Abstract
Objective
To explore biologic correlates to age at onset in patients with juvenile idiopathic arthritis (JIA) using peripheral blood mononuclear cell (PBMC) gene expression analysis.
Methods
PBMCs were isolated from 56 healthy controls and 104 patients with recent-onset JIA (39 with persistent oligoarticular JIA, 45 with rheumatoid factor–negative polyarticular JIA, and 20 with systemic JIA). RNA was amplified and labeled using NuGEN Ovation, and gene expression was assessed with Affymetrix HG-U133 Plus 2.0 GeneChips.
Results
A total of 832 probe sets revealed gene expression differences (false discovery rate 5%) in PBMCs from children with oligoarticular JIA whose disease began before age 6 years (early-onset disease) compared with those whose disease began at or after age 6 years (late-onset disease). In patients with early-onset disease, there was greater expression of genes related to B cells and less expression of genes related to cells of the myeloid lineage. Support vector machine analyses identified samples from patients with early- or late-onset oligoarticular JIA (with 97% accuracy) or from patients with early- or late-onset polyarticular JIA (with 89% accuracy), but not from patients with systemic JIA or healthy controls. Principal components analysis showed that age at onset was the major classifier of samples from patients with oligoarticular JIA and patients with polyarticular JIA.
Conclusion
PBMC gene expression analysis reveals biologic differences between patients with early-and late-onset JIA, independent of classification based on the number of joints involved. These data suggest that age at onset may be an important parameter to consider in JIA classification. Furthermore, pathologic mechanisms may vary with age at onset, and understanding these processes may lead to improved treatment of JIA.
Juvenile idiopathic arthritis (JIA) encompasses the majority of childhood arthritides. Six subtypes of JIA are distinguished largely on the basis of clinical and laboratory features present in the first 6 months of disease, with a seventh category reserved for individuals who meet insufficient criteria or cannot be unambiguously classified. There is increasing evidence for heterogeneity within the defined subtypes, as well as commonalities between subtypes, that are not currently accounted for in the JIA classification system (1). Genome-level technologies that provide comprehensive assessments of gene sequence and expression offer unprecedented opportunities to further define JIA subtypes based on molecular phenotypes and to advance understanding of disease mechanisms that will improve therapeutic approaches.
The JIA classification system, proposed and subsequently revised by the International League of Associations for Rheumatology (ILAR) (2, 3), was intended to define relatively homogeneous and mutually exclusive categories of childhood arthritis for research purposes. The ILAR classification system has become generally accepted (4) and is currently used to define specific JIA phenotypes for genome-wide association studies aimed at defining genetic polymorphisms that create susceptibility to childhood arthritis. Evidence based on gene expression signatures found early in disease indicates that there are biologic differences between controls and the major JIA subtypes including persistent oligoarticular, rheumatoid factor (RF) negative, polyarticular, enthesitis-related, and systemic arthritis (5).
Despite support for a biologic basis for the ILAR classification, there is also emerging evidence for significant heterogeneity within JIA subtypes (6–8). For example, Fall et al showed heterogeneity based on high ferritin levels and a distinct gene expression pattern in a subset of systemic arthritis patients with active or occult macrophage activation syndrome, a potentially life-threatening complication of certain rheumatic diseases (6). In addition, following up on previous observations (5), Griffin et al showed that polyarticular JIA appears to include subsets of patients with a prominent monocyte activation signature or varying proportions of a counter-regulatory gene expression signature (7).
One clinical parameter that has been reported to have biologic implications for disease, but that is not used for JIA classification except for enthesitis-related arthritis, is the age at which symptoms attributable to JIA begin, also known as age at onset. The involvement of HLA genes has been implicated in age at onset (9–11), and a recent study using high-resolution HLA typing identified additional genetic differences that may influence age at onset in JIA (12). In addition, patients with early-onset JIA have a different Ig κ light chain repertoire (13) and are more likely to be antinuclear antibody (ANA) positive (14).
The current study represents an ongoing multicenter investigation into molecular features of peripheral blood mononuclear cells (PBMCs) from patients with recent-onset JIA prior to treatment with disease-modifying antirheumatic drugs (DMARDs) or biologics (5–7). In the current study, substantial peripheral blood gene expression differences were identified in patients with early-onset persistent oligoarticular JIA compared with patients with late-onset disease. Clustering revealed biologic themes related to age at onset that extended to patients with RF-negative polyarticular JIA but not to healthy controls or patients with systemic arthritis. Results of principal components analyses (PCAs) are consistent with the view that age at onset may be an important characteristic for classification of certain JIA subtypes and perhaps may be more biologically relevant than the number of joints involved. Differential gene expression patterns together with known and emerging genetic and antibody repertoire differences suggest that pathologic mechanisms may differ between patients with early-onset disease and those with late-onset disease, with important implications for the treatment of patients with JIA.
