Abstract
We recently demonstrated that vaccinated rhesus macaques controlled viral replication of a heterologous SIV challenge. Here, we analyzed anamnestic SIV-specific CD4+ T-cell responses expanding immediately after challenge and show that successful vaccinees consistently targeted a short region of the Gag-p27 Capsid (amino acids 249-291). We have also defined the Major Histocompatibility Complex Class II (MHC-II) restricting alleles for several of these responses and show that DQ-restricted CD4+ T-cells depend on unique combinations of both the DQA and DQB alleles. Analysis of SIV-specific CD4+ T-cell responses elicited by a successful vaccine may have important implications in the understanding of vaccine design.
Keywords: HIV, SIV, CD4+ T-cells, Mamu-MHC, Major Histocompatibility Complex, Rhesus macaques
The role of CD4+ T-cells in controlling HIV/SIV viral replication is controversial. Evidence indicates that CD4+ T-cells are an important factor in controlling other pathogens that establish chronic infections (Matloubian et al. 1994; Hasenkrug et al. 1998; Grakoui et al. 2003; Virgin et al. 2009). In HIV and SIV infection, the presence of antigen-specific CD4+ T-cells correlates with control of viral replication (Kalams and Walker 1998; Letvin et al. 2006; Kawada et al. 2007). Elite Controllers (ECs), who reduce viral replication to undetectable levels in the chronic phase, often have high frequency antigen-specific CD4+ T-cell responses (Rosenberg et al. 1997; Friedrich et al. 2007), suggesting that these cells might be important in the maintenance of control. In addition, although HIV and SIV may preferentially infect antigen-specific CD4+ T-cells (Douek et al. 2002; Brenchley et al. 2006), the frequency of this preferential infection in vivo is low (Douek et al. 2002; Virgin and Walker 2010). Evidence may also suggest that high frequency effector-memory SIV-specific CD4+ T-cell responses elicited through vaccination associate with reduced viral replication following viral challenge (Hansen et al. 2009). These studies suggest that HIV/SIV-specific CD4+ T-cells may play a direct role in controlling viral replication. However, we only have a limited knowledge of what successful HIV/SIV-specific CD4+ T-cell responses should target in the virus. A better understanding of the role these CD4+ T-cells have in controlling HIV replication should be a high priority (Virgin and Walker 2010).
We recently vaccinated eight Indian rhesus macaques with DNA/Ad5 encoding Gag, Tat, Rev, Nef, Pol, Vif, Vpr and Vpx and mucosally challenged them with a low dose heterologous SIVsmE660 swarm virus (Wilson et al. 2009). Seven of the vaccinees clearly became infected with the challenge virus, while one vaccinee may not have been productively infected and was not included in this analysis. Five of these seven vaccinated animals had reduced peaks of viremia and controlled viral replication to less than 50 viral RNA/copies of plasma during the chronic phase of infection. These animals also mounted several high frequency SIV-specific CD4+ T-cell responses that expanded during peak viremia after SIVsmE660 challenge. The remaining two vaccinees had high viral loads; one was indistinguishable from the unvaccinated naïve controls (Wilson et al. 2009). Here we investigated the anamnestic SIVmac239-specific CD4+ T-cell responses detected after vaccination in the five animals that controlled viral replication to less than 50 viral RNA copies/ml of plasma. Previous studies have analyzed the CD4+ T-cell response directed against the entire viral proteome (Kaufmann et al. 2004) in HIV-infected humans and in SIV-infected EC Indian rhesus macaques (Friedrich et al. 2007). However few studies to date have analyzed the antigen-specific anamnestic CD4+ T-cell responses that expand during the peak of viremia in the setting of a successful vaccine.
