Abstract
Background
The barley-Puccinia hordei (barley leaf rust) pathosystem is a model for investigating partial disease resistance in crop plants and genetic mapping of phenotypic resistance has identified several quantitative trait loci (QTL) for partial resistance. Reciprocal QTL-specific near-isogenic lines (QTL-NILs) have been developed that combine two QTL, Rphq2 and Rphq3, the largest effects detected in a recombinant-inbred-line (RIL) population derived from a cross between the super-susceptible line L94 and partially-resistant line Vada. The molecular mechanism underpinning partial resistance in these QTL-NILs is unknown.
Results
An Agilent custom microarray consisting of 15,000 probes derived from barley consensus EST sequences was used to investigate genome-wide and QTL-specific differential expression of genes 18 hours post-inoculation (hpi) with Puccinia hordei. A total of 1,410 genes were identified as being significantly differentially expressed across the genome, of which 55 were accounted for by the genetic differences defined by QTL-NILs at Rphq2 and Rphq3. These genes were predominantly located at the QTL regions and are, therefore, positional candidates. One gene, encoding the transcriptional repressor Ethylene-Responsive Element Binding Factor 4 (HvERF4) was located outside the QTL at 71 cM on chromosome 1H, within a previously detected eQTL hotspot for defence response. The results indicate that Rphq2 or Rphq3 contains a trans-eQTL that modulates expression of HvERF4. We speculate that HvERF4 functions as an intermediate that conveys the response signal from a gene(s) contained within Rphq2 or Rphq3 to a host of down-stream defense responsive genes. Our results also reveal that barley lines with extreme or intermediate partial resistance phenotypes exhibit a profound similarity in their spectrum of Ph-responsive genes and that hormone-related signalling pathways are actively involved in response to Puccinia hordei.
Conclusions
Differential gene expression between QTL-NILs identifies genes predominantly located within the target region(s) providing both transcriptional and positional candidate genes for the QTL. Genetically mapping the differentially expressed genes relative to the QTL has the potential to discover trans-eQTL mediated regulatory relays initiated from genes within the QTL regions.
Background
Plants have evolved complex mechanisms to defend against pathogen attack. Two types of immunity have been described: Pathogen-Associated Molecular Pattern (PAMP)-Triggered Immunity (PTI) and Effector-Triggered Immunity (ETI). PTI is induced at an early stage when PAMPs are recognized by Pattern Recognition Receptors (PRRs), whereas ETI is induced by direct or indirect association of a Resistance (R) protein with a pathogen-derived effector [1-4]. The outcomes of the two immune systems appear to be partial or quantitative resistance and non-host resistance (PTI), and qualitative resistance (ETI). Recently, Niks and Marcel [5] proposed that the varying efficacy of PTI suppression by pathogen effectors may explain partial resistance. In cereal crops, the barley-Puccinia hordei Otth (barley leaf rust) pathosystem is a model for investigating partial and non-host resistance. Microscopic studies on resistance levels in relation to the pathogen developmental phases has indicated plant cell wall penetration and haustorium formation by P. hordei as critical phases determining the success or failure of the infection [6]. Pre-haustorial resistance reduces the chance of successful haustorium formation by the fungal pathogen in the host cells. Failed attempts are typically associated with cell wall appositions [6-10]. Such pre-haustorial basal host defence is a typical reaction to Ph-infection in most (if not all) barley lines exhibiting partial resistance [6]. Post-haustorial resistance is usually due to R gene-mediated hypersensitive response after haustorium formation [9].
These two types of resistance have strategic significance in plant breeding for resistance to diseases. Quantitative or partial resistance has become increasingly important because of its broader spectrum and higher durability compared to R-gene mediated race-specific resistance. Many of the genes underlying partial resistance have plant developmental stage-dependent effectiveness [11]. Currently, over 20 quantitative trait loci (QTL) for quantitative basal resistance to leaf rust from five different mapping populations have been mapped to barley chromosomes [11-16]. They are named Rphq genes [Resistance to Puccinia hordei (quantitative)]. Of these, 10 were effective during the seedling stage, and were detected by QTL analysis of the latency period exhibited by the rust fungus on seedling leaves [15]. Considerable effort has been expended in an attempt to identify the genes underlying these QTL. Notably, a set of NILs and reciprocal NILs have been developed that contain single (Rphq2, 3, 4) or combined (Rphq2+3) introgressed segment(s) carrying resistance and susceptibility QTL allele(s) that were identified in an L94 × Vada RIL population [11,16,17]. L94 is an Ethiopian landrace and highly susceptible to barley leaf rust. Vada is a Dutch cultivar expressing a high level of partial resistance. Following a positional cloning strategy, Marcel et al. [18] have fine-mapped Rphq2, the QTL with largest effect, to an interval of 0.11 cM corresponding to less than 200 kb in physical length.
Microarray technology is being widely used to address various biological, biochemical and genetic questions. Microarray-based gene expression studies can be generally grouped into two major categories. The first aims to address specific biological questions by monitoring the differential expression of genes under contrasting conditions or over time. The most common studies in this field are the investigations on host-pathogen interactions. Profiling changes in genome-wide expression in response to pathogen challenge has identified a large spectrum of genes that are responsive to pathogen attack or are associated with plant resistance in various pathosystems (reviewed by Wise et al. [19]). The second category is based on the more recently emerged concept of 'genetical genomics' [20] or expression QTL (eQTL) mapping that combines highly parallel gene expression studies with the power of genetic segregation. eQTL studies have been performed on maize, eucalyptus and Arabidopsis [21]. eQTL analyses in barley have addressed the global genetic architecture of transcript abundance in
[22], the phenomenon of limited pleiotropy [23] and as an approach to identify the causal or candidate genes underlying partial resistance to fungal diseases [24,25]. While both categories of microarray studies are based on variation in transcript abundance, eQTL analysis provides a genetic dimension that can differentiate cis-from trans-regulation and the genetic locations of a large number of genes through the co-location of high LOD eQTL (i.e. highly differentially expressed) and their structural genes [26]. This is particularly valuable for a crop with large and unsequenced genome like barley.
Here, using a previously reported Agilent 15 k custom array [25], we performed differential expression analysis of QTL-NILs and their recurrent parental lines at 18 hours post-inoculation (hpi) with Puccinia hordei. Our major objective was to identify candidate genes for Rphq2 and Rphq3. In addition, transcript profiles between Ph-infected parents and their respective mock-inoculated controls allowed the establishment of transcriptomic signatures for each line in response to Ph-infection. Our results indicate that transcriptional differentiation between QTL-NILs and their respective recurrent parents reveals components of a regulatory transcriptional relay induced in response to Ph-infection. The datasets generated offer a basis for further studies on defence signalling in relation to partial resistance to P. hordei in barley.
