Abstract
We have analysed a family with nine congenital neutropenia patients in four generations, several of which we have studied in a long-term follow-up of over 25 years. The patients were mild to severe neutropenic and suffered from various recurrent bacterial infections. Mutations in the genes ELANE, CSF3R and GFI1 have been reported in patients with autosomal dominant congenital neutropenias. Using a small-scale linkage analysis with markers around the ELANE, CSF3R, CSF3 and GFI1 genes, we were able to determine that the disease segregated with markers around the ELANE gene. We identified a novel mutation in the ELANE gene in all of the affected family members that was not present in any of the healthy family members. The mutation leads to an A28S missense mutation in the mature protein. None of these patients developed leukaemia. This is the first truly multigenerational family with mutations in ELANE as unambiguous cause of severe congenital neutropenia SCN.
Keywords: Severe congenital neutropenia, ELANE, Neutrophil elastase, Mutation, ELA2
Introduction
Congenital neutropenia is a disorder of myelopoiesis characterised by impaired neutrophil differentiation with maturation arrest at the promyelocyte stage. Absolute blood neutrophil counts (ANC) in neutropenia are below 0.5 × 109/l (severe), 0.5–1 × 109/l (moderate) or 1–1.5 × 109/l (mild). A normal ANC is on average 4 × 109/l. The major clinical problem associated with neutropenia is recurrence of bacterial infections.
Several forms of congenital neutropenia can be distinguished: autosomal dominant forms such as severe congenital neutropenia (SCN) [1], cyclic neutropenia [2], myelokathexis [3], as well as recessive forms like Shwachman–Diamond’s syndrome [4]. In patients with autosomal dominant SCN, mutations have been found in three genes: CSF3R, which encodes the granulocyte colony-stimulating factor receptor (G-CSFR); GFI1, which encodes the growth factor independent 1 transcriptional repressor (GFI-1); and ELANE (previously known as ELA2), which encodes neutrophil elastase. In 83 sporadic SCN patients, mutations have been found in CSF3R [5–17], and at least 32 of these have developed leukaemia. Mutations in GFI1 in SCN patients have been described in one father and his two young children [18] while transient somatic GFI1 mutations were detected in a boy that also had mosaic tetraploidy [19]. In 141 SCN patients, mutations have been found in ELANE (mainly missense mutations in exon 4) [17, 20–35]; 24 of these patients developed leukaemia. One promoter polymorphism in ELANE is also thought to contribute to SCN [36, 37]. All mutations in ELANE, CSFR3 and GFI1 were present heterozygously in the patients, as new mutations in the sporadic cases, or inherited in a dominant fashion in the familial cases.
The occurrence of leukaemia in part of the SCN patients has given rise to doubts as to whether the mutations in ELANE and CSF3R are causal or the effect of a (pre-) leukaemic disorder [38]. Only eight small families with ELANE mutations were found, in which only a parent and one or two children were affected [20, 23, 32] and the same ELANE mutation was found in five children fathered by the same sperm donor [28]. The lack of families or families with three or more generations affected, or with more affected branches, has, however, cast doubt on the causal effect of the mutations. To complicate the picture, six SCN/leukaemia patients have been found to have mutations in both the CSF3R and ELANE genes [17, 35, 39].
We have studied a four-generation family with nine congenital neutropenia patients in which the disease is inherited in an autosomal dominant fashion. Most of these patients suffer from recurrent bacterial infections, including gingivitis in all. Several of the patients have had appendicitis; none has developed leukaemia. The granulocyte counts in these patients qualify them as mild to severe neutropenic. We performed linkage analysis in the core family using highly polymorphic genetic markers around the most likely candidate genes, ELANE, CSF3R, GFI1 and CSF3. This analysis indicated a clear linkage between the ELANE gene and the disorder. Sequence analysis revealed a mutation in exon two: r.169G>T that results in the substitution of an alanine by a serine at amino acid 28 in the mature protein (A28S). The mutation is present in all patients tested but not in healthy family members. This is the first multigenerational family with mutations in ELANE as unambiguous cause of SCN.