PATIENTS AND METHODS
Patients and controls
Prior to treatment with DMARDs or biologics, 104 patients with recent-onset JIA (Table 1) were enrolled at 5 clinical centers (46 at Cincinnati Children’s Hospital Medical Center [CCHMC], 12 at Children’s Hospital of Philadelphia, 13 at Children’s Hospital of Wisconsin, 28 at Schneider Children’s Hospital, and 5 at Toledo Children’s Hospital). Using cumulative clinical and laboratory information from the 2-year study period (including nonstudy visits when appropriate), subjects were classified according to ILAR criteria (3) (Table 1). For simplicity, patients with persistent oligoarticular disease and those with RF-negative polyarticular disease are hereafter referred to as having oligoarticular JIA and polyarticular JIA, respectively. The 56 controls were apparently healthy children from the Cincinnati area. Study subjects have been described previously (5–7).
Table 1.
Characteristics of the study subjects*
| Controls (n = 56) | Patients with persistent oligoarticular JIA (n = 39) | Patients with RF-negative polyarticular JIA (n = 45) | Patients with systemic JIA (n = 20) | |
|---|---|---|---|---|
| No. with early/late disease onset | –† | 23/16 | 18/27 | 14/6 |
| Age at onset, mean ± SD years | NA | 5.9 ± 4.2 | 7.4 ± 4.7 | 5.3 ± 4.7 |
| Disease duration at sampling, mean ± SD years | NA | 0.4 ± 0.3 | 0.7 ± 0.6 | 0.3 ± 0.5 |
| Age at sampling, mean ± SD years | 9.1 ± 5.1 | 6.3 ± 4.3 | 8.1 ± 4.7 | 5.6 ± 4.6 |
| Female sex, % | 57 | 69 | 87 | 35 |
| ANA positive, no./no. tested | ND | 16/37 | 17/34 | 2/5 |
JIA = juvenile idiopathic arthritis; RF = rheumatoid factor; NA = not applicable; ANA = antinuclear antibody; ND = not determined.
In the case of controls, their age at the time of sampling was used (20 were age <6 years and 36 were age ≥6 years).
Sample preparation
Peripheral blood was collected into acid citrate dextrose tubes as described previously (5). PBMCs were isolated by Ficoll (GE Healthcare) gradient centrifugation at the site of collection, and RNA was immediately stabilized in TRIzol (Invitrogen). Samples were processed as rapidly as possible and stored at −80°C. Samples were shipped on dry ice to CCHMC, where RNA was isolated, purified over RNeasy columns (Qiagen), and stored in water at −80°C.
RNA processing and analysis
RNA quality was assessed using standard protocols adopted by the CCHMC Gene Expression Microarray Core, using an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA (100 ng per sample) was labeled using NuGEN Ovation Version 1 (NuGEN Technologies). Labeled samples were hybridized to Affymetrix HG-U133 Plus 2.0 GeneChips and scanned with an Agilent G2500A GeneArray scanner. This array has 54,681 probe sets measuring more than 47,000 transcripts; thus, many transcripts are interrogated by more than 1 probe set (e.g., STAT1 is represented by 7 probe sets). In addition, many probe sets are annotated with more than 1 transcript identifier, so the actual number of transcripts (and thus the actual number of differentially expressed genes) is difficult to specify. We have attempted to apply the terms gene and probe set accurately, but they are occasionally used interchangeably.
Statistical analysis
Affymetrix GeneChip data were imported into GeneSpring GX 7.3.1 (Agilent Technologies) and preprocessed using robust multichip analysis (RMA) (15) followed by normalization of each probe set to the median of all samples. Distance-weighted discrimination was used to align centroids of predefined groups to adjust for batch-to-batch variation (16). Statistical analysis was performed using the t-test with a Benjamini and Hochberg–corrected false discovery rate (FDR) of 5%. Comparisons of samples from patients were made based on the age at which symptoms attributable to JIA began, commonly referred to as age at onset, and for controls based on the age at which samples were acquired. Patients were enrolled very early in the disease course, and consequently the difference between age at onset and age at sample acquisition is small (<4–8 months depending on JIA subtype) (Table 1).
Hierarchical clustering was performed using Pearson’s correlation. Principal components and support vector machine analyses were performed on samples. Support vector machine analysis is a class prediction method that uses only gene expression patterns to determine class membership. This analysis was performed as cross-validation rather than as a training set/test set methodology. With this method, the expected accuracy is determined as the percentage of the group with the highest population, since the simplest classification is to put all samples into a single group. For example, the expected prediction accuracy for samples from patients with early-onset oligoarticular disease is 23/(23 + 16), or 59%, where 23 is the number of samples in the group with the largest population and 23 + 16 is the total number of samples from patients with persistent oligoarticular disease. Functional analysis of the data was performed using Ingenuity Pathways Analysis (Ingenuity Systems; www.ingenuity.com). GeneChip data are available from the National Center for Biotechnology Information Gene Expression Omnibus, series accession GSE20307.
RESULTS
Gene expression patterns in oligoarticular JIA based on age at onset
PBMCs were obtained for global gene expression analysis from patients with recent-onset oligoarticular JIA prior to treatment with DMARDs or biologics (Table 1). Comparing patients with early disease onset (age at onset <6 years; n = 23) and patients with late disease onset (age at onset ≥6 years; n = 16) revealed 832 probe sets detecting differentially expressed genes (FDR 5%) (see Supplemental Table 1, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home). Clustering of samples using these 832 probe sets is shown in Figure 1A. Using support vector machine analysis, 38 of 39 samples from patients with oligoarticular disease (97%) were classified correctly as being from patients with early-onset or late-onset disease (Table 2).
