Abstract
Comprehensive characterization of human tissues promises novel insights into the biological architecture of human diseases and traits. We assessed metabonomic, transcriptomic, and genomic variation for a large population-based cohort from the capital region of Finland. Network analyses identified a set of highly correlated genes, the lipid–leukocyte (LL) module, as having a prominent role in over 80 serum metabolites (of 134 measures quantified), including lipoprotein subclasses, lipids, and amino acids. Concurrent association with immune response markers suggested the LL module as a possible link between inflammation, metabolism, and adiposity. Further, genomic variation was used to generate a directed network and infer LL module's largely reactive nature to metabolites. Finally, gene co-expression in circulating leukocytes was shown to be dependent on serum metabolite concentrations, providing evidence for the hypothesis that the coherence of molecular networks themselves is conditional on environmental factors. These findings show the importance and opportunity of systematic molecular investigation of human population samples. To facilitate and encourage this investigation, the metabonomic, transcriptomic, and genomic data used in this study have been made available as a resource for the research community.
Keywords: bioinformatics, biological networks, integrative genomics, metabonomics, transcriptomics
Introduction
Our understanding of the genetic basis of complex disease has recently been transformed by genome-wide association studies, leading to the identification and cataloging of hundreds of genomic loci associated with human disease (Hindorff et al, 2009). In parallel, current technologies have brought systematic functional investigation of the underlying disease pathways within reach. Building upon previous work to integrate genetic and transcriptional profiles to uncover disease genes (Hubner et al, 2005; Mehrabian et al, 2005), Chen et al (2008) and Emilsson et al (2008) constitute two recent large-scale studies, which led to the identification of novel candidate genes for obesity and the characterization of the macrophage-enriched metabolic network module, a subnetwork enriched for genes in inflammatory processes and metabolic syndrome. These studies added to the growing body of evidence linking inflammation and the immune response with systemic metabolism and metabolic disorders (Hotamisligil, 2006).
We recently characterized the lipid–leukocyte (LL) module, a cluster of tightly correlated co-expressing genes, that harbored key basophil and mast cell (BMC)-specific mediators of the immune response and strongly associated with total measures of serum triglycerides (TGs), high-density lipoprotein (HDL) cholesterol (C), and apolipoprotein B (APOB) (Inouye et al, 2010). However, lipid metabolism is heterogeneous and lipoprotein subpopulations are presently utilized to both appreciate the various counteracting metabolic phenomena and more accurately assess the risk for various vascular outcomes (Chasman et al, 2009). Further, lipids comprise only a fraction of the metabolic compounds in the circulatory system; therefore, a comprehensive assessment is essential to understand potentially causative and reactive relationships between serum metabolites and molecular networks (Holmes et al, 2008). To this end, the serum metabonomes of 518 individuals from a population-based cohort, the Dietary, Lifestyle, and Genetic determinants of Obesity and Metabolic syndrome (DILGOM) study, were determined by proton nuclear magnetic resonance spectroscopy (NMR). The DILGOM study consists of unrelated individuals, 240 males and 278 females, aged 25–74 years, sampled from the capital region of Finland (Supplementary Figure 1). Combined with the genome-wide profiles of genetic and transcriptional variation from blood leukocytes of the same individuals, this information represents a powerful resource for large-scale functional investigation, thus we have made the data publically available for researchers (see Data Availability section of Materials and methods). In this study, we provide both a proof of concept and roadmap for integrated analysis of variation at the genetic, transcriptional, and metabonomic levels. In doing so, we also comprehensively characterize the role of LL module as a network linking metabolic compounds and the immune response.
Results and Discussion
Blood serum is a site of systemic metabolism in the human body. However, the vast array of molecular compounds in serum makes their identification and quantification a challenging task. Here, we partitioned serum compounds into three molecular windows using a unique NMR metabonomics platform (Soininen et al, 2009) with an optimized measurement and analysis protocol providing absolute quantitative information on 134 metabolic measures, including 14 lipoprotein subclasses (see Materials and methods and Supplementary Figure 2), various low-molecular-weight metabolites, and also individual lipid molecules together with their degree of (poly)(un)saturation (Supplementary Table 1).
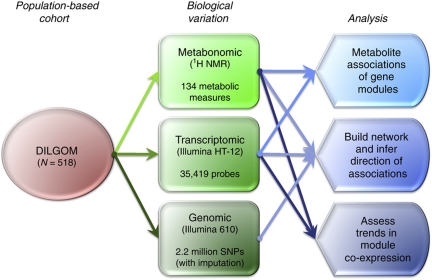
An overview of the data integration and the different analysis in the study is given in Figure 1.
Figure 1.
Overview of data integration and analyses. A high-level view of the study design and analysis employed.
A gene network associated with blood metabolites
Across all metabolites, 1341 significant gene expression associations were observed; however, the degree of intercorrelation between metabolites was strong (Supplementary Figure 3). Genes belonging to the core LL module (HDC, FCER1A, GATA2, CPA3, MS4A2, SPRYD5, and SLC45A3) were not only strongly associated with lipoprotein and lipid levels but were also among the strongest signals for glycoproteins, isoleucine, and 3-hydroxybutyrate. Glycoproteins are known to have potentially allergenic effects (Altmann, 2007); however, the associations with isoleucine (an essential amino acid) and creatine (a nitrogenous organic acid) suggest LL module's involvement in pathways outside of lipid metabolism and a potential molecular clue as to how BMC-mediated inflammation may arise from various serum compounds.
Network analysis of gene co-expression showed the clustering of LL module genes (Supplementary Table 2) and the association of the core module genes with metabolites (Supplementary Figure 6) (Horvath and Dong, 2008; Langfelder and Horvath, 2008). A Spearman's rank correlation of the LL module expression profile (first principal component) with metabolite distributions offered fine-scale detail of potentially causative/reactive effects (Table I, see Supplementary Figures 7–16). After correction for the 21 modules tested (a Bonferroni-corrected significance level of 2.4 × 10−3), expression of LL module was, as expected from univariate analysis, positively associated with glycoprotein (P=1.83 × 10−8) and creatine levels (P=9.02 × 10−4), and negatively associated with isoleucine (P=4.03 × 10−16). Our previous findings showed association of LL module expression with total HDL and APOB but not with total low-density lipoprotein (LDL) (Inouye et al, 2010). The deeper lipoprotein phenotypes obtained by serum NMR metabonomics allowed for division of the APOB–lipoprotein cascade into 10 subclasses: six for the very-low-density lipoprotein (VLDL) fraction, the intermediate-density lipoprotein (IDL), and three LDL subclasses. All VLDL subclasses had a strong correlation with LL module expression, indicating dominance of the VLDL fraction in the APOB association (Table I), as could be expected (Lusis et al, 2008). The current results further validated that, at current detection power, there remained little evidence for association with total LDL-C (P=0.07), while a few measures related to the medium and small LDL subclasses did associate weakly with LL module expression (Supplementary Figure 14).