Materials and methods
Patients
Patient II-6 presented with recurrent infections, such as mouth ulcers, gingivitis, conjunctivitis and otitis media, and complaints of arthralgia and chronic fatigue. She had an adenotonsillectomy at age 6 and jaundice at age 16. She died 1 year after being diagnosed with a diffuse large B cell non-Hodgkin’s lymphoma in the brain at the age of 72. Patient III-1, a 50-year-old male, has had mastoiditis, leading to a double mastoidectomy in infancy. In addition, he had otorrhea and repeated skin and mouth infections (paradontitis, gingivitis), bronchopneumonia and infectious complications after operation(s). Patient III-3, a 49-year-old female, had very painful, recurrent mouth ulcers and skin folliculitis, myalgia and general fatigue. She had a mastoidectomy at the age of 16 after repeated mastoiditis and an acute appendicitis led to an appendectomy at the age of 18. She has further had a bout of severe gastroenteritis at the age of 28 and several wound infections after operations and after giving birth. Patient III-5 is a 43-year-old male with general fatigue, arthralgia and stiff muscles after exercise. The patient has further had mouth and skin infections, recurrent upper airway infections in his youth and had an adenotonsillectomy at the age of 4 and an appendectomy for an acute appendicitis at age 13. Patient III-7 is a 38-year-old male who has not suffered from recurrent infections although small wounds healed only slowly. Although actively playing sports, he did complain of fatigue. Patient IV-1 is a 20-year-old woman and patient IV-4 is a 14-year-old girl with recurrent infections, mainly mild gingivitis. Patient IV-6 is an 8-year-old boy; he was given antibiotic prophylaxis and remains as of yet free of severe infections. Patient I-2 died (committed suicide) at the age of 75 and is known to have suffered from recurrent infections although we have not investigated this. No material was available from this patient.
DNA and RNA isolation
Blood was collected in heparin tubes (Becton Dickinson, USA). Genomic DNA was isolated from white blood cells by a salting out method and dissolved in 10 mM Tris and 0.1 mM EDTA, pH 7.5. RNA was isolated using Trizol (GibcoBRL) according to manufacturer’s instructions and dissolved in 10 mM Tris and 0.1 mM EDTA, pH 7.5.
Analysis of polymorphic markers
A limited linkage analysis around the ELANE, CSF3R, CSF3 and GFI1 genes was performed with the following markers: AFMa302yb5, AFMa312zc9 (CSF3), AFM260zg5, AFM214yc7, AFM077xg (CSF3R), KB9, AFMa310wd9, D19S814 (ELANE), AFMa205xd5 and AFMa109wh5 (GFI1). Primer sequences are available on request. Primers were ordered at Biosource Europe, and one of each set was 5′ FAM-labelled. PCRs were typically performed on 50 ng of genomic DNA in 10 mM Tris-HCl (pH 9.6), 50 mM NaCl, 2.5 mM MgCl2, 200 μg/ml BSA, 200 μM dNTPs, 10 pmol primers and 0.5 U AmpliTaq® DNA polymerase (Applied Biosystems) in a total volume of 25 μl. Thirty to 35 cycli were performed with an annealing temperature varying from 58°C to 65°C. PCR products were analysed on gel before analysis of 0.1–1 μl of the PCR product diluted in HiDi formamide and 0.25 μl ROX-400HD size standard (Applied Biosystems) on an ABI PRISM®3700 DNA analyzer (Applied Biosystems). Allele sizes of the markers were analysed using GeneScan 3.5 and Genotyper (Applied Biosystems) software.
Mutational analysis
Exons 1, 2, 4 and 5 of ELANE and all five exons of CSF3 along with at least 20 bases of flanking intron or UTR were sequenced on genomic DNA (primers are available upon request). PCRs were performed on 200 ng of genomic DNA in 10 mM Tris-HCl (pH 9.6), 50 mM NaCl, 2.5 mM MgCl2, 200 μg/ml BSA, 200 μM dNTPs, 10 pmol primers and 0.8 U AmpliTaq DNA polymerase (Applied Biosystems) in a total volume of 50 μl. Amplification was done in 30 cycles with an annealing temperature of 55°C to 60°C depending on the primer pair. PCR products were analysed on gel and purified using a QIAquick PCR purification kit (Qiagen). Products were sequenced using the BigDye Terminators v 2.0 Cycle Sequencing kit (Applied Biosystems) and an ABI PRISM®3700 DNA analyzer (Applied Biosystems). To verify the presence/absence of the mutation in the family, exon 2 PCR products were AcyI (Roche) digested following the manufacturer’s instructions.
