Abstract
An essential question in human biology is how cells and tissues differ in gene and protein expression and how these differences delineate specific biological function. Here, we have performed a global analysis of both mRNA and protein levels based on sequence-based transcriptome analysis (RNA-seq), SILAC-based mass spectrometry analysis and antibody-based confocal microscopy. The study was performed in three functionally different human cell lines and based on the global analysis, we estimated the fractions of mRNA and protein that are cell specific or expressed at similar/different levels in the cell lines. A highly ubiquitous RNA expression was found with >60% of the gene products detected in all cells. The changes of mRNA and protein levels in the cell lines using SILAC and RNA ratios show high correlations, even though the genome-wide dynamic range is substantially higher for the proteins as compared with the transcripts. Large general differences in abundance for proteins from various functional classes are observed and, in general, the cell-type specific proteins are low abundant and highly enriched for cell-surface proteins. Thus, this study shows a path to characterize the transcriptome and proteome in human cells from different origins.
Keywords: cell lines, expression, human, proteome, transcriptome
Introduction
The human body displays a complex array of biological functions mediated by the expression of mRNA and proteins. The construction of complex organs, such as the kidney or the brain, is far from understood and there is a need to dissect in a systematic manner the expression of genes and proteins using quantitative methods. The complete sequences of human genomes (Lander et al, 2001; Venter et al, 2001) have facilitated such studies and opened up the possibility for whole-genome analysis on both the RNA and the protein levels. The ultimate goal of such an endeavor is to define the quantitative levels of the transcriptome and the proteome in various cell types in human tissues and organs.
We have recently described an antibody-based immunohistochemistry analysis of 48 human organs and tissues (Ponten et al, 2009) based on proteins corresponding to one third of all human genes, showing that a large portion of the analyzed proteins were detected across the tissues in a ubiquitous manner. This led to the suggestion that tissue specificity is achieved by precise regulation of protein levels in space and time, and that different tissues in the body acquire their unique characteristics by controlling not which proteins are expressed but how much of each is produced (Ponten et al, 2009). Similarly, a detailed study of 1% of the human genome showed that chromosomes are ubiquitously transcribed and that the majority of all bases are included in primary transcripts (Birney et al, 2007). These results have recently been supported by deep sequencing, demonstrating that a majority of the transcripts can be detected in a human cell line (Sultan et al, 2008) and that a large fraction (75%) of the human protein-coding genes are expressed in most tissues (Ramskold et al, 2009).
Most analyses of whole proteomes by mass spectrometry have so far been performed on yeast cells (Ghaemmaghami et al, 2003; de Godoy et al, 2008; Picotti et al, 2009). Analysis of the human proteome using mass spectrometry has so far only been performed on a moderate fraction of the complete proteome. The proteomic limitation combined with the limitation of quantification accuracy in array-based methods for RNA analysis have resulted in relatively low correlations between RNA and protein levels (de Sousa Abreu et al, 2009), as exemplified by studies on yeast (Griffin et al, 2002; Greenbaum et al, 2003) and human cancers (Chen et al, 2002). Recent technological advances in the field of mass spectrometry make it possible to perform deep proteome analysis also of complex organisms (Cox and Mann, 2007) with quantitative mass spectrometry methods such as the stable isotope-based SILAC method (Ong et al, 2002; Mann, 2006). The technological developments of RNA-seq together with accurate SILAC quantification enable a global comparative analysis of RNA and protein levels and changes in higher eukaryotes such as humans.
A complication in global comparisons of RNA and protein levels in tissues and organs is the multitude of cell types and developmental stages present in most tissues. We have therefore decided to compare the human transcriptome and the proteome with quantitative methods in three established human cell lines of different functional origins allowing an analysis of relatively homogenous cellular populations. Although caution needs to be taken due to the artificial nature of cell lines grown in vitro, the aim of the study was to categorize all the protein-coding genes based on their cell specificity and expression levels. The analysis was performed using deep sequencing of mRNA, proteomics analysis using triple isotope labeled SILAC mass spectrometry and antibody-based confocal microscopy (Barbe et al, 2008; Berglund et al, 2008). The RNA-seq data give the absolute number of reads per kilobase for each gene. The triple-SILAC MS data accurately determine the relative abundance of each of the proteins in the three cell lines. Furthermore, the summed peptide intensities roughly estimate the absolute amount of each of the identified proteins. Since the cells were harvested during exponential growth, steady-state levels of RNA and proteins could be analyzed and compared. The study further allowed quantitative analysis of changes on the RNA and protein levels between the three cell lines, and allowed us to annotate the expression of all genes across these functionally different cells.