Table 1. Significant LL module associations with serum metabolite concentrations and immune response markers.
| Spearman's correlation | P-value | |
|---|---|---|
| Metabolite | ||
| Concentration of chylomicrons and extremely large VLDL particles | −0.464 | 4.46E−29 |
| Triglycerides in medium VLDL | −0.442 | 3.46E−26 |
| Concentration of large VLDL particles | −0.441 | 4.51E−26 |
| Free cholesterol in large VLDL | −0.441 | 4.89E−26 |
| Triglycerides in large VLDL | −0.441 | 5.20E−26 |
| Triglycerides in VLDL | −0.439 | 7.25E−26 |
| Concentration of very large VLDL particles | −0.439 | 8.88E−26 |
| Total cholesterol in large VLDL | −0.438 | 1.00E−25 |
| Triglycerides in very large VLDL | −0.438 | 1.09E−25 |
| Phospholipids in large VLDL | −0.437 | 1.43E−25 |
| Total lipids in large VLDL | −0.436 | 1.98E−25 |
| Triglycerides in VLDL (Lipido) | −0.434 | 3.77E−25 |
| Concentration of medium VLDL particles | −0.43 | 8.80E−25 |
| Cholesterol esters in large VLDL | −0.43 | 9.68E−25 |
| Total lipids in very large VLDL | −0.429 | 1.32E−24 |
| Total lipids in medium VLDL | −0.428 | 1.59E−24 |
| Serum total triglycerides | −0.427 | 2.53E−24 |
| Phospholipids in medium VLDL | −0.423 | 6.17E−24 |
| Triglycerides in small VLDL | −0.421 | 1.27E−23 |
| Total triglycerides | −0.418 | 2.87E−23 |
| Mobile lipids −CH2− | −0.417 | 3.25E−23 |
| Free cholesterol in medium VLDL | −0.417 | 3.26E−23 |
| Phospholipids in very large VLDL | −0.414 | 6.36E−23 |
| Ratio of triglycerides to phosphoglycerides | −0.41 | 2.14E−22 |
| Total cholesterol in medium VLDL | −0.405 | 7.78E−22 |
| Concentration of small VLDL particles | −0.394 | 1.18E−20 |
| Cholesterol esters in medium VLDL | −0.383 | 1.36E−19 |
| Phospholipids in chylomicrons and extremely large VLDL | −0.381 | 2.56E−19 |
| Total lipids in chylomicrons and extremely large VLDL | −0.376 | 7.91E−19 |
| Triglycerides in small HDL | −0.375 | 1.00E−18 |
| Total lipids in small VLDL | −0.374 | 1.18E−18 |
| Triglycerides in chylomicrons and extremely large VLDL | −0.372 | 2.03E−18 |
| Phospholipids in small VLDL | −0.362 | 1.70E−17 |
| Mobile lipids −CH3 | −0.35 | 2.11E−16 |
| Isoleucine | −0.347 | 4.03E−16 |
| Unsaturated lipids | −0.343 | 9.41E−16 |
| Free cholesterol in small VLDL | −0.341 | 1.43E−15 |
| Triglycerides in very small VLDL | −0.337 | 2.97E−15 |
| ω-9 and saturated fatty acids | −0.329 | 1.61E−14 |
| Total fatty acids | −0.316 | 1.66E−13 |
| Total cholesterol in IDL (Lipido) | −0.307 | 9.13E−13 |
| Apolipoprotein B by apolipoprotein A-I | −0.299 | 3.90E−12 |
| Total cholesterol in small VLDL | −0.278 | 1.18E−10 |
| Free cholesterol in large HDL | 0.273 | 2.59E−10 |
| ω-6 and -7 fatty acids | −0.269 | 5.02E−10 |
| Apolipoprotein B | −0.263 | 1.17E−09 |
| Average number of methylene groups per a double bond | −0.263 | 1.24E−09 |
| Ratio of bisallylic groups to total fatty acids | 0.263 | 1.29E−09 |
| Ratio of bisallylic groups to double bonds | 0.255 | 4.16E−09 |
| Total cholesterol in large HDL | 0.253 | 5.15E−09 |
| Total lipids in large HDL | 0.245 | 1.57E−08 |
| Phospholipids in large HDL | 0.244 | 1.77E−08 |
| Glycoproteins | −0.244 | 1.83E−08 |
| Cholesterol esters in large HDL | 0.243 | 2.26E−08 |
| Concentration of large HDL particles | 0.241 | 2.87E−08 |
| Average number of double bonds in a fatty acid chain | 0.24 | 3.39E−08 |
| Leucine | −0.236 | 5.58E−08 |
| Total cholesterol in HDL2 (Lipido) | 0.235 | 5.94E−08 |
| Ratio of ω-9 and saturated fatty acids to total fatty acids | −0.234 | 6.82E−08 |
| Phospholipids in very large HDL | 0.221 | 4.00E−07 |
| Total cholesterol in HDL3 (Lipido) | −0.211 | 1.30E−06 |
| Concentration of very large HDL particles | 0.205 | 2.56E−06 |
| Triglycerides in IDL | −0.199 | 5.24E−06 |
| Total lipids in very large HDL | 0.198 | 5.49E−06 |
| Ratio of ω-6 and -7 fatty acids to total fatty acids | 0.194 | 8.94E−06 |
| Total cholesterol in HDL | 0.191 | 1.17E−05 |
| Free cholesterol in very large HDL | 0.186 | 1.96E−05 |
| Concentration of very small VLDL particles | −0.182 | 3.23E−05 |
| Cholesterol esters in very large HDL | 0.177 | 5.08E−05 |
| 3-Hydroxybutyrate | 0.171 | 9.48E−05 |
| Concentration of small LDL particles | −0.17 | 1.02E−04 |
| Total cholesterol in very large HDL | 0.168 | 1.25E−04 |
| 18:2, Linoleic acid | −0.154 | 4.48E−04 |
| Concentration of small HDL particles | −0.154 | 4.48E−04 |
| Total phosphoglycerides | −0.151 | 5.78E−04 |
| Total lipids in small LDL | −0.147 | 7.87E−04 |
| Other polyunsaturated fatty acids than 18:2 | −0.146 | 8.30E−04 |
| Creatine | 0.145 | 9.02E−04 |
| Total lipids in very small VLDL | −0.145 | 9.13E−04 |
| Description of average fatty acid chain length (not carbon number) | 0.141 | 1.26E−03 |
| Phospholipids in medium LDL | −0.141 | 1.31E−03 |
| Triglycerides in very large HDL | −0.141 | 1.33E−03 |
| Immune response marker | ||
| Interleukin-1 receptor antagonist | −0.203 | 3.14E−06 |
| High-molecular-weight adiponectin | 0.189 | 1.55E−05 |
