Abstract
Background
The Nile tilapia is the second most important fish in aquaculture. It is an excellent laboratory model, and is closely related to the African lake cichlids famous for their rapid rates of speciation. A suite of genomic resources has been developed for this species, including genetic maps and ESTs. Here we analyze BAC end-sequences to develop comparative physical maps, and estimate the number of genome rearrangements, between tilapia and other model fish species.
Results
We obtained sequence from one or both ends of 106,259 tilapia BACs. BLAST analysis against the genome assemblies of stickleback, medaka and pufferfish allowed identification of homologies for approximately 25,000 BACs for each species. We calculate that rearrangement breakpoints between tilapia and these species occur about every 3 Mb across the genome. Analysis of 35,000 clones previously assembled into contigs by restriction fingerprints allowed identification of longer-range syntenies.
Conclusions
Our data suggest that chromosomal evolution in recent teleosts is dominated by alternate loss of gene duplicates, and by intra-chromosomal rearrangements (~one per million years). These physical maps are a useful resource for comparative positional cloning of traits in cichlid fishes. The paired BAC end sequences from these clones will be an important resource for scaffolding forthcoming shotgun sequence assemblies of the tilapia genome.
Background
Tilapia (Oreochromis spp.) are among the most important species in aquaculture and a primary source of animal protein for millions of people in the developing world [1]. Only limited efforts have been made toward genetic improvement of these species [2]. The sequence of the tilapia genome will be a fundamental resource used for genetic selection, on traits such as growth performance and disease resistance, to create strains of fish optimized for the unique culture conditions of each country.
Tilapia and other closely related species of African cichlid fishes are also widely used in basic research. Because of their intimate physiological relationship with the environment, tilapia are ideal for studies of ion regulation [3,4], the accumulation of heavy metals [5], and detoxification of biotoxins [6]. Nile tilapia expressing a humanized insulin gene are being studied as a source of islet cells which might be transplanted into humans for control of type I diabetes [7]. Tilapia are also an important model for studying environmental influences on sex differentiation [8]. The closely related haplochromine cichlids of the East African lakes are a model system for studying the genetic basis of behavior [9] and evolutionary processes of adaptation and speciation [10].
Cichlid genomics
Considerable progress has been made in developing genomic resources for tilapia and other East African cichlid fishes. Genetic maps have been published for tilapia [11], Lake Malawi haplochromines [12], and Astatotilapia burtoni [13]. There are also extensive collections of ESTs for Lake Victoria haplochromines [14,15], A. burtoni [16,17] and Nile tilapia [18]. Several BAC libraries have been constructed for Nile tilapia [19], and fingerprinted to construct a physical map [20]. BAC libraries have been constructed also for haplochromine cichlids from lakes Malawi [21], Victoria [22] and Tanganyika [23].
Comparative physical maps
Comparative maps have been a useful intermediate resource for many agricultural species before complete genome sequences were available [24-26]. Most often these comparative maps have relied on mapping homologous gene markers in radiation hybrid panels [27], but comparative maps have also been based on analysis of BAC end sequences [28,29]. Until a complete genome sequence is available for tilapia, comparative maps to the genome sequences of model fish species will provide the best organization of the partial sequence data for cichlid fishes.
The utility of a comparative map is proportional to the extent to which synteny exists between the two genomes. Useful comparative maps have been constructed between cattle and human (100MY divergence)[30]. The divergence among many fish lineages is much older, creating the potential for more extensive genome rearrangements. The Ostariophysi (e.g. zebrafish) and Acanthopterygii (e.g. medaka) diverged ~300MY ago [31]. Divergence among Percomorph groups (e.g. Tilapia and pufferfish) occurred more than 100MY ago [32]. The utility of comparative maps across these greater evolutionary distances is not yet clear.
Early research suggested that the rate of chromosome evolution is relatively low in non-mammalian vertebrates [33]. Recently it has been suggested that the rate of chromosomal rearrangement increases immediately after episodes of whole-genome duplication [34]. Teleost fishes experienced an additional round of whole genome duplication about 300 MY ago [35], and recent papers have suggested that fishes continue to have a high rate of chromosomal rearrangement [36]. However, the more extensive inter-chromosomal rearrangements detected in the zebrafish genome may be due to unique evolutionary processes in that lineage, and there appear to have been no major inter-chromosomal rearrangements in the medaka genome during the last 300MY [37]. The green pufferfish shows relatively little inter-chromosomal rearrangement since divergence from the ancestral bony vertebrate [38]. Most of the changes in the pufferfish lineage represent fusions that reduced the chromosome number after whole genome duplications.