The SIV-specific CD4+ T-cell response in vaccinated animals primarily targeted Gag
We used groups of peptide-pools (ten 15-mer peptides overlapping by 11 amino acids) that spanned the entire SIVmac239 proteome to determine the targets of these responses using Enzyme-linked immunosorbent spot assay (ELISPOT) (Wilson et al. 2009). Most of these responses were directed against Gag (Fig. 1A). Since the majority of these CD4+ T-cell responses were observed after vaccination (Wilson et al. 2009) and because the expansion in frequency occurred soon after infection, these responses are likely anamnestic vaccine-induced SIVmac239-specific CD4+ T-cell responses. Thus, the anamnestic vaccine-induced responses recognized epitopes from the heterologous SIVsmE660 challenge virus. These broad Gag-specific responses continued into the chronic phase of infection (Fig. 1B). The Gag protein from HIV and SIV has previously been shown to be a major immunodominant target for virus-specific CD4+ T-cells in HIV (Kaufmann et al. 2004) and SIV (Friedrich et al. 2007; Giraldo-Vela et al. 2008; Sacha et al. 2009). Interestingly, Gag- and Nef-specific CD4+ T-cells increased in frequency when SIV replication resumed after in vivo CD8-depletion of ECs, despite the high viral loads present in the peripheral blood (Friedrich et al. 2007). Similarly, SIV-specific CD4+ T-cells also increased in frequency following SIVsmE660 challenge (Wilson et al. 2009), implicating these cells in the control of viral replication.
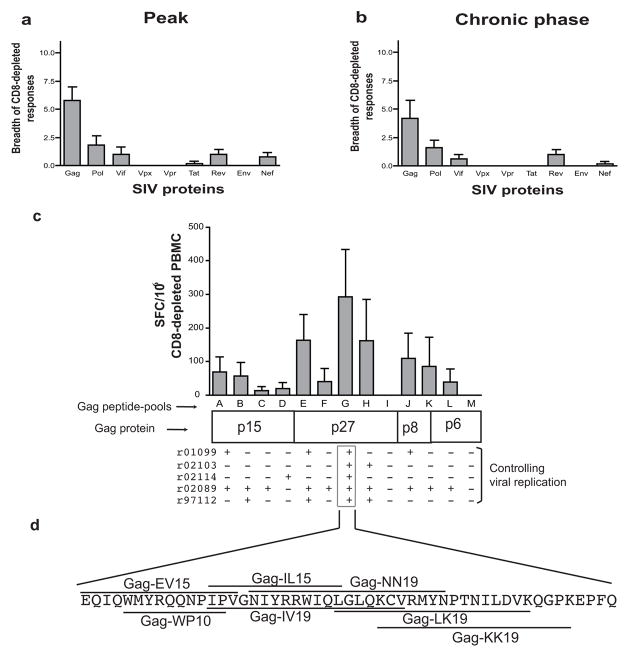
Fig. 1. Breadth and frequency of anamnestic SIV-specific CD4+ T-cell responses.
The breath of SIV-specific CD8-depleted PBMC responses present in five vaccinated animals that controlled viral replication targeted mainly SIV Gag during the peak of viremia after SIVmE660 challenge (A) (Wilson et al. 2009). Some of the responses detected during the peak of viremia were still present in the chronic phase of infection (six months post-infection) (B). Several of the highest frequency responses in CD8-depleted PBMC from vaccinated animals that controlled viral replication were directed against the Gag G peptide-pool (amino acids 241-291), located within the SIV Capsid (Gag-p27), during the chronic phase of infection (C). A “+” sign in panel (C) represents an anamnestic IFNγ positive CD8-depleted PBMC response detected against a particular region of Gag. The SIV-specific CD4+ T-cell responses directed against the Gag G peptide-pool (amino acids 241-291) located within SIV Capsid (Gag-p27) are shown underlying or overlying the amino acid sequence (D). The bars in panels (A), (B) and (C) represent the mean and the standard deviation
Some of the highest frequency CD4+ T-cell responses were elicited by a short piece of the SIV Capsid (Gag-p27, amino acids 249-291, Gag G peptide-pool; Fig. 1C). All of the vaccinated animals that controlled viral replication had strong anamnestic CD8-depleted PBMC responses directed against this short region of the SIV Capsid (amino acids 249-291) during the peak of viremia (Wilson et al. 2009) and maintained them during the chronic phase of infection (Fig. 1C). We mapped seven different anamnestic SIV-specific CD4+ T-cell responses targeting the Gag G-peptide-pool (amino acids 241-291). All of these targeted epitopes overlapped each other (Fig. 1D). It had been shown previously that certain discrete regions of HIV can be presented to CD4+ T-cells more readily than others acting as a “hotspot” for the generation of epitopes (Surman et al. 2001). These “hotspots” have also been reported to occur in the Chlamydia trachomatis major outer protein (Kim and DeMars 2001). Here, this discrete region of SIV Capsid (amino acids 241-291) also appeared to be a “hotspot” for the generation of epitopes recognized by SIV-specific CD4+ T-cells. More importantly, however, all of the vaccinees that controlled viral replication consistently targeted this region after vaccination and following SIVsmE660 challenge (Gag G peptide-pool, amino acids241-291; Table 1).