| C-reactive protein | −0.16 | 2.62E−04 |
Of particular interest were the opposite metabolic correlations observed for HDL subclasses both in terms of LL module association and overall (Supplementary Figure 3). For example, the smallest HDL subclass behaved similarly to the VLDL subclasses and had a negative correlation with the larger HDL subclasses. This finding has also been observed by others (Chasman et al, 2009) and suggests that the serum HDL fraction does not have a coherent physiological function and that the smallest HDL particles are a metabolically distinct entity, tightly positively connected with TG metabolism and the APOB lipoprotein cascade. Thereby, the smallest HDL particles appear potentially proatherogenic in contradiction to the larger particles thought to have a role in the reverse C transport (Tall, 1998). The capacity of total HDL-C measurement to describe the functional aspects of HDL metabolism is dependent on the relative concentrations of circulating HDL subclasses; the more large particles the better the measure will be, but if there is a preponderance of smaller HDL particles, its physiological meaning is confounded. The size-dependent metabolic behavior of HDL particles clearly represents the inadequacy of current total lipoprotein measures used in clinical practice but also offers an opportunity to delve more deeply into the (patho)physiology of lipoprotein metabolism and the immune response.
The LL module is associated with interleukin-1 receptor antagonist, high-molecular-weight adiponectin, and C-reactive protein
Given the gene composition of LL module, serum markers of the immune response were measured for all individuals in the DILGOM study. These included total serum immunoglobulin E (IgE), interleukin-1 receptor antagonist (IL-1ra), C-reactive protein (CRP), and high-molecular-weight adiponectin (HMWA). As with blood metabolites, the expression of LL module was tested against these new variables (Table I). LL module exhibited strong negative association with IL-1ra and CRP levels (P=3.1 × 10−6 and 2.6 × 10−4, respectively) and strong positive association with HMWA levels (P=1.6 × 10−5). Total serum IgE levels did not show significant association with LL module (P=0.60) nor with FCER1A expression itself (P=0.42), findings which were consistent with previous observations (Weidinger et al, 2008).
Associations with IL-1ra, CRP, and HMWA represent intriguing insights into LL module's potential physiological role. The IL-1 system is important in insulin resistance, glucose homeostasis, and inflammation. IL-1ra is a cytokine, which inhibits the activation of IL-1 receptor, in turn dampening the inflammatory response. The modulation of IL-1ra is a compensatory phenomenon to the levels of proinflammatory IL-1β; however, recombinant human IL-1ra has shown promise as a treatment for type 2 diabetes (Larsen et al, 2007). Interestingly, IL-1β has also been shown to induce degranulation of mast cells (Haak-Frendscho et al, 1988). Adiponectin is a hormone secreted by adipocytes. It is inversely related to incidence of obesity and diabetes and was recently shown to be the strongest protective factor in a set of novel biomarkers for type 2 diabetes (Salomaa et al, 2010). However, its association with cardiovascular disease is controversial (Sattar et al, 2008). CRP is an acute phase reactant and inflammatory marker. It has been shown to be induced by IL-1 (Ganapathi et al, 1991) and inhibited by adiponectin (Devaraj et al, 2008).
A directed network of blood metabolites and the LL module
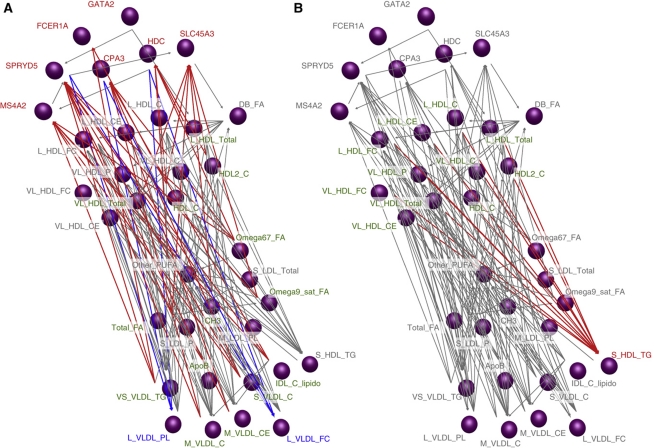
While we have considered undirected network models for the association of LL module expression with metabolite concentrations, the use of genetic variants allows for the construction of an edge-oriented network to infer gene/gene or gene/metabolite causality (Schadt et al, 2005; Li et al, 2006; Aten et al, 2008). Using Network Edge Orientation (NEO) conditional correlation analysis (Aten et al, 2008), we inferred an edge-oriented network of core LL module gene expression with all metabolites that (a) were significantly associated with LL module expression and (b) displayed the genetic component needed for edge orientation (at least one SNP associated with P<5.0 × 10−7). This led to a network with 36 nodes (the 7 core LL genes plus 29 metabolites) and 137 causal edges (Figure 2 and Supplementary Table 3). Core LL module was largely reactive to fatty acids and high/low/intermediate-density lipoprotein fractions; however, some lipoprotein components (notably the free C and phospholipids in large VLDL) contradicted this trend and appeared to be driven by expression of HDC, SPRYD5, and CPA3. Interestingly, the TGs present in the smallest HDL particles (those which showed the strongest negative relationship with larger HDL subclasses, see Supplementary Figure 3) were strongly driven by the concentration of large/medium HDL.