The goal of the present study was to construct a comparative physical map between tilapia and the latest sequence assemblies for three other percomorph species: stickleback, medaka and pufferfish. From this comparative map we estimate the extent of chromosomal rearrangement during the recent evolution of these species.
Results and Discussion
New BAC library
The BAC library (VMRC-44) constructed at the Benaroya Research Institute consists of 73,728 clones (192 384-well plates) with an average insert size of 150 kb. This represents a total of 11 Gbp or approximately 10× coverage of the tilapia genome. The methods used to prepare this library are presented in Additional file 1.
Sequencing statistics
Genoscope
The construction of the BAC libraries sequenced at Genoscope was reported previously [19]. A total of 35,000 clones from these libraries (average insert 182 kb, ~5.6× genome coverage), have been restriction fingerprinted and assembled into 3,600 contigs [20]. Genoscope end sequenced a total of 40,704 clones (52 plates from library 3 and 54 plates from library 4). From 37,383 clones, a total of 68,032 end sequences were obtained, representing 6.8× clone coverage of the genome. The mean trimmed length of the sequences was 562.6 bp, for a total dataset of 38,272,386 bp representing 3.8% sequence coverage of the genome.
Broad
The Broad Institute end sequenced 73,728 clones (192 plates) from the Benaroya library, obtaining a sequence for at least one end of 68,876 clones, representing 10.0× clone coverage of the genome. Multiple attempts were made to sequence some clones and therefore, a total of 153,216 end sequences were finally submitted to GenBank. The mean length of the sequences was 757.3 bp, for a dataset of 116,029,366 bp. After quality trimming and vector removal with Lucy, a total of 124,995 sequences remained, with a mean length of 527.3 bp, for a total of 65,912,624 bp, representing 6.6% sequence coverage of the genome. These sequences were previously analyzed for their repeat content [39].
Microsatellites
Microsatellite motifs were identified in 7,230 (3.7%) of the 193,027 sequences. These included 5,027 dinucleotide, 1,250 trinucleotide, and 953 tetranucleotide repeats (Additional file 2Table S1). Over half of the repeats (3,887) were AC dinucleotides. AT and AG dinucleotides were also abundant. AAT was the most frequent trinucleotide. These microsatellites could be exploited to develop new genetic markers and could be used to anchor the FPC-based physical map [20] to the genetic map [11].
Genes
A total of 16,636 (8.6%) repeat-masked sequences had a significant (1e-5) BLASTx hit to the Uniprot database. We found that 38,020 (19.7%) of the repeat-masked sequences had a significant (1e-50) BLASTn hit to the 116,899 Nile tilapia EST set [40]. Therefore, 49,823 (25.8%) of the sequences had either a significant BLASTx hit to Uniprot or a significant BLASTn hit to the Nile tilapia ESTs. There were 4,833 (2.5%) sequences that had a significant hit to both Uniprot and the Nile tilapia ESTs.
Comparative mapping
A total of 193,027 BAC end sequences were BLASTed against the genome assemblies of stickleback, medaka and pufferfish. The results are summarized in Table 1. The proportion of sequences that had hits with e-values less than e-10 ranged from 11 percent against pufferfish, 15 percent against medaka and 17 percent against stickleback. Twenty-eight percent of the BACs had at least one hit to the stickleback genome assembly.
Table 1.