Results
Transcriptomic signatures of response to P. hordei (Ph infected vs. Mock inoculated)
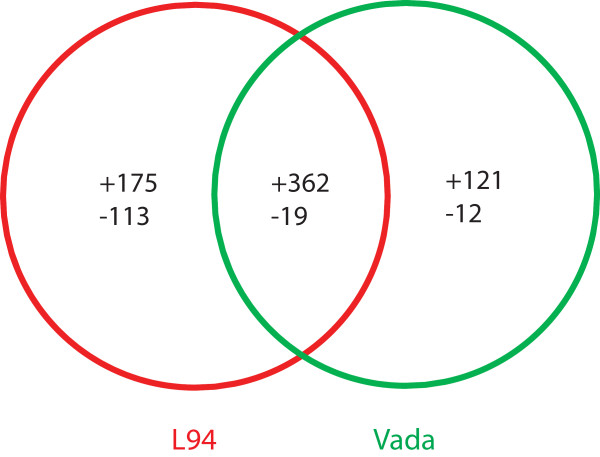
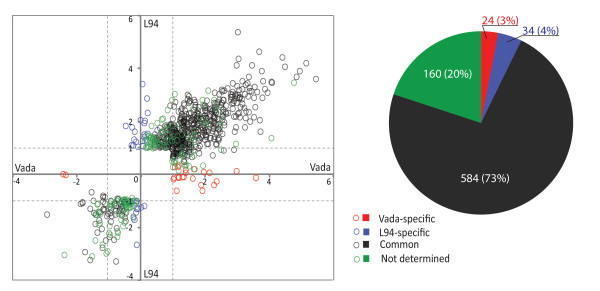
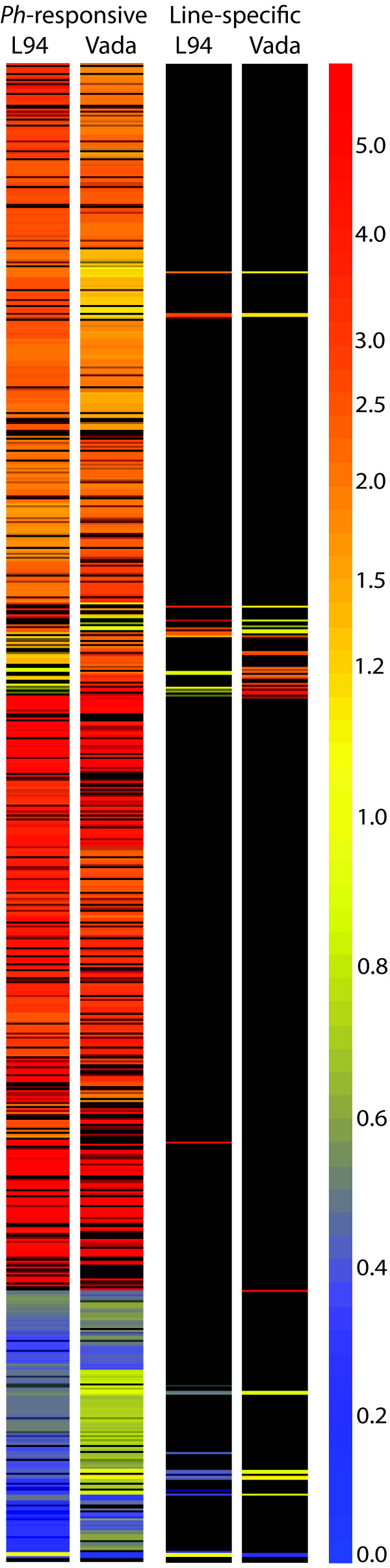
Plant defence responses involve transcriptional activation of a plethora of specific genes and regulation of their temporal and spatial expression [27]. To investigate the genome-wide transcriptional signatures of susceptible and partially resistant barley lines L94 and Vada respectively in response to P. hordei infection, we compared Ph-infected with mock-inoculated leaf material. A stringent threshold with fold change >2 and false discovery rate (FDR) <0.05 was adopted for declaring significant differences. At this threshold, 669 and 514 genes were respectively identified in L94 and Vada as 'significantly differentially expressed' with 381 (362 up + 19 down) overlapping between the two lines while 421 (L94 175 up +113 down and Vada 121 up + 12 down) were present in only one of the two parents (Figure 1). This yielded a total of 802 genes which we considered 'Ph-responsive'. Close examination of the expression data of the 421 'Ph-responsive' genes from both parents showed that while a substantial number failed to meet the stringent thresholds applied (fold change >2, FDR <0.05) they still exhibited statistically significant differential expression in both parents. Therefore, a relaxed threshold ignoring the fold changes was adopted for the follow-up analysis on the commonality and specificity of response to Ph-infection between the resistant and susceptible lines using all 802 Ph-responsive genes. We plotted the log-transformed expression ratios of Vada against L94 and classified them into four groups. Genes that showed the same expression patterns (up- or down-regulation) and expression changes at p < 0.05 in both lines were defined as being common to both lines (Figure 2, black empty circles), whereas, those that showed significant expression changes in one line but no significant expression changes (p > 0.5) or a contrasting expression pattern in the other line were considered as being line-specific (Figure 2, red empty circles for Vada and blue for L94). The remaining genes that had no strong evidence to suggest either commonality or specificity were grouped into 'not determined' (Figure 2, green empty circles). There were a total of 584, 24, 34 and 160 genes that appeared to be in common, Vada- or L94-specific or 'not determined' representing 73%, 3%, 4% and 20% of the 802 Ph-responsive genes respectively. Figure 3 shows a colour-coded heat map that was converted from the relative expression ratios (signal intensity from Ph-infected vs. mock-inoculated controls) of the 802 genes and the 58 (24 + 34) line-specific genes showing the overall similarity and specificity of gene expression in L94 and Vada. Full expression information of the 802 genes and the line-specific genes is given in the Additional File 1 (Table S1) and 2 (Table S2) respectively.
Figure 1.
Venn diagram showing number of Ph-responsive genes (fold change >2, FDR <0.05) identified in L94 and Vada. '+' and '-' represent up- and down-regulation respectively.
Figure 2.
Scatter plot of log ratios (ratio of signal intensity Ph-infected/Mock control) of the 802 Ph-responsive genes from Vada (horizontal axis) and L94 (vertical axis). Colour-coded circles represent genes in different groups with proportions shown in the pie chart. Log ratios >0 or <0 indicates up- or down-regulation respectively, dashed lines set at 1 and -1 corresponding to 2× fold change in expression.
Figure 3.
A heat map illustrating expression patterns of the 802 Ph-responsive genes identified in L94 and Vada. Genes are organized by 'gene tree' hierarchical clustering implemented in GeneSpring based on overall similarity in expression patters (the gene tree has been omitted for clarity). The color bar indicates the expression ratios of the two treatments (Ph-infection vs. mock-inoculated controls). Red and blue represent up- and down-regulation respectively, whereas yellow represent no significant alteration. Left panel shows 802 genes that were significantly (FC >2, FDR <0.05) altered in at least one of the two lines; right panel shows the 58 line-specific genes that were only significantly (FC >2, FDR <0.05) altered in one line but not the other.