Figure 2.
Edge-directed network of LL module and serum metabolites. Genetic variation was used to infer a causal network of core LL module expression and metabolites (see Supplementary Methods and Supplementary Table 1 for abbreviations). Core LL module genes are represented by purple nodes in the upper left, whereas metabolites significantly associated with genetic variation and LL module are denoted by all other nodes. Arrows denote directed edges. (A) Shows core LL module (red) reactivity to most lipoprotein subclass levels (green); however, two components of large VLDL (blue) were predicted downstream of CPA3, SPRYD5, and HDC expression. (B) Shows that triglycerides in the small HDL subclass (red) are predicted to be largely reactive to concentrations of the larger HDL subclasses (green). Source data is available for this figure at www.nature.com/msb.
Conditional co-expression: dependency on metabolite environment
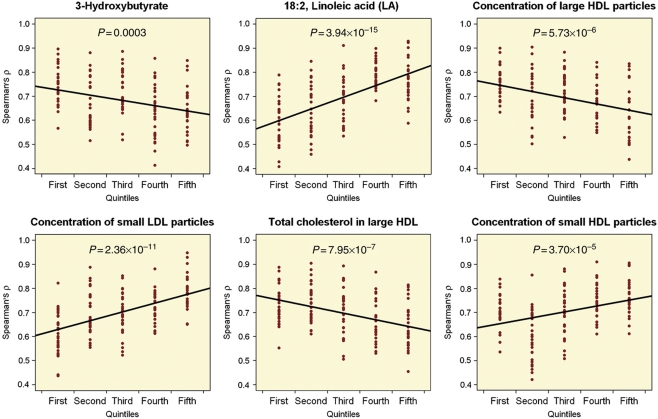
The large number of unrelated individuals in the data set allowed for the investigation of the metabolite dependencies underlying the co-expression in LL module. For example, if module co-expression is reduced in the face of extreme metabolite levels, then this offers potential biological insight into how cells might regulate gene expression when faced with different extracellular environments. For each metabolite the log2-normalized expression values were partitioned into quintiles of metabolite level, and the co-expression of all core LL module gene pairs were measured via a Spearman's rank correlation coefficient. A linear model was fitted to all co-expression pairs across the quintiles in order to identify trends in co-expression. We refer to this fitted model as co-expression trend. If there is no co-expression trend, then the slope of the fitted line should be zero (or not significant). In our data, there were 63 metabolites, which showed both a significant association (P<2.4 × 10−3) with LL module expression and a significant co-expression trend (P<0.05). For all of these metabolites, there was an inverse relationship between the direction of association with LL module expression and the direction of co-expression trend (Figure 3). To put it in another way, if both LL module expression and metabolite levels increase (as is the case with HDL-C), then the connectivity of the core genes in LL module will decrease. This raises the hypothesis that the functionality of LL module is reduced at increasing level of metabolites, such as large/medium HDL and 3-hydroxybutyrate, and increased by metabolites such as TGs, glycoproteins, and small HDL. As the LL module contains key genes specific to mast cells and basophils, this represents an intriguing prediction as to how a gene network in these cells might respond to extracellular metabolites.
Figure 3.
Conditional co-expression of LL module. Core LL module expression was partitioned into quintiles based on metabolite concentration rank. For each quintile (x axis, lowest to highest metabolite concentration), co-expression of all non-redundant pairs was calculated using Spearman's rank correlation coefficient (y axis). Fitting a linear model to all co-expression pairs suggests that if LL module expression is negatively correlated with metabolite concentration, then co-expression among genes belonging to LL module increases and vice versa.
Conclusions
As the quantification of human tissues acquires more dimensions, our understanding of gene function and interaction will become immensely enriched. In this report, we provide a proof of concept for the integration of metabonomic, transcriptomic, and genomic data. In doing so, we elucidate functional aspects of LL module within and outside lipid pathways by showing its strong correlations with lipoprotein particle size, organic acids, and immune response markers. We also show the dependency of the network connections on the concentrations of circulating metabolites. Building upon our knowledge of genetic variation associated with lipid levels, we construct an edge-oriented graph to infer causality, providing population-based evidence that LL module is reactive to both HDL and small VLDL subclasses and that components of the smallest HDL subclasses are negatively correlated and reactive to larger HDL subclasses.
From our systems analysis of a population-based cohort, we have shown that LL module implicates mast cells and basophils as having a key role in systemic metabolism. This study dramatically increases LL module's blood metabolite associations to 83 from 3 (Inouye et al, 2010) and, through the gene content of LL module, offers a firm basis for biochemical studies targeting metabolite-stimulated mast cell and basophil secretion of histamine and proteases, which can in turn act on many tissues including vasculature and inflammatory. Further, associations with IL-1ra, HMWA, and CRP represent potentially fruitful research avenues in the connection of adiposity, inflammation and, through lipoproteins, atherogenesis (Lyon et al, 2003).
Taken together, these observations offer a comprehensive view of interactions between an immune response subnetwork and serum metabolic compounds and the inadequacy of current total cholesterol measures. This novel combination of metabonomic, transcriptomic, and genomic profiling illustrates the immense potential for integrative genomics in epidemiology.
Materials and methods
Sample collection, genotyping, and expression
Samples were collected as part of the Dietary, Lifestyle, and Genetic determinants of Obesity and Metabolic syndrome (DILGOM) study. Study participants were aged 25–74 years and were drawn from the Helsinki/Vantaa area of southern Finland. Participants were asked to fast overnight for a period of at least 10 h before giving a blood sample. Blood samples were left at room temperature for 45 min before the serum and plasma were separated via centrifugation and subsequent storage in a −70°C freezer.
Extraction, purification, and quantification of DNA were performed as previously described (Inouye et al, 2010) before genotyping on the Illumina 610-Quad SNP array (Illumina Inc., San Diego, CA, USA). RNA protocols for the Illumina HT-12 expression array were also previously described (Inouye et al, 2010). Technical replicates for each sample were done.