BLAST statistics against three fish genome assemblies.
| # BACs one end | # BACs both ends | Sequences w/hit | BACs w/hit | Type 1* | Type 2* | Type 3* | Type 4* | 3/(2+3) | (3+4)/(2+3+4) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Stickleback | ||||||||||
| Genoscope | 6,734 | 30,649 | 11,229 | 10,048 | 7,987 | 797 | 54 | 153 | 0.063 | 0.206 |
| Broad | 12,758 | 56,118 | 21,754 | 19,510 | 14,405 | 1,416 | 142 | 633 | 0.054 | 0.286 |
| Combined | 19,492 | 86,767 | 33,053 | 29,558 | 22,392 | 2,213 | 196 | 786 | 0.057 | 0.259 |
| Medaka | ||||||||||
| Genoscope | 6,734 | 30,649 | 9,764 | 9,278 | 7,087 | 469 | 43 | 226 | 0.084 | 0.364 |
| Broad | 12,758 | 56,118 | 19,699 | 17,943 | 13,907 | 886 | 51 | 624 | 0.054 | 0.432 |
| Combined | 19,492 | 86,767 | 29,463 | 27,221 | 20,994 | 1,355 | 94 | 850 | 0.065 | 0.410 |
| Tetraodon | ||||||||||
| Genoscope | 6,734 | 30,649 | 6,931 | 6,386 | 3,879 | 265 | 14 | 62 | 0.050 | 0.222 |
| Broad | 12,758 | 56,118 | 14,260 | 13,227 | 7,279 | 503 | 19 | 188 | 0.036 | 0.291 |
| Combined | 19,492 | 86,767 | 21,191 | 19,613 | 11,158 | 768 | 33 | 250 | 0.041 | 0.269 |
*Numbers for type 1, 2, 3 & 4 do not include hits to the contigs in the 'unordered chromosome' of each genome assembly. Type 2 hits were scored when the two end sequences of a clone hit within 300 kb of each other in the target genome.
We classified the BACs into one of four types, according to the pattern of BLAST hit. Type 1 clones are those for which only a single sequence produced a hit in the target genome. Type 2 clones are those in which the sequences from the two ends of the BAC hit in the appropriate opposing orientation within 300 kb in the target genome. Type 3 clones are those in which the two end sequences of a BAC hit the same chromosome in the target genome outside of the 300 kb range. Type 4 BACs are those in which the two sequences hit different chromosomes in the target genome.
Since the average BLAST hit rate against the stickleback genome is 17%, we expected the proportion of clones with hits on both ends would be 2.9%. In fact we observed a slightly greater proportion (3.7%), possibly reflecting a clustering of conserved sequences in the genomes. When both ends of a BAC had BLAST hits, they were most often found within 300 kb on the same chromosome in the target genome (type 2). A much smaller proportion (3-5%) were found at larger distances on the same chromosome in the target genome (type 3).
Conservation of gene order
We can use the ratios of type 2, 3 and 4 hits (Table 1) to estimate the number of rearrangements between genomes. Across the three species, 27-41% of double hit clones are type 3 or 4. If the BAC clone inserts average 150 kb, and every third clone has a break in synteny, it would suggest a breakpoint every 3 × 150 kb = 450 kb across the genome. This is equivalent to more than 2000 breakpoints across the genome, or about 100 breakpoints per chromosome. We suspect this simple statistic overestimates the true number of chromosomal rearrangements.
The best estimate of intra-chromosomal rearrangements is the number of type 3 BACs relative to the number of type 2 + type 3 BACs. This proportion is between 3 and 6%, suggesting an intra-chromosomal rearrangement every 20 × 150 kb = 3 Mb. If the average chromosome is 48 Mb, this suggests about 16 breakpoints (e.g. 8 inversions) per chromosome. We detected a mean of 2.1 breakpoints per chromosome, with at least one rearrangement on each stickleback chromosome (Additional file 3Table S2). The observed breakpoints were spanned by an average of 3.5 BAC clones. Unfortunately, the relatively low clone coverage of the type 3 BACs does not allow us to identify all of the likely breakpoints, or precisely map their locations. Still, the high end of these estimates (8 inversions/chromosome) suggests there have been only 160 inversions since the divergence of tilapia and stickleback. The type 3 hits are visualized in Circos plots in Additional files 4, 5, 6, Figures S1-S3.
