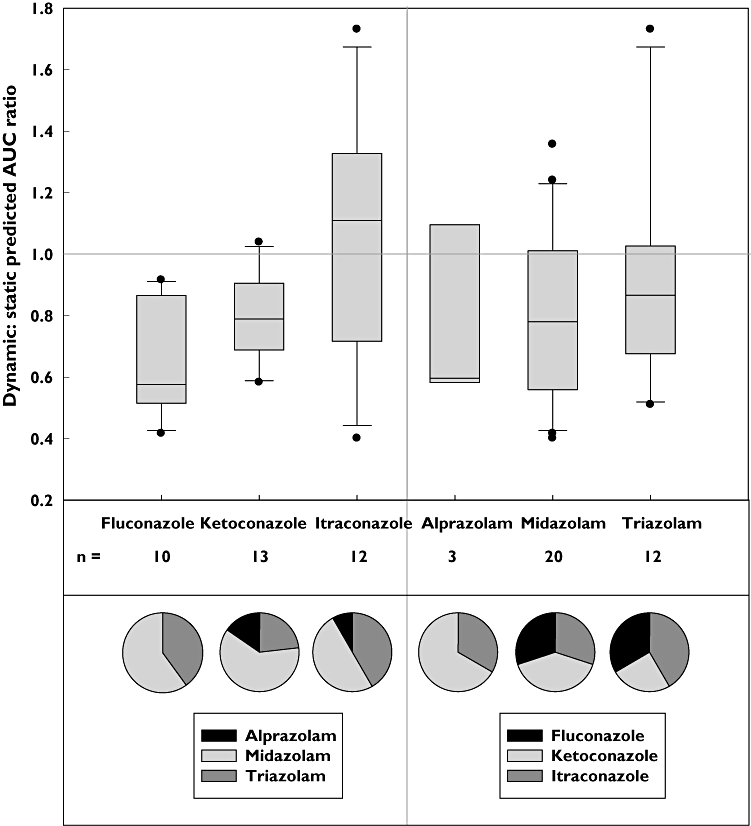
Comparison of the predicted AUC ratio for 39 DDIs performed using either the time-based dynamic or static model in Simcyp. The median and inter-quartile ranges are represented by the black line and box boundaries, respectively, whiskers represent 10–90% ranges and outliers are represented by •. The predictions are classified according to the inhibitor or victim drug and the pie charts underneath display the proportion of DDIs for each
Table 3.
Comparison of predicted AUC ratios by the dynamic and static models according to the inhibitor and potency of DDI investigated. Weak, moderate and strong DDIs refer to interactions with fold increase in victim drug AUC <2, 2–5 and >5-fold, respectively
| Inhibitor | Weak DDI | Moderate DDI | Strong DDI | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Dynamic : static predicted AUC ratio | >1 | <1 | >1 | <1 | >1 | <1 | >1 | <1 |
| Fluconazole | – | 1/10 | – | 9/10 | – | – | – | 10/10 |
| Itraconazole | – | – | 3/12 | 4/12 | 4/12 | 1/12 | 7/12 | 5/12 |
| Ketoconazole | – | 1/13 | – | 2/13 | 2/13 | 8/13 | 2/13 | 11/13 |
| Subtotal | – | 2/35 | 3/35 | 15/35 | 6/35 | 9/35 | 9/35 | 26/35 |
| Total | 2/35 | 18/35 | 15/35 | 35/35 | ||||
DDI prediction accuracy from the static and dynamic model
Predicted : observed AUC ratios from virtual trials obtained using either the static or dynamic model are shown in Figure 2. For 35 DDIs, 71% and 77% were predicted within 2-fold using the dynamic and static model, respectively, using the parameters detailed in Table 2. The static model also resulted in higher precision and lower bias across the range of DDI potency (Table 4), although the difference was not significant (P > 0.1, Student's t-test). Fluconazole DDIs showed good prediction accuracy as either all or 9/10 studies were predicted within 2-fold when using the static or dynamic model, respectively, although all of these DDIs were either weak or moderate interactions, i.e. AUC ratio <5. A clear under-prediction trend was observed with increasing potency of DDIs with over 40% of strong DDIs (AUC increase >5-fold) under-predicted regardless of the model used. The prediction of ketoconazole DDIs particularly showed this trend, with 40–50% of strong DDIs under-predicted using the dynamic or static model. However, overall prediction accuracy for ketoconazole DDIs was comparable between the models with 54–62% of studies within a 2-fold margin (Table 4). There was no difference observed in prediction accuracy between fluconazole DDIs resulting from steady-state inhibitor concentrations (five studies) or single dosing (five studies) as nearly all (4/5 or 5/5) were predicted within 2-fold, respectively. In the case of ketoconazole, two of the 13 DDIs with this inhibitor used a single inhibitor dose. However, those DDIs were predicted within 2-fold regardless of the model used, whereas either 5 or 6 of the remaining 11 multiple dosing studies were predicted within 2-fold using the dynamic or static model, respectively. Itraconazole DDIs with steady-state inhibitor concentrations (seven studies) or with single dosing regimens (five studies), were predicted 86% and 60% within 2-fold, respectively, when using the dynamic model (regardless of metabolite inclusion). However, a difference was observed between the dosing scenarios when using the static model with 57% of steady-state and 100% of single dosing studies predicted within 2-fold. Overall, 83 and 100% of the 12 single dose studies, and 65 and 61% of the 23 multiple dose studies were predicted within 2-fold using the dynamic and static models, respectively.
Figure 2.
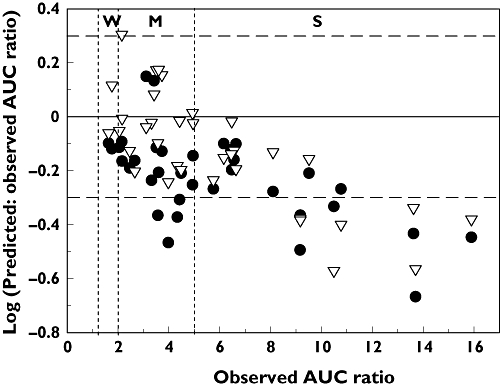
Comparison of predicted : observed and observed AUC ratios for 35 DDIs with azole inhibitors. Predictions were performed using either the time-based dynamic (•) or static (▿) model in Simcyp. Horizontal dashed lines represent the 2-fold margins, and vertical dashed lines represent the boundaries between weak (W), moderate (M) and strong (S) drug–drug interactions
Table 4.