Serum NMR metabonomics
We have recently introduced a high-throughput serum NMR metabonomics platform with an optimized measurement and analysis protocol, providing absolute quantitative information on ∼140 metabolic measures (Ala-Korpela et al, 2009; Mora et al, 2009; Soininen et al, 2009). The methodology is based on three molecular windows, two of which (LIPO and low-molecular-weight molecule (LMWM)) are applied to native serum and one for serum lipid extracts (LIPID). The LIPO window gives information, e.g., on the lipoprotein subclass distribution and lipoprotein particle concentrations for 14 lipoprotein subclasses, and the LMWM window on various low-molecular-weight metabolites such as amino acids, 3-hydroxybutyrate, and creatinine. The combination of these two molecular windows is likely to contain most of the metabolic information available by 1H NMR metabonomics of native serum (Ala-Korpela, 1995, 2008). The LIPID window is run from serum lipid extracts to obtain detailed molecular information on various serum lipid constituents, such as free and esterified C, sphingomyelin, (poly)(un)saturation and ω-3 fatty acids (Tukiainen et al, 2008).
In our setup, the NMR data are measured using a Bruker AVANCE III spectrometer operating at 500.36 MHz (1H observation frequency; 11.74 T) and equipped with an inverse selective SEI probehead, including an automatic tuning and matching unit and a z-axis gradient coil for automated shimming. A BTO-2000 thermocouple serves to temperature stabilization, the sample at the level of approximately 0.01°C. Notably, very stable and high-performance electronics is also a prerequisite for some of the implemented concepts, including metabolite quantification without per sample chemical referencing or double tube systems (Soininen et al, 2009).
The serum samples are stored in a freezer at −80°C. Before sample preparation, the frozen samples are first slowly thawed in a refrigerator (+4°C) overnight. The samples are then mixed gently and spun in a centrifuge at 3400 × g to remove possible precipitate. Aliquots of each sample (300 μl) are mixed with 300 μl of sodium phosphate buffer (75 mM Na2HPO4 in 80%/20% H2O/D2O, pH 7.4; including also 0.08% sodium trimethylsilyl [2,2,3,3-D4]propionate and 6.2 mM sodium azide). The sample preparation is done automatically with a Gilson Liquid Handler 215 to 5-mm-outer-diameter standard NMR tubes. In the automatic preparation procedure, 300 μl of buffer is first transferred into the NMR tubes, after which 300 μl of serum is added to the buffer and the resulting solution is mixed thoroughly by aspirating three times.
The LIPO window represents a conventional spectrum of human serum showing broad overlapping resonances arising mainly from different lipid molecules in various lipoprotein particles. The LIPO data are now recorded with 80 k data points after four dummy scans using eight transients acquired with an automatically calibrated 90° pulse and applying a Bruker noesypresat pulse sequence with mixing time of 10 ms and irradiation field of 25 Hz to suppress the water peak. The acquisition time is 2.7 s and the relaxation delay 3.0 s. The 90° pulse is calibrated automatically for each sample. A constant receiver gain setting is used for all the samples.
The LMWM spectrum is dominated by numerous glucose resonances and shows signals from various LMWMs. In addition, some lipid resonances are still visible in the LMWM spectrum, representing the most mobile −CH3 and (−CH2−)n groups of lipid molecules in the lipoprotein particles. The LMWM data are acquired with such spectrometer settings (using a T2-relaxation-filtered pulse sequence) that suppress most of the broad macromolecule and lipoprotein lipid signals and in that way enhance the detection of rapidly tumbling smaller solutes, i.e., with 64 k data points using 24 transients acquired after four steady-state scans with a Bruker 1D CPMG pulse sequence with water peak suppression and a 78 ms T2-filter with a fixed echo delay of 403 μs to minimize diffusion and J-modulation effects. The acquisition time is 3.3 s and the relaxation delay 3.0 s. Both LIPO and LMWM data are processed and phase corrected in an automated manner. Before Fourier transformations to spectra, the FIDs (both LIPO and LMWM) are zero-filled to 128 k data points and then multiplied with an exponential window function with a 1.0 Hz line broadening.
Serum lipid extraction is carried out by adding 5 ml of methanol, 10 ml dichloromethane, and 15 ml 0.15 M sodium chloride solution to the serum samples (including 300 μl of serum and 300 μl of the NMR buffer). The samples are shaken and centrifuged at 2400 × g for 20 min at 4°C. The organic phase is recovered and the aqueous phase is then extracted again with 10 ml of dichloromethane to enhance the yield. After shaking and centrifuging (as before), the organic phase is recovered. The separated organic phases are combined and evaporated to dryness with pressurized air. For the NMR measurements, the lipid extracts are dissolved into 0.6 ml of CDCl3 containing 0.03% of tetramethylsilane to serve as a chemical shift reference. The 64 k LIPID data are acquired using 32 transients after four dummy scans. A relaxation delay of 3.0 s and an acquisition time of 3.3 s are used. The measured data are zero-filled to 128 k and multiplied with an exponential window function with a line broadening of 0.5 Hz.
1H NMR is inherently a quantitative technique allowing for absolute metabolite quantification. Although the multi-metabolic nature of serum inevitably causes signal overlap, the metabolite content and concentrations can be extracted by appropriate experimental settings and advanced computational techniques (Ala-Korpela, 1995). In the case of the heavily overlapping data of lipoprotein lipids in the LIPO window, it is our experience that regression modeling performs more reliably than line fitting-based approaches. Therefore, we have now implemented several lipoprotein subclasses (e.g., VLDL subclasses), as well as other lipoprotein (e.g., HDL-C), and serum lipid (e.g., TGs) quantification models using regression modeling. These models can be directly applied to the LIPO spectra in an automated manner and the computation of the measures is instant (as the analysis with existing models is non-iterative). A simplified and computationally more efficient modification of the approach presented recently in Vehtari et al (2007) is used. All the models used are cross-validated against NMR-independent lipid data. Before applying the predictive models, we verify that the new input spectrum lies within the limits (±10%) of the training data set; otherwise, the particular sample is rejected from the subclass analyses with no outcome.