Type 4 BACs are possible evidence of inter-chromosomal rearrangements, and represent 24-37% of the two-hit BACs. This might suggest more than 100 breakpoints in synteny for each chromosome. However, we do not think this statistic is an indication of a large number of inter-chromosomal transfers of genes. Rather, it probably includes many instances in which one of the BLAST matches is to a paralog on a second chromosome. For example, if the syntenic copy of the gene has been lost, BLAST will identify a paralog on another chromosome as the best hit. This kind of gene loss is a common feature of fish genomes, which underwent a whole-genome duplication about 300MY ago. Alternate loss of even a small proportion of genes from these duplicated regions would be sufficient to create the pattern. There are about 1,250 genes/chromosome, and if only 5% of them (60 genes/chromosome) were deleted after the whole genome duplication, it would be sufficient to create the pattern we see in the BAC data. The fact that type 4 BLAST hits have much lower e-values than type 2 BLAST hits (Figure 1) tends to reinforce this view.
Figure 1.
Distribution of BLAST scores for type 2, 3 and 4 hits of the Broad Institute tilapia BAC end sequences on the stickleback genome assembly. The distribution was truncated at e-10.
We mapped the rearrangements onto a phylogeny of the four species. The results suggest that approximately 15-20 rearrangements have occurred on each lineage since they diverged from their common ancestor. There is no indication that the rate of rearrangement is higher in one lineage than another.
Comparative physical maps
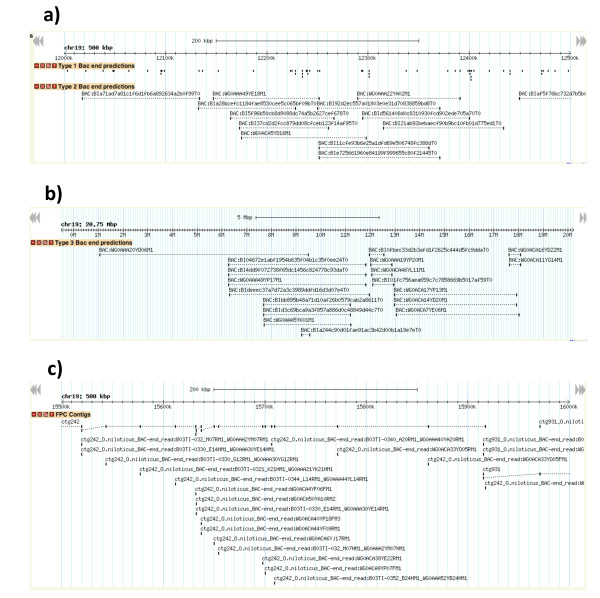
These BLAST results are displayed in a GBrowse interface at http://www.BouillaBase.org (Figure 2). Separate tracks display the type 1, 2, 3 and 4 BLAST hits. An additional track displays the BLAST hits from each of the fingerprint contigs in the previous physical map [20]. Because these FPC contigs contain multiple BAC clones, they help to tie the physical map together at larger scales than the end sequences of individual clones.
Figure 2.
Comparative mapping of tilapia data mapped on the stickleback genome assembly displayed in GBrowse at http://www.BouillaBase.org. a) Type 1 and 2 BLAST hits, b) Type 3 BLAST hits, c) FPC contigs.
Conclusions
End-sequencing of these BAC libraries was a key step in preparing the tilapia genome for shotgun sequencing. Together with the BAC fingerprint database, these sequences will provide long-range structure for scaffolding the contigs of genome assemblies to construct a golden path across the genome.
Recent molecular phylogenies appear to have reached a consensus that cichlids are more closely related to medaka than to either pufferfish or stickleback [40-42]. Nevertheless, a higher number of the tilapia BAC end sequences hit stickleback (33,053) than either medaka (29,463) or pufferfish (21,191). This discrepancy might be due to variation in the quality of each assembly, or it might support an alternative phylogenetic reconstruction. Regardless, it appears that the stickleback sequence is currently the best reference sequence for building comparative maps of tilapia [43].
Finally, these data suggest that chromosomal evolution in recent teleosts is dominated by alternate loss of gene duplicates, and by intra-chromosomal rearrangements. The rate of these rearrangements is relatively slow, on the order of one per million years. So the prospects are good for building useful comparative maps between sequenced genomes and the large number of as yet unsequenced teleost species of commercial or scientific importance.