Figure 1.
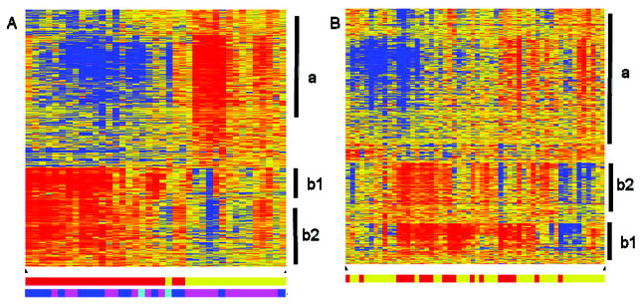
Heat maps of samples using age at onset–related probe sets. Eight hundred thirty-two probe sets reflecting differentially expressed genes were identified between samples from patients with early-onset persistent oligoarticular disease and from patients with late-onset persistent oligoarticular disease (false discovery rate 5%). Hierarchical clustering of the probe sets was used to identify patterns in the data from patients with persistent oligoarticular disease (A) or from control subjects (B). Vertical bars indicate clusters of probe sets with similar expression patterns related to cells of the myeloid lineage (a), immunoglobulins (b1), or B cells (b2). In the clustering diagram, red indicates increased expression relative to the median of all samples, blue indicates decreased expression relative to the median of all samples, and yellow indicates the median of all samples. Bottom in A and B, Bars indicate samples from patients with early-onset disease (younger patients) (red) or samples from patients with late-onset disease (older patients) (yellow). Bottom in A only, Bars indicate antinuclear antibody status (blue indicates positive, magenta indicates negative, cyan indicates not tested).
Table 2.
Results of support vector machine analyses*
| Subject classification | Prediction accuracy, % | Expected by chance, %† |
|---|---|---|
| Oligoarticular JIA | 97 | 59 |
| RF-negative polyarticular JIA | 89 | 62 |
| Systemic JIA | 75 | 70 |
| Controls | 70 | 64 |
See Table 1 for definitions.
See Patients and Methods for details.
A readily identifiable group of probe sets with a high proportion related to immunoglobulins was apparent in the clustering (Figure 1A, cluster b1). In order to ensure that this group of probe sets representing a small number of genes did not unduly bias the analysis, the 106 probe sets corresponding to these genes were removed from the list and the samples were reclustered, resulting in similar separations (data not shown). All 832 probe sets were used in later analyses in order to include as much biologic information as possible.
Biologic meaning of the differentially expressed genes
Clustering the samples from patients with oligoarticular JIA using the 832 probe sets identified 2 major gene expression patterns (Figure 1A, clusters a and b). Gene cluster a consisted of 508 probe sets detecting genes with greater expression in patients with late-onset oligoarticular disease. Using Ingenuity Pathway Analysis, the top-ranked network was related to antigen presentation and cell-mediated immune response (data not shown). Inspection of the 508 probe sets revealed an enrichment of genes expressed in cells of the myeloid lineage including platelets (glycoprotein Ib, platelet, alpha polypeptide; glycoprotein Ib, platelet, beta polypeptide; glycoprotein IX, platelet; glycoprotein VI, platelet; glycoprotein IIIa, platelet [CD61]; platelet-derived growth factor alpha; platelet-derived growth factor C; platelet endothelial aggregation receptor 1; platelet factor 4 [CXCL4]; proplatelet basic protein [CXCL7]). Other biologic themes included coagulation (coagulation factor XIII, A1 peptide; thrombospondin 1; tissue factor pathway inhibitor) and innate immunity (clusterin; complement factor H).
Gene cluster b (Figure 1A; separated into b1 and b2) comprised 324 probe sets indicating genes with increased expression in samples from patients with early-onset oligoarticular disease compared with that in samples from patients with late-onset oligoarticular disease. Ingenuity Pathway Analysis of this group of probe sets identified a network related to humoral immunity (data not shown). Further analysis of the individual genes showed that cluster b1 contained 106 probe sets, many of which are annotated as immunoglobulins or immunoglobulin-related genes (as noted above). Cluster b2 contained 218 probe sets with a strong representation of genes related to B cells. They included the prototypical B cell marker, CD19, and other B cell–related markers (CD22, CD38, CD40, CD72, CD79A, CD79B, CD200). Additionally, expression differences were observed for transcription factors (TCF3, EBF, EIF5B) and other B cell proteins (BCL11A, BCNP1, BLNK, BRDG1, BTLA, BANK1, SP110) (Table 3).
Table 3.