The method of choice to quantify the low-molecular-weight metabolites (as well as the residual lipoprotein lipids resonances) in the LMWM spectra and the lipid signals in the LIPID spectra is iterative lineshape fitting analysis. A simple singlet-multiplet approach works well for the majority of the low-molecular-weight metabolites. A model lineshape-based approach needs to be adopted in the case of the LIPID spectra to enable quantitative analysis of severely overlapping peaks and to increase the quantification accuracy (Mierisova and Ala-Korpela, 2001). Intensity ratios of well-defined multiplets are used as constraints, and, in some cases, the known coupling constants or relative line widths are also constrained. A number of constraints were created based on model spectra that were acquired from pure lipid compounds. This kind of analysis is often termed as ‘the use of biochemical prior knowledge’ and its use is recommended to decrease the mathematical uncertainties with overlapping resonances (Mierisova and Ala-Korpela, 2001). This sophisticated methodology allows us to get information on the amounts of several lipid components, e.g., sphingomyelin and ω-3 fatty acids. Also, the average degree of (poly)(un)saturation can be calculated from these variables. Absolute quantification cannot be directly established in the lipid extraction procedure due to some experimental variation in the lipid acquisition. Therefore, each LIPID spectrum is scaled (via the fitted C resonances) according to the serum total C as estimated from the corresponding LIPO spectrum using a regression model. The PERCH NMR software (Perch Solutions Ltd, Kuopio, Finland) is used for all the lineshape fitting analyses (Soininen et al, 2005).
Lipoprotein measures and subclass quantification
Lipoprotein profiling by 1H NMR spectroscopy is currently well established (Ala-Korpela, 1995, 2008) and is incorporated in the serum NMR metabonomics platform (Soininen et al, 2009). Regarding the quantifications, we applied here a computationally more efficient modification of the approach presented recently in Vehtari et al (2007). Serum concentrations of total TGs and total C, as well as, for example, VLDL-TG, IDL-C, LDL-C, and HDL-C, were determined. Incorporation of the NMR measures into the recently introduced computational approach also enabled estimation of apolipoprotein A-I and APOB (Niemi et al, 2009). In addition, total lipid and particle concentrations in 14 lipoprotein subclasses were obtained, as well as various subclass-specific lipid concentrations as total C, in all the LDL subclasses. The subclasses referred here were calibrated via high-performance liquid chromatography (Okazaki et al, 2005) and are as follows: chylomicrons and extremely large VLDL particles (with particle diameters from ∼75 nm upwards), five different VLDL subclasses, namely, very large VLDL (average particle diameter of 64.0 nm), large VLDL (53.6 nm), medium VLDL (44.5 nm), small VLDL (36.8 nm), and very small VLDL (31.3 nm); IDL (28.6 nm), three LDL subclasses as large LDL (25.5 nm), medium LDL (23.0 nm), and small LDL (18.7); and four HDL subclasses as very large HDL (14.3 nm), large HDL (12.1 nm), medium HDL (10.9 nm), and small HDL (8.7 nm).
Immune response marker quantification
Serum concentrations of IgE were analyzed at the National Institute for Health and Welfare by Quantia IgE immunoturbidimetric assay (Biokit, S. A., Barcelona, Spain) using analyzer Architect c8000 (Abbott Laboratories, Abbott Park, IL, USA). The analytical range of the IgE method was from 25 to 1000 IU/ml. Because nearly half of the results were below 25 IU/ml, we assumed that there was a linear correlation between the measured absorbance and the concentration of IgE. According to the absorbance at 25 IU/ml, we calculated a factor that we used to generate semiquantitative results for stratifying samples below 25 IU/ml. The interassay coefficient of variation (CV) of IgE varied from 2.4% (high level control, 283 IU/ml) to 13% (low level control, 37 IU/ml).
IL-1ra and HMWA were determined at the National Institute of Health and Welfare laboratory in Turku. Intra-assay CVs were 5.3, 2.2, and 2.2% for IL-1ra at nominal concentration levels of 170, 400, and 1485 pg/ml, and 8.5 and 6.3% for HMWA at nominal concentration levels of 3000 and 6800 ng/ml. Interassay CVs were 11.9, 10.3, 9.9, and 9.0% for IL-1ra at nominal concentration levels of 145, 290, 520, and 1290 pg/ml, and 18.1, 18.7, and 19.5% for HMWA at nominal concentration levels of 3300, 4400, and 9500 ng/ml.
Data quality, processing, and normalization
As metabolite distributions did not achieve normality, Box-Cox power transformations were implemented for each metabolite and the resulting distributions were corrected for age, gender, and the first 10 principal components by taking the standardized residuals from a multiple linear regression with the above as covariates. The principal components are calculated directly from the SNP genotypes and as such capture genetic variation among the samples. We control for this variation in the metabolite levels because we wish to remove metabolite variation which is attributable to genetic population structure in Finland. Outliers of >4 s.d.s from the mean were removed. See Supplementary Figure 3 for Spearman's rank correlations between metabolites.
Processing of the expression data was performed as previously described (Inouye et al, 2010). Briefly, all arrays were quantile normalized at the strip-level and technical replicates were combined via a bead-count weighted average. Technical replicates were removed from further analysis if their Pearson's correlation coefficient was <0.94 or their Spearman's rank correlation coefficient was <0.60. Probes were removed if they mapped to a non-autosomal chromosome, erythrocyte globin components, or more than one genomic position. Genotypes were called and imputed using the Illuminus and IMPUTE algorithms (Marchini et al, 2007; Teo et al, 2007), respectively, and underwent stringent filtering for low-quality SNPs, samples, population structure and relatedness.
Association analysis
All modeling was performed using the R-statistical computing language (http://www.r-project.org/). After normalization and correction for confounding variables, each metabolite concentration was tested for association with the expression values of each gene probe using linear regression. For each metabolite and gene probe, Yi is the standardized residual for individual i, Xi is the probe's log2-normalized expression value for the individual, and εi is a normally distributed random variable with mean equal to zero and constant variance:
Nominal P-values were calculated for the test of no association, b=0. A strict significance level for each metabolite was employed using Bonferroni's correction. With 35 419 tests for each metabolite, this gives a significance level of 1.41 × 10−6.
Network analysis
Network analysis was carried out using the R-packages, WGCNA (Langfelder and Horvath, 2008) and NEO (Aten et al, 2008).