RT-PCR
The sequence of exon 3 of ELANE was analysed in RNA from blood. One microgram of RNA was reverse-transcribed in a reaction containing 5 pmol oligo-dT primer, RT buffer (Invitrogen), 50 μM DTT, 250 μM dNTPs and 24 U RNAsin (Promega) using 200 U SuperScriptTM reverse transcriptase (Invitrogen). One microlitre of cDNA was used in a PCR reaction in 10 mM Tris-HCl (pH 9.6), 50 mM NaCl, 2.5 mM MgCl2, 200 μg/ml BSA, 200 μM dNTPs, 10 pmol primers and 0.8 U AmpliTaq DNA polymerase (Applied Biosystems) in a total volume of 50 μl. Primer sequences are available upon request. PCR products were analysed on agarose gels. PCR products were sequenced as described above.
Neutrophil elastase ELISA
Neutrophil elastase levels were measured using an enzyme-linked immunosorbent assay (ELISA) that detects both free and α1-anti-trypsin inhibitor complexed neutrophil elastase, with a lower limit of detection of 1.5 ng/ml [40]. The ELISA is specific for human neutrophil elastase.
α1-Anti-trypsin phenotyping
α1-Anti-trypsin phenotypes were analysed by standard isoelectric focusing.
LL-37 ELISA
LL-37 in plasma was measured with an ELISA kit (Hycult Biotechnology, Uden, The Netherlands).
Neutrophil purification and migration
Granulocytes were isolated from heparinized venous blood as described [41, 42]. Purity was always greater than 95%. Neutrophil migration was assessed by means of the FluoroBlok inserts (Falcon; Becton Dickinson). Cells (5 × 106/ml) were labelled with 1 μM calcein-AM (Molecular Probes) for 30 min at 37°C, washed twice and resuspended in N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer at a concentration of 2 × 106/ml. Chemoattractant solution (platelet-activating factor (PAF), interleukin 8 (IL-8) and complement 5a (C5a), all at 10 nM) or medium alone (0.8 ml/well) was placed in a 24-well plate, and 0.3-ml cell suspension was delivered to the inserts (3-μm pore size) and placed in the 24-well plate. Cell migration was assessed by measuring fluorescence in the lower compartment at 2.5-min intervals for 45 min with the HTS7000+ plate reader (Perkin Elmer). Maximal slope of migration was estimated over a 10-min interval.
Results
Haematology
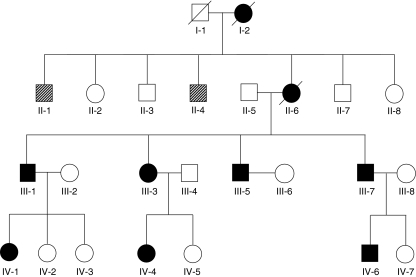
A pedigree of the family was assembled (Fig. 1). Haematological data of patients II-2, III-3 and III-6 have been collected since 1983 and are summarised in Table 1. Haematological data for most other affected individuals and some unaffected family members are presented in Table 1 as well. Based on these data, there is a striking heterogeneity in the neutropenia in this family. Individuals III-1, III-3, III-5, IV-4 and IV-6 can be classified as severely neutropenic. Individual II-6 can be classified as moderately neutropenic although she has had more often severe (n = 8) than mild episodes (n = 5). Individual III-7 can be classified as mildly neutropenic. Although neutrophil counts varied over time in these patients, we did not observe a cyclic effect in neutrophil counts of patient II-6 or III-3 when followed at 2- to 3-day intervals for several weeks.
Fig. 1.
The congenital neutropenia family. Solid symbol, individual affected with congenital neutropenia. Open symbol, unaffected individual. Hatched symbol, status unknown. Individuals I-1, I-2, II-1 and II-4 were not available for genetic analyses
Table 1.