Results and discussion
Antibody-based protein profiling
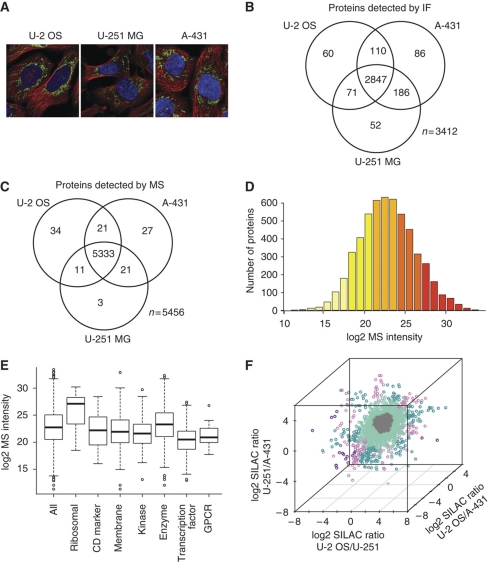
Three functionally different human cell lines were analyzed in the study: a bone osteosarcoma (U-2 OS), an epidermoid squamous cell carcinoma (A-431) and a brain glioblastoma (U-251 MG) (Figure 1A). A subset of the proteome, corresponding to 3877 genes, was studied using antibody-based immunofluorescence confocal microscopy (IF) as part of the Human Protein Atlas (HPA) project, as described earlier (Barbe et al, 2008; Berglund et al, 2008). Similar to the conclusion from a previous study (Ponten et al, 2009), we found that out of the detected proteins (n=3412), the majority (83%) were found in all three cell lines and few (6%) of the proteins were expressed exclusively in only one of the three cell lines (Figure 1B).
Figure 1.
The protein profiles in the three human cell lines based on confocal microscopy and mass spectrometry analysis. The numbers of analyzed and detected genes for each platform are shown in Supplementary Table 1. (A) The three cell lines used in the study: U-2 OS, U-251 MG and A-431 illustrated by confocal immunofluorescent images, where the microtubules (red), nuclei (blue) and an antibody (HPA024087) toward the human TUFM are staining mitochondria (green). (B) Venn diagram showing the number of proteins (%) detected by confocal IF in each cell line and the overlap between the cell lines. (C) Venn diagram showing the number of proteins detected by MS in each cell line and the overlap between the cell lines. (D) Distribution of log2 MS intensity for all detected proteins (n=5405) in U-2 OS. Bars are colored according to MS intensity, ranging from light yellow (low MS intensity) to dark red (high MS intensity). (E) The distribution of log2 MS intensity values for the protein categories (see Supplementary Table 2 for details) in U-2 OS. (F) Three-dimensional plot of log2 protein (SILAC) ratios between two cell lines performed for all three combinations. The genes are colored by the variation of expression between the three cell lines as: similar (less than two-fold difference between all cell lines in gray), slightly changed (two- to four-fold difference between two cell lines in light turquoise or between all cell lines in light purple) or substantially changed (more than four-fold difference between two cell lines in dark turquoise or between all cell lines in dark purple).