WGCNA was used to generate an undirected network model of genes associated with blood metabolite concentrations. We constructed the network using the top 10% of expression signals across all metabolites (3520 unique probes). Pearson's correlation coefficients were used to construct the correlation matrix, and a soft power threshold of seven (Supplementary Figure 4) was used to generate the adjacency matrix. The matrix was then hierarchically clustered and a dynamic tree cut with a minimum module size of 10 probes was used to identify clusters of tightly correlated genes (i.e., modules or subnetworks). Individual modules can also be intercorrelated; therefore, we used each module's summary expression profile (first singular vector) to merge tightly correlated modules at a dendrogram height of <0.20 (Supplementary Figure 5). Metabolite concentrations were tested for association with each module using the summary expression profile. A t-test of Spearman's rank correlation was employed and the associations presented in heatmaps for each metabolite window (Supplementary Figures 7–16). In the heatmaps, the LL module corresponds to module A.
NEO was used to predict the directedness of the network using causal SNPs as anchors. We only included metabolites that were associated with LL module expression, not ratios, and, as edge orienting depends on a sufficient genetic basis for both linked traits, have at least one SNP association with P-value <5.0 × 10−7. This leads to the inclusion of 29 metabolites in a directed network with core LL module. For an SNP to be considered as a causal anchor in the network, it needed to satisfy one of the following: (a) have association P-value <5.0 × 10−7 for one or more LL module-associated metabolites, (b) be in cis to a core LL module gene and have a expression association P-value <5.0 × 10−3. To assign SNPs as causal anchors, we adopted the automatic SNP selection approach implemented in NEO, which uses both greedy and forward stepwise regression (Aten et al, 2008). An oriented edge was considered causal according to three criteria: (i) if its NEO score (in this case the local SEM-based Edge Orienting Next Best Orthogonal Causal Anchor, LEO.NB.OCA score) was >0.30, (ii) if its causal model P-value was >0.10, and (iii) if its A → B path coefficient was a Z-score test statistic >1.96 or <−1.96. See Supplementary Table 3 for a full list of network statistics. The robustness of the SNP selection as causal anchors was evaluated for all inferred causal edges using the robustness analysis in NEO (Supplementary Figure 17). The combined greedy and forward stepwise selection approach was used to select the top 1, 2, 3, 4, and 5 most highly correlated SNPs, as causal anchors for metabolites and expressed genes, and, for each step, edge-orientation scores were calculated for the originally inferred edge orientation, i.e., that which was based on the whole SNP set. This analysis showed that core LL module's reactivity to blood metabolites was robust to SNP selection.
Conditional co-expression
It is important to understand how the LL module's core co-expression changes with metabolite levels. For each core gene, log2-normalized expression values were partitioned into quintiles. In order to make quintile numbers consistent, the last quintile (highest metabolite concentration) consisted of 0–4 more individuals than other quintiles. Changing the quintile with 0–4 individuals did not affect downstream analysis. For each quintile, co-expression was assessed by calculating Spearman's rank correlation coefficient between all possible non-redundant pairs (N=28) of the LL module core genes. A linear model was fitted to all co-expression pairs across all quintiles in order to test the hypothesis that co-expression of module is unaffected by metabolite concentration, i.e., the fitted linear function has zero slope (Figure 3). See Supplementary Figure 18 for all metabolites.
Data availability
We have made the metabonomic, transcriptomic, and genomic data used in this study publically available. The metabonomic measures are available as Supplementary Table 4, the raw and normalized gene expression intensities have been deposited in the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-TABM-1036, and the genotype data has been deposited in the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under the accession number EGAS00000000086. Both ArrayExpress and EGA are hosted by the European Bioinformatics Institute.
Supplementary Material
Supplementary tables S1–3, Supplementary figures S1–18
Normalized metabonomic data for all individuals used in the study
Acknowledgments
This manuscript is dedicated in memory of Prof LP. We thank the participants of the DILGOM study. MI has been supported by the Wellcome Trust and an NHMRC Biomedical Australian Training Fellowship (no. 637400). VS was supported by the Finnish Foundation for Cardiovascular Research, Sigrid Jusélius Foundation and the Academy of Finland, Grant number 129494. PJ was supported by the Academy of Finland (Grant number 118065 for the DILGOM project). This work has been supported by the Academy of Finland (MJS, MAK), the Finnish Cardiovascular Research Foundation (MJS, MAK), the Jenny and Antti Wihuri Foundation (AJK), and the Sigrid Jusélius Foundation (MJS). We also thank the comments of three anonymous peer reviewers, who have substantially improved the original paper.
Author contributions: The study was conceived and designed by LP, VS, AP, MP, MAK, and MI. Metabonomics was performed by MAK, PS, LSK, AJK, and MJS. Experimental work was performed by KS, EH, PJ, SM, AJ, and JL. Analyses were performed by MI, JK, and SR. MI and MAK wrote the paper with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ala-Korpela M (1995) 1H NMR spectroscopy of human blood plasma. Prog Nucl Magn Reson Spectr 27: 475–554 [Google Scholar]
- Ala-Korpela M (2008) Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med 46: 27–42 [DOI] [PubMed] [Google Scholar]
- Ala-Korpela M, Soininen P, Savolainen MJ (2009) Letter by Ala-Korpela et al regarding article, ‘Lipoprotein particle profiles by nuclear magnetic resonance compared with standard lipids and apolipoproteins in predicting incident cardiovascular disease in women’. Circulation 120: e149; author reply e150 [DOI] [PubMed] [Google Scholar]
- Altmann F (2007) The role of protein glycosylation in allergy. Int Arch Allergy Immunol 142: 99–115 [DOI] [PubMed] [Google Scholar]
- Aten JE, Fuller TF, Lusis AJ, Horvath S (2008) Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst Biol 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, Ordovas JM, Ripatti S, Parker AN, Miletich JP, Ridker PM (2009) Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet 5: e1000730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA et al. (2008) Variations in DNA elucidate molecular networks that cause disease. Nature 452: 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S, Torok N, Dasu MR, Samols D, Jialal I (2008) Adiponectin decreases C-reactive protein synthesis and secretion from endothelial cells: evidence for an adipose tissue-vascular loop. Arterioscler Thromb Vasc Biol 28: 1368–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U et al. (2008) Genetics of gene expression and its effect on disease. Nature 452: 423–428 [DOI] [PubMed] [Google Scholar]