Table 1.
Epitopes recognized by anamnestic SIVmac239-specific CD4+ T-cell responses in vaccinated animalsa
| SIV Proteinb | Peptide Pool & aa positionc | Epitope & aa position (abbreviation)d | Epitope Sequencee | MHC-II restricting allelef | Animal(s) targeting the epitopeg | |
|---|---|---|---|---|---|---|
| Gag | Matrix (p15) | A1-51 | EL17-31 (EL15) | EKIRLRPNGKKKYML | DRB1*0303 | r02089 |
| KA27-37 (KA11) | KKYMLKHVVWA | DRB1*1007 | r02089 | |||
| LL41-51 (LL11) | LDRFGLAESLL | DQB1*1708 | r01099 | |||
| DQA1*2402 | ||||||
| B41-91 | SL49-67 (SL19) | SLLENKEGCQKILSVLAPL | - | r97112 | ||
| KP59-69 (KP11) | KILSVLAPLVP | DPB1*06 | r02089 | |||
| SE77-91 (SE15) | SLYNTVCVIWCIHAE | - | r97112 | |||
| C81-131 | KH93-107 (KH15) | KVKHTEEAKQIVQRH | - | r02089 | ||
| Capsid (p27) | D121-171 | TE153-163 (TL11) | TLNAWVKLIEE | DPB1*04 | r02114 | |
| E161-211 | CA181-199 (CA19) | CTPYDINQMLNCVGDHQAA | DRB1*0309 | r97112 | ||
| DI185-203 (DI19) | DINQMLNCVGDHQAAMQII | DQB1*1801 | r02089 | |||
| DQA1*2601 | ||||||
| VA193-211 (VA19) | VGDHQAAMQIIRDIINEEA | - | r02089 & r01099 | |||
| F201-251 | AY237-251 (AY15) | AGTTSSVDEQIQWMY | - | r02089 | ||
| G241-291 | WP249-258 (WP10) | WMYRQQNPIP | DQB1*1801 | r02089 | ||
| DQA1*2601 | ||||||
| EV245-259 (EV15) | EQIQWMYRQQNPIPV | - | r01099 | |||
| IL257-271 (IL15) | IPVGNIYRRWIQLGL | DPB1*02 | r01099 | |||
| IV257-275 (IV19) | IPVGNIYRRWIQLGLQKCV | DRB*w303 | r02114 | |||
| NN261-279 (NN19) | NIYRRWIQLGLQKCVRMYN | DQB1*1801 | r02089 | |||
| DQA1*2601 | ||||||
| LK269-287 (LK19) | LGLQKCVRMYNPTNILDVK | - | r02089 | |||
| KK273-291 (KK19) | KCVRMYNPTNILDVKQGPK | DRBw*201 | r97112, r02103 & r02114 | |||
| H281-331 | RT300-310 (RT11) | RFYKSLRAEQT | DRB1*0303 | r02089 | ||
| QA309-327 (QA19) | QTDAAVKNWMTQTLLIQNA | - | r02103 | |||
| WC317-331 (WC15) | WMTQTLLIQNANPDC | DRB1*1007 | r02089 & r97112 | |||
| (p2) & (p8) | J361-411 | PI376-391 (PI16) | PIPFAAAQQRGPRKPI | DQB1*1801 | r02089 | |
| DQA1*2601 | ||||||
| RA385-403 (RA19) | RGPRKPIKCWNCGKEGHSA | - | r01099 | |||
| K401-451 | KD419-429 (KD11) | KMDHVMAKCPD | DPB1*06 | r02089 | ||
| (p1) & (p6) | L451-491 | KV443-452 (KV10) | KPRNFPMAQV | DQB1*1801 | r02089 | |
| DQA1*2601 | ||||||
| LK469-483 (LK15) | LLKNYMQLGKQQREK | - | r02089 | |||
| Pol | RT (p51) | G199-250 | AP200-218 (AP19) | ALREICEKMEKDGQLEEAP | - | r97112 |
| MP208-226 (MP19) | MEKDGQLEEAPPTNPYNTP | - | r02114 | |||
| FI228-442 (FI15) | FAIKKKDKNKWRMLI | DRB1*0303 | r02089 | |||
| KV231-250 KV19) | KKDKNKWRMLIDFRELNRV | - | r97112 | |||
| I280-330 | DY280-294 (DY15) | DAYFSIPLDEEFRQY | - | r02089 | ||
| M440-490 | GE440-458 (GE19) | GIKTKHLCRLIRGKMTLTE | DPB1*06 | r02089 | ||
| EY471-486 (EY15) | ENKIILSQEQEGCYY | - | r02089 | |||