Selected B cell related genes*
| Probe ID no.‡ | Gene symbol‡ | p§ | Controls | Fold change† | ||
|---|---|---|---|---|---|---|
| Patients with persistent oligoarticular JIA | Patients with RF-negative polyarticular JIA | Patients with systemic JIA | ||||
| 1558662_s_at | BANK1 | 0.003 | 1.17 | 1.74 | 1.71 | 2.24 |
| 222891_s_at | BCL11A | 0.004 | 1.18 | 1.43 | 1.24 | 1.73 |
| 1553369_at | BCNP1 | 0.009 | 1.13 | 1.97 | 1.67 | 2.39 |
| 207655_s_at | BLNK | 0.005 | 1.23 | 1.65 | 1.59 | 2.1 |
| 1554343_a_at | BRDG1 | 0.01 | 1.18 | 1.68 | 1.61 | 2.28 |
| 236226_at | BTLA | 0.04 | 1.03 | 1.62 | 1.6 | 1.52 |
| 206398_s_at | CD19 | 0.004 | 1.21 | 2.16 | 2.07 | 2.75 |
| 204581_at | CD22 | 0.008 | 1.07 | 1.83 | 1.53 | 2.05 |
| 205692_s_at | CD38 | 0.003 | 1.5 | 1.61 | 1.48 | 1.44 |
| 215346_at | CD40 | 0.03 | 1.13 | 1.38 | 1.22 | 1.46 |
| 215925_s_at | CD72 | 0.03 | 1.17 | 2.01 | 2.23 | 3.14 |
| 205049_s_at | CD79A | 0.04 | 1.24 | 1.62 | 1.56 | 2.23 |
| 205297_s_at | CD79B | 0.03 | 1.25 | 1.62 | 1.59 | 1.95 |
| 209583_s_at | CD200 | 0.04 | 1 | 1.74 | 1.5 | 1.71 |
| 232204_at | EBF | 0.008 | 1.15 | 1.81 | 1.4 | 1.86 |
| 201025_at | EIF5B | 0.02 | 0.97 | 1.2 | 1.1 | 0.97 |
| 209762_x_at | SP110 | 0.03 | 1.17 | 1.24 | 1.1 | 1.19 |
| 210776_x_at | TCF3 | 0.003 | 1.02 | 1.17 | 1.14 | 1.08 |
See Table 1 for definitions.
Ratio of geometric mean of expression in patients with early-onset disease:geometric mean of expression in patients with late-onset disease, or ratio of geometric mean of expression in younger controls (age <6 years at time of sampling):geometric mean of expression in older controls (age ≥ 6 years at time of sampling).
Ratio of geometric mean of expression in patients with early-onset disease:geometric mean of expression in patients with late-onset disease, or ratio of geometric mean of expression in younger controls (age <6 years at time of sampling):geometric mean of expression in older controls (age ≥ 6 years at time of sampling).
Affymetrix HG-U133 Plus 2.0 GeneChips probe set ID numbers and gene symbols.
By false discovery rate adjusted t-test comparing samples from patients with early- and late-onset oligoarticular disease.
CD19+ B cells do not differ between patients with early-onset oligoarticular JIA and patients with late-onset oligoarticular JIA
PBMC samples from a subset of the patients with oligoarticular JIA were analyzed by flow cytometry. The average percentage of CD19+ B cells was 1.3-fold higher in patients with early-onset disease than in patients with late-onset disease (n = 18 and n = 17, respectively), but this difference was not statistically significant. This finding is consistent with the finding of a previous study (17) that showed a slight, but not statistically significant, increase in CD19+ B cells in patients with oligoarticular JIA; however, Wouters et al identified a significant increase (<1.8-fold) in CD5+ B cells between the same groups. Unfortunately, CD5+ B cells were not tested in the current study.
Analysis of gene expression patterns relative to patient characteristics
Variables other than age at onset were investigated as a possible explanation for the differentially expressed genes. Clustering was not related to the sex of the patient, the duration of disease prior to sampling (mean ± SD 0.5 ± 0.3 years for early onset and 0.3 ± 0.3 years for late onset), or the clinical center at which the samples were collected. The mean number of active joints did not differ between patients with early-onset disease and patients with late-onset disease (1.4 and 1.1, respectively; P = 0.74).
Considering ANA status according to our most recent information (as reported by the clinical laboratory at each center, with up to 2 years of followup), 14 of 22 patients with early-onset oligoarticular disease (1 patient not tested) were ANA positive, while 2 of 15 patients with late-onset oligoarticular disease (1 patient not tested) were ANA positive. A direct comparison of samples (by t-test; FDR 5%) from ANA-positive and ANA-negative patients did not identify differentially expressed genes. However, more ANA-positive patients had the early-onset signature as determined by membership in the early-onset cluster (14 of 22 patients with early-onset disease) compared with membership in the late-onset cluster (2 of 15 patients with late-onset disease) (χ2 = 9.2, P ≤ 0.003).