Haematological data
| Average blood counts (>20 years) | Blood counts at last checkup | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II-6 | III-3 | III-7 | II-2 | II-3 | II-7 | II-8 | III-1 | III-5 | III-8 | IV-1 | IV-4 | IV-6 | IV-7 | |
| Leucocytes, in 109/l | 3.6 | 3.9 | 4.8 | 8.8 | 6.4 | 6.7 | 7 | 4.1 | 2.8 | 8.3 | 3.2 | 6.0 | 3.9 | 6.0 |
| Eosinophils (%) | 6.8 | 7.6 | 2.4 | 5.4 | 4.2 | 4.2 | 5.9 | 5.6 | 4.3 | 2.5 | 3.5 | 17.0 | 3.3 | 2.0 |
| Basophils (%) | 1.6 | 1.8 | 0.9 | 0.8 | 0.6 | 0.5 | 0.9 | 2.5 | 2.5 | 0.2 | 1.9 | 1.0 | 1 | 0.5 |
| Neutrophils (%) | 16 | 7.3 | 33 | 59 | 60 | 54 | 56 | 0.3 | 3.4 | 73 | 18 | 6.0 | 11 | 54 |
| Lymphocytes (%) | 55 | 60 | 47 | 27 | 22 | 32 | 29 | 58 | 59 | 19 | 58 | 59 | 65 | 32 |
| Monocytes (%) | 19 | 20 | 14 | 8.6 | 13 | 9.5 | 8.2 | 33 | 31 | 5.4 | 18.4 | 17.0 | 19 | 11.6 |
| l.u.c. (%) | 5.3 | 6.0 | 3.3 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 0.0 | N.D. | N.D. |
| Erythrocytes, in 1012/l | 4.2 | 4.4 | 4.9 | 4.1 | 4.8 | 4.9 | 4.7 | 4.2 | 5.2 | 3.4 | 4.4 | 4.8 | 4.3 | 4.3 |
| Thrombocytes, in 109/l | 371 | 299 | 246 | 253 | 239 | 222 | 273 | 220 | 341 | 193 | 262 | 375 | 227 | N.D. |
| Neutrophils, in 109/l | 0.8 | 0.3 | 1.6 | 5.2 | 3.9 | 3.6 | 3.9 | 0.01 | 0.09 | 6 | 0.6 | 0.4 | 0.4 | 3.2 |
Patients are indicated in bold; healthy family members are indicated in normal font. Average blood counts were based on 27 measurements for II-6 (in nine of which only leucocytes, erythrocytes and neutrophils were counted), 27 measurements for III-3 and 20 measurements for III-7 (in two of which only leucocytes, erythrocytes and neutrophils were counted)
l.u.c. large undifferentiated cells, N.D. not determined
Neutrophil development
Neutropenia can result from reduced production of neutrophils in the bone marrow or from increased apoptosis in the blood and/or from increased clearance from the peripheral tissues. To study the production of neutrophils in the bone marrow, biopsies were taken from individuals II-6, III-1, III-3 and III-5 in 1983 and of II-6 and III-3 again in 2003. The biopsies were analysed with the following result: In III-1 and III-5, the bone marrow material contained few cells, and although all cell types were present, a typical pattern of maturation arrest was observed of neutrophil precursors at the promyelocyte–myelocyte stage with a relative increase in eosinophil cells. The bone marrow from individuals II-6 and III-3 appeared at both occasions rich in cells with all cell types present but again with a typical pattern of maturation arrest of neutrophil precursors at the promyelocyte–myelocyte stage with a relative increase in eosinophil cells.
Neutrophil migration and function
In SCN patients, neutrophil migration has been reported to be decreased [43]. We tested neutrophil function of a severe patient (III-3) and a mild patient (III-7) by measuring directed cell motility (chemotaxis) towards neutrophil-specific stimuli (i.e. C5a, IL-8 or PAF) acting via different members of the chemotaxin receptor superfamily. In this test, the neutrophils did not show abnormalities (data not shown).
Genetic analysis
Since in autosomal dominant-inherited neutropenia mutations have been described in several genes, we decided to perform a small-scale linkage analysis around the ELANE, CSF3R, GFI1 and CSF3 genes. The individuals analysed were II-5, II-6, III-1 to III-5, III-7, III-8 and IV-1 to IV-5. Only the markers around the ELANE gene followed the inheritance pattern of the disorder in this family. We subsequently sequenced all five exons (including at least 20 bases of flanking introns or untranslated regions) from genomic DNA or RNA of patients II-6, III-3 and IV-1. All sequences were wild type except for a heterozygous r.169G>T mutation [44] in exon 2 in the three patients. The presence or absence of the mutation in the complete family was then tested using an AcyI digest on exon 2 PCR products. In patients II-6, III-1, III-3, III-5, III-7, IV-1, IV-4 and IV-6, the mutation was present heterozygously. All healthy family members analysed were homozygous for the wild-type allele. The mutation r.169G>T results in a substitution of an alanine (A) for a serine (S) at amino acid 28 of the mature protein.