| (p15) | S676-726 | IK688-706 (IK19) | IEEMIKKSEIYVAWVPAHK | - | r02089 | |
| Integrase (p31) | V796-846 | GQ808-822 (GQ19) | GFIEAEVIPQETGRQ | DRBw*201 | r97112 | |
| PH816-838 (PH23) | PQETGRQTALFLLKLAGRWPITH | - | r97112 | |||
| X880-934 | TR900-914 (TR15) | TIVLMAVHCMNFKRR | - | r02113 | ||
| Y924-978 | EK924-942 (EK19) | ERLINMITTEQEIQFQQSK | - | r02089 | ||
| Vif | A1-51 | MI1-15 (MI15) | MEEEKRWIAVPTWRI | - | r02114 | |
| EY17-31 (EY15) | ERLERWHSLIKYLKY | - | r02089 | |||
| C81-131 | AA105-119 (AA15) | ADILLHSTYFPCFTA | - | r02089 | ||
| Rev | A1-51 | EL9-20 (EL12) | ELRKRLRLIHLL | DRB1*0318 | r01099 | |
| RT13-23 (RT11) | RLRLIHLLHQT | DPB1*06 | r02089 & r97112 | |||
| B40-90 | KY40-58 (KY19) | KRRWRRRWQQLLALADRIY | - | r02089 | ||
| AD72-86 (AD15) | AIQQLQNLAIESIPD | - | r02089 & r97112 | |||
| Nef | E166-216 | EY209-223 (EY15) | EVLAWKFDPTLAYTY | DRB1*0309 | r02103 | |
We mapped the SIV-specific CD4+ T-cell responses from five vaccinated animals (r01099, r02103, r02114, r02089, r97112), and generated 23 CD4+ T-cell clones specific for 23 different epitopes to determine MHC-II restriction.
Only the proteins that elicited an anamnestic SIV-specific CD4+ T-cell response were included in the table.
Peptide-pools were comprised of ten peptides of 15 amino acids in length that overlapped by 11 amino acids. We used peptide-pools that span the entire SIVmac239 proteome to detect anamnestic SIV-specific CD4+ T-cell immune responses. The amino acid (aa) position for each peptide-pool in SIVmac239 is written in subscript next to each letter.
Epitopes are named after their two flanking amino acids (aa) and their position in the SIVmac239 protein.
Epitopes highlighted in gray denote epitopes that were mapped to a core epitope.
The alpha chain is only denoted for the DQ alpha alleles. A dash (−) indicates that the restricting MHC-II allele for the targeted epitope was not determined.
Animals with SIV-specific CD4+ T-cell responses targeting the same epitope are in bold.
Although this short region of SIV Capsid (amino acids 241-291) elicited some of the highest frequency responses in CD4+ T-cells from vaccinated animals that controlled viral replication, this region may not be as effective at inducing the same type of responses in CD8+ T-cells. Only the SIV-derived CD8+ T-cell epitopes QI9 (Sacha et al. 2008; Valentine et al. 2009) and the rarely detected YL9 (Sacha et al. 2008; Valentine et al. 2009) have been described for this region of SIV Capsid. These two epitopes only elicit subdominant responses in SIV-infected animals (Sacha et al. 2008; Valentine et al. 2009). This difference between epitopes recognized by CD8+ and CD4+ T-cells likely occurs because epitopes presented to these two cell types are derived from different antigen-processing pathways.