Genome-wide Ph-responsive genes have previously been investigated [25] using Steptoe (St) and Morex (Mx), two barley cultivars with similar, intermediate levels of resistance to P. hordei (leaf materials were prepared from the same experiment as the current study with L94 and Vada). We therefore compared the data from L94/Vada with those from St/Mx. At exactly the same thresholds (i.e. FC >2, FDR <0.05) a total of 1154 genes were identified as Ph-responsive in St/Mx [25]. Applying exactly the same criteria as described above, we identified 913 (79%), 21 (1.8%), and 19 (1.6%) genes that were common, St-specific and Mx-specific respectively. We then explored the common genes in each of these categories between the two experiments (Table 1). 75.4% (605) of the 802 genes detected with L94/Vada were also detected with St/Mx and more than half of the genes (466) were significant in all four lines, highlighting the similarity of response to Ph-infection across genotypes. Of the 24 and 34 genes that were specifically detected in Vada and L94 respectively, 13 and six of these were reproducibly identified as Ph-responsive in Steptoe or Morex (Additional File 3, Table S3). All of the 13 Vada (resistant)-specific genes were up-regulated in Vada and St or Mx. Ten of these genes showed significant differential expression (p < 0.05) between St and Mx. Of the six L94 (susceptible)-specific genes, only one up-regulated gene (unigene21775) showed significant differential expression between St and Mx (Additional File 3, Table S3).
Table 1.
Number of overlapping genes (shown in matrix) in different categories detected in two experiments with St/Mx and Vada/L94
| Ph-responsive (1154) | Common (913) | St-specific (21) | Mx-specific (19) | |
|---|---|---|---|---|
| Ph-responsive (802) | 605 | 532 | 9 | 5 |
| Common (584) | 506 | 466 | 5 | 3 |
| Vada-specific (24) | 13 | 6 | 3 | 2 |
| L94-specific (34) | 6 | 3 | 0 | 1 |
Note: Ph-responsive genes were selected on criteria with fold change >2 and FDR <0.05; genes with similar patterns were selected from Ph-responsive gene on p < 0.05 without considering fold changes.
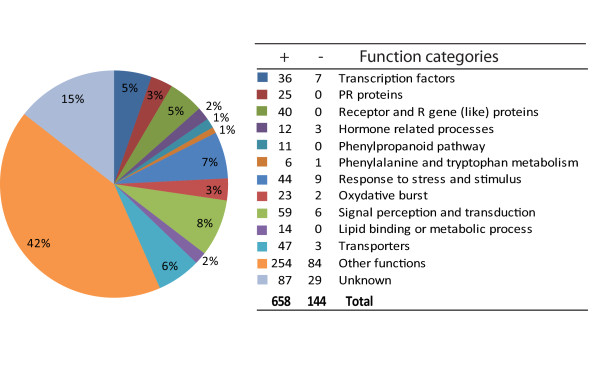
To further characterise the biological processes represented by the 802 Ph-responsive genes, we performed gene ontology (GO) analysis by classifying the Ph-responsive genes into functional biological categories based on GO terms retrieved from their rice homologues through the rice database at http://rice.plantbiology.msu.edu/annotation_pseudo_goslim.shtml. The Ph-responsive genes were associated with a broad range of biological processes. The primary category was related to defence response. We further classified these genes into 11 major functional categories following the GO terms in 'biological process' with all remaining genes grouped into 'other functions' or 'unknown'. The results are shown in Figure 4. They indicate that at the sampling time point of 18 hpi, the plants had responded to defend against the Ph-infection.
Figure 4.
Functional classification of the 802 Ph-responsive genes. Number of up (+) or down (-) regulated genes are shown in the table (see Additional File 1, Table S1 for details).
Differential expression analysis between Ph-infected recurrent parents (L94 vs. Vada)
We performed genome-wide differential expression analysis by comparing expression differences between Ph-infected L94 and Ph-infected Vada at 18 hpi. A total of 1411 genes were identified as being differentially expressed (FC >2, FDR <0.05), of which 247 were Ph-responsive genes as described above. The majority (1164) represent genome-wide, genotype-specific differences in gene expression. The detailed information of these genes regarding their expression ratios, p-values and functional annotations is presented in Additional File 4 (Table S4).
Differential expression between Ph-infected QTL-NILs and Ph-infected recurrent parents
To identify QTL-specific and differentially expressed genes accounted for by genetic differences in the QTL regions, the two reciprocal QTL-NILs were compared with their respective recurrent parents: L94 vs. L94-Rphq2+3 and Vada vs. Vada-Rphq2+3. A total of 94 genes were identified as significant (FC >2, FDR <0.05) in at least one comparison. Of these, 39 genes showed a significant difference in one recurrent parent/QTL-NIL comparison but not with the other. We attribute these observations to the different size and incomplete overlap of the introgressed segments in the two recurrent parent/QTL-NIL pairs. These genes were, therefore, not pursued further. The remaining 55 genes showed expression differences at p < 0.05 in both comparisons and were, therefore, considered potentially relevant to the QTL regions. This suggests that differential expression results from genetic factors differing specifically within the QTL regions (Additional File 5, Table S5, and Figure 5). Of these 55 genes, 50 were present on the list of 1411 differentially expressed genes between Ph-infected Vada and Ph-infected L94. The remaining five genes (Table S5, underlined) did not fulfil the criteria (FC >2, FDR <0.05) set for the differential expression between the two Ph-infected recurrent parents, but their expression differences were still statistically significant (p < 0.05).
Figure 5.
Heat map of the genes significantly and differentially expressed in the three comparisons. 'L' and 'V' on top of the heat map refer to L94 and Vada respectively. Roman numerals represent the four biological replicates. Colour coding represents the transcript abundance ratios. The two comparisons involving NILs were performed on microarray slide 3 and showed reversed colouring reflecting the reciprocal features of the NILs in their genetic background. Comparison between the two parents was conducted on microarray slide 2 with transcript abundance being calculated as L94/Vada. The genes (rows) and treatment groups (columns) are clustered through gene tree generation by GeneSpring program on distance (gene tree has been omitted for clarity).
Transcription of QTL-specific and differentially expressed genes in response to Ph-infection
To identify whether the 55 QTL-specific and differentially expressed genes were also Ph-responsive, expression data from the Ph-infected vs. Mock-inoculated experiment was re-investigated (Table S5). Six genes showed changes that fulfilled the criteria (fold change >2, FDR <0.05) set for defining Ph-responsive genes. Twelve genes did not fully meet the criteria, but their level of differential expression was still statistically significant (p < 0.05) in at least one of the lines. The others were not statistically significant.