In this study, patients were enrolled and PBMCs were obtained as early in the disease course as possible; therefore, there was an average of only 4–8 months between disease onset and sampling (Table 1). In order to ensure that our findings related to age at onset rather than age at sampling, samples from healthy controls were analyzed. Comparing samples obtained from control subjects at age <6 years (n = 20) with those obtained from control subjects at age ≥6 years (n = 36) identified only 22 probe sets detecting differentially expressed genes. Of these 22 probe sets, 11 were also found in the list of 832 probe sets from samples from patients with persistent oligoarticular disease. To further explore possible age-related effects independent of disease, samples from the 56 normal controls were subjected to hierarchical clustering using the 832 probe sets. This resulted in a nearly random distribution of control samples regardless of age at sampling (Figure 1B), in sharp contrast to the result with early-onset versus late-onset oligoarticular JIA (Figure 1A). Prediction accuracy using support vector machine analysis was only 70%, compared with 64% expected by chance (Table 2). These findings show that the age of the patient does not contribute significantly to the large number of differentially expressed genes found in early-onset JIA versus late-onset JIA. Instead, the differences are due to disease processes modified by the age at which disease begins.
Since the list of 832 probe sets was derived by comparing patients with early-onset disease and patients with late-onset disease, we wanted to determine if there were differences between patients and controls. Since the B cell signature was most prominent in early-onset disease and the myeloid signature was prominent in patients in whom disease started later, we compared patients with controls in each age group using a gene expression index (7, 18) (geometric mean of the expression of the 218 B cell probe sets or 508 myeloid probe sets). The B cell signature was present in 9 of 23 samples from patients with early-onset oligoarticular JIA compared with 2 of 20 samples from healthy controls (χ2 = 4.8, P < 0.029), while the myeloid signature was present in 15 of 16 samples from patients with late-onset oligoarticular JIA but in only 5 of 20 samples from older controls (χ2 = 19.3, P < 0.00002). In contrast, the B cell signature was not prominent in patients with late-onset oligoarticular disease, nor was the myeloid signature found in patients with early-onset oligoarticular disease. This demonstrates that subsets of the gene expression differences distinguishing early-onset and late-onset oligoarticular JIA also distinguish patients with arthritis from control subjects, depending on their age.
Gene expression patterns similar to those in oligoarticular JIA are seen in early-onset polyarticular JIA, but not in systemic JIA
To determine if these gene expression differences could be extended to other JIA subtypes, the 832 probe sets were used to cluster samples from patients with polyarticular JIA or systemic JIA (Figure 2A). Hierarchical clustering showed that the differentially expressed genes were able to separate samples from patients with early- and late-onset polyarticular JIA, with only 3 outliers. Support vector machine analysis indicated that these 832 probe sets could correctly predict the age at onset category of patients with polyarticular JIA with 89% accuracy, compared with 62% expected by chance (Table 2).
Figure 2.
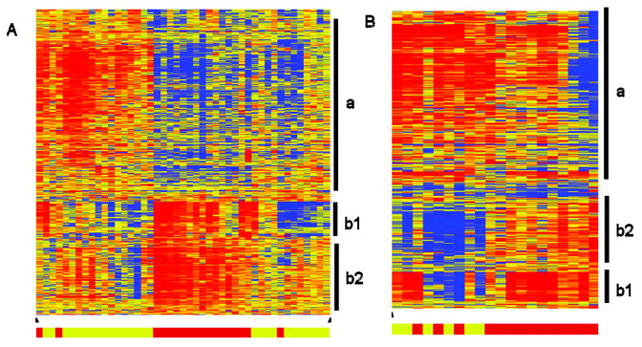
Evaluation of 832 age at onset–related probe sets in additional subtypes of juvenile idiopathic arthritis (JIA). Eight hundred thirty-two probe sets reflecting differentially expressed genes between samples from patients with early-onset persistent oligoarticular disease and from patients with late-onset persistent oligoarticular disease were used to cluster samples from patients with rheumatoid factor–negative polyarticular JIA (A) or from patients with systemic JIA (B). Vertical bars indicate clusters of probe sets with similar expression patterns related to cells of the myeloid lineage (a), immunoglobulins (b1), or B cells (b2). In the clustering diagram, red indicates increased expression relative to the median of all samples, blue indicates decreased expression relative to the median of all samples, and yellow indicates the median of all samples. Bottom, Bars indicate samples from patients with early-onset disease (younger patients) (red) or samples from patients with late-onset disease (older patients) (yellow).
Clustering of samples from patients with systemic JIA showed expression patterns that differed from those in samples from patients with either oligoarticular or polyarticular JIA (Figure 2B). It was noted that the myeloid-related probe sets had increased expression in most samples from patients with systemic JIA, including those from patients with early-onset disease. This was in contrast to decreased expression in samples from patients with early-onset oligoarticular or early-onset polyarticular disease. Additionally, some of the samples from patients with late-onset systemic disease showed a difference in expression of the immunoglobulin-related probe sets compared with that of the B cell–related probe sets, a pattern that differed from that of the other subtypes of JIA. Support vector machine analysis prediction accuracy of samples from patients with systemic JIA was only 75%, compared with 70% expected by chance (Table 2). Gene expression data were also available from patients with enthesitis-related arthritis (n = 29), RF-positive polyarticular JIA (n = 15), psoriatic arthritis (n = 8), and extended oligoarticular JIA (n = 7) but were not included in our analysis due to small numbers of patients with early-onset disease in each group.