Besides the SIV Capsid region, vaccinated animals that controlled viral replication targeted several different regions of Gag, Pol, Vif, Rev and Nef. We found 47 distinct anamnestic SIV-specific CD4+ T-cells responses. Interestingly, only five of the identified epitopes were targeted in more than one animal. Most animals appeared to have an exclusive set of anamnestic SIV-specific CD4+ T-cell responses (Table 1). The fine mapping of all of these CD4+ T-cell responses suggested that the targeted epitopes were mostly clustered together; however, the region of the SIV Capsid in the Gag G peptide-pool (amino acids 241-291) appeared to contain an unusual number of tightly clustered CD4+ T-cell epitopes (Table 1).
Vaccinated animals shared MHC-II alleles
To identify the MHC-II alleles expressed by the five vaccinated animals controlling viral replication, we generated CD4+ T-cell clones and cDNA libraries as previously described (Giraldo-Vela et al. 2008). All of the alleles appeared to be part of haplotypes that have been previously described (Table 2) (Doxiadis et al. 2000; Doxiadis et al. 2003; de Groot et al. 2004). Notably, animal r02089 seemed to be homozygous for a common MHC-II haplotype (DRB1*0303, DRB1*1007, DRA1*0105, DQB1*1801, DQA1*2601, DPB1*06, DPA1*0202) (Table 2) (Doxiadis et al. 2003). MHC-II homozygosity has previously been associated with faster disease progression in SIV (Sauermann et al. 2000) and HIV (Zijenah et al. 2002). Interestingly, this homozygous animal made CD4+ T-cell responses against 27 epitopes, whereas other vaccinated animals that controlled viral replication made CD4+ T-cell responses to fewer than 11 on average (Table 1). We also found that rhesus macaque r02089 shared what appeared to be an entire MHC-II haplotype with r97112. In addition, r97112 shared several MHC-II alleles with r02103 and r02114 (Table 2). However, we found no common MHC-II allele expressed among all of the five vaccinated animals controlling viral replication. Surprisingly, despite all of the MHC-II alleles we determined to be shared, the SIV-specific CD4+ T-cell responses elicited in these animals targeted mostly different epitopes (Table 1).
Table 2.
| Allele group | r02089 | r97112 | r02103 | r02114 | r01099 |
|---|---|---|---|---|---|
| DRB | DRB1*0303 | DRB1*0303 | - | - | - |
| DRB1*1007 | DRB1*1007 | - | - | - | |
| - | DRB1*0309 | DRB1*0309 | DRB1*0309 | - | |
| - | DRB*w20101 | DRB*w20101 | DRB*w20101 | - | |
| - | - | DRB*w307 | - | - | |
| - | - | DRB*w702 | - | - | |
| - | - | DRB1*0404 | - | - | |
| - | - | - | DRB*w401+ | - | |
| - | - | - | DRB*w303 | - | |
| - | - | - | - | DRB*w2002 | |
| - | - | - | - | DRB*w2501 | |
| - | - | - | - | DRB*w603 | |
| - | - | - | - | DRB*w604+ | |
| - | - | - | - | DRB1*0318+ | |
| DRA | DRA1*0105 | DRA1*0105 | - | - | - |
| - | DRA1*01024 | DRA1*01024 | DRA1*01024 | - | |
| - | - | DRA1*01025 | - | - | |
| - | - | - | DRA1*01021 | DRA1*01021 | |
| - | - | - | - | DRA1*0103 | |
| DQB | DQB1*1801 | DQB1*1801 | - | - | - |
| - | DQB1*0601 | DQB1*0601 | DQB1*0601 | - | |
| - | - | DQB1*1811 | - | - | |
| - | - | - | - | DQB1*1808 | |
| - | - | - | - | DQB1*1708 | |
| DQA | DQA1*2601 | DQA1*2601 | - | - | - |
| - | DQA1*0104 | DQA1*0104 | DQA1*0104 | - | |
| - | - | DQA1*2602 | - | - | |
| - | - | - | - | DQA1*2402 | |
| DPB | DPB1*06 | DPB1*06 | - | - | DPB1*06 |
| - | DPB1*12 | - | - | - | |
| - | - | DPB1*01 | - | - | |
| - | - | DPB1*13 | DPB1*13 | - | |
| - | - | - | DPB1*04 | - | |
| - | - | - | - | DPB1*02 | |
| DPA | DPA1*0202 | DPA1*0202 | - | - | DPA1*0202 |
| - | DPA1*0208 | - | - | - | |
| - | - | DPA1*0204 | - | - | |
| - | - | - | DPA1*0601 | - | |
| - | - | - | - | DPA1*0203 | |
Vaccinated animals that controlled viral replication of the challenge virus SIVsmE660 to lower than 50 viral RNA copies/ml of plasma during the chronic phase of infection.