Identification of positional candidates for Rphq2 and Rphq3
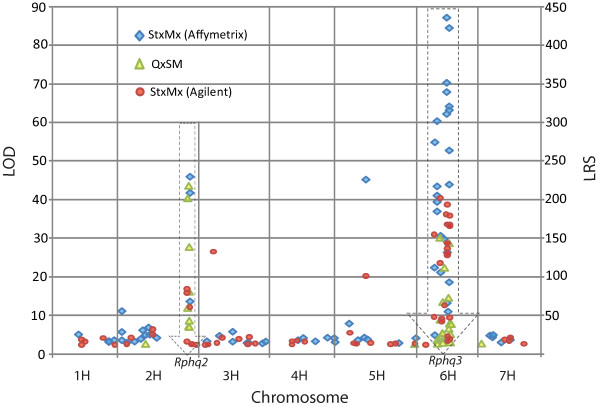
To determine the map position of the 55 QTL-specific and differentially expressed genes, we took advantage of available datasets previously generated in three different eQTL studies (germinating embryos [22]; P. tritici infected leaves http://genenetwork.org, R. Wise, unpublished data) and Ph-infected seedling leaves [25]. 52 of the 55 genes had one or more eQTL in at least one of these three experiments, yielding 163 eQTL in total. The distribution of these eQTL was investigated by plotting their map positions against their LOD/LRS values (Figure 6). 40 genes with eQTL mapped to within the QTL regions (nine at Rphq2 and 31 at Rphq3) (Figure 6, Table S5), of which 33 (83%) had LOD >10 or LRS >50 suggesting they are cis-eQTL (i.e. their structural genes map to the same locus as the eQTL). We then explored three available gene-based mapping datasets: Illumina OPA-SNPs [28], Single Feature Polymorphisms (R. Wise, unpublished data) and TDMs [22] to help assign genetic map positions to the 55 genes. This allowed four and nine genes to be placed within the confidence intervals of Rphq2 and Rphq3 respectively. All of these genes overlapped with the eQTL except two (unigene7920 and 2826) for which no eQTL was detected in the three eQTL studies (Table S5). Rphq2 and Rphq3 on chromosome 2H and 6H are syntenic to regions on rice chromosomes Os04 and Os02 respectively. Conservation of synteny allowed us to infer the approximate map positions of an additional 15 genes to within the QTL regions (Table S5). Thus, of the 55 QTL-specific differentially expressed genes, the map location of nine and 34 fell within Rphq2 and Rphq3 respectively, whilst the 11 others remain unknown. Of note was the observation that one gene (unigene6636), encoding an Ethylene-Responsive Transcription Factor 4 (HvERF4) (rice orthologue Os05g41780.1), has been mapped as Illumina OPA-SNP marker 11_10686 to chromosome 1H at position 71 cM [28]. This map position is consistent with a location based on conservation of synteny between rice Os05 and barley chromosome 1H, suggesting that differential expression of this gene is the consequence of trans-regulation by a gene located within either Rphq2 or Rphq3.
Figure 6.
Distribution of the 163 eQTL detected from three experiments for the 52 genes (3 genes without eQTL detected) differentially expressed in QTL-specific NILs. Blue diamond, red dots and green triangles represent eQTL identified by Potokina et al. [22], Chen et al. [25] and Wise et al. (unpublished results) respectively. eQTL co-located with Rphq2 and Rphq3 were framed with dash-lined arrows. Significance levels of eQTL detected in the St/Mx population refer to LOD score, those with 'Q × SM' population refer to LRS.
Discussion
In this study, we performed differential expression analysis of two reciprocal QTL-NILs and compared them with their respective recurrent parents. As QTL-NILs differ genetically from their recurrent parent only in the selected QTL regions, we would anticipate that genetic polymorphism between these QTL regions would account for any differential expression observed. However, due to the complexity of gene regulation, differentially expressed genes may not necessarily be located in the introgressed QTL regions, which may themselves contain regulatory genes affecting the expression of other genes spread throughout the genome. We therefore established the map positions of differentially expressed genes by exploiting previously generated gene mapping datasets. Of the 55 genes highlighted in our comparisons between NILs and recurrent parents, 40 detected eQTL in at least one of the three previous eQTL studies and co-located at the QTL regions, most (83%) having high LOD/LRS scores [22,25] (Table S5). eQTL with high LOD scores have been demonstrated previously to be almost always cis-eQTL [22,25,26] placing these genes within the Rphq2 or Rphq3 QTL regions. The observation that so many significantly differentially expressed genes appeared to be regulated in cis- is in agreement with previous studies [25,29]. An exception was unigene 6636, encoding HvERF4. This gene mapped to 71 cM on chromosome 1H, consistent with the position of its rice homologue Os05g41780.1 predicted by conservation of synteny [28]. This observation raises the possibility that the introgressed regions at either Rphq2 or Rphq3 contain a polymorphic trans-acting regulator that differentially modulates expression of HvERF4. No eQTL for HvERF4 was detected at the regions corresponding to Rphq2 or Rphq3 in the St/Mx DH mapping population, consistent with the fact that it does not segregate for Rphq2 or Rphq3.
HvERF4 is a member of a family of plant transcription factors functionally involved in defence signalling pathways related to ethylene, jasmonic acid and abscisic acid. Over-expression of Arabidopsis AtERF4 represses the expression of pathogenesis-related (PR) genes such as basic chitinase and beta-1,3-glucanase genes and genes containing a GCC-box [30], the core sequence element of promoters required for responsiveness to ethylene [31]. In our previous experiment with Steptoe and Morex, cultivars with similar but intermediate levels of partial resistance to leaf rust, we also observed that HvERF4 was significantly up-regulated by Ph-infection but no differential expression (p > 0.2) was observed between the parents [25]. Here, HvERF4 was induced in Ph-infected L94 (susceptible) (FC = 4.42) and Vada (partially resistant) (FC = 2.42) as compared to mock-inoculated controls (Table S5), and the expression level of the Vada allele was only a third (FC = 0.34) of that of the L94 allele after induction. The association of resistance/susceptibility with lower/higher expression of HvERF4 appears to be in agreement with the negative regulatory role of HvERF4 on the expression of PR and other defence responsive genes. However, consistent association of higher expression of PR genes with resistance was not observed in Vada and L94. This may reflect the general complexity of natural resistance response coupled with allelic variation at PR genes between these two lines. While this train of inference highlights HvERF4 as potentially important in this specific defense interaction, none of the so far reported 20 QTL for partial resistance to leaf rust, nor any of the QTL for resistance to heterologous rusts is co-located with HvERF4 at 71 cM on chromosome 1H [11-16]. Thus, HvERF4 is not a positional candidate for any of the reported QTL. However, of direct relevance is a previously highlighted eQTL hotspot for
genes that was associated with OPA-SNP 11_20157 [25] at 70 cM (98 cM on the consensus map [28]) on chromosome 1H spanning the region containing HvERF4. This hotspot comprised 127 eQTL in less than a 10 cM interval and contained genes primarily involved in defence response [25]. Given its known role in PR-protein regulation, we speculate that HvERF4 represents a key regulatory relay component of the signalling pathway that controls expression of at least a portion of the genes with eQTL located at the hotspot on chromosome 1H. Considering these observations together we hypothesise that the causal genetic polymorphism at either Rphq2 or Rphq3 differentially regulates HvERF4 in trans (possibly through direct or indirect modulation of ethylene, jasmonic acid or abscisic acid levels, known in Arabidopsis to alter levels of AtERF4 expression [30]), the consequence of which is differential regulation of down-stream defence responses. In this scenario, the candidate genes for Rphq2 or Rphq3 would be those acting up-stream rather than down-stream of HvERF4 and possibly involved directly or indirectly in plant hormone signalling pathways. While we did not find such a candidate from the annotated functions of the QTL-specific and differentially expressed candidates for Rphq2 and Rphq3, the gene controlling expression of HvERF4 may, however, not be differentially expressed between L94 and Vada, may not be on our expression platform (which probably contains less than half of the barley genes) or may not be at the orthologous position in rice. An alternative to identifying the causal gene for Rphq2 or Rphq3 could be through map-based cloning of the trans-eQTL for HvERF4.