Based on the heterogeneity in presentation of the patients in this family, ranging from mild to severe neutropenic, we hypothesised that a variation in another gene might be modifying the clinical outcome of disease severity in these patients. We had observed in the small-scale linkage analysis around ELANE, CSF3R, CSF3 and GFI1 that the severely affected individuals III-1, III-3 and III-5 had inherited a different CSF3 haplotype from their father than the mildly affected III-7. We therefore sequenced the CSF3 gene from individuals II-5, II-6, III-1, III-3, III-5 and III-7. We identified only one variation, r.555G>A in exon 5, that does not affect the amino acid sequence since it is in the third base of the codon for a leucine (CTG) at position 185 in the protein. This variation was present in the mother (II-6) and III-3 but not in the other individuals.
α1-Anti-trypsin phenotype
α1-Anti-trypsin is the major serum inhibitor of neutrophil elastase. Li and Horwitz [21] have shown, using constructs of various ELANE mutations, that the inhibition by α1-anti-trypsin remains similar to that observed for the wild type. However, α1-anti-trypsin deficiency could influence the disease severity between the various patients in this family, which ranges from mild to severe. We therefore analysed the α1-anti-trypsin phenotype in serum from patients III-3 (severe) and III-7 (mild). Both patients were homozygous for the common M variant that is associated with normal α1-anti-trypsin levels.
LL-37 analysis
It was recently reported that LL-37 plasma levels can be used for differential diagnosis of neutropenic disorders [45]. LL-37 levels were measured in plasma from 14 members of this family and 14 unrelated controls (Table 2). In healthy family members and controls, the LL-37 levels ranged from undetectable to 113 ng/ml with an average value of 18 ng/ml (21 individuals analysed). In the granulocytopenia patients, the LL-37 levels ranged from undetectable (in three individuals) to 239 ng/ml with an average value of 53 ng/ml (seven patients analysed). No correlation between severity and LL-37 concentrations was detected.
Table 2.
LL-37 concentration in plasma
| Neutrophil counta (×109/l) | LL-37 in ng/ml | |
|---|---|---|
| Patient III-1 | 0.01 | <10 |
| Patient III-5 | 0.09 | <10 |
| Patient III-3 | 0.3 | 239 |
| Patient IV-1 | 0.6 | 24 |
| Patient II-6 | 0.8 | 24 |
| Patient III-7 | 1.6 | <10 |
| Patient IV-4 | 0.4 | 86 |
| Mean of above 7 patients | 0.5 | 53c |
| 21 healthy controls | N.D.b | <10 to 113 (mean 18)c |
N.D. not determined
aFrom Table 1
bAverage normal neutrophil count is 4 × 109/l
cFor calculating, average <10 was set to 0
Response to G-CSF treatment
G-CSF treatment is known to reverse neutropenia in SCN patients with ELANE mutations. It has, however, been reported that the very low (<20 ng/ml) neutrophil elastase in plasma from SCN patients does not improve upon treatment with G-CSF [46]. Similarly, neutrophil elastase in neutrophils was also observed not to increase upon G-CSF treatment [31]. It has been suggested that expression of granule-associated proteins, such as neutrophil elastase, and whether G-CSF has any effect on this expression may depend on the particular ELANE mutation [47]. We therefore initially administered two different doses of G-CSF (Neupogen) with 3-week interval to patient III-3 to monitor the effect on neutrophil elastase. We determined the neutrophil elastase circulating in the plasma just before, 1 day after and 2 days after subcutaneous G-CSF injection. Before the first G-CSF treatment, circulating neutrophil elastase level in patient III-3 was 14.3 ng/ml; this increased to 25.7 ng/ml 1 day after injection of 300 μg G-CSF and to 28.6 ng/ml at 2 days after injection. Three weeks later, neutrophil elastase levels were 22.5 ng/ml before injection, which increased to 35.1 ng/ml 1 day after injection with 600 μg G-CSF and to 37.4 ng/ml 2 days after injection. We also measured neutrophil elastase concentrations twice in plasma from the mildly neutropenic brother (III-7) who did not receive G-CSF treatment. In his plasma, an average of 49.4 ng/ml neutrophil elastase was present. In healthy controls, neutrophil elastase concentration in plasma is about 300 ng/ml [46].
Patients II-6, III-3 and IV-4 have been receiving 300 μg G-CSF twice weekly for over 6 years, and patients III-1 and II-5 have received 300 μg G-CSF twice weekly for 2 years without adverse effects. They have remained free of major infections and have since had neutrophil counts ranging from 0.6 to 32.8 × 109/l (Table 3). Patients III-7, IV-1 and IV-6 have not received G-CSF treatment.
Table 3.