- Ganapathi MK, Rzewnicki D, Samols D, Jiang SL, Kushner I (1991) Effect of combinations of cytokines and hormones on synthesis of serum amyloid A and C-reactive protein in Hep 3B cells. J Immunol 147: 1261–1265 [PubMed] [Google Scholar]
- Haak-Frendscho M, Dinarello C, Kaplan AP (1988) Recombinant human interleukin-1 beta causes histamine release from human basophils. J Allergy Clin Immunol 82: 218–223 [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK (2008) Metabolic phenotyping in health and disease. Cell 134: 714–717 [DOI] [PubMed] [Google Scholar]
- Horvath S, Dong J (2008) Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 4: e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS (2006) Inflammation and metabolic disorders. Nature 444: 860–867 [DOI] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Müller A, Cook SA, Kurtz TW, Whittaker J et al. (2005) Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37: 243–253 [DOI] [PubMed] [Google Scholar]
- Inouye M, Silander K, Hamalainen E, Salomaa V, Harald K, Jousilahti P, Mannisto S, Eriksson J, Saarela J, Ripatti S, Perola M, van Ommen GJB, Taskinen MR, Palotie A, Dermitzakis ET, Peltonen L (2010) An immune response network associated with blood lipid levels. PLoS Genet 6: e1001113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CM, Faulenbach M, Vaag A, Vølund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY (2007) Interleukin-1 receptor antagonist in type 2 diabetes mellitus. N Engl J Med 356: 1517–1526 [DOI] [PubMed] [Google Scholar]
- Li R, Tsaih SW, Shockley K, Stylianou IM, Wergedal J, Paigen B, Churchill GA (2006) Structural model analysis of multiple quantitative traits. PLoS Genet 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Attie AD, Reue K (2008) Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet 9: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon CJ, Law RE, Hsueh WA (2003) Minireview: adiposity, inflammation, and atherogenesis. Endocrinology 144: 2195–2200 [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906–913 [DOI] [PubMed] [Google Scholar]
- Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, Castellani LW, Suh M, Armour C, Edwards S, Lamb J, Lusis AJ, Schadt EE (2005) Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet 37: 1224–1233 [DOI] [PubMed] [Google Scholar]
- Mierisova S, Ala-Korpela M (2001) MR spectroscopy quantitation: a review of frequency domain methods. NMR Biomed 14: 247–259 [DOI] [PubMed] [Google Scholar]
- Mora S, Otvos JD, Rifai N, Rosenson RS, Buring JE, Ridker PM (2009) Lipoprotein particle profiles by nuclear magnetic resonance compared with standard lipids and apolipoproteins in predicting incident cardiovascular disease in women. Circulation 119: 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi J, Makinen VP, Heikkonen J, Tenkanen L, Hiltunen Y, Hannuksela ML, Jauhiainen M, Forsblom C, Taskinen MR, Kesaniemi YA, Savolainen MJ, Kaski K, Groop PH, Kovanen PT, Ala-Korpela M (2009) Estimation of VLDL, IDL, LDL, HDL(2), apoA-I, and apoB from the Friedewald inputs-apoB and IDL, but not LDL, are associated with mortality in type 1 diabetes. Ann Med 41: 451–461 [DOI] [PubMed] [Google Scholar]
- Okazaki M, Usui S, Ishigami M, Sakai N, Nakamura T, Matsuzawa Y, Yamashita S (2005) Identification of unique lipoprotein subclasses for visceral obesity by component analysis of cholesterol profile in high-performance liquid chromatography. Arterioscler Thromb Vasc Biol 25: 578–584 [DOI] [PubMed] [Google Scholar]
- Salomaa V, Havulinna A, Saarela O, Zeller T, Jousilahti P, Jula A, Muenzel T, Aromaa A, Evans A, Kuulasmaa K, Blankenberg S (2010) Thirty-one novel biomarkers as predictors for clinically incident diabetes. PLoS One 5: e10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar N, Wannamethee SG, Forouhi NG (2008) Novel biochemical risk factors for type 2 diabetes: pathogenic insights or prediction possibilities? Diabetologia 51: 926–940 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A et al. (2005) An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37: 710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen P, Kangas AJ, Wurtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Jarvelin MR, Kahonen M, Lehtimaki T, Viikari J, Raitakari OT, Savolainen MJ, Ala-Korpela M (2009) High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 134: 1781–1785 [DOI] [PubMed] [Google Scholar]
- Soininen P, Haarala J, Vepsäläinen J, Niemitz M, Laatikainen R (2005) Strategies for organic impurity quantification by 1H NMR spectroscopy: constrained total-line-shape fitting. Analytica Chimica Acta 542: 178–185 [Google Scholar]
- Tall AR (1998) An overview of reverse cholesterol transport. Eur Heart J 19(Suppl A): A31–A35 [PubMed] [Google Scholar]
- Teo YY, Inouye M, Small KS, Gwilliam R, Deloukas P, Kwiatkowski DP, Clark TG (2007) A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 23: 2741–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T, Tynkkynen T, Makinen VP, Jylanki P, Kangas A, Hokkanen J, Vehtari A, Grohn O, Hallikainen M, Soininen H, Kivipelto M, Groop PH, Kaski K, Laatikainen R, Soininen P, Pirttila T, Ala-Korpela M (2008) A multi-metabolite analysis of serum by 1H NMR spectroscopy: early systemic signs of Alzheimer′s disease. Biochem Biophys Res Commun 375: 356–361 [DOI] [PubMed] [Google Scholar]
- Vehtari A, Makinen VP, Soininen P, Ingman P, Makela SM, Savolainen MJ, Hannuksela ML, Kaski K, Ala-Korpela M (2007) A novel Bayesian approach to quantify clinical variables and to determine their spectroscopic counterparts in 1H NMR metabonomic data. BMC Bioinformatics 8 (Suppl 2): S8. doi: 10.1186/1471-2105-8-S2-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, Gohlke H, Wagenpfeil S, Ollert M, Ring J, Behrendt H, Heinrich J, Novak N, Bieber T, Krämer U, Berdel D, von Berg A, Bauer CP, Herbarth O, Koletzko S et al. (2008) Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 4: e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables S1–3, Supplementary figures S1–18
Normalized metabonomic data for all individuals used in the study
Data Availability Statement
We have made the metabonomic, transcriptomic, and genomic data used in this study publically available. The metabonomic measures are available as Supplementary Table 4, the raw and normalized gene expression intensities have been deposited in the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-TABM-1036, and the genotype data has been deposited in the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under the accession number EGAS00000000086. Both ArrayExpress and EGA are hosted by the European Bioinformatics Institute.