Accuracy of 35 DDI predictions using either the time-based dynamic or static model in Simcyp, classified according to either inhibitor or DDI potency (moderate and strong indicating 2–5 and >5-fold increase in AUC of the victim drug, respectively)
| Model | Number of studies within 2-fold (%) | Number of studies within 1.5-fold (%) | afe | rmse |
|---|---|---|---|---|
| Dynamic | ||||
| Inhibitor | ||||
| Fluconazole (n = 10) | 90 | 50 | 1.52 | 1.29 |
| Ketoconazole (n = 13) | 54 | 31 | 2.07 | 5.60 |
| Itraconazole (n = 12) | 75 | 50 | 1.46 | 2.64 |
| Potency | ||||
| Moderate (n = 18) | 78 | 50 | 1.51 | 1.54 |
| Strong (n = 15) | 60 | 27 | 1.97 | 5.57 |
| Overall* (n = 35) | 71 | 43 | 1.68 | 3.81 |
| Static | ||||
| Inhibitor | ||||
| Fluconazole (n = 10) | 100 | 100 | 1.06 | 0.96 |
| Ketoconazole (n = 13) | 62 | 54 | 1.63 | 4.95 |
| Itraconazole (n = 12) | 75 | 33 | 1.43 | 3.26 |
| Potency | ||||
| Moderate (n = 18) | 94 | 72 | 1.04 | 1.15 |
| Strong (n = 15) | 53 | 40 | 1.89 | 5.36 |
| Overall* (n = 35) | 77 | 60 | 1.34 | 3.61 |
Accuracy of the two weak DDIs is not displayed in the analysis according to the DDI potency but was included in the overall analysis.
The impact of active metabolite inclusion on DDI prediction accuracy
The inclusion of the most potent and abundant active metabolite of itraconazole (hydroxy-itraconazole) into the dynamic model resulted in an increase in the predicted AUC ratio. Although this did not result in a difference in prediction accuracy for moderate DDIs, a significantly higher prediction accuracy was observed for strong DDIs (80% within 2-fold compared with 40% without the metabolite). The dynamic model (including metabolite) also showed improved prediction accuracy for strong DDIs when compared with the static model (80% within 2-fold compared with 60%, respectively), and had an increased overall number of studies within 1.5-fold (50% compared with 25–33% observed with other two scenarios). The overall trend of under-prediction at higher potency DDIs can be observed in Figure 3 for the predictions made using the static and dynamic (without metabolite) model. Overall, the dynamic (with metabolite) and static model result in comparable numbers of studies predicted within 2-fold (9/12).
Figure 3.
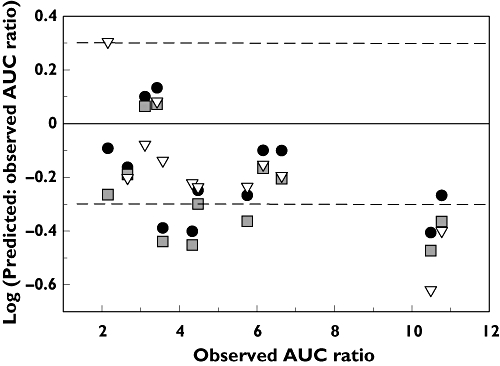
Comparison of predicted : observed and observed AUC ratios for 12 itraconazole DDIs. Predictions were performed in Simcyp using either the time-based dynamic model in the presence (•) or absence ( ) of the itraconazole metabolite (hydroxy-itraconazole) or using the static model (▿). The horizontal dashed lines represent the 2-fold margins
) of the itraconazole metabolite (hydroxy-itraconazole) or using the static model (▿). The horizontal dashed lines represent the 2-fold margins
Impact of properties of the victim drug on dynamic DDI prediction accuracy
Figure 4 shows the comparison of predicted and observed AUC ratios according to the victim drugs, obtained using either the dynamic or static model. From the 23 DDIs with midazolam or alprazolam, 90 and 67% predictions obtained by dynamic model were within 2-fold of the in vivo value, respectively. Bias in prediction of midazolam and alprazolam was comparable (afe of 1.18 and 1.29, respectively). However, prediction of midazolam DDIs showed higher imprecision (rmse of 3.39). In the case of triazolam, bias and imprecision were higher (afe and rmse of 1.67 and 4.29, respectively). In addition, only 42% of triazolam DDIs were predicted within 2-fold of the observed AUC ratio regardless of inhibitor. None of the interactions within 2-fold was in the class of strong DDIs and only 4/7 of the moderate DDIs were predicted successfully. Similar precision and bias was observed per substrate for predictions using the static model compared with the dynamic model.
Figure 4.
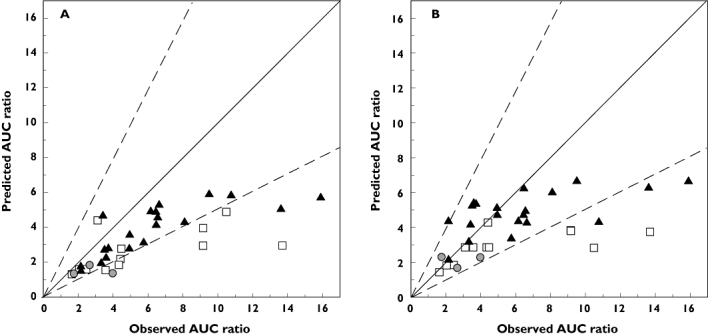
Predicted and observed AUC ratios for azole DDIs classified according to the victim drug where  represents alprazolam, ▴ midazolam and □ triazolam. Predictions used the dynamic model (plus metabolite for itraconazole DDIs) (A) or the static model (B). The solid and dashed lines represent the line of unity and 2-fold margins respectively. Error bars have been removed for clarity
represents alprazolam, ▴ midazolam and □ triazolam. Predictions used the dynamic model (plus metabolite for itraconazole DDIs) (A) or the static model (B). The solid and dashed lines represent the line of unity and 2-fold margins respectively. Error bars have been removed for clarity
The impact of altered dosing times of the three victim drugs (from t = −10 to +24 h) on the predicted AUC ratio was assessed using a DDI involving itraconazole (Figure 5A). A similar trend was observed for all three substrates but it was more pronounced for midazolam and triazolam DDIs with a maximum difference in the predicted AUC ratio of 4.2 and 2.8, respectively, depending on the time of administration. In contrast, the impact on the lower potency DDI with alprazolam was negligible, with a maximum 11.3% difference in AUC ratio seen across the different dosing times. Prediction was particularly sensitive to the dosing time of −2 to 0 h, with an increase in AUC ratio of up to 2.5-fold for midazolam. A 16% decline in AUC ratio was observed when midazolam was administered 1 h after the itraconazole dose rather than simultaneously (Figure 5B). This was in contrast to simulations with ketoconazole reported by Zhao et al. [41] where only a minimal change in AUC ratio was observed during the same time period. Limited observed data were available to assess directly prediction success related to the dose timing. Hence, proportional decreases in the AUC ratio were investigated. In the case of interactions involving midazolam, a 43 ± 4% (n = 3) decrease in AUC ratio was observed between dosing at 1 or 2 h after the inhibitor dose. This decrease was also seen in simulations, but accounted for only 13%. In the case of triazolam, an increase in observed AUC ratio was noted with substrate dosing from 0–3 h after inhibitor dose (based on [38]), whereas simulated results display a 34% decrease. The subsequent 20% decrease observed in vivo from 3 to 24 h was over-predicted in simulations by 38%.
Figure 5.