Methods
Sequence trimming
Both trimmed and untrimmed quality scores and FASTA sequences for the Genoscope library 4 sequences were available, whereas only trimmed FASTA sequences for the Genoscope library 3 were available. To achieve essentially the same level of trimming for both Genoscope libraries and the Broad library, the Genoscope library 4 data was used to determine a set of parameters that trimmed the data in the same way as had been done for both the Genoscope libraries. The following Lucy 1.20p [44] settings were used: -error 0.025 0.02, -bracket 10 0.005, -window 50 0.08 10 0.12, and -vector with the FASTA sequence of the pBAC-Lac cloning vector [45] for the Genoscope libraries and the FASTA sequence of the pCC1BAC cloning vector (Epicentre Biotechnologies) for the Broad library.
Annotation
Identification of microsatellites
We scanned the BAC end sequences for microsatellites that might be useful for genetic mapping. We used the Tandem Repeats Finder http://tandem.bu.edu/trf/trf.html[46] to identify microsatellite motifs. The BAC ends containing microsatellites have been color-coded in the annotation tracks in the GMOD browser.
Identification of genes
The BAC end sequences were masked with RepeatMasker version open-3.2.8 [47] against a combination of the Repbase [48] RepeatMasker libraries, release 20090604 and tilapia specific repeats [39]. The sequences were then aligned to the Uniprot database (release-2010_05) using BLASTx, and a database of 116,899 Sanger ESTs from Nile tilapia [18] using BLASTn. Significant hits were defined with an e-value threshold of 1e-5 for Uniprot, or 1e-50 for the ESTs.
BLAST analysis
Comparative mapping was performed by running BLASTn against the pufferfish, stickleback and medaka genome assemblies. The genomes were downloaded from the UCSC Genome Browser http://hgdownload.cse.ucsc.edu/downloads.html. The following versions were used for the respective genomes: Feb. 2004 (Genoscope 7/tetNig1), Feb. 2006 (Broad/gasAcu1), and Oct. 2005 (NIG/UT MEDAKA1/oryLat2). FASTA sequences were downloaded and formatted into BLAST databases for use with the NCBI BLASTall tool and scripts utilizing BioPerl were used to parse the results. Type 2 hits were defined as mate pairs that hit the target genome in opposing orientation at a distance of 300 kb or less. Type 3 hits were defined as mate pairs that hit the same chromosome, regardless of orientation. Type 4 hits were defined as mate pairs that hit different chromosomes. The positions of the BLAST hits were visualized with Circos [49].
Online access to the resource
We used the GMOD browser http://www.gmod.org to develop a comparative genome server for fishes that maps tilapia ESTs and BAC end-sequences onto the genome assemblies of stickleback, medaka and pufferfish. This server can be accessed through our www site http://www.BouillaBase.org.
Additional data files
The Benaroya/Broad BAC end sequences are available in the NCBI Trace Archive under Center_Project 'G1447'. The Genoscope sequences are available as accession numbers FQ242537 - FQ280267.
Authors' contributions
LS and MAC carried out the bioinformatic analyses. TK, AEH and BYL constructed and prepared the BAC libraries for sequencing at Genoscope. CA and AS constructed the BAC library that was sequenced at the Broad Institute. CD and JP sequenced the BAC libraries at Genoscope. JJ, FDP and KLT organized the sequencing at the Broad Institute. JFB, HDC, COC and TDK prepared the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental Methods. Description of methods used in constructing the BAC library.
Table S1 Microsatellite motifs identified in the BAC end sequences.
Table S2 Number of type 3 BACs spanning potential recombination breakpoints in the comparative map to stickleback.
Figure S1 Circos plot of the type 3 BLAST hits on the stickleback genome. The chromosomes of the stickleback genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.
Figure S2 Circos plot of the type 3 BLAST hits on the medaka genome. The chromosomes of the medaka genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.
Figure S3 Circos plot of the type 3 BLAST hits on the Tetraodon genome. The chromosomes of the Tetraodon genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.
Contributor Information
Lucile Soler, Email: lucile.soler@cirad.fr.
Matthew A Conte, Email: mconte@umd.edu.
Takayuki Katagiri, Email: takakata@kaiyodai.ac.jp.
Aimee E Howe, Email: soxfan2626@hotmail.com.
Bo-Young Lee, Email: bylee613@snu.ac.kr.