Mass spectrometry-based protein profiling
A deep proteomic analysis of the three cell lines was performed using a triple-SILAC method in which the three cell lines were cultivated with amino acids with different isotopes and then analyzed by mass spectrometry. The triple-SILAC method was chosen since it is particularly well suited for comparative quantification of protein levels between different cell lines by generating triple peak patterns for each protein. Specifically in this setup, identification of one of the peptides automatically yields the identity of the other two. Therefore, sampling biases due to the LC–MS/MS method do not influence the measured diversity of the analyzed proteins. A majority of the detected proteins were found across all the three cell lines (Figure 1C). Approximately 5500 proteins were quantified using the triple-SILAC method and the dynamic range as scored by the mass spectrometry intensity scores was ∼106 (Figure 1D). It is noteworthy that the coverage of the proteome is not complete and that even within the estimated 106-fold technical abundance range, there may be undetected proteins. A comparison of some selected protein classes reveals that, in general, the ribosomal proteins were most abundant in all three cell lines as judged from the MS intensity scores and the transcription factors and G-protein coupled receptors (GPCRs) were the least abundant (Figure 1E; Supplementary Table 2). A quantitative analysis of the differences between SILAC ratios (Figure 1F) shows that most (65%) of the detected proteins have similar expression levels (less than two-fold differences) in the three cell lines (gray), suggesting a ‘house-keeping' role with common functions in the three cell lines. Obviously, the proteins (one third) with differential expression are interesting starting points for further studies regarding the cell-specific functions in the various cell types.
Sequencing-based transcript profiling
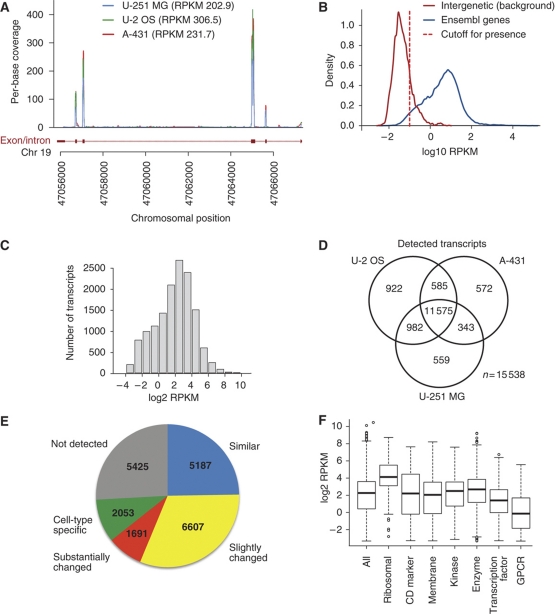
The RNA levels in the three cell lines were analyzed using digital RNA-seq based on >20 million separate reads for each cell line and the RPKM values (reads per kilobase of exon model per million mapped reads) were calculated for each gene. This approach has been shown to be very sensitive and specific when it comes to estimating expression levels, with Pearson's correlation values between RNA-seq and RT–qPCR of ∼0.95 (Cloonan et al, 2008; Mortazavi et al, 2008; Sandberg et al, 2008; Wang et al, 2008). A RPKM threshold value of 0.1 was set to detect the presence of a transcript for a particular gene, which corresponds to a false-discovery rate (FDR) and false-negative rate of 5% (Supplementary Figure S1B). An example of RPKM values (Figure 2A) across the exons and introns of a gene on chromosome 19 shows that the reads are predominately localized to the predicted exons, suggesting relatively low background from non-functional transcripts. Similarly, a plot showing the exon/intron ratios for 9630 genes, where both exons and introns were detected (Supplementary Figure S1A), demonstrates that a majority of the genes have considerably more reads in the exons as compared with the corresponding introns, with a trend toward higher significance for high abundant transcripts, confirming that most transcripts have been generated through specific splicing events. This conclusion is supported by a comparison of RPKM values from known coding and non-coding regions of the chromosomes (Figure 2B), showing that the false-positive and false-negative rates are relatively low (5%) using a cutoff value of 0.1 RPKM. The number of transcripts across the genome (Figure 2C) suggests a dynamic range of 104 from the highest abundant transcripts to the cutoff value of 0.1 RPKM. This is considerably lower than the observed dynamic range of proteins as estimated by the MS intensity values.
Figure 2.
The transcript profiles in the three human cell lines based on RNA sequencing (RNA-seq). (A) RNA-seq reads mapping to a part of the RPS19 gene (ENSG00000105372). The reads align almost exclusively to the exons of the gene. (B) Distribution of RPKM values for all protein-coding genes (blue) and a set of ≈3000 intergenic regions defined as a genomic region at least 1 kb away from a known gene or EST. To detect present genes, we used a RPKM cutoff value of 0.1 (red dashed line), which corresponds to false-positive and false-negative rates of 5% (Supplementary Figure S1). (C) Distribution of log2 RPKM values for all detected transcripts (n=14 064) in U-2 OS. (D) Venn diagram showing the number of transcripts detected by RNA-seq in each cell line and the overlap between the cell lines. (E) The number of genes present in the categories ‘similar', ‘slightly changed', ‘substantially changed', ‘cell-type specific' and ‘not detected'. (F) Distribution of log2 RPKM values for all transcripts detected in protein categories (see Supplementary Table 3 for details) in U-2 OS.