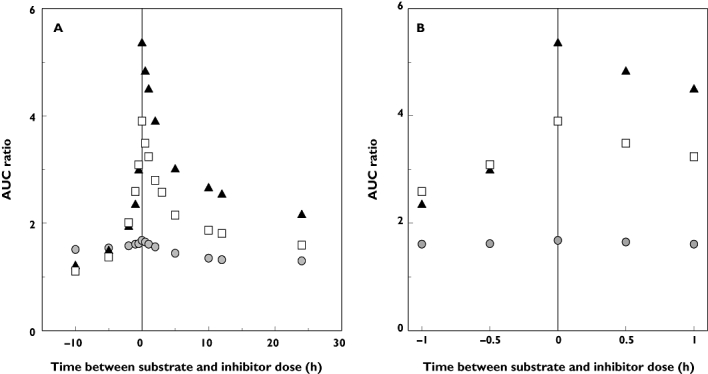
Impact of dosing time from −10 to 24 h on the predicted AUC ratio in the DDI between itraconazole as the inhibitor and the three substrates (A). Focus on the dosing time from −1 to 1 h (B). The increase in AUC was predicted using the dynamic model (with the inclusion of the hydroxy-itraconazole active metabolite) and with alprazolam ( ), midazolam (▴) or triazolam (□) as the victim drugs, with dosing schedules as defined in the methods
), midazolam (▴) or triazolam (□) as the victim drugs, with dosing schedules as defined in the methods
Use of a range of itraconazole ka values between 0.3–1 h−1 had a minimal impact on the predicted AUC ratio for all victim drugs investigated (data not shown), resulting in a maximal difference in AUC ratios of 11% for triazolam. Maximal differences for alprazolam and midazolam were <1% in both cases.
Assessment of inter-individual variability via the dynamic model
Figure 6 shows the median and distribution of predicted AUC ratios from ketoconazole : triazolam (Figure 6A) and itraconazole : triazolam (Figure 6B) DDIs in each virtual trial compared with the actual data reported in nine individuals [40]. In the case of ketoconazole, an under-prediction trend was apparent, as illustrated by the predicted median AUC ratio of 3.7 (obtained from 10 virtual trials, interquartile range of 2.9–4.7) in comparison with the observed median AUC ratio of 8.96 (interquartile range of 6.1–11.3). However, the coefficient of variation was consistent across the virtual trials (29–57%) and with the variation observed in the actual study (50%). Fold difference between maximum and minimum predicted AUC ratios across 10 virtual trials ranged from 2.7–5.9, whereas 6.4-fold was observed in the actual study. In the case of itraconazole, predicted median AUC ratio was 4.7 (obtained from 10 virtual trials, interquartile range of 4.0–5.6) in comparison with the observed median AUC ratio of 6.4 (interquartile range of 6.3–8.9). However, virtual trials estimated only 23–39% coefficient of variation, consistent with the 2.9-fold difference between maximum and minimum predicted AUC ratio across 10 virtual trials. This was in contrast to 105% coefficient of variation and 11-fold difference between maximum and minimum AUC ratio observed in the clinical study due to one significant outlier. If the analysis is performed without this outlier then the fold difference between the maximum and minimum AUC ratio is reduced to 2.7. Coefficient of variation is also significantly reduced (to 28%) and is therefore consistent with the observed data (Figure 6). In both cases, up to three CYP3A5*1/*1 subjects were included per virtual trial and the variation in CYP3A4 liver and intestinal abundance in each virtual trial is displayed in Figures 6C and D, respectively. The mean CYP3A4 abundance in the liver is fairly constant across the virtual trials, with a mean value of 8200 nmol, ranging from 633 to 62 212 nmol, with coefficient of variation of 105.8%. Similarly, the CYP3A4 abundance in the gut has a consistent mean of 60 nmol, but has an approximately 15-fold range and a coefficient of variation of 54.7%.
Figure 6.
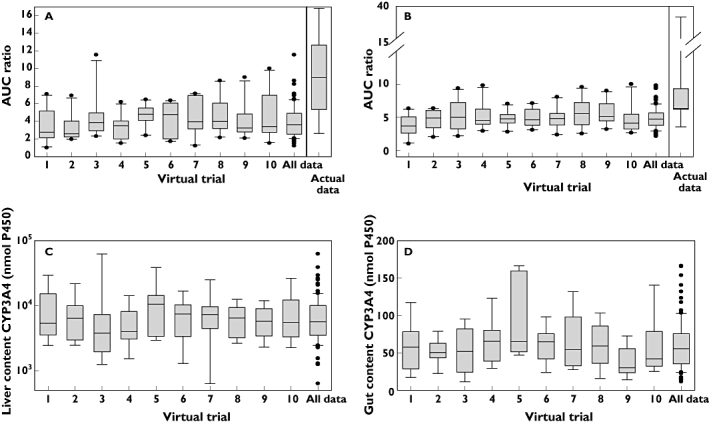
Predicted AUC ratios obtained in 10 individual and combined virtual trials for the ketoconazole-triazolam (A) or itraconazole-triazolam (B) DDI compared with the actual distribution of the data from the nine individual AUC ratios reported by [40]. Predictions used triazolam CLint from HLM ([52, 66–69]) in the dynamic model and ketoconazole and itraconazole parameters as detailed in Table 2. The variation in liver (C) and gut (D) content of CYP3A4 in each trial is additionally displayed (data were the same in both DDI simulations). Box and whisker plots illustrate the distribution in the prediction success; the black line represents the median value, the box represents the inter-quartile range boundaries, lower and upper whiskers represent 10–90% range and outliers are represented by •
Discussion
Simcyp is an ADME simulator with the ability to include inter-individual variability within DDI prediction [7, 9, 15]. Previously, the utility of Simcyp to predict DDIs has been investigated across a range of inhibitors and victim drugs, also assessing the impact of the time course of inhibition and induction [31]. However, the majority of comparisons of DDI prediction between the static and dynamic model used the static model equation [4], or the static model in Simcyp compared with an alternative dynamic model [72] rather than both models within Simcyp. Therefore, the parameters utilized have not been consistent, which confounds the direct comparison between the two models. In addition, some of the studies focus on only one inhibitor [41, 73, 74] or one victim drug [31, 75] and conclusions may therefore be specific to that drug without the ability to ascertain more general trends. In contrast, the current study performs predictions for both the dynamic and static models in Simcyp, allowing consistency in parameters and direct assessment of the impact of the time course. This study comprehensively assessed the prediction accuracy of reversible DDIs of azole inhibitors and different victim drugs, including a representative number of weak, medium and strong interactions. Despite a plethora of research available with these combinations there is still no consensus on certain practices. In addition, a sufficient number of studies per inhibitor or victim drug allowed identification of trends and individual assessment of parameters in the current study. The impact of different parameters including active metabolites of inhibitor, dose timing and ka was investigated, as well as the ability of Simcyp to predict inter-individual variability in the extent of DDIs.