If animals shared MHC-II alleles, these alleles were located on the same row. If an MHC-II allele detected in an animal was not present in another, a dash (−) is used. A positive (+) sign next to an MHC-II allele indicates that the allele was not isolated with cDNA libraries but it was presumed to be part of the haplotype (Doxiadis et al. 2000; Doxiadis et al. 2003; de Groot et al. 2004). The presence of MHC-II alleles was determined by capture with cDNA libraries or MHC-II typing and comparing the results to MHC-II haplotypes previously described (Doxiadis et al. 2000; Doxiadis et al. 2003; de Groot et al. 2004).
MHC-II DQ beta and DQ alpha alleles are both involved in CD4+ T-cell-epitope restriction
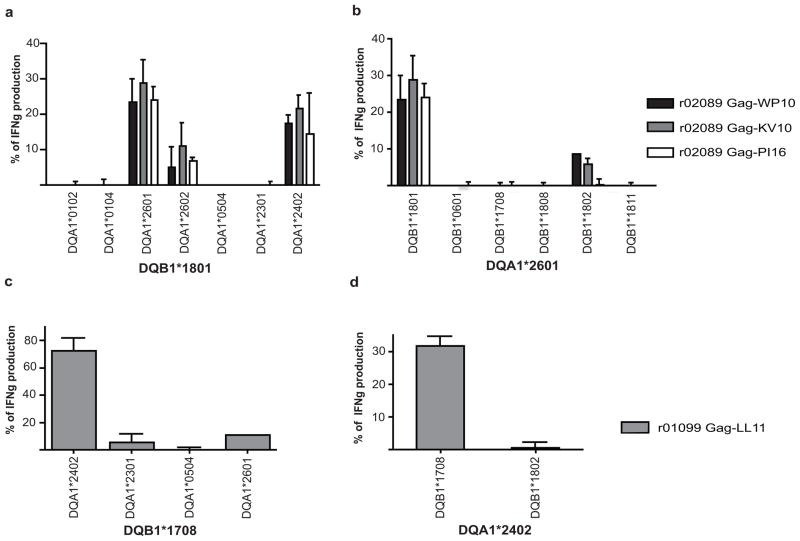
Unlike MHC-I complexes, in which a single chain forms the peptide-binding site, the peptide-binding grove for MHC-II is formed by the pairing of two distinct alpha and beta chains. In the case of MHC-II DR molecules, the amino acid variability of the DR alpha chain is restricted to regions outside the peptide-binding groove (Doxiadis et al. 2000; Doxiadis et al. 2003; de Groot et al. 2004); consequently, it is the MHC-II DR beta chain that determines the specificity for peptide binding. On the contrary, for MHC-II DQ molecules we found that restriction of SIV-specific CD4+ T-cells depended on both the DQ alpha and the DQ beta alleles (Fig. 2). We tested three different SIV-specific CD4+ T-cell clones targeting three different epitopes, Gag-WP10, Gag-KV10 and Gag-PI16 (Table 1), all derived from r02089. We observed that only DQB1*1801 paired with DQA1*2601 or DQA1*2402 elicited optimal IFNγ secretion (Fig. 2A). By contrast, the allele DQB1*1801 paired with DQA1*2602 elicited little IFNγ secretion from the three clones tested (Fig. 2A). Four other DQ alpha alleles did not elicit any IFNγ secretion from the three clones tested (Fig. 2A). We then paired DQA1*2601 with several DQ beta alleles and again tested clones specific for Gag-WP10, Gag-KV10 and Gag-PI16. We found that only the allele pair DQA1*2601- DQB1*1801 induced the secretion of IFNγ (Fig. 2B). The DQA1*2601 allele paired with DQB1*1802 allele did not induce a significant secretion of IFNγ. In addition, another SIV-specific CD4+ T-cell clone (Gag-LL11, derived from r01099, Table 1) was specific only for the alleles DQA1*2402 and DQB1*1708 (Fig. 2C). We then paired the allele DQA1*2402 with another DQB allele (DQB1*1802) and found that it did not elicit IFNγ secretion from this Gag-LL11 clone (Fig. 2D). We found no difference in IFNγ secretion of SIV-specific CD4+ T-cell clones restricted by MHC-II DR and DP alleles using different combinations of DR alpha and DP alpha alleles respectively (not shown). To analyze different DQA and DQB pairings, as well as to determine the MHC-II restriction of each SIV-specific CD4+ T-cell clone, we used RM3 cells. These Epstein-Barr virus-transformed B-cells do not express MHC-II molecules (Calman and Peterlin 1987) and can be used to express MHC-II proteins (Giraldo-Vela et al. 2008). After transfection of RM3 cells with single MHC-II alpha and beta chains and pulsing them with peptides, we used them as antigen presenting cells for SIV-specific CD4+ T-cells (Giraldo-Vela et al. 2008). Thus, when defining which MHC-II DQ alleles restrict SIV-specific CD4+ T-cell epitopes, both the DQ alpha and the DQ beta alleles must be identified.