Defining the transcriptome of the cell lines
The sequence-based transcriptome analysis suggests that the majority (74%) of the detected transcripts are expressed in all the three cell lines (Figure 2E) and only 13% of the detected genes are found exclusively in one of the cell lines. Global analysis of the transcriptome allowed us to estimate the fraction of genes that are cell specific, expressed at similar or different levels in the cell lines (Figure 2E). The study revealed that 25% (n=5187) of all genes show no or little (less than two-fold) change in transcript levels between all of the cells, while an additional 40% show at least two-fold changes between the two or more cell lines. Approximately 10% showed a cell-type specific pattern and a quarter (26%) of the predicted genes were not found in any of the three cell lines, suggesting that they constitute genes expressed in other cell types than the ones analyzed here or alternatively are genes no longer needed, and therefore turned off, when grown in vitro in cell cultures.
Gene ontology enrichment analyses
Gene ontology (GO)-based enrichment analyses were carried out for the proteins identified as similarly or differentially expressed by the quantitative analysis of SILAC ratios (Figure 1F). The results show that proteins with similar expression are enriched in functions related to intracellular maintenance, such as organelles, RNA processing, metabolic process, etc (Supplementary Table 4), while proteins that are slightly (Supplementary Table 5) or substantially (Supplementary Table 6) changed in expression in the cell lines are enriched for proteins expressed on the surface of the cell, such as plasma membrane, cell junction, cell adhesion, etc. Similarly, a GO-based enrichment analysis was also carried out for the various categories of the transcriptome analysis (Figure 2E) and the result confirms the analysis by proteomics. The ubiquitously expressed genes are predominantly coding for intracellular proteins, that is organellar or metabolic proteins and proteins of the cytoplasm and nucleus, while the cell-type specific transcripts are highly enriched for genes coding for plasma membrane proteins and other membrane-bound proteins (Supplementary Tables 7–14). The brain-derived U-251 MG cell line also expresses cell-specific transcripts annotated as nervous system development (Supplementary Table 13), which fits well with its neurological origin.
RNA abundance analysis
We further investigated the mRNA abundance within each of the categories defined in Figure 2E. The analysis for U-2 OS (Table I) demonstrates that a majority (94%) of the genes expressed in a cell-type specific manner are present at low levels, while the genes coding for proteins with similar levels in the three cell lines are present at medium or high levels (38 and 51%, respectively). The analysis for the other cell lines (Supplementary Tables 15 and 16) shows similar trends and the results suggest that proteins with differential expression in the cell lines in general are less abundant as compared with the gene products present at similar levels in the three cell lines.
Table 1. The mRNA abundance in the cell line U-2 OS for the different categories.