The ability of Simcyp to incorporate inter-individual variability and to generate a range of output values gives an advantage over the equation-based static model (equation 1). It also allows a successful level of prediction when using the unbound drug concentrations, therefore conforming to the free drug hypothesis. This is in contrast to some previous publications advocating the use of total concentration based on the prediction success and number of studies estimated within the 2-fold margin [1, 6]. In the current analysis, the Simcyp static model gave consistently higher results than the dynamic model (Figure 1), as the ratio of the predicted AUC ratios by dynamic : static model were predominantly <1. This is not surprising considering the consistently higher average inhibitor concentration used in static predictions. A comparable number of studies was found within 2-fold of the observed AUC ratio regardless of the model used, with similar bias (afe of 1.68 and 1.34) and precision (rmse of 3.81 and 3.61) for the dynamic or static model, respectively. Use of the weighting according to the number of subjects in the clinical studies did not affect bias and precision of predictions performed in the dynamic model (afe and rmse of 1.64 and 3.61, respectively). Any benefit from incorporating the time-course of the inhibitor in the dynamic model was relatively minor for ketoconazole despite its short half-life (approximately 3.3 ± 1 h). The largest differences between static and dynamic model predictions were observed for interactions involving itraconazole or triazolam. The difference observed with itraconazole studies was likely to be explained by the inclusion of the active metabolite, where studies with a longer time scale included the additional action of hydroxy-itraconazole. The pie charts in Figure 1 indicate that the data are not skewed by a disproportionate amount of DDI studies involving any particular inhibitor or victim drug.
Good overall prediction of DDIs investigated was observed using both the dynamic and static models with 71 and 77% of studies within 2-fold of the observed AUC ratio, respectively. However, a trend for under-prediction was observed (using the parameter input values detailed in Table 2 rather than Simcyp default values) particularly involving the strong interactions (with ketoconazole and itraconazole), regardless of the model used. Five studies (12 data points) were included in the subset where DDIs were reported after single dosing of the inhibitor. One of the assumptions behind the prediction model shown in equation 1 is that the inhibitor concentrations are at steady-state. As demonstrated by the simulations performed by Zhao et al. [41], this would result in significant differences in the DDI magnitude for substrates with a long t1/2. The single dose studies in this subset only involve midazolam or triazolam which both have relatively short half-lives. Predictions from virtual trials for these victim drugs support the findings by Zhao et al. [41] and were not affected by the single dosing regimen of the inhibitor. However, the limited number of clinical studies in this range confounds any firm conclusions. No clear trend was observed to support a difference between static and dynamic DDI predictions or the accuracy against the observed clinical AUC ratio in terms of the bioavailability (0.32–0.88 for midazolam and alprazolam, respectively) [76] or t1/2 (1.5–12 h for triazolam and alprazolam, respectively) of the victim drugs in the dataset. This lack of trend was still apparent when other victim drugs were included in the assessment (i.e. simvastatin and ciclosporin, t1/2 of 2 and 6 h, respectively, and bioavailability (F) < 0.3).
The inclusion of the most abundant and potent active metabolite of itraconazole resulted in improved prediction accuracy in the dynamic model, demonstrating the importance of this parameter in DDI predictions (Figure 3). Simcyp currently allows the inclusion of one metabolite into DDI prediction, excluding the other two potentially relevant itraconazole active metabolites, keto- and N-desalkyl-itraconazole. N-desalkyl-itraconazole has a particularly long t1/2 and a comparable ratio of steady-state average unbound concentration and Ki to the hydroxy metabolite [27]. Therefore, it may contribute to the persistence of inhibition after itraconazole administration. The variation in itraconazole ka of approximately 2-fold of the clinical range in the dynamic model resulted in marginal difference in predicted AUC ratio (<11%). The impact may be seen for the larger range in ka (e.g. at 10-fold as simulated in Zhao et al. [41]), but assuming that the range reported in the clinical study was proportional to the overall patient population, this 10-fold range is unlikely to be observed.
Good prediction accuracy was observed for DDIs involving alprazolam or midazolam as the victim drug, with the majority of these DDIs predicted within 2-fold of the observed value. However, there was an under-prediction trend observed for triazolam DDIs regardless of the model used. Therefore, the impact of alternative in vitro clearance and permeability data used as input parameters was assessed. Minimal improvement in AUC ratio prediction accuracy was seen when triazolam in-house permeability data (from either Caco-2 or MDCK-MDR1 cells) [67] were used in the Qgut model to estimate triazolam FG values, which were consequently used in DDI prediction. The triazolam CLint utilized in AUC ratio predictions represented a mean value from four different studies performed in human liver microsomes. However, in one of these studies [52], a 20-fold difference in CLint was observed across the six donors, resulting in a maximum 5-fold difference (range of 2.20 to 9.54) in AUC ratio in the Simcyp simulations. This AUC ratio difference is not observed in individual trials, but can be observed across the whole virtual population. This difference could be due to other confounding factors rather than a direct result of the CLint variability. Differences in predicted AUC ratio resulting from the use of CLint from human liver microsomes and the default recombinant values in Simcyp were minor in the case of triazolam.
In the current study impact of different dosing regimens (staggered dosing) of the inhibitor on DDI prediction was assessed using an itraconazole interaction. In addition to midazolam (previously studied in a ketoconazole-midazolam interaction pair [41]), triazolam and alprazolam were also included in the assessment. Investigation found that the most pronounced difference based solely on virtual trials was observed with midazolam, and the maximum AUC ratio was observed with simultaneous dosing. However, the same trends were not observed when compared with the observed clinical data with different dosing schedules. For example, the maximum AUC ratio in one study with triazolam [38] was actually observed with substrate dosing 3 h after the inhibitor dose. However, this observed difference may be a result of the study design, considering that food intake was included with the inhibitor dose in this scenario and not when the inhibitor and substrate were simultaneously dosed. The decreases in the AUC for the dosing time >3 h for both midazolam and triazolam was over-estimated in the simulations in comparison with the observed AUC ratios [38]. However, limited availability of clinical data for different dosing regimens and the wide range of DDI magnitude observed for the same dosing regimen confounds any clear trends. The differential impact of dosing time between previous work with ketoconazole [41] and current analysis with itraconazole cannot be explained by the approximately 4-fold longer t1/2 of itraconazole (21 ± 6 h) in comparison with ketoconazole [77].
The prediction of inter-individual variability in DDI magnitude was assessed in comparison with the one study where AUC values of the victim drug ± inhibitor were reported for each individual [40]. In the case of ketoconazole, the ratio of median predicted : observed values across the 10 virtual trials ranged from 0.29–0.52 highlighting the under-prediction trend. However, predicted inter-individual variability in the AUC ratio reflected observed variability, as the coefficient of variation across the trials (45%) was consistent with the actual study (50%). A similar under-prediction trend was observed in the case of itraconazole, but to a lesser extent, as the ratio of median predicted/observed values was 0.74. However, variability estimated across trials for this interaction pair (32% coefficient of variation) was considerably lower in comparison with the observed data with this inhibitor (105%), associated with one significant outlier. If this subject was excluded from the analysis of the itraconazole DDI with triazolam, the predicted variability and differences between minimum and maximum AUC ratio are comparable to the observed data.
Assessment of different physiological, demographic and genetic characteristics of the individual subjects in the virtual population was performed in order to rationalize the variability seen in virtual subjects' AUC ratio, although corresponding data from in vivo trials were not available for direct comparison. This included simulated CYP3A4 content, based on a meta-analyses of liver data [9] or intestinal data from 31 individuals [78], which could only partially explain the inter-individual variability in AUC ratios. The prediction success of DDIs can only be validated against the reported in vivo values. However, it is advisable to consider these data very critically, considering that variability can be introduced as a result of inconsistency in study designs and data analysis. The small size of the population observed in this DDI study (nine subjects) or overall in the studies investigated (mean 10.0 ± 2.76, range 4–20 subjects) may result in a reduced ability to predict accurately population values, either by limiting the variability observed, or increasing the power of one subject if a significantly different result is observed, as illustrated in the case of itraconazole-triazolam.