Chris Amemiya, Email: camemiya@benaroyaresearch.org.
Andrew Stuart, Email: astuart@benaroyaresearch.org.
Carole Dossat, Email: cdossat@genoscope.cns.fr.
Julie Poulain, Email: poulain@genoscope.cns.fr.
Jeremy Johnson, Email: jjohnson@broadinstitute.org.
Federica Di Palma, Email: fdipalma@broadinstitute.org.
Kerstin Lindblad-Toh, Email: kersli@broad.mit.edu.
Jean-Francois Baroiller, Email: baroiller@cirad.fr.
Helena D'Cotta, Email: dcotta@cirad.fr.
Catherine Ozouf-Costaz, Email: ozouf@mnhn.fr.
Thomas D Kocher, Email: tdk@umd.edu.
Acknowledgements
This work was supported by a grant from the USDA-NRICGP (#2006-04830) to TDK. BAC end-sequences from Genoscope have been produced through the Project "BAC end-sequencing for comparative genomics and assembly of the genome of tilapia Oreochromis niloticus" funded by CNS. We thank Elodie Pepey for her help with the robotics at CIRAD. The authors would like to thank Gaëtan Droc from the Cirad joint unit UMR DAP (Plant Development and Genetic Improvement) for help with the Circos diagrams. Thanks also to the Broad Institute Sequencing Platform for sequencing the Benaroya/Broad BAC library and making the data available.
References
- Coward K, Little DC. Culture of the 'aquatic chicken': present concerns and future prospects. Biologist (London) 2001;48:12–6. [PubMed] [Google Scholar]
- Hulata G. Genetic manipulations in aquaculture: a review of stock improvement by classical and modern technologies. Genetica. 2001;111:155–73. doi: 10.1023/A:1013776931796. [DOI] [PubMed] [Google Scholar]
- Fiol DF, Chan SY, Kültz D. Regulation of osmotic stress transcription factor 1 (Ostf1) in tilapia (Oreochromis mossambicus) gill epithelium during salinity stress. J Exp Biol. 2006;209:3257–65. doi: 10.1242/jeb.02352. [DOI] [PubMed] [Google Scholar]
- Breves JP, Hasegawa S, Yoshioka M, Fox BK, Davis LK, Lerner DT, Takei Y, Hirano T, Grau EG. Acute salinity challenges in Mozambique and Nile tilapia: Differential responses of plasma prolactin, growth hormone and branchial expression of ion transporters. Gen Comp Endocrinol. 2010;167:135–142. doi: 10.1016/j.ygcen.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Wang F, Leung AO, Wu SC, Yang MS, Wong MH. Chemical and ecotoxicological analyses of sediments and elutriates of contaminated rivers due to e-waste recycling activities using a diverse battery of bioassays. Environ Pollut. 2009;157:2082–90. doi: 10.1016/j.envpol.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Prieto AI, Jos A, Pichardo S, Moreno I, de Sotomayor MA, Moyano R, Blanco A, Cameán AM. Time-dependent protective efficacy of Trolox (vitamin E analog) against microcystin-induced toxicity in tilapia (Oreochromis niloticus) Environ Toxicol. 2009;24:563–79. doi: 10.1002/tox.20458. [DOI] [PubMed] [Google Scholar]
- Wright JR Jr, Pohajdak B. Cell therapy for diabetes using piscine islet tissue. Cell Transplant. 2001;10:125–43. [PubMed] [Google Scholar]
- Baroiller JF, D'Cotta H, Saillant E. Environmental effects on fish sex determination and differentiation. Sex Dev. 2009;3:118–35. doi: 10.1159/000223077. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–98. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Lee BY, Lee WJ, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A, Stern JE, Terai Y, Kocher TD. A second-generation genetic linkage map of tilapia (Oreochromis spp.) Genetics. 2005;170:237–44. doi: 10.1534/genetics.104.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc Natl Acad Sci USA. 2003;100:5252–7. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanetra M, Henning F, Fukamachi S, Meyer A. A microsatellite-based genetic linkage map of the cichlid fish, Astatotilapia burtoni (Teleostei): a comparison of genomic architectures among rapidly speciating cichlids. Genetics. 2009;182:387–97. doi: 10.1534/genetics.108.