| Category | Description (gene/protein expression) | Number of genes | Low | Medium | High |
|---|---|---|---|---|---|
| The mRNA abundance has been divided into three similar sized fractions: low (RPKM between 0.1 and 2), medium (RPKM between 2 and 8) and high (RPKM>8) and the number of genes and the percent of genes within each category are shown. | |||||
| Similar levels | At the same level in all three cell lines (at most two-fold change between all cell lines) | 5187 | 548 (11%) | 1976 (38%) | 2663 (51%) |
| Slightly changed levels | Moderately up- or down-regulated in at least one of the cell lines (two- to four-fold change between two or more cell lines) | 6607 | 2257 (35%) | 2243 (35%) | 1877 (29%) |
| Substantially changed levels | Prominently up- or down-regulated in at least one of the cell lines (at least four-fold change between all differentially expressed cell lines) | 1691 | 814 (52%) | 485 (31%) | 279 (18%) |
| Cell-type specific | Exclusively expressed in one cell line | 922 | 863 (94%) | 52 (6%) | 7 (1%) |
Comparison between the mRNA and protein abundance
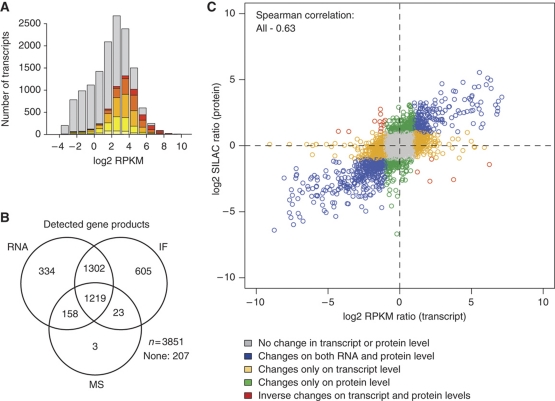
An analysis of the mRNA abundance for the various protein classes (Figure 2F) in U-2 OS shows a remarkably similar profile as compared with the MS analysis (Figure 1E). This is reassuring since it demonstrates that protein classes with highly abundant mRNA also have high levels of proteins and vice versa. The only disagreement between the two abundance plots is the class of GPCRs, which is higher in average abundance for proteins as compared with the mRNA data and relative to the other protein classes (Supplementary Table 3). This is probably due to the fact that very few GPCR proteins (n=7) were detected by the proteome analysis as compared with the RNA analysis (n=159), most likely due to the limit of detection achieved in this study that made it difficult to detect very low abundant proteins. A comparison of the genes detected by the transcriptome and the proteome analyses (Figure 3A; comparing Figures 1D and 2D) shows that most of the high abundant mRNA genes are detected by the MS analysis, while many low abundant transcripts were not detected on the protein level. This is expected and shows the remarkable sensitivity of sequence-based transcript profiling using next generation sequencing instruments. Although neither the MS intensity scores nor the RPKM values are strictly quantitative, our comparison suggests that the dynamic range of proteins is higher than the dynamic range of the transcripts. This might indicate large differences in translational efficiency (Man and Pilpel, 2007; Spruill and McDermott, 2009) or half lives (Ciechanover, 2005) for high abundant proteins as compared with low abundant proteins and that this phenomenon is less accentuated for the RNA molecules. This is supported by a recent study showing that sequence features related to translation and protein degradation account for as much as one third of the protein abundance variation (Vogel et al, 2010). These observations regarding differences in dynamic range between the transcriptome and the proteome should be further explored by absolute quantification of both the RNA and the protein levels for selected genes of various abundances.
Figure 3.
Comparative analysis of RNA and protein profiles. (A) Comparative analysis of the number of proteins detected by MS and RNA-seq in U-2 OS, respectively. Transcripts also detected at the protein level are colored according to the MS intensity as shown in Figure 1D. The data for A-431 and U-251 MG are presented in Supplementary Figures S2 and S3, respectively. (B) Venn diagram showing the overlap in gene products between the three methods for all genes studied by all three methods (n=3851) in U-251 MG cells. The overlap for the three methods in A-431 and U-2 OS cells is presented in Supplementary Figure S4. (C) Correlation between changes on protein (log2 SILAC ratio) and transcript levels (log2 RPKM ratio) for U-2 OS over A-431 cells. A two-fold change between the cell lines was used as cutoff for up-/down-regulation on RNA and protein levels. Data for the changes between the other cell lines are presented in Supplementary Figure S6. The Spearman correlation for randomized RNA and protein pairs is zero for all cell lines (Supplementary Figure S7). The correlation between RPKM and MS intensity levels are presented in Supplementary Figure S8.
Overlap between the three-assay platforms
A comparative analysis was carried out for the genes with data from all three platforms (n=3851) to investigate the overlap between transcripts (RNA-seq) and proteins (IF and MS). The analysis shows that approximately one third of the proteins were detected by all three methods (Figure 3B). Another third of the genes were only detected by the RNA-based and antibody-based methods, suggesting that these are below the detection limit for the MS analysis performed here. Note that among the proteins detected by MS, <1% were not detected by RNA-seq or IF, suggesting very few false positives for the MS analysis. The RNA and MS intensities for the various classes show that the genes detected by all platforms are more abundant, both as determined by RNA-seq and MS and that these targets also have more reliable western blot (WB) results for their corresponding antibodies (Supplementary Figure S5), while proteins detected exclusively by the antibody-based method (9%) show less reliable WB results. This suggests that some of the proteins detected only by the antibody-based method are false positives, resulting from cross-reactivity to related proteins and thus it is possible to use the mass spectrometry and the RNA analysis as a tool for validation of the specificity of the antibodies.