Overall, this study has reinforced the value of Simcyp as a successful platform to assess the prediction of DDIs in a population based model, as high prediction accuracy was observed for 35 DDIs. An under-prediction trend was observed in the current dataset for strong DDIs, with no overall significant difference in the prediction accuracy between the static and dynamic model. This lack of major difference between the two models can be attributed to the fact that concentrations of azoles are substantially higher in comparison with their Ki regardless of the approach used. However, this cannot be extrapolated to less potent or inhibitors administered at a low dose. Prediction accuracy was related to the particular victim drug in the DDI, with high prediction accuracy observed for midazolam DDIs and lower prediction accuracy seen for triazolam DDIs. Incorporation of the time course of inhibition was mainly beneficial for itraconazole, due to the ability to include the contribution of the active metabolite in the prediction model. An increasing number of compounds with multiple interaction mechanisms or contributing inhibitory metabolites [20, 22, 31] emphasize the importance of the dynamic model approach. Predicted inter-individual variability in DDI magnitude reflected the variability observed in vivo despite some under-prediction of mean, and highlighted the importance of critical evaluation of both the input parameters used in the simulations and the clinical data used for validation of the prediction success.
Acknowledgments
The work was funded by a consortium of pharmaceutical companies (GlaxoSmithKline, Lilly, Novartis, Pfizer and Servier) within the Centre for Applied Pharmacokinetic Research at the University of Manchester. The research was also funded by a grant from Simcyp Ltd. E.J.G was partly financially supported by a Simcyp studentship.
Glossary
Abbreviations
- ADME
absorption, disposition, metabolism and excretion
- afe
average fold error
- AUC
area under the concentration-time curve
- CLint
intrinsic clearance
- DDI
drug-drug- interaction
- F
bioavailability
- FDA
Food & Drug Administration
- FG
fraction escaping metabolism in the intestine
- fmCYP
fraction metabolized by a specific Cytochrome P450 enzyme
- fup
fraction unbound in plasma
- HLM
human liver microsomes
- [I]
inhibitor concentration
- ka
absorption rate constant
- Ki
inhibition constant
- mse
mean squared error
- Qgut
hybrid parameter of enterocytic blood flow and drug permeability
- rmse
root mean squared error
- SD
single dose
- t1/2
half-life of the drug
- Tmax
time at which maximal drug plasma concentration occurs.
Competing Interests
A.R-H. is seconded part time to Simcyp Limited from the University of Manchester, and A.R-H., K.R-Y and G.T.T. are shareholders of the company. K.R.-Y. and G.T.T. are employed by Simcyp Ltd. J.B.H. is on the Simcyp Limited Scientific Advisory Board.
REFERENCES
- 1.Brown HS, Galetin A, Hallifax D, Houston JB. Prediction of in vivo drug-drug interactions from in vitro data. Clin Pharmacokinet. 2006;45:1035–50. doi: 10.2165/00003088-200645100-00006. [DOI] [PubMed] [Google Scholar]
- 2.Brown HS, Ito K, Galetin A, Houston JB. Prediction of in vivo drug-drug interactions from in vitro data: impact of incorporating parallel pathways of drug elimination and inhibitor absorption rate constant. Br J Clin Pharmacol. 2005;60:508–18. doi: 10.1111/j.1365-2125.2005.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galetin A, Hinton LK, Burt H, Obach RS, Houston JB. Maximal inhibition of intestinal first-pass metabolism as a pragmatic indicator of intestinal contribution to the drug-drug interactions for CYP3A4 cleared drugs. Curr Drug Metab. 2007;8:685–93. doi: 10.2174/138920007782109805. [DOI] [PubMed] [Google Scholar]
- 4.Einolf HJ. Comparison of different approaches to predict metabolic drug-drug interactions. Xenobiotica. 2007;37:1257–94. doi: 10.1080/00498250701620700. [DOI] [PubMed] [Google Scholar]
- 5.Huang SM, Temple R, Throckmorton DC, Lesko LJ. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther. 2007;81:298–304. doi: 10.1038/sj.clpt.6100054. [DOI] [PubMed] [Google Scholar]
- 6.Ito K, Hallifax D, Obach RS, Houston JB. Impact of parallel pathways of drug elimination and multiple cytochrome P450 involvement on drug-drug interactions: CYP2D6 paradigm. Drug Metab Dispos. 2005;33:837–44. doi: 10.1124/dmd.104.003715. [DOI] [PubMed] [Google Scholar]
- 7.Jamei M, Marciniak S, Feng K, Barnett A, Tucker G, Rostami-Hodjegan A. The Simcyp® population-based ADME simulator. Expert Opin Drug Metab Toxicol. 2009;5:211–23. doi: 10.1517/17425250802691074. [DOI] [PubMed] [Google Scholar]
- 8.Obach RS, Walsky RL, Venkatakrishnan K, Gaman EA, Houston JB, Tremaine LM. The utility of in vitro cytochrome P450 inhibition data in the prediction of drug-drug interactions. J Pharmacol Exp Ther. 2006;316:336–48. doi: 10.1124/jpet.105.093229. [DOI] [PubMed] [Google Scholar]
- 9.Rostami-Hodjegan A, Tucker GT. Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov. 2007;6:140–8. doi: 10.1038/nrd2173. [DOI] [PubMed] [Google Scholar]
- 10.Ito K, Brown HS, Houston JB. Database analyses for the prediction of in vivo drug-drug interactions from in vitro data. Br J Clin Pharmacol. 2004;57:473–86. doi: 10.1111/j.1365-2125.2003.02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanamitsu S, Ito K, Sugiyama Y. Quantitative prediction of in vivo drug-drug interactions from in vitro data based on physiological pharmacokinetics: use of maximum unbound concentration of inhibitor at the inlet to the liver. Pharm Res. 2000;17:336–43. doi: 10.1023/a:1007509324428. [DOI] [PubMed] [Google Scholar]
- 12.Houston JB, Galetin A. Methods for predicting in vivo pharmacokinetics using data from in vitro assays. Curr Drug Metab. 2008;9:940–51. doi: 10.2174/138920008786485164. [DOI] [PubMed] [Google Scholar]