Marcel et al. [18] narrowed down the genetic interval for Rphq2 to 0.11 cM corresponding to a physical length of 183 kb in barley (Marcel and Niks unpublished data) and a 69.7 kb syntenic region on rice chromosome 4. Inspection of all predicted genes in the Rphq2 syntenic interval in rice identified a cluster of six peroxidase genes and a MAP3K gene [18] as potential candidates because of their functional involvement in defence responses. In this study, we identified four barley genes at Rphq2 that were differentially expressed and had homologues located in the syntenic region in rice (Table S5, unigene1852 (no.7), unigene2111 (no.9), unigene13865 (no. 8) and unigene8521 (no. 46)). Unigene1852 and 2111 both encode peroxidases and are within the 0.11 cM interval containing Rphq2. The other two, according to the fine mapping data of Marcel et al. [18] fell just outside the candidate interval. However, given the frequent breakdown in conservation of synteny, positional candidate gene identification using this approach alone remains problematic. Differential expression in the QTL-NILs identified an additional five candidate genes (Figure 5 and Table S5: no. 2, 3, 19, 21and 51) that were not apparently present in the syntenic region of rice. Two of these encode proteins that are functionally involved in signal transduction (Additional file 5, Table S5, no.19 and 21 encoding a kinase and a receptor-like kinase respectively), one PR protein, and one with homology to human NIPA1, implicated in Prader-Willi/Angelman syndrome 1 [32]. One gene showed no homology to known genes. Thus, these five genes, together with the two peroxidase genes, are potential positional candidates for Rphq2. Further refinement of the candidate gene list will require knowledge of the role of these genes in defence response and correlation of transcript levels with resistance/susceptibility. Many more genes (i.e. 31) were identified as being differentially expressed and located at the Rphq3 region. This is expected given the larger interval of the QTL (28 cM for Rphq3 vs. 4 cM for Rphq2) and Rphq3 may, therefore, account for differential expression of most of the genes with unassigned map positions. Functionally, none of the differentially expressed genes at Rphq2 or Rphq3 appear to be obvious candidates for a regulator of HvERF4.
Many defence genes encoding PR proteins and components of the phenylpropanoid pathway such as phenylalanine ammonia lyase (PAL) were, as would be expected, Ph-responsive. PR genes encoding beta-1,3-glucanases, chitinases and thaumatin-like proteins exhibit strong in vitro anti-fungal activity [33] and numerous studies have shown that transgenic plants expressing PR-proteins have significant improvement of disease resistance [34-37]. PAL, the first committed enzyme in the phenylpropanoid pathway, is involved in synthesis of both phytoalexins and lignin. Phytoalexins are antimicrobial while lignin synthesis contributes to formation of papillae, which are physical barriers against cell wall penetration by the pathogen [38]. As part of the general response to pathogen infection, few of the genes fell into these categories co-located at the two QTL for partial resistance. One exception, unigene25195, encoding a chitinase (PR3), co-located at Rphq2, was Ph-responsive and differentially expressed between Rphq2 and Rphq2. However, it was not prioritized as a candidate for Rphq2 since the higher level of gene expression was associated with the susceptibility allele Rphq2. Whereas a number of defence genes were activated in response to Ph-infection, none was found to be a promising candidate for Rphq2 or Rphq3. Our results support the notion that components of the general defence response have incremental, rather than deterministic, roles in the outcome of an interaction between a plant and a pathogen [39]. Many attempts to identify genes for disease resistance have highlighted those involved in signal transduction [40,41] or physiological and cellular functions [42,43] rather than defence per se [44,45].
Ph-infection triggers a broad range of biological responses with defence response genes being significantly over-represented. Of note is a set of genes encoding receptor-like kinase (RLK), receptor-like proteins (RLP), WRKY, MAPK and PR proteins (Additional File 1, Table S1), which form a complete and well-explored defence signalling cascade starting with the perception of PAMPs, activation of WRKY transcription factors and the subsequent induction of PR proteins [3,46]. Our results also suggest that, in the absence of cognate R genes to P. hordei, plants still mount reactions similar to R-gene mediated responses as indicated by the significant up-regulation of genes coding for R gene (-like) proteins and marker genes for oxidative burst such glutathione S-transferase and peroxidase. Although no obvious R genes were identified as candidates for the QTL in this study, R gene-like mediated responses may contribute to basal resistance as a complementary mechanism to PAMP-triggered defence responses. Support for this is provided by observations that resistance QTL are often coincident with the location of R-gene homologues [47-51] and that mutated R genes can induce a resistance phenotype similar to quantitative resistance controlled by multiple genes [52-54].
One striking characteristic of the responses to Ph-infection was the activation of signalling pathways related to a broad range of plant hormones including ethylene, gibberellins, auxin, and brassinosteroid as indicated by the up-regulation of genes encoding ethylene-responsive transcription factors, ACC oxidase, auxin-responsive proteins, brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1), gibberellin receptors and a DELLA protein (Additional File 1, Table S1). All of these hormones have been reported to be involved in plant defence responses [55-57] and various defence pathways are interconnected through hormone-mediated signalling pathways forming complex regulatory networks [55,56,58-60]. Here, the identification of the ethylene-responsive factor HvERF4 as a putative link between pathogen perception and response is consistent with a role for differential hormone signalling in partial resistance. Understanding the role of Rphq2 of Rphq3 in initiating and coordinating the response requires further work.
Substantial overlap of Ph-responsive genes was identified in super-susceptible (L94) and partially-resistant (Vada) lines. Over 70% of Ph-responsive genes were detected in both L94 and Vada and had the same expression patterns (up- or down-regulation) in both lines. An even higher percentage of overlapping Ph-responsive genes (79%) was discovered in both Steptoe and Morex, two cultivars with similar and intermediate level of partial resistance. Given that these lines are genetically diverse, we conclude that barley lines without known cognate R genes to P. hordei exhibit similar responses at the transcriptional level, and that observed differences are largely quantitative. Similar findings have been observed in the comparison between compatible and incompatible interactions [61-63]. A small proportion (7%) of Ph-responsive genes in this study did appear to be resistant/susceptible line-specific and it may be that they determine part of the observed phenotypic differences between lines. However, in Ph-infected leaves we found no evidence for their differential expression in the comparisons between the two QTL-NILs and their respective recurrent parents. Therefore, if the variation in resistance, accounted for by Rphq2 or Rphq3, is regulated at the transcriptional level, these are not strong candidate genes.
We generated a robust expression data set in reciprocal Rphq2/Rphq3 QTL-NILs at 18 hpi, which is the timepoint previously described as being the most critical during P. hordei invasion in barley [25]. However, we realise that transcriptional re-programming in response to pathogen infection is a dynamic and complex process and that defence-associated genes respond to input stimuli with different timing and amplitude. A limitation of our experiment is, therefore, that defence response scenarios constructed on the transcriptional profiles of the 802 Ph-responsive genes identified here is simply a snapshot of a dynamic process, at the point when infection hyphae have just attempted penetration of the host cells forming haustoria [25]. To extend our understanding of the complex regulatory mechanisms occurring during defence against P. hordei, a more comprehensive investigation would involve sampling at multiple timepoints covering the whole infection period.
Conclusions
Differential expression with QTL-NILs identifies genes predominantly located at the target region(s) providing both transcriptional and positional candidate genes underlying the QTL. Positional analysis of the differentially expressed genes relative to the QTL has the potential to discover regulatory relays initiated from genes within the QTL.