Comparison of changes in mRNA and protein levels between the cell lines
Since this global analysis has been performed on three cell lines, it was possible to analyze the differences of transcript and protein expression between the cell lines, with the advantage that transcript or peptide detection efficiency of individual gene products does not influence the analysis. The changes in expression on transcript (log2 RPKM ratios) and protein (log2 SILAC ratios) levels were plotted for all genes detected by MS and the Spearman's rank correlation coefficients of the entire data set, depending on the cell lines analyzed, vary between 0.58 and 0.63 (Figure 3C; Supplementary Figure S6). A linear relationship between RNA and protein changes can be observed, suggesting that transcript changes between cell lines are accompanied with similar changes on the protein level. GO-based enrichment analyses confirmed that surface-expressed proteins are over-represented for the differentially expressed genes, while intracellular and organelle proteins are frequent in the group of non-varied proteins (Supplementary Tables 17–21). Together, these results demonstrate a high correlation between changes in transcript and protein levels on an individual gene product basis in the three cell lines.
Conclusions
In summary, the global analysis of human cells with deep sequencing of RNA complemented with quantitative SILAC-based proteomics and antibody-based confocal microscopy has allowed a rough estimate of the fraction of the proteome that is cell specific, similarly or differentially expressed in three selected cell lines of functionally different origins, as shown in Figure 2E. The majority of all genes were shown to be expressed in the three cell lines. This high conservation of expression could in part be due to the fact that all analyzed samples were of similar origin, that is cell lines grown in vitro; however, recent studies of both RNA and protein profiles in human tissues from many origins have reported similar conclusions (Ponten et al, 2009; Ramskold et al, 2009). Our study suggests that more than half of the genes are expressed in all the three cell lines with small (up to four-fold) changes in expression levels, while close to 20% are substantially either up- or down-regulated (more than four-fold) or not present at all in one or two of the cell lines. These latter groups of proteins are interesting for further in depth studies to understand cell-specific functions and differences between cells and tissues. Interestingly, our data suggest that the cell-type specific proteins, in general, are low abundant and highly enriched for cell-surface proteins (Table I; Supplementary Tables 4–11), although more in depth studies are needed to rule out that some of these proteins have not been scored due to difficulties to measure quantitative levels for very low abundant gene products. Twenty-five percent of the genes predicted from the genome sequence were not present in any of the three cell lines and these are also interesting for studies of human tissue specificity, although some of these genes might be silent genes or pseudo-genes not actually translated into proteins. Interestingly, the correlation of changes between the cell lines on RNA and protein levels for individual gene products was high, even though our results indicate that the genome-wide dynamic range of protein expression is considerably larger than that of the transcriptome. The human protein-coding genes stratified into the various categories are listed in a searchable format on the HPA (http://www.proteinatlas.org) along with the expression data from the transcript and proteome analysis. This study demonstrates the power of comparative transcriptome and proteome studies to define protein expression across human cell lines and to explore the relationship between RNA and protein expression both on an individual gene level and across the whole genome.
Materials and methods
Cell cultivation
The glioblastoma cell line U-251 MG (Professor Bengt Westermark, Uppsala University), the epidermoid carcinoma cell line A-431 (DSMZ) and the osteosarcoma cell line U-2 OS (ATCC-LGC) were grown at 37°C in a 5% CO2 environment in media suggested by the provider. For the subsequent analysis, cells were harvested during logarithmic growth (at 70–80% confluency). For the MS analysis, cells were SILAC labeled by cultivation in DMEM media where the natural lysine and arginine were replaced by stable isotope labeled arginine and lysine. A-431 cells were cultured with light forms of these amino acids (Arg0, Lys0), U-251 MG cells were cultured with medium forms of the amino acids (Arg6-L-13C6 and Lys4-L-2H4) and U-2 OS were labeled with heavy amino acids (Arg10-L-13C615N4 and Lys8-L-13C615N2).