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kobayashi N, Shin-i T, Horiike T, Tateno Y, Kohara Y, Okada N. Extensive analysis of ORF sequences from two different cichlid species in Lake Victoria provides molecular evidence for a recent radiation event of the Victoria species flock: identity of EST sequences between Haplochromis chilotes and Haplochromis sp. "Redtailsheller". Gene. 2004;343:263–9. doi: 10.1016/j.gene.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Watanabe M, Horiike T, Kohara Y, Okada N. Extensive analysis of EST sequences reveals that all cichlid species in Lake Victoria share almost identical transcript sets. Gene. 2009;441:187–91. doi: 10.1016/j.gene.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Renn SC, Aubin-Horth N, Hofmann HA. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics. 2004;5:42. doi: 10.1186/1471-2164-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Renn SC, Steinke D, Braasch I, Hofmann HA, Meyer A. Annotation of expressed sequence tags for the East African cichlid fish Astatotilapia burtoni and evolutionary analyses of cichlid ORFs. BMC Genomics. 2008;9:96. doi: 10.1186/1471-2164-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Howe AE, Conte MA, D'Cotta H, Pepey E, Baroiller JF, di Palma F, Carleton KL, Kocher TD. An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genomics. 2010;11:278. doi: 10.1186/1471-2164-11-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Asakawa S, Minagawa S, Shimizu N, Hirono I, Aoki T. Construction and characterization of BAC libraries for three fish species; rainbow trout, carp and tilapia. Anim Genet. 2001;32:200–4. doi: 10.1046/j.1365-2052.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Kidd C, Tomasino E, Davis JT, Wishon C, Stern JE, Carleton KL, Howe AE, Kocher TD. A BAC-based physical map of the Nile tilapia genome. BMC Genomics. 2005;6:89. doi: 10.1186/1471-2164-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma F, Kidd C, Borowsky R, Kocher TD. Construction of bacterial artificial chromosome libraries for the Lake Malawi cichlid (Metriaclima zebra), and the blind cavefish (Astyanax mexicanus) Zebrafish. 2007;4:41–7. doi: 10.1089/zeb.2006.9996. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kobayashi N, Fujiyama A, Okada N. Construction of a BAC library for Haplochromis chilotes, a cichlid fish from Lake Victoria. Genes Genet Syst. 2003;78:103–5. doi: 10.1266/ggs.78.103. [DOI] [PubMed] [Google Scholar]
- Lang M, Miyake T, Braasch I, Tinnemore D, Siegel N, Salzburger W, Amemiya CT, Meyer A. A BAC library of the East African haplochromine cichlid fish Astatotilapia burtoni. J Exp Zool B Mol Dev Evol. 2006;306:35–44. doi: 10.1002/jez.b.21068. [DOI] [PubMed] [Google Scholar]
- Everts-van der Wind A, Larkin DM, Green CA, Elliott JS, Olmstead CA, Chiu R, Schein JE, Marra MA, Womack JE, Lewin HA. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc Natl Acad Sci USA. 2005;102:18526–31. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kirkness EF, Nefedov M, McWilliam S, Ratnakumar A, Barris W, Zhao S, Shetty J, Maddox JF, O'Grady M, Nicholas F, Crawford AM, Smith T, de Jong PJ, McEwan J, Oddy VH, Cockett NE. the International Sheep Genomics Consortium. Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biol. 2007;8:R152. doi: 10.1186/gb-2007-8-7-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KM, Chaves LD, Mendoza KM. An integrated and comparative genetic map of the turkey genome. Cytogenet Genome Res. 2007;119:113–26. doi: 10.1159/000109627. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Gustafson-Seabury A, Durkin K, Wagner ML, Goh G, Seabury CM, Brinkmeyer-Langford C, Lee EJ, Agarwala R, Stallknecht-Rice E, Schäffer AA, Skow LC, Tozaki T, Yasue H, Penedo MC, Lyons LA, Khazanehdari KA, Binns MM, MacLeod JN, Distl O, Guérin G, Leeb T, Mickelson JR, Chowdhary BP. A 4,103 marker integrated physical and comparative map of the horse genome. Cytogenet Genome Res. 2008;122:28–36. doi: 10.1159/000151313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatcheva MB, Chen K, Larkin DM, Meyers SN, Marron BM, He W, Schook LB, Beever JE. Piggy-BACing the human genome I: constructing a porcine BAC physical map through comparative genomics. Anim Biotechnol. 2008;19:28–42. doi: 10.1080/10495390701807634. [DOI] [PubMed] [Google Scholar]