Methods
Plant materials
The plant materials used in this study included both recurrent parental lines L94 (highly susceptible to P. hordei) and Vada (high level of partial resistance to P. hordei) and the QTL-NIL named L94-Rphq2+3 and Vada-Rphq2+3 according to the introgressed resistance/susceptibility QTL alleles. Gene symbol 'Rphq' refers to the resistance allele of the QTL, i.e. the allele contributed by Vada, and 'rphq' refers to the susceptibility L94 allele. Neither of these cultivars carries a cognate R-gene to P. hordei. The NIL 'L94-Rphq2+3' was previously developed through a marker-assisted backcross programme by incorporating leaf rust resistance alleles Rphq2 and Rphq3 from Vada into L94 susceptible genetic background, whereas the NIL Vada-rphq2+3 was generated by reciprocally incorporating the corresponding susceptibility QTL alleles rphq2 and rphq3 from L94 into Vada genetic background [18]. The resulting resistance levels (relative latency period in hours) of the NILs are 120 ± 1.77 for L94-Rphq2+3, 106 ± 2.54 for Vada-Rphq2+3, as compared to 100 ± 1.77 for L94 and 127 ± 1.80 for Vada [18]. The genetic lengths of the two introgression segments on chromosome 2H were 4.6 cM for Rphq2 and 4.4 cM for Rphq2; the two QTL segments on chromosome 6H were 22.6 cM for Rphq3 and 45.8 cM for rphq3 [18].
Plant growth and leaf inoculations were performed as previously described [25]. The parental lines L94 and Vada and their QTL-NILs, each with 10 seedlings were grown in one tray (37 × 39 cm) in two rows 30 cm apart. A total of eight trays were prepared, with four each used as biological replicates for pathogen inoculation and mock inoculation. The plant growth conditions were as described by Chen et al. [25].
Pathogen inoculation
Inoculation with P. hordei isolate 1.2.1 was performed on 9-day old seedlings when the first leaf was fully developed and the second leaf was emerging. Leaves were laid horizontal and gently fixed over the soil prior to inoculation. The inoculation was described in Chen et al. [25]. Per plant tray, 8 mg of urediospores plus 32 mg of Lycopodium spores (added as a carrier) were thoroughly mixed by vortexing and applied to the adaxial sides of the seedling leaves using a settling tower inoculation facility. This amount of spores corresponds to a deposition of about 500 spores per cm2. Mock inoculation of parental lines was carried out using 40 mg of Lycopodium spores only. All trays were transferred to a dark dew chamber at 18°C and 100% relative humidity for 10 hours overnight, before being placed in the glasshouse for infection development.
Leaf sampling
At 18 hpi, both pathogen- and mock-inoculated leaf blades of each replicate and treatment were collected separately into falcon tubes and immediately flash-frozen in liquid nitrogen before being stored at -80°C until use.
RNA isolation, labelling and microarray platform
RNA isolation was done using the TRIZOL® reagent according to the manufacturer's protocol. cDNA synthesis, labeling and hybridization were performed following the optimized protocol developed by the Sequencing & Microarray Facility at SCRI. The Agilent 8 × 15 k format custom array system was used as the platform for RNA profiling. Detailed protocols are described in Chen et al. [25].
Sample layout on the 8 × 15 k Agilent arrays
The Agilent platform may be used as a two-colour microarray system allowing two differentially-labeled samples to be tested on a single array. We used three different sample layouts depending upon the biological questions to be addressed: 1) RNA samples from Ph-infected parents and mock-inoculated controls (four replicates) were hybridized onto single arrays to identify Ph-responsive genes (array slide 1 in Table 2); 2) RNA samples from Ph-infected L94 and Vada (four replicates) were hybridized onto single arrays to test genome-wide differential expression (slide 2 in Table 2); 3) RNA samples from Ph-infected L94 and L94-Rphq2+3 or Vada and Vada-Rphq2+3 were put on single arrays with four replicates (8 arrays) (slide 3 in Table 2) to compare expression levels of parental lines with their respective NILs. In all sample layouts, a balanced dye swap strategy was applied as indicated in the Table 2.
Table 2.
Microarray experimental design:
| Array slide | Replicate | Sample pairs | |||||
|---|---|---|---|---|---|---|---|
| Name | Treatment | Label | Name | Treatment | Label | ||
| 1 | I | L94 | Mock | C3 | L94 | Ph-infected | C5 |
| 1 | I | Vada | Mock | C3 | Vada | Ph-infected | C5 |
| 1 | II | L94 | Mock | C3 | L94 | Ph-infected | C5 |
| 1 | II | Vada | Mock | C3 | Vada | Ph-infected | C5 |
| 1 | III | L94 | Mock | C5 | L94 | Ph-infected | C3 |
| 1 | III | Vada | Mock | C5 | Vada | Ph-infected | C3 |
| 1 | IV | L94 | Mock | C5 | L94 | Ph-infected | C3 |
| 1 | IV | Vada | Mock | C5 | Vada | Ph-infected | C3 |
| 2 | I | L94 | Ph-infected | C3 | Vada | Ph-infected | C5 |
| 2 | II | L94 | Ph-infected | C5 | Vada | Ph-infected | C3 |
| 2 | III | L94 | Ph-infected | C3 | Vada | Ph-infected | C5 |
| 2 | IV | L94 | Ph-infected | C5 | Vada | Ph-infected | C3 |
| 3 | I | L94 | Ph-infected | C3 | L94-Rphq2+3 | Ph-infected | C5 |
| 3 | I | Vada | Ph-infected | C3 | Vada- Rphq2+3 | Ph-infected | C5 |
| 3 | II | L94 | Ph-infected | C5 | L94- Rphq2+3 | Ph-infected | C3 |
| 3 | II | Vada | Ph-infected | C5 | Vada- Rphq2+3 | Ph-infected | C3 |
| 3 | III | L94 | Ph-infected | C3 | L94- Rphq2+3 | Ph-infected | C5 |
| 3 | III | Vada | Ph-infected | C3 | Vada- Rphq2+3 | Ph-infected | C5 |
| 3 | IV | L94 | Ph-infected | C5 | L94- Rphq2+3 | Ph-infected | C3 |
| 3 | IV | Vada | Ph-infected | C5 | Vada- Rphq2+3 | Ph-infected | C3 |
Array slide 1: Ph-infected vs. mock-inoculated controls for Ph-responsive genes; Array slide 2: Ph-infected L94 vs. Ph-infected Vada for differentially expressed genes; Array slide 3: Ph-infected parents vs. Ph-infected QTL-NILs for QTL specific and differentially expressed genes.
Deposition of microarray data
The raw microarray data and relevant experimental metadata, which are MIAME (Minimum Information About a Microarray Experiment) compliant, are deposited at the ArrayExpress microarray data archive http://www.ebi.ac.uk/microarray-as/ae/ at the European Bioinformatics Institute (accession numbers: E-TABM-980).
Data extraction, normalisation and significance criteria for differential expression
Data extraction and normalisation were done independently for the three different experiments with GeneSpring (v.7.3) software as described previously [25]. Briefly, dye swap was corrected in relevant samples, followed by Lowess (LOcally WEighted polynomial regreSSion) normalisation to minimize differences in dye incorporation efficiency in a two-channel microarray platform [64]. Differentially expressed genes were first selected on fold change >2 followed by a Students t-test on log-transformed normalised ratio data, setting the False Discovery Rate (FDR) to 0.05.