RNA-seq
Total RNA was prepared from frozen cells using RNeasy mini kit (Qiagen). A measure of 10 μg of total RNA was depleted of ribosomal RNA (5S, 5.8S, 18S and 28S) using the RiboMinus Eukaryote kit for RNA-seq (Invitrogen) after which libraries were constructed using SOLiD Whole Transcriptome kit rev C. The 50-bp sequencing was carried out on the SOLiD3 platform. The fragments were aligned using BWA (Li and Durbin, 2009) after which only unique matches were retained for calculation of expression levels. Briefly, the RPKM value was calculated by dividing the number of reads mapping to each gene by the length of the gene and number of reads from the library to compensate for slightly different read depths for different samples. This generates RPKM values that are estimations of expression values for each gene. In order to determine what RPKM value to use to detect present genes, we used the approach described by Ramskold et al (2009), where a collection of intergenic regions were used to estimate the background over which genes can be called present with high confidence. In our case, the threshold value of 0.1 was set to detect the presence of a transcript for a particular gene, which corresponds to a FDR and false-negative rate of 5% (Supplementary Figure S1B). The raw sequence data have been deposited to the NCBI short read archive with accession number SRA012517.
Mass spectrometry
Cells were lysed with SDS buffer containing 0.1 M Tris/HCl pH 7.6, 0.1 M DTT and 4% SDS. Protein digestion was performed once by in-gel digestion (Shevchenko et al, 2006), and twice according to the FASP protocol (Wisniewski et al, 2009b). For in-gel digest, the protein lysates of the three cell lines were combined at a ratio of 1:1:1, and were separated by SDS–PAGE. The gel was excised to 15 slices and trypsin digestion was performed according to the standard protocol (Shevchenko et al, 2006). Alternatively, the combined lysates were digested with trypsin using the FASP procedure (Wisniewski et al, 2009b). After FASP, digestion peptides were separated to seven fractions using strong anion exchange (SAX) in a StageTip format (Wisniewski et al, 2009a). After peptide purification on StageTips (Rappsilber et al, 2007), eluted peptides were separated on a reverse phase C18 column (75 μm i.d., 3 μm beads, Dr Maisch) using the EASY-nLC system (Proxeon Biosystems now Thermo Fisher Scientific). MS analysis was performed on the LTQ-Orbitrap XL instrument (Thermo Fisher Scientific). In-gel fractions were analyzed using 100 min gradients, and the SAX fractions were analyzed using 180 min gradients. Data were acquired in data-dependent mode. Survey scan acquisition was performed in the 300–1700 m/z range (R=60 000 and target value of 1 000 000). Data were processed using MaxQuant software (version 1.0.14.3) (Cox and Mann, 2008). MS/MS spectra were searched with the MASCOT search engine against the decoy IPI-human database (forward and reverse sequences). The search included variable modifications of N-terminal acetylation and methionine oxidation, and fixed modification of cysteine carbamidomethylation. Analysis was limited to peptides of six or more amino acids and maximum two mis-cleavages. We set the protein and peptide FDR to 0.01. In case the identified peptides were shared by two proteins (homologs or isoforms), they were reported by MaxQuant as one protein group. We required a minimum of two quantification events to determine the protein ratio. The MS data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash:
aOW7DUnjIVHTrK6zlezv28jbseXTXR+cbUFOw/bbyZMUyXCDkckblUCpZaZwIt0SjBimMbAX9gQAUDP5bJjcISBfdXcAAAAAAAAbSg=.
Immunofluorescence microscopy
The expression of 3877 proteins was analyzed in the three cell lines using antibodies generated in the HPA project (Uhlen et al, 2005; Berglund et al, 2008) and confocal microscopy as previously described in the study of ∼500 proteins (Barbe et al, 2008). The confocal images and annotations of the protein subcellular distribution are available in version 7.0 of HPA database (http://www.proteinatlas.org).