- 13.Ito K, Chiba K, Horikawa M, Ishigami M, Mizuno N, Aoki J, Gotoh Y, Iwatsubo T, Kanamitsu S, Kato M, Kawahara I, Niinuma K, Nishino A, Sato N, Tsukamoto Y, Ueda K, Itoh T, Sugiyama Y. Which concentration of the inhibitor should be used to predict in vivo drug interactions from in vitro data? AAPS PharmSci. 2002;4:E25. doi: 10.1208/ps040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obach RS. Predicting drug-drug interactions from in vitro drug metabolism data: challenges and recent advances. Curr Opin Drug Discov Devel. 2009;12:81–9. [PubMed] [Google Scholar]
- 15.Rostami-Hodjegan A, Tucker G. ‘In silico’ simulations to assess the ‘in vivo’ consequences of ‘in vitro’ metabolic drug-drug interactions. Drug Discov Today. 2004;1:441–8. doi: 10.1016/j.ddtec.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Galetin A, Burt H, Gibbons L, Houston JB. Prediction of time-dependent CYP3A4 drug-drug interactions: impact of enzyme degradation, parallel elimination pathways, and intestinal inhibition. Drug Metab Dispos. 2006;34:166–75. doi: 10.1124/dmd.105.006874. [DOI] [PubMed] [Google Scholar]
- 17.Obach RS, Walsky RL, Venkatakrishnan K. Mechanism-based inactivation of human cytochrome P450 enzymes and the prediction of drug-drug interactions. Drug Metab Dispos. 2007;35:246–55. doi: 10.1124/dmd.106.012633. [DOI] [PubMed] [Google Scholar]
- 18.Brown HS, Chadwick A, Houston JB. Use of isolated hepatocyte preparations for cytochrome P450 inhibition studies: comparison with microsomes for Ki determination. Drug Metab Dispos. 2007;35:2119–26. doi: 10.1124/dmd.107.017095. [DOI] [PubMed] [Google Scholar]
- 19.Brown HS, Griffin M, Houston JB. Evaluation of cryopreserved human hepatocytes as an alternative in vitro system to microsomes for the prediction of metabolic clearance. Drug Metab Dispos. 2007;35:293–301. doi: 10.1124/dmd.106.011569. [DOI] [PubMed] [Google Scholar]
- 20.Hinton LK, Galetin A, Houston JB. Multiple inhibition mechanisms and prediction of drug-drug interactions: status of metabolism and transporter models as exemplified by gemfibrozil-drug interactions. Pharm Res. 2008;25:1063–74. doi: 10.1007/s11095-007-9446-6. [DOI] [PubMed] [Google Scholar]
- 21.Ogilvie BW, Zhang D, Li W, Rodrigues AD, Gipson AE, Holsapple J, Toren P, Parkinson A. Glucuronidation converts gemfibrozil to a potent, metabolism-dependent inhibitor of CYP2C8: implications for drug-drug interactions. Drug Metab Dispos. 2006;34:191–7. doi: 10.1124/dmd.105.007633. [DOI] [PubMed] [Google Scholar]
- 22.Isoherranen N, Hachad H, Yeung CK, Levy RH. Qualitative analysis of the role of metabolites in inhibitory drug-drug interactions: literature evaluation based on the metabolism and transport drug interaction database. Chem Res Toxicol. 2009;22:294–8. doi: 10.1021/tx800491e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galetin A, Ito K, Hallifax D, Houston JB. CYP3A4 substrate selection and substitution in the prediction of potential drug-drug interactions. J Pharmacol Exp Ther. 2005;314:180–90. doi: 10.1124/jpet.104.082826. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Quinney SK, Gorski JC, Jones DR, Hall SD. Semiphysiologically based pharmacokinetic models for the inhibition of midazolam clearance by diltiazem and its major metabolite. Drug Metab Dispos. 2009;37:1587–97. doi: 10.1124/dmd.109.026658. [DOI] [PubMed] [Google Scholar]
- 25.Isoherranen N, Kunze KL, Allen KE, Nelson W. L, Thummel KE. Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab Dispos. 2004;32:1121–31. doi: 10.1124/dmd.104.000315. [DOI] [PubMed] [Google Scholar]
- 26.Quinney SK, Galinsky RE, Jiyamapa-Serna VA, Chen Y, Hamman MA, Hall SD, Kimura RE. Hydroxyitraconazole, formed during intestinal first-pass metabolism of itraconazole, controls the time course of hepatic CYP3A inhibition and the bioavailability of itraconazole in rats. Drug Metab Dispos. 2008;36:1097–101. doi: 10.1124/dmd.108.020644. [DOI] [PubMed] [Google Scholar]
- 27.Templeton IE, Thummel KE, Kharasch ED, Kunze KL, Hoffer C, Nelson WL, Isoherranen N. Contribution of itraconazole metabolites to inhibition of CYP3A4 in vivo. Clin Pharmacol Ther. 2008;83:77–85. doi: 10.1038/sj.clpt.6100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo SD, Kang E, Jun H, Shin BS, Lee KC, Lee KH. Absorption, first-pass metabolism, and disposition of itraconazole in rats. Chem Pharm Bull. 2000;48:798–801. doi: 10.1248/cpb.48.798. Tokyo. [DOI] [PubMed] [Google Scholar]
- 29.Polasek TM, Polak S, Doogue MP, Rostami-Hodjegan A, Miners JO. Assessment of inter-individual variability in predicted phenytoin clearance. Eur J Clin Pharmacol. 2009;65:1203–10. doi: 10.1007/s00228-009-0703-y. [DOI] [PubMed] [Google Scholar]
- 30.Tucker GT, Houston JB, Huang S-M. Optimizing drug development: strategies to assess drug metabolism/transporter interaction potential – towards a consensus. Br J Clin Pharmacol. 2001;52:107–17. doi: 10.1046/j.0306-5251.2001.temp.1441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahmi OA, Hurst S, Plowchalk D, Cook J, Guo F, Youdim K, Dickins M, Phipps A, Darekar A, Hyland R, Obach RS. Comparison of different algorithms for predicting clinical drug-drug interactions, based on the use of CYP3A4 in vitro data; predictions of compounds as precipitants of interaction. Drug Metab Dispos. 2009;37:1658–66. doi: 10.1124/dmd.108.026252. [DOI] [PubMed] [Google Scholar]
- 32.Rowland-Yeo K, Jamei M, Yang J, Tucker GT, Rostami-Hodjegan A. Physiologically-based mechanistic modelling to predict complex drug-drug interactions involving simultaneous competitive and time-dependent enzyme inhibition by parent compound and its metabolite in both liver and gut – the effect of diltiazem on the time-course of exposure to triazolam. Eur J Pharm Sci. 2010;39:298–309. doi: 10.1016/j.ejps.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang YH. Confidence assessment of the Simcyp time-based approach and a static mathematical model in predicting clinical drug-drug interactions for mechanism-based CYP3A inhibitors. Drug Metab Dispos. 2010;38:1094–104. doi: 10.1124/dmd.110.032177. [DOI] [PubMed] [Google Scholar]