- Liu H, Jiang Y, Wang S, Ninwichian P, Somridhivej B, Xu P, Abernathy J, Kucuktas H, Liu Z. Comparative analysis of catfish BAC end sequences with the zebrafish genome. BMC Genomics. 2009;10:592. doi: 10.1186/1471-2164-10-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin DM, Everts-van der Wind A, Rebeiz M, Schweitzer PA, Bachman S, Green C, Wright CL, Campos EJ, Benson LD, Edwards J, Liu L, Osoegawa K, Womack JE, de Jong PJ, Lewin HA. A cattle-human comparative map built with cattle BAC-ends and human genome sequence. Genome Res. 2003;13:1966–72. doi: 10.1101/gr.1560203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Diogo R, He S. In: The Timetree of Life. Hedges SB, Kumar S, editor. Oxford: Oxford University Press; 2009. Teleost fishes (Teleostei) pp. 335–338. [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Sarich VM, Maxson LR. The importance of gene rearrangement in evolution: evidence from studies on rates of chromosomal, protein, and anatomical evolution. Proc Natl Acad Sci USA. 1974;71:3028–3030. doi: 10.1073/pnas.71.8.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. Rearrangement rate following the whole-genome duplication in teleosts. Mol Biol Evol. 2007;24:860–7. doi: 10.1093/molbev/msm003. [DOI] [PubMed] [Google Scholar]
- Hurley IA, Mueller RL, Dunn KA, Schmidt EJ, Friedman M, Ho RK, Prince VE, Yang Z, Thomas MG, Coates MI. A new time-scale for ray-finned fish evolution. Proc R Soc B. 2007;274:489–98. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18:544–50. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–9. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Shirak A, Grabherr M, Di Palma F, Lindblad-Toh K, Hulata G, Ron M, Kocher TD, Seroussi E. Identification of repetitive elements in the genome of Oreochromis niloticus: tilapia repeat masker. Mar Biotechnol (NY) 2010;12:121–5. doi: 10.1007/s10126-009-9236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Kumazawa Y, Miya M, Mabuchi K, Nishida M. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol Biol. 2008;8:215. doi: 10.1186/1471-2148-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dettaï A, Cruaud C, Couloux A, Desoutter-Meniger M, Lecointre G. RNF213, a new nuclear marker for acanthomorph phylogeny. Mol Phylogenet Evol. 2009;50:345–63. doi: 10.1016/j.ympev.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Chen W-J, Mayden RL. A phylogenomic perspective on the new era of ichthyology. Bioscience. 2010;60:421–432. doi: 10.1525/bio.2010.60.6.6. [DOI] [Google Scholar]
- Sarropoulou E, Nousdili A, Magoulas A, Kotoulas G. Linking the genomes of nonmodel teleosts through comparative genomics. Mar Biotechnol. 2008;10:227–233. doi: 10.1007/s10126-007-9066-5. [DOI] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N. Human BAC library: construction and rapid screening. Gene. 1997;191:69–79. doi: 10.1016/S0378-1119(97)00044-9. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996-2010. http://www.repeatmasker.org
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogen Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods. Description of methods used in constructing the BAC library.
Table S1 Microsatellite motifs identified in the BAC end sequences.
Table S2 Number of type 3 BACs spanning potential recombination breakpoints in the comparative map to stickleback.
Figure S1 Circos plot of the type 3 BLAST hits on the stickleback genome. The chromosomes of the stickleback genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.
Figure S2 Circos plot of the type 3 BLAST hits on the medaka genome. The chromosomes of the medaka genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.
Figure S3 Circos plot of the type 3 BLAST hits on the Tetraodon genome. The chromosomes of the Tetraodon genome are represented on the circle. The position of BAC mate pair BLAST hits are indicated with red arcs.