Polyarticular JIA, but not systemic JIA, identifies age at onset–related probe set differences
Separate analyses were performed using samples from patients with polyarticular JIA or systemic JIA. A comparison of samples from patients with early-onset (n = 17) or late-onset (n = 28) polyarticular JIA identified 531 probe sets detecting differentially expressed genes, of which 224 (42%) overlapped with the 832 probe sets defined using samples from patients with oligoarticular JIA. The overlapping genes included many related to B cells (CD19, CD22, CD72, BANK1, BCL11A, BCNP1, BLNK, BRDG1, BTLA) and immunoglobulin-related probe sets. A similar analysis of samples from patients with early-onset (n = 14) and late-onset (n = 6) systemic JIA (FDR 10% to allow for the smaller numbers of samples) did not identify any differentially expressed probe sets. These findings support the conclusion that age at onset is an important characteristic for patients with polyarticular JIA but not for those with systemic JIA.
Age at onset is more informative than the number of joints involved for certain JIA subtypes
Since samples from patients with oligoarticular or polyarticular JIA could distinguish them as having early-onset or late-onset disease using the same probe sets, the question arose as to whether the expression patterns were the same in both JIA subtypes. To address this, PCA (a statistical method that identifies and visually displays the key underlying patterns in a multidimensional data set) was performed on samples from these subtypes, using the 832 age at onset–related probe sets (Figure 3). The first component explained 41% of the variance and corresponded to the age at onset of the patients (Figure 3A), while none of the first 3 components related to JIA subtype (Figure 3B). This result suggests that age at onset is more informative than disease subtype for distinguishing JIA patients based on PBMC gene expression in the context of the identified signature.
Figure 3.
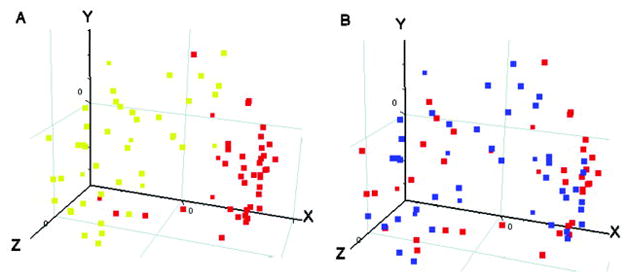
Principal components analysis (PCA) for 832 age at onset–related probe sets. Samples from patients with persistent oligoarticular juvenile idiopathic arthritis (JIA) and patients with rheumatoid factor (RF)–negative polyarticular JIA were arranged according to PCA using the 832 age at onset–related probe sets. The first 3 principal components are shown, comprising 41% (x-axis), 11% (y-axis), and 6% (z-axis) of the total variation observed. Each point represents 1 sample. A, Red indicates early-onset disease, and yellow indicates late-onset disease. B, Red indicates persistent oligoarticular disease, and blue indicates RF-negative polyarticular disease.
DISCUSSION
he current study was undertaken to explore heterogeneity within the subgroup of JIA patients currently classified as having persistent oligoarticular disease and ended up also identifying commonalities between polyarticular and oligoarticular subtypes. Since some clinical observations and genetic studies (9–11) suggest that the age at which disease begins may have biologic implications, we investigated whether the age at onset is reflected by the molecular phenotype of the disease. The rationale for choosing age 6 years as the cutoff was based on genetic studies showing that some major histocompatibility complex (MHC) genes operative in JIA appear to have a “window-of-effect” during which time they may contribute to risk of disease, but that they may be neutral or even protective at other times (9–11). For many of the MHC genes, this window is limited to the first 6 years of life.
The comparison of PBMC gene expression profiles obtained from oligoarticular JIA patients with age at onset <6 years versus those with age at onset ≥6 years identified 832 probe sets representing differentially expressed genes. Hierarchical clustering of these 832 probe sets revealed 2 main clusters distinguishing the 2 groups of patients. One cluster (cluster a) comprised genes with higher levels of expression in patients with late-onset oligoarticular JIA and was enriched for the genes related to cellular immunity and myeloid cell lineage (Figure 1A). This cluster overlaps with the monocyte signature we previously reported in a subset of older patients with polyarticular JIA (7).
The other cluster (cluster b) was highly enriched for genes related to humoral immunity encoding for immunoglobulins, other B lymphocyte–related cell surface markers, and B cell–specific proteins including several transcription factors (Figure 1). Additional evidence that cluster b is derived from B lymphocytes comes from a comparison of the list of probe sets in this cluster with B cell signatures generated by other groups. Of 324 probe sets in cluster b (Figure 1A), 97 overlapped with a list of probe sets representing differentially expressed genes in purified B cells (19). In contrast, there was no overlap with the signatures of other purified cell types, including T cells and granulocytes (19).