Fig. 2. MHC-II DQ restriction of SIV-specific CD4+ T-cell responses is allele specific.
Three CD4+ T-cell clones specific for the Gag-WP10, Gag-KV10 and Gag-PI16 epitopes derived from animal r02089 were restricted by the DQB1*1801 allele and only the DQA1*2601 or DQA1*2402 alleles (A). DQA1*2601 paired with other DQ beta molecules and specific peptide, did not elicit IFNγ secretion from the same three clones (B). The CD4+ T-cell clone specific for the Gag-LL11 epitope derived from r01099 and restricted by DQB1*1708 also required the DQA1*2402 allele for IFNγ secretion (C). A different DQ beta allele besides DQB1*1708 paired with DQA1*2402 did not elicit IFNγ secretion from the same clone (D). Bars in the four panels show the mean and the standard deviation of results from one representative experiment. Secretion of IFNγ was normalized to each SIV-specific CD4+ T-cell clone when stimulated with homologous BLCLs and relevant peptide. Single MHC-II allele pairs were expressed using RM3 cells
Here we showed that a particular region of the SIV Capsid is consistently targeted by SIV-specific CD4+ T-cells in vaccinated animals that control viral replication. This region contained several overlapping epitopes that were restricted by different MHC-II alleles. We also mapped several anamnestic SIV-specific CD4+ T-cell responses that expanded immediately after challenge. The study of cellular immune responses that correlate with a successful immune response, in this case elicited by a successful vaccine, may be important in HIV vaccine design.
Acknowledgments
We thank Matija Peterlin for supplying us with the RM3 cell line. We thank Shari Piaskowski and Jessica R. Furlott for assistance with ELISPOT and Debra L Fisk for MHC-II typing. Lara Vojnov, Mauricio Martins and Nick Maness provided helpful discussions. We thank Bridget Giraldo-Vela for the helpful support and comments given during the completion of this manuscript.
This research was funded by NIH grants R24-RR015371, R24-RR016038, R01-AI049120, R01-AI052056 and NIAID contract number HHSN 266200400088C to D.I.W. In addition, this work was supported by NCRR grant P51-RR000167 to the Wisconsin National Primate Research Center, University of Wisconsin-Madison. This research was conducted in part at a facility constructed with support from the Research Facilities Improvement grants RR15459-01 and RR020141-01 (WNPRC).