Abbreviations
Ph: Puccinia hordei; QTL: quantitative trait loci; eQTL: expression QTL; QTL-NIL: QTL-specific nearly isogenic line; RIL: recombinant inbred line, PAMP: pathogen-associated molecular pattern; PTI: PAMP-triggered immunity; ETI: effector-triggered immunity, FDR: false discovery rate; GO: gene ontology; LOD: log of odds; LRS: likelihood ratio statistics; FC: fold change; SFP: single feature polymorphism; TDM: transcript derived marker.
Authors' contributions
Conceived and designed the experiments: XC, REN, AD and RW; performed the experiments: XC; wrote the paper: XC and RW. Pathogen infection and sampling: REN, XC, TCM and AV. Microarray and data deposition: PH and JM. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Expression information of Ph-reponsive genes identified on L94 and Vada (Ph-infected vs. mock control).
Table S2. Expression information of resistant/susceptible line-specific and Ph-responsive genes.
Table S3. Expression of the resistant/susceptible line-specific genes (upper/lower panel) reproduced as Ph-responsive genes in St and Mx.
Table S4. Genome-wide differentially expressed genes in Ph-infected seedlings between Vada and L94.
Table S5. List of the 55 differentially expressed genes showing expression ratios and p-values in different comparisons and map position of eQTL and corresponding genes from different sources.
Contributor Information
Xinwei Chen, Email: xinwei.chen@scri.ac.uk.
Rients E Niks, Email: rients.niks@wur.nl.
Peter E Hedley, Email: pete.hedley@scri.ac.uk.
Jenny Morris, Email: jenny.morris@scri.ac.uk.
Arnis Druka, Email: arnis.druka@scri.ac.uk.
Thierry C Marcel, Email: thierry.marcel@versailles.inra.fr.
Anton Vels, Email: anton.vels@wur.nl.
Robbie Waugh, Email: robbie.waugh@scri.ac.uk.
Acknowledgements
We gratefully acknowledge J McNicol, C Hackett and D Roberts for valuable discussions concerning the experimental and custom array design; F Yeo, A Gonzalez, Z Kohutova, F Meijer-Dekens, R Aghnoum, M Macaulay and K McLean for their kind help with sampling; and Drs A Newton and G Bryan for their critical review of the manuscript. Funding for this experiment was provided by the European Union Bioexploit Grant No. 513959 (FOOD) to RW and RN http://www.bioexploit.net and by Scottish Government Rural and Environment Research and Analysis Directorate (RERAD) Programme 1, Work Package 1 http://www.programme1.net/programmes. We thank Dr R Wise for access to unpublished eQTL data uploaded in the GeneNetwork.
References
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature Immunology. 2005;6:973–79. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiology. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annual Review of Phytopathology. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Current Opinion in Plant Biology. 2007;10:335–341. doi: 10.1016/j.pbi.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Niks RE, Marcel TC. Nonhost and basal resistance: how to explain specificity? New Phytologist. 2009;182:817–828. doi: 10.1111/j.1469-8137.2009.02849.x. [DOI] [PubMed] [Google Scholar]
- Niks RE. Failure of haustorial development as a factor in slow growth and development of Puccinia hordei in partially resistant barley seedlings. Physiological and Molecular Plant Pathology. 1986;28:309–322. doi: 10.1016/S0048-4059(86)80073-X. [DOI] [Google Scholar]
- O'Connell RJ, Panstruga R. Tete a tete inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist. 2006;171:699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- Heath MC. Cellular interactions between biotrophic fungal pathogens and host or nonhost plants. Canadian Journal of Plant Pathology. 2002;24:259–264. doi: 10.1080/07060660209507007. [DOI] [Google Scholar]
- Collins NC, Niks RE, Schulze-Lefert P. Resistance to cereal rusts at the plant cell wall - what can we learn from other host-pathogen systems? Australian Journal of Agricultural Research. 2007;58:476–489. doi: 10.1071/AR06065. [DOI] [Google Scholar]
- Hardham AR, Jones DA, Takemoto D. Cytoskeleton and cell wall function in penetration resistance. Current Opinion in Plant Biology. 2007;10:342–348. doi: 10.1016/j.pbi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Qi X, Niks RE, Stam P, Lindhout P. Identification of QTLs for partial resistance to leaf rust (Puccinia hordei) in barley. Theoretical and Applied Genetics. 1998;96:1205–1215. doi: 10.1007/s001220050858. [DOI] [Google Scholar]
- Qi X, Jiang G, Chen W, Niks RE, Stam P, Lindhout P. Isolate-specific QTLs for partial resistance to Puccinia hordei in barley. Theoretical and Applied Genetics. 1999;99:877–884. doi: 10.1007/s001220051308. [DOI] [Google Scholar]
- Jafary H, Szabo LJ, Niks RE. Innate nonhost immunity in barley to different heterologous rust fungi is controlled by sets of resistance genes with overlapping specificities. Molecular Plant-Microbe Interaction. 2006;19:1270–1279. doi: 10.1094/MPMI-19-1270. [DOI] [PubMed] [Google Scholar]
- Jafary H, Albertazzi G, Marcel TC, Niks RE. High diversity of genes for nonhost resistance of barley to heterologous rust fungi. Genetics. 2008;178:2327–2339. doi: 10.1534/genetics.107.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel TC, Varshney RK, Barbieri M, Jafary H, de Kock MJD, Graner A, Niks RE. A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theoretical and Applied Genetics. 2007;114:487–500. doi: 10.1007/s00122-006-0448-2. [DOI] [PubMed] [Google Scholar]
- Marcel TC, B Gorguet M, Truong Ta, Kohutova Z, Vels A, Niks RE. Isolate-specificity of quantitative trait loci for partial resistance of barley to Puccinia hordei confirmed in mapping populations and near-isogenic lines. New Phytologist. 2008;177:743–755. doi: 10.1111/j.1469-8137.2007.02298.x. [DOI] [PubMed] [Google Scholar]
- van Berloo R, Aalbers H, Werkman A, Niks RE. Resistance QTL confirmed through development of QTL-NILs for barley leaf rust resistance. Molecular Breeding. 2001;8:187–195. doi: 10.1023/A:1013722008561. [DOI] [Google Scholar]
- Marcel TC, Aghnoum R, Durand J, Varshney RK, Niks RE. Dissection of the barley 2L1.0 region carrying the 'Laevigatum' quantitative resistance gene to leaf rust using near-isogenic lines (NIL) and subNIL. Molecular Plant-Microbe Interactions. 2007;20:1604–1615. doi: 10.1094/MPMI-20-12-1604. [DOI] [PubMed] [Google Scholar]
- Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA. Transcript profiling in host-pathogen interactions. Annual Review of Phytopathology. 2007;45:329–369. doi: 10.1146/annurev.phyto.45.011107.143944. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends in Genetics. 2001;17:388–391. doi: 10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Jiang N, Chen X, Kearsey M, Waugh R. eQTL analysis in Plants. Plant Biotechnology Journal. 2009;8:10–27. doi: 10.1111/j.1467-7652.2009.00460.x. [DOI] [PubMed] [Google Scholar]
- Potokina E, Druka A, Luo Z, Wise R, Waugh R, Kearsey MJ. Gene expression quantitative trait locus analysis of 16 000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. The Plant Journal. 2008;53:90–101. doi: 10.1111/j.1365-313X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Potokina E, Druka A, Luo Z, Moscou M, Wise R, Waugh R, Kearsey MJ. Tissue-dependent limited pleiotropy affects gene expression in barley. The Plant Journal. 2008;56:287–296. doi: 10.1111/j.1365-313X.2008.03601.x. [DOI] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Bonar N, Druka I, Zhang L, Marshall DF, Kearsey M, Waugh R. Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp tritici in barley. Theoretical and Applied Genetics. 2008;117:261–272. doi: 10.1007/s00122-008-0771-x. [DOI] [PubMed] [Google Scholar]