A separate study by Hystad et al (20) used microarray analyses to identify gene expression signatures associated with various stages of B cell differentiation (i.e., hematopoietic stem cells, early B cells, pro–B cells, pre–B cells, and immature B cells). Of the 324 probe sets in cluster b, many overlap with genes that are expressed predominantly in immature B cells, including several transcription factors (TCF3, EBF, EIF5B). The overlap also includes many immunoglobulin-related genes as well as genes encoding cell surface markers CD19 and CD79A. Combined with the absence of transcripts from the gene encoding CD27, a marker of memory B cells, it appears that cluster b is derived from relatively early stage transitional and perhaps mature naive B cells. This is consistent with a recent report describing detectable expansion of transitional B cells in peripheral blood of patients with oligoarticular JIA (21).
Martini previously suggested that 1 goal of the ILAR classification, to define homogeneous subgroups, might be better served by combining patients with oligoarticular, RF-negative polyarticular, or psoriatic arthritis and exploring the value of variables such as age at onset, symmetry of arthritis, and ANA status to define subsets (22). This concept was supported by a subsequent study showing that ANA-positive patients were similar in terms of age at onset, sex ratio, frequency of symmetric arthritis, and frequency of iridocyclitis (14). In addition, it was shown that lymphoid neogenesis and plasma cell infiltration of the synovium were more frequent in ANA-positive JIA patients rather than being related to disease activity or severity (23). The results in the current study are supportive of patient regrouping, since we identified a B cell signature and a high preponderance of ANA-positive patients in early-onset oligoarticular JIA. Further, the B cell signature was present in patients with early-onset RF-negative polyarticular JIA. In the same manner, the myeloid signature was present in patients with late-onset oligoarticular and those with late-onset RF-negative polyarticular disease. Taken together, the signatures identified in this study suggest that early-onset JIA and late-onset JIA have similarities that cross the boundaries proscribed by the JIA classification, the derivation of which was based mainly on the number of active joints.
In our study, we enrolled patients as early in the disease course as possible, and therefore age at onset and age at sampling were similar. In general, healthy younger children have a slightly higher proportion of B lymphocytes in their peripheral circulation (24), and the first 6 years of life are characterized by a particularly steep increase in the levels of serum immunoglobulins. Therefore, we considered age-related normal physiologic changes as a possible explanation for the observed differences between patients with early-onset disease and those with late-onset disease. To address this possibility, samples from healthy controls were analyzed to assess the contribution of age. Using the same statistical methods and similar sample sizes, we identified only 22 probe sets detecting differentially expressed genes between samples from control subjects age <6 years and samples from control subjects age ≥6 years. Of these, 11 overlapped with the 832 probe sets from samples from patients with persistent oligoarticular disease. One caveat, however, is that fewer subjects in the control group than in the patient group were age <2 years. Nevertheless, additional evidence that the observed gene expression differences are a feature of disease rather than simply a reflection of age is the ability of the patterns from the 832 probe sets to separate samples from younger and older patients with oligoarticular disease, but not from younger and older control subjects, by either hierarchical clustering or support vector machine analyses.
The study was expanded to assess whether differences related to age at onset could be identified in other subtypes of JIA. Patterns similar to those in oligoarticular JIA were found in patients with RF-negative polyarticular JIA but not in patients with systemic arthritis, indicating disease subtype specificity. It was also apparent using PCA that the age at onset was a more important characteristic than the JIA subtype, which is defined to a large extent by the number of affected joints.
In our study, the B cell signature was present in patients with disease onset at age <6 years. The fact that healthy children in this age group have a physiologic increase in the activity of B cells is intriguing. One might speculate that this early “physiologic hyperactivity” of B cells during development contributes, in the context of additional genetic and environmental factors, to susceptibility to JIA. Interestingly, a similar age-dependent effect seems to play a role in the development of B cell autoimmunity to a myelin surface antigen in pediatric multiple sclerosis (25).
Overall, the differential gene expression patterns that distinguish patients with early- or late-onset oligoarticular or polyarticular JIA suggest that different pathologic mechanisms may be active depending on the age at disease onset. Moreover, the expression differences related to age at onset appear to be more robust than the number of joints involved at classifying samples from patients with oligoarticular and polyarticular JIA subtypes. Considering other genetic and antibody repertoire differences, we propose that age at onset should be considered in future efforts to refine JIA classification.
Acknowledgments
We acknowledge and appreciate the contributions of Dr. Jarek Meller (CCHMC) for discussions regarding the statistical analysis, the CCHMC Gene Expression Microarray Core for working to ensure the production of high-quality data in a timely manner, and the CCHMC division of Biomedical Informatics for providing sufficient computing capacity and support for this data analysis.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Barnes had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Barnes, Grom, Thompson, Griffin, Colbert, Glass.
Acquisition of data. Barnes, Grom, Thompson, Griffin, Luyrink, Colbert, Glass.
Analysis and interpretation of data. Barnes, Grom, Thompson, Griffin, Colbert, Glass.