References
- Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol. 2006;80(14):6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calman AF, Peterlin BM. Mutant human B cell lines deficient in class II major histocompatibility complex transcription. J Immunol. 1987;139(7):2489–2495. [PubMed] [Google Scholar]
- de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE. Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol. 2004;172(10):6152–6157. doi: 10.4049/jimmunol.172.10.6152. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, de Groot NG, de Groot N, Rouweler AJ, Noort R, Verschoor EJ, Bontjer I, Bontrop RE. Evolutionary stability of MHC class II haplotypes in diverse rhesus macaque populations. Immunogenetics. 2003;55(8):540–551. doi: 10.1007/s00251-003-0590-9. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, de Groot NG, Noort R, Bontrop RE. Unprecedented polymorphism of Mhc-DRB region configurations in rhesus macaques. J Immunol. 2000;164(6):3193–3199. doi: 10.4049/jimmunol.164.6.3193. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, Leon EJ, Soma T, Napoe G, Capuano SVr, Wilson NA, Watkins DI. Subdominant CD8+ T-Cell Responses Are Involved in Durable Control of AIDS Virus Replication. J Virol. 2007;81(7):3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, Piaskowski SM, Furlott JR, Morales-Martinez JP, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;82(2):859–870. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302(5645):659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak MJ, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15(3):293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkrug KJ, Brooks DM, Dittmer U. Critical role for CD4(+) T cells in controlling retrovirus replication and spread in persistently infected mice. J Virol. 1998;72(8):6559–6564. doi: 10.1128/jvi.72.8.6559-6564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalams SA, Walker BD. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J Exp Med. 1998;188(12):2199–2204. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, Frahm N, Brander C, Sette A, Walker BD, Rosenberg ES. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78(9):4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M, Tsukamoto T, Yamamoto H, Takeda A, Igarashi H, Watkins DI, Matano T. Long-term control of simian immunodeficiency virus replication with central memory CD4+ T-cell preservation after nonsterile protection by a cytotoxic T-lymphocyte-based vaccine. J Virol. 2007;81(10):5202–5211. doi: 10.1128/JVI.02881-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, DeMars R. Epitope clusters in the major outer membrane protein of Chlamydia trachomatis. Curr Opin Immunol. 2001;13(4):429–436. doi: 10.1016/s0952-7915(00)00237-5. [DOI] [PubMed] [Google Scholar]
- Letvin NL, Mascola JR, Sun Y, Gorgone DA, Buzby AP, Xu L, Yang ZY, Chakrabarti B, Rao SS, Schmitz JE, Montefiori DC, Barker BR, Bookstein FL, Nabel GJ. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312(5779):1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68(12):8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278(5342):1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- Sacha JB, Giraldo-Vela JP, Buechler MB, Martins MA, Maness NJ, Chung C, Wallace LT, Leon EJ, Friedrich TC, Wilson NA, Hiraoka A, Watkins DI. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. Proc Natl Acad Sci U S A. 2009;106(24):9791–9796. doi: 10.1073/pnas.0813106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha JB, Reynolds MR, Buechler MB, Chung C, Jonas AK, Wallace LT, Weiler AM, Lee W, Piaskowski SM, Soma T, Friedrich TC, Wilson NA, Watkins DI. Differential antigen presentation kinetics of CD8+ T-cell epitopes derived from the same viral protein. J Virol. 2008;82(18):9293–9298. doi: 10.1128/JVI.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauermann U, Stahl-Hennig C, Stolte N, Muhl T, Krawczak M, Spring M, Fuchs D, Kaup FJ, Hunsmann G, Sopper S. Homozygosity for a conserved Mhc class II DQ-DRB haplotype is associated with rapid disease progression in simian immunodeficiency virus-infected macaques: results from a prospective study. J Infect Dis. 2000;182(3):716–724. doi: 10.1086/315800. [DOI] [PubMed] [Google Scholar]
- Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, White SW, Doherty PC, Hurwitz JL. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci U S A. 2001;98(8):4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, Loffredo JT, Bean AT, Leon EJ, MacNair CE, Beal DR, Piaskowski SM, Klimentidis YC, Lank SM, Wiseman RW, Weinfurter JT, May GE, Rakasz EG, Wilson NA, Friedrich TC, O’Connor DH, Allison DB, Watkins DI. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J Virol. 2009;83(22):11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Walker BD. Immunology and the elusive AIDS vaccine. Nature. 2010;464(7286):224–231. doi: 10.1038/nature08898. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, Liang X, Wang F, Thoryk E, Heidecker GJ, Citron MP, Huang L, Lin J, Vitelli S, Ahn CD, Kaizu M, Maness NJ, Reynolds MR, Friedrich TC, Loffredo JT, Rakasz EG, Erickson S, Allison DB, Piatak MJ, Lifson JD, Shiver JW, Casimiro DR, Shaw GM, Hahn BH, Watkins DI. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83(13):6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijenah LS, Hartogensis WE, Katzenstein DA, Tobaiwa O, Mutswangwa J, Mason PR, Louie LG. Association of high HIV-1 RNA levels and homozygosity at HLA class II DRB1 in adults coinfected with Mycobacterium tuberculosis in Harare, Zimbabwe. Hum Immunol. 2002;63(11):1026–1032. doi: 10.1016/s0198-8859(02)00684-5. [DOI] [PubMed] [Google Scholar]