- Chen X, Hackett CA, Niks RE, Hedley PE, Booth C, Druka A, Marcel TC, Vels A, Bayer M, Milne I, Morris J, Ramsay L, Marshall D, Cardle L, Waugh R. An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE. 2010;5:e8598. doi: 10.1371/journal.pone.0008598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZW, Potokina E, Druka A, Wise R, Waugh R, Kearsey MJ. SFP genotyping from Affymetrix arrays is robust but largely detects cis-acting expression regulators. Genetics. 2007;176:789–800. doi: 10.1534/genetics.106.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis: 2001 status. Current Opinion in Plant Biology. 2001;4:301–308. doi: 10.1016/S1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao S, Varshney RK, Szűcs P, Sato K, Hayes PM, Matthews DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BG, Halkier BA, Kliebenstein DJ. Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends in Plant Science. 2008;13:72–77. doi: 10.1016/j.tplants.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M. Identification of an ethylene-responsive region in the promoter of tobacco class I chitinase gene. Plant Molecular Biology. 1995;27:923–932. doi: 10.1007/BF00037020. [DOI] [PubMed] [Google Scholar]
- Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) American Journal of Human Genetics. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Hadwiger LA, Boller T. Antifungal hydrolases in pea tissue. 1. Purification and characterization of two chitinases and β-1,3-glucanase differentially regulated during development and in response to fungal infection. Plant Physiology. 1988;87:325–333. doi: 10.1104/pp.87.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglie K, Chet I, Holliday M, Cressman R, Biddle Ph, Knowlton S, Mauvais CJ, Broglie R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb C. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology. 1994;12:807–812. doi: 10.1038/nbt0894-807. [DOI] [Google Scholar]
- Lin W, Anuratha CS, Datta K, Potrykus I, Muthukrishnan S, Datta SK. Genetic engineering of rice for resistance to sheath blight. Bio/Technology. 1995;13:686–691. doi: 10.1038/nbt0795-686. [DOI] [Google Scholar]
- Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theoretical and Applied Genetics. 1999;98:1138–1145. doi: 10.1007/s001220051178. [DOI] [Google Scholar]
- Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- Collinge DB, Jensen MK, Lyngkjaer MF, Rung J. How can we exploit functional genomics approaches for understanding the nature of plant defences? Barley as a case study. European Journal of Plant Pathology. 2008;121:257–266. doi: 10.1007/s10658-008-9271-8. [DOI] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Panstruga R, Schulze-Lefer P. Live and let live: insights into powdery mildew disease and resistance. Molecular Plant Pathology. 2002;3:495–502. doi: 10.1046/j.1364-3703.2002.00145.x. [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Jordan F, Osbourn A. First encounters-Deployment of defence-related natural products by plants. New Phytologist. 2006;172:193–207. doi: 10.1111/j.1469-8137.2006.01863.x. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Current Opinion in Plant Biology. 2007;10:335–341. doi: 10.1016/j.pbi.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Wagner C, Schweizer G, Kraemer M, Dehmer-Badani AG, Ordon F, Friedt W. The complex quantitative barley-Rhynchosporium secalis interaction: newly identified QTL may represent already known resistance genes. Theoretical and Applied Genetics. 2008;118:113–122. doi: 10.1007/s00122-008-0881-5. [DOI] [PubMed] [Google Scholar]
- Tan MYA, Hutten RCB, Celis C, Park TH, Niks RE, Visser RGF, van Eck HJ. The RPi-mcd1 locus from Solanum microdontum involved in resistance to Phytophthora infestans, causing a delay in infection, maps on potato chromosome 4 in a cluster of NBS-LRR genes. Molecular Plant-Microbe Interactions. 2008;21:909–918. doi: 10.1094/MPMI-21-7-0909. [DOI] [PubMed] [Google Scholar]
- Xiao W, Zhao J, Fan S, Li L, Dai J, Xu M. Mapping of genome-wide resistance gene analogs (RGAs) in maize (Zea mays L.) Theoretical and Applied Genetics. 2007;115:501–508. doi: 10.1007/s00122-007-0583-4. [DOI] [PubMed] [Google Scholar]
- Zimnoch-Guzowska E, Marczewski W, Lebecka R, Flis B, Schäfer-Pregl R, Salaminin F, Gebhardt C. QTL analysis of new sources of resistance to Erwinia carotovora ssp. atroseptica in potato done by AFLP, RFLP, and resistance-gene-like markers. Crop Science. 2000;40:1156–1167. doi: 10.2135/cropsci2000.4041156x. [DOI] [Google Scholar]
- Pflieger S, Lefebvre V, Caranta C, Blattes A, Goffinet B, Palloix A. Disease resistance gene analogs as candidates for QTLs involved in pepper-pathogen interactions. Genome. 1999;42:1100–1110. doi: 10.1139/gen-42-6-1100. [DOI] [PubMed] [Google Scholar]
- Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS, Whalen MC, Ronald P. Xa21D encodes a receptor-like molecule with a leucine-rich repeat that determines race-specific recognition and is subject to adaptative evolution. The Plant Cell. 1998;10:765–779. doi: 10.2307/3870663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG. Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. The Plant Cell. 1997;9:641–651. doi: 10.2307/3870513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. The Plant Cell. 1997;9:521–532. doi: 10.2307/3870504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009;69:473–88. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends in Plant Science. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host & Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. Brassinosteroid-mediated stress responses. Journal of Plant Growth Regulation. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT. Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.) Plant Molecular Biology. 1999;41:443–454. doi: 10.1023/A:1006372612574. [DOI] [PubMed] [Google Scholar]
- Muessig C, Lisso J, Coll-Garcia D, Altmann T. Molecular analysis of brassinosteroid action. Plant Biology. 2006;8:291–296. doi: 10.1055/s-2005-873043. [DOI] [PubMed] [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends in Plant Science. 2005;10:71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nature Genetics. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Wise RP. Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. The Plant Cell. 2004;16:2514–2528. doi: 10.1105/tpc.104.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30:4e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Expression information of Ph-reponsive genes identified on L94 and Vada (Ph-infected vs. mock control).
Table S2. Expression information of resistant/susceptible line-specific and Ph-responsive genes.
Table S3. Expression of the resistant/susceptible line-specific genes (upper/lower panel) reproduced as Ph-responsive genes in St and Mx.
Table S4. Genome-wide differentially expressed genes in Ph-infected seedlings between Vada and L94.
Table S5. List of the 55 differentially expressed genes showing expression ratios and p-values in different comparisons and map position of eQTL and corresponding genes from different sources.