References
- 1.Thompson SD, Barnes MG, Griffin TA, Grom AA, Glass DN. Heterogeneity in juvenile idiopathic arthritis: impact of molecular profiling based on DNA polymorphism and gene expression patterns [commentary] Arthritis Rheum. 2010;62:2611–5. doi: 10.1002/art.27561. [DOI] [PubMed] [Google Scholar]
- 2.Petty RE, Southwood TR, Baum J, Bhettay E, Glass DN, Manners P, et al. Revision of the proposed classification criteria for juvenile idiopathic arthritis: Durban, 1997. J Rheumatol. 1998;25:1991–4. [PubMed] [Google Scholar]
- 3.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 4.Duffy CM, Colbert RA, Laxer RM, Schanberg LE, Bowyer SL. Nomenclature and classification in chronic childhood arthritis: time for a change? [commentary] Arthritis Rheum. 2005;52:382–5. doi: 10.1002/art.20815. [DOI] [PubMed] [Google Scholar]
- 5.Barnes MG, Grom AA, Thompson SD, Griffin TA, Pavlidis P, Itert L, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:2102–12. doi: 10.1002/art.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 7.Griffin TA, Barnes MG, Ilowite NT, Olson JC, Sherry DD, Gottlieb BS, et al. Gene expression signatures in polyarticular juvenile idiopathic arthritis demonstrate disease heterogeneity and offer a molecular classification of disease subsets. Arthritis Rheum. 2009;60:2113–23. doi: 10.1002/art.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes MG, Aronow BJ, Luyrink LK, Moroldo MB, Pavlidis P, Passo MH, et al. Gene expression in juvenile arthritis and spondyloarthropathy: pro-angiogenic ELR+ chemokine genes relate to course of arthritis. Rheumatology (Oxford) 2004;43:973–9. doi: 10.1093/rheumatology/keh224. [DOI] [PubMed] [Google Scholar]
- 9.Hall PJ, Burman SJ, Laurent MR, Briggs DC, Venning HE, Leak AM, et al. Genetic susceptibility to early onset pauciarticular juvenile chronic arthritis: a study of HLA and complement markers in 158 British patients. Ann Rheum Dis. 1986;45:464–74. doi: 10.1136/ard.45.6.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass D, Litvin D, Wallace K, Chylack L, Garovoy M, Carpenter CB, et al. Early-onset pauciarticular juvenile rheumatoid arthritis associated with human leukocyte antigen-DRw5, iritis, and antinuclear antibody. J Clin Invest. 1980;66:426–9. doi: 10.1172/JCI109872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis–associated HLA alleles. Arthritis Rheum. 1999;42:1843–53. doi: 10.1002/1529-0131(199909)42:9<1843::AID-ANR8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis Rheum. 2010;62:1781–91. doi: 10.1002/art.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morbach H, Richl P, Faber C, Singh SK, Girschick HJ. The κ immunoglobulin light chain repertoire of peripheral blood B cells in patients with juvenile rheumatoid arthritis. Mol Immunol. 2008;45:3840–6. doi: 10.1016/j.molimm.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Ravelli A, Felici E, Magni-Manzoni S, Pistorio A, Novarini C, Bozzola E, et al. Patients with antinuclear antibody–positive juvenile idiopathic arthritis constitute a homogeneous subgroup irrespective of the course of joint disease. Arthritis Rheum. 2005;52:826–32. doi: 10.1002/art.20945. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marron JS, Todd MJ, Ahn J. Distance weighted discrimination. J Am Stat Assoc. 2007;102:1267–71. doi: 10.1198/jasa.2010.tm08487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wouters CH, Ceuppens JL, Stevens EA. Different circulating lymphocyte profiles in patients with different subtypes of juvenile idiopathic arthritis. Clin Exp Rheumatol. 2002;20:239–48. [PubMed] [Google Scholar]
- 18.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hystad ME, Myklebust JH, Bo TH, Sivertsen EA, Rian E, Forfang L, et al. Characterization of early stages of human B cell development by gene expression profiling [published erratum appears in J Immunol 2009;182:5882] J Immunol. 2007;179:3662–71. doi: 10.4049/jimmunol.179.6.3662. [DOI] [PubMed] [Google Scholar]
- 21.Corcione A, Ferlito F, Gattorno M, Gregorio A, Pistorio A, Gastaldi R, et al. Phenotypic and functional characterization of switch memory B cells from patients with oligoarticular juvenile idiopathic arthritis. Arthritis Res Ther. 2009;11:R150. doi: 10.1186/ar2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martini A. Are the number of joints involved or the presence of psoriasis still useful tools to identify homogeneous disease entities in juvenile idiopathic arthritis? J Rheumatol. 2003;30:1900–3. [PubMed] [Google Scholar]
- 23.Gregorio A, Gambini C, Gerloni V, Parafioriti A, Sormani MP, Gregorio S, et al. Lymphoid neogenesis in juvenile idiopathic arthritis correlates with ANA positivity and plasma cells infiltration. Rheumatology (Oxford) 2007;46:308–13. doi: 10.1093/rheumatology/kel225. [DOI] [PubMed] [Google Scholar]
- 24.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112:973–80. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin KA, Chitnis T, Newcombe J, Franz B, Kennedy J, McArdel S, et al. Age-dependent B cell autoimmunity to a myelin surface antigen in pediatric multiple sclerosis. J Immunol. 2009;183:4067–76. doi: 10.4049/jimmunol.0801888. [DOI] [PMC free article] [PubMed] [Google Scholar]