Western blotting
WB analysis was performed on U-251 MG cell lysate using 3860 antibodies generated in the HPA project (Uhlen et al, 2005; Berglund et al, 2008). Annotation of WB results was performed on a 7-level scale ranging from level 1 (single band corresponding to the right size in kDa ±20%) to level 7 (only band(s) not corresponding to the predicted size) as previously described (Bjorling and Uhlen, 2008). The WBs are available in version 7.0 of HPA database (http://www.proteinatlas.org).
Data analysis
All data were mapped to gene identifiers obtained from Ensembl (Hubbard et al, 2009) version 57.37 (21 248 protein-coding genes) and analyzed using the R statistical programming environment (R Development Core Team, 2009) and the scatterplot3d package (Ligges and Mächler, 2003). DAVID (Huang da et al, 2009) was used for a GO (Ashburner et al, 2000)-based enrichment analysis of gene lists. Genes were grouped into the following five categories based on the RNA-seq data for all the three cell lines. Genes not detected in any cell line or only in one were categorized as ‘not present' or ‘cell-type specific', respectively. The ‘similar level' category was defined as at most a two-fold change in RPKM levels between all pair wise combinations of cell lines for a gene. All genes with at least a two-fold change between RPKM levels in two or more cell lines were considered differentially expressed, and were classified as either ‘slightly' or ‘substantially' changed. The latter group was constructed to find genes with great differences in abundance between all three cell lines. Hence, the category of ‘substantially changed' genes consisted of all genes with at least a four-fold change between all differentially expressed cell lines, whereas the ‘slightly changed' genes had at least a two-fold change between two or more cell lines. For the sake of simplicity, these arbitrary expression classification windows were set to two- and four-fold, respectively, based on the statistical analysis of the fraction of reads detected when comparing two cell lines (Supplementary Figure S1C).
Supplementary Material
Consists of 8 figures and 21 tables.
IF expression data
MS expression data
RNA-seq expression data
Acknowledgments
We acknowledge Martin Dodel, Fredrik Pontèn, Sophia Hober, Per-Åke Nygren and Caroline Kampf for valuable contributions. Funding was provided by the Knut and Alice Wallenberg Foundation and PROSPECTS, a 7th Framework grant by the European Directorate (grant agreement HEALTH-F4-2008-201648/PROSPECTS). Author contributions: MU and MM conceived and designed the study. LF, DK and EL performed most of the bioinformatics analysis. DK and JL performed the RNA sequencing. IM, JC, TG and MM performed the MS analysis. CÄ performed the western blot analysis. EL performed the IF analysis. MU and EL wrote the paper. LF and MM contributed analysis, text and comments to the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H (2008) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7: 499–508 [DOI] [PubMed] [Google Scholar]
- Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7: 2019–2027 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorling E, Uhlen M (2008) Antibodypedia, a portal for sharing antibody and antigen validation data. Mol Cell Proteomics 7: 2028–2037 [DOI] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG (2002) Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 1: 304–313 [DOI] [PubMed] [Google Scholar]
- Ciechanover A (2005) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ 12: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ, Grimmond SM (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2007) Is proteomics the new genomics? Cell 130: 395–398 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O′Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 323–333 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K et al. (2009) Ensembl 2009. Nucleic Acids Res 37: D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligges U, Mächler M (2003) Scatterplot3d—an R package for visualizing multivariate data. J Stat Softw 8: 1–20 [Google Scholar]
- Man O, Pilpel Y (2007) Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet 39: 415–421 [DOI] [PubMed] [Google Scholar]
- Mann M (2006) Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7: 952–958 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten F, Gry M, Fagerberg L, Lundberg E, Asplund A, Berglund L, Oksvold P, Bjorling E, Hober S, Kampf C, Navani S, Nilsson P, Ottosson J, Persson A, Wernerus H, Wester K, Uhlen M (2009) A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol 5: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- R development Core Team (2009) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Spruill LS, McDermott PJ (2009) Role of the 5′-untranslated region in regulating translational efficiency of specific mRNAs in adult cardiocytes. FASEB J 23: 2879–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O′Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML (2008) A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920–1932 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M et al. (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Vogel C, de Sousa Abreu R, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M (2009a) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 8: 5674–5678 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009b) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consists of 8 figures and 21 tables.
IF expression data
MS expression data
RNA-seq expression data