- 34.Chien JY, Lucksiri A, Ernest CS, Gorski JC, Wrighton SA, Hall SD. Stochastic prediction of CYP3A-mediated inhibition of midazolam clearance by ketoconazole. Drug Metab Dispos. 2006;34:1208–19. doi: 10.1124/dmd.105.008730. [DOI] [PubMed] [Google Scholar]
- 35.Ahonen J, Olkkola KT, Neuvonen PJ. Effect of route of administration of fluconazole on the interaction between fluconazole and midazolam. Eur J Clin Pharmacol. 1997;51:415–9. doi: 10.1007/s002280050223. [DOI] [PubMed] [Google Scholar]
- 36.McCrea J, Prueksaritanont T, Gertz BJ, Carides A, Gillen L, Antonello S, Brucker MJ, Miller-Stein C, Osborne B, Waldman S. Concurrent administration of the erythromycin breath test (EBT) and oral midazolam as in vivo probes for CYP3A activity. J Clin Pharmacol. 1999;39:1212–20. doi: 10.1177/00912709922012015. [DOI] [PubMed] [Google Scholar]
- 37.Kharasch ED, Walker A, Hoffer C, Sheffels P. Sensitivity of intravenous and oral alfentanil and pupillary miosis as minimally invasive and noninvasive probes for hepatic and first-pass CYP3A activity. J Clin Pharmacol. 2005;45:1187–97. doi: 10.1177/0091270005280077. [DOI] [PubMed] [Google Scholar]
- 38.Neuvonen PJ, Varhe A, Olkkola KT. The effect of ingestion time interval on the interaction between itraconazole and triazolam. Clin Pharmacol Ther. 1996;60:326–31. doi: 10.1016/S0009-9236(96)90059-4. [DOI] [PubMed] [Google Scholar]
- 39.Olkkola KT, Ahonen J, Neuvonen PJ. The effects of the systemic antimycotics, itraconazole and fluconazole, on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Anesth Analg. 1996;82:511–6. doi: 10.1097/00000539-199603000-00015. [DOI] [PubMed] [Google Scholar]
- 40.Varhe A, Olkkola KT, Neuvonen PJ. Oral triazolam is potentially hazardous to patients receiving systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;56:601–7. doi: 10.1038/clpt.1994.184. [DOI] [PubMed] [Google Scholar]
- 41.Zhao P, Ragueneau-Majlessi I, Zhang L, Strong JM, Reynolds KS, Levy RH, Thummel KE, Huang S-M. Quantitative evaluation of pharmacokinetic inhibition of CYP3A substrates by ketoconazole: a simulation study. J Clin Pharmacol. 2009;49:351–9. doi: 10.1177/0091270008331196. [DOI] [PubMed] [Google Scholar]
- 42.Varhe A, Olkkola KT, Neuvonen PJ. Fluconazole, but not terbinafine, enhances the effects of triazolam by inhibiting its metabolism. Br J Clin Pharmacol. 1996;41:319–23. doi: 10.1046/j.1365-2125.1996.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varhe A, Olkkola KT, Neuvonen PJ. Effect of fluconazole dose on the extent of fluconazole-triazolam interaction. Br J Clin Pharmacol. 1996;42:465–70. doi: 10.1046/j.1365-2125.1996.45111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenblatt DJ, Wright CE, von Moltke LL, Harmatz JS, Ehrenberg BL, Harrel LM, Corbett K, Counihan M, Tobias S, Shader RI. Ketoconazole inhibition of triazolam and alprazolam clearance: differential kinetic and dynamic consequences. Clin Pharmacol Ther. 1998;64:237–47. doi: 10.1016/S0009-9236(98)90172-2. [DOI] [PubMed] [Google Scholar]
- 45.Schmider J, Brockmoller J, Arold G, Bauer S, Roots I. Simultaneous assessment of CYP3A4 and CYP1A2 activity in vivo with alprazolam and caffeine. Pharmacogenetics. 1999;9:725–34. doi: 10.1097/01213011-199912000-00007. [DOI] [PubMed] [Google Scholar]
- 46.Eap CB, Buclin T, Cucchia G, Zullino D, Hustert E, Bleiber G, Golay KP, Aubert AC, Baumann P, Telenti A, Kerb R. Oral administration of a low dose of midazolam (75 µg) as an in vivo probe for CYP3A activity. Eur J Clin Pharmacol. 2004;60:237–46. doi: 10.1007/s00228-004-0762-z. [DOI] [PubMed] [Google Scholar]
- 47.Lam YW, Alfaro CL, Ereshefsky L, Miller M. Pharmacokinetic and pharmacodynamic interactions of oral midazolam with ketoconazole, fluoxetine, fluvoxamine, and nefazodone. J Clin Pharmacol. 2003;43:1274–82. doi: 10.1177/0091270003259216. [DOI] [PubMed] [Google Scholar]
- 48.Olkkola KT, Backman JT, Neuvonen PJ. Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;55:481–5. doi: 10.1038/clpt.1994.60. [DOI] [PubMed] [Google Scholar]
- 49.Tsunoda SM, Velez RL, von Moltke LL, Greenblatt DJ. Differentiation of intestinal and hepatic cytochrome P450 3A activity with use of midazolam as an in vivo probe: effect of ketoconazole. Clin Pharmacol Ther. 1999;66:461–71. doi: 10.1016/S0009-9236(99)70009-3. [DOI] [PubMed] [Google Scholar]
- 50.Lee J-I, Chaves-Gnecco D, Amico JA, Kroboth PD, Wilson JW, Frye RF. Application of semisimultaneous midazolam administration for hepatic and intestinal cytochrome P450 3A phenotyping. Clin Pharmacol Ther. 2002;72:718–28. doi: 10.1067/mcp.2002.129068. [DOI] [PubMed] [Google Scholar]
- 51.Chung E, Nafziger AN, Kazierad DJ, Bertino JS., Jr Comparison of midazolam and simvastatin as cytochrome P450 3A probes. Clin Pharmacol Ther. 2006;79:350–61. doi: 10.1016/j.clpt.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 52.von Moltke LL, Greenblatt DJ, Harmatz JS, Duan SX, Harrel LM, Cotreau-Bibbo MM, Pritchard GA, Wright CE, Shader RI. Triazolam biotransformation by human liver microsomes in vitro: effects of metabolic inhibitors and clinical confirmation of a predicted interaction with ketoconazole. J Pharmacol Exp Ther. 1996;276:370–9. [PubMed] [Google Scholar]
- 53.Yasui N, Kondo T, Otani K, Furukori H, Kaneko S, Ohkubo T, Nagasaki T, Sugawara K. Effect of itraconazole on the single oral dose pharmacokinetics and pharmacodynamics of alprazolam. Psychopharmacology. 1998;139:269–73. doi: 10.1007/s002130050715. [DOI] [PubMed] [Google Scholar]
- 54.Ahonen J, Olkkola KT, Neuvonen PJ. Effect of itraconazole and terbinafine on the pharmacokinetics and pharmacodynamics of midazolam in healthy volunteers. Br J Clin Pharmacol. 1995;40:270–2. [PMC free article] [PubMed] [Google Scholar]
- 55.Backman JT, Kivisto KT, Olkkola KT, Neuvonen PJ. The area under the plasma concentration-time curve for oral midazolam is 400-fold larger during treatment with itraconazole than with rifampicin. Eur J Clin Pharmacol. 1998;54:53–8. doi: 10.1007/s002280050420. [DOI] [PubMed] [Google Scholar]
- 56.Ito K, Ogihara K, Kanamitsu S, Itoh T. Prediction of the in vivo interaction between midazolam and macrolides based on in vitro studies using human liver microsomes. Drug Metab Dispos. 2003;31:945–54. doi: 10.1124/dmd.31.7.945. [DOI] [PubMed] [Google Scholar]
- 57.Yang J, Jamei M, Yeo KR, Tucker GT, Rostami-Hodjegan A. Prediction of intestinal first-pass drug metabolism. Curr Drug Metab. 2007;8:676–84. doi: 10.2174/138920007782109733. [DOI] [PubMed] [Google Scholar]
- 58.Gibbs MA, Thummel KE, Shen DD, Kunze KL. Inhibition of cytochrome P-450 3A (CYP3A) in human intestinal and liver microsomes: comparison of Ki values and impact of CYP3A5 expression. Drug Metab Dispos. 1999;27:180–7. [PubMed] [Google Scholar]
- 59.Galetin A, Brown C, Hallifax D, Ito K, Houston JB. Utility of recombinant enzyme kinetics in prediction of human clearance: impact of variability, CYP3A5, and CYP2C19 on CYP3A4 probe substrates. Drug Metab Dispos. 2004;32:1411–20. doi: 10.1124/dmd.104.000844. [DOI] [PubMed] [Google Scholar]
- 60.Kilford PJ, Stringer R, Sohal B, Houston JB, Galetin A. Prediction of drug clearance by glucuronidation from in vitro data: use of combined cytochrome P450 and UDP-glucuronosyltransferase cofactors in alamethicin-activated human liver microsomes. Drug Metab Dispos. 2009;37:82–9. doi: 10.1124/dmd.108.023853. [DOI] [PubMed] [Google Scholar]
- 61.Klieber S, Hugla S, Ngo R, Arabeyre-Fabre C, Meunier V, Sadoun F, Fedeli O, Rival M, Bourrie M, Guillou F, Maurel P, Fabre G. Contribution of the N-glucuronidation pathway to the overall in vitro metabolic clearance of midazolam in humans. Drug Metab Dispos. 2008;36:851–62. doi: 10.1124/dmd.107.019539. [DOI] [PubMed] [Google Scholar]
- 62.Ito K, Iwatsubo T, Kanamitsu S, Ueda K, Suzuki H, Sugiyama Y. Prediction of pharmacokinetic alterations caused by drug-drug interactions: metabolic interaction in the liver. Pharmacol Rev. 1998;50:387–412. [PubMed] [Google Scholar]
- 63.Barone JA, Moskovitz BL, Guarnieri J, Hassell AE, Colaizzi JL, Bierman RH, Jessen L. Enhanced bioavailability of itraconazole in hydroxypropyl-β-cyclodextrin solution versus capsules in healthy volunteers. Antimicrob Agents Chemother. 1998;42:1862–5. doi: 10.1128/aac.42.7.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galetin A, Houston JB. Intestinal and hepatic metabolic activity of five cytochrome P450 enzymes: impact on prediction of first-pass metabolism. J Pharmacol Exp Ther. 2006;318:1220–9. doi: 10.1124/jpet.106.106013. [DOI] [PubMed] [Google Scholar]
- 65.Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes P450: role of CYP3A4 and CYP3A5. Drug Metab Dispos. 2003;31:938–44. doi: 10.1124/dmd.31.7.938. [DOI] [PubMed] [Google Scholar]
- 66.Patki KC, Von Moltke LL, Harmatz JS, Hesse LM, Court MH, Greenblatt DJ. Effect of age on in vitro triazolam biotransformation in male human liver microsomes. J Pharmacol Exp Ther. 2004;308:874–9. doi: 10.1124/jpet.103.059311. [DOI] [PubMed] [Google Scholar]
- 67.Gertz M, Harrison A, Houston JB, Galetin A. Prediction of human intestinal first-pass metabolism of 25 CYP3A substrates from in vitro clearance and permeability data. Drug Metab Dispos. 2010 doi: 10.1124/dmd.110.032649. (in press); doi: 10.1124/dmd.110.032649. [DOI] [PubMed] [Google Scholar]
- 68.Kuypers DR, de Jonge H, Naesens M, Vanrenterghem Y. Effects of CYP3A5 and MDR1 single nucleotide polymorphisms on drug interactions between tacrolimus and fluconazole in renal allograft recipients. Pharmacogenet Genomics. 2008;18:861–8. doi: 10.1097/FPC.0b013e328307c26e. [DOI] [PubMed] [Google Scholar]
- 69.Yu KS, Cho JY, Jang IJ, Hong KS, Chung JY, Kim JR, Lim HS, Oh DS, Yi SY, Liu KH, Shin JG, Shin SG. Effect of the CYP3A5 genotype on the pharmacokinetics of intravenous midazolam during inhibited and induced metabolic states. Clin Pharmacol Ther. 2004;76:104–12. doi: 10.1016/j.clpt.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 70.Obach RS, Baxter JG, Liston TE, Silber BM, Jones BC, MacIntyre F, Rance DJ, Wastall P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J Pharmacol Exp Ther. 1997;283:46–58. [PubMed] [Google Scholar]
- 71.Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm. 1981;9:503–12. doi: 10.1007/BF01060893. [DOI] [PubMed] [Google Scholar]
- 72.Perdaems N, Blasco H, Vinson C, Chenel M, Whalley S, Cazade F, Bouzom F. Predictions of metabolic drug-drug interactions using physiologically based modelling: two cytochrome P450 3A4 substrates coadministered with ketoconazole or verapamil. Clin Pharmacokinet. 2010;49:239–58. doi: 10.2165/11318130-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 73.Rakhit A, Pantze MP, Fettner S, Jones HM, Charoin JE, Riek M, Lum BL, Hamilton M. The effects of CYP3A4 inhibition on erlotinib pharmacokinetics: computer-based simulation (SimCYP) predicts in vivo metabolic inhibition. Eur J Clin Pharmacol. 2008;64:31–41. doi: 10.1007/s00228-007-0396-z. [DOI] [PubMed] [Google Scholar]
- 74.Youdim KA, Zayed A, Dickins M, Phipps A, Griffiths M, Darekar A, Hyland R, Fahmi O, Hurst S, Plowchalk DR, Cook J, Guo F, Obach RS. Application of CYP3A4 in vitro data to predict clinical drug-drug interactions; predictions of compounds as objects of interaction. Br J Clin Pharmacol. 2008;65:680–92. doi: 10.1111/j.1365-2125.2007.03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyland R, Dickins M, Collins C, Jones H, Jones B. Maraviroc: in vitro assessment of drug-drug interaction potential. Br J Clin Pharmacol. 2008;66:498–507. doi: 10.1111/j.1365-2125.2008.03198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galetin A, Gertz M, Houston JB. Potential role of intestinal first-pass metabolism in the prediction of drug-drug interactions. Expert Opin Drug Metab Toxicol. 2008;4:909–22. doi: 10.1517/17425255.4.7.909. [DOI] [PubMed] [Google Scholar]
- 77.Thummel KE, Shen DD, Isoherranen N, Smith HE. Appendix II: Design and Optimization of Dosage Regimens: Pharmacokinetic Data. Goodman & Gilman's The Pharmacological Basis of Therapeutics. Medical Publishing Division: McGraw-Hill; 2008. [Google Scholar]
- 78.Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 ‘pie’. Drug Metab Dispos. 2006;34:880–6. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]