G-CSF treatment response
| Range of blood countsa | |||||
|---|---|---|---|---|---|
| II-6 | III-1 | III-3 | III-5 | IV-4 | |
| Leucocytes, in 109/l | 5.0–11.6 | 6.3–7.4 | 4.2–12.1 | 4.0–7.9 | 5.5–41.7 |
| Neutrophils, in 109/l | 1.7–7.1 | 1.7–2.1 | 0.4–6.6 | 1.0–3.2 | 0.8–32.8 |
aData are minimum and maximum from nine (II-6), four (III-1), 14 (III-3), three (III-5) and six (IV-4) measurements
Discussion
We investigated a multigenerational family with SCN and found the cause of the disease to be a mutation in the ELANE gene. This is the first unambiguous multigenerational family with autosomal dominant inheritance of SCN. In addition, the inheritance of the disease in this family proves that the ELANE mutation in this family is not a result of a pre-leukemic disorder but the cause of the congenital neutropenia.
ELANE mutations in multigenerational families are common in cyclic neutropenia but have not been found in SCN. Thus far, the vast majority of ELANE mutations in SCN were found in sporadic cases and occasionally a small nuclear family was found in which the mutation was present in one parent and one or two children. In two of the families reported, the parent was found to be mosaic for the ELANE mutation [23, 38]. In one family, the father was neutropenic and developed myelodysplasia, but in the remaining four families, haematological and clinical data of the parents are not discussed, and it remains unclear whether these parents are SCN patients [20]. The authors suggest these familial cases can be either due to dominant inheritance or the result of a new germline mutation [20] but have not proceeded to test for mosaicism.
The mutation found in this family leads to a missense mutation, A28S in the mature protein, that has not been identified before. A similar mutation (A28T) has been described in a sporadic patient with SCN [20]. Serine and threonine are both hydrophilic amino acids that can be phosphorylated while alanine is a hydrophobic amino acid that is not phosphorylated. Li and Horwitz have shown, by expressing constructs of various ELANE mutations in RBL-1 cells, that the A28T mutant protein exerts reduced proteolytic activity and a dominant negative effect similar to the other missense mutations tested [21]. The A28S mutation will likely have a similar effect on the function of neutrophil elastase.
SCN is known to be a heterogeneous disease in which severity and symptoms can widely vary. Here we show that also within one family the neutropenia can vary from mild to severe. We have attempted to identify factors that modify the disease in this family by analysing variations in other candidate genes, in α1-anti-trypsin and in LL-37 concentration but have been unable to find an explanation: A paternal CSF3 haplotype appeared to segregate with severe neutropenia, but no variation in the paternal CSF3 gene was found that could account for the phenotype. The mild and severe patient analysed for α1-anti-trypsin had the same M phenotype. Finally, LL-37 concentrations were not correlated with disease severity.
In contrast to the results reported by Karlsson et al. [45] who found LL-37 concentrations to be invariably higher in controls than in SCN patients, we did not observe this in our samples. The mean LL-37 concentration was even higher in our patients than in controls. Six of the 15 patients in the report by Karlsson et al. had ELANE mutations, and it is unclear from the report which of these were included in the LL-37 measurements; in addition, the mutations in the six patients were different from the mutation in our family. The difference in LL-37 concentrations between Karlsson’s SCN patients and our patients may be due to a number of factors, the most obvious being that we both studied only a limited number of patients with different mutations that may not allow generalisations.
Severe neutropenic patients can be treated with G-CSF to increase the number of neutrophils, reduce the number of infections and increase survival [48]. Unfortunately, long-term treatment with G-CSF does result in an increase in the incidence of leukaemia [49]. It has been suggested in the past that the increase in leukaemia may be due to the G-CSF treatment, but it now appears more likely to be due to increased survival of pre-leukaemic patients [50, 51]. Most of the patients described here are being treated with G-CSF and have not suffered from major infections since. No leukaemia has been reported thus far. One complication that is likely not related to G-CSF therapy was the development of a lethal diffuse large B cell non-Hodgkin’s lymphoma in the brain in patient II-6. Brain lymphomas have not been reported before in the course of congenital neutropenia, or as possible side effect of G-CSF treatment.
G-CSF treatment in patients with ELANE mutations does in general resolve neutropenia but does not always correct the deficiency in granule-associated proteins that is seen during neutropenia [31, 46, 47]. This may be dependent on the type of ELANE mutation in the patient. We observed that neutrophil elastase concentrations in the plasma did not drastically increase in a pilot with either a single or a double dose of G-CSF.
We investigated the first multigenerational family with SCN and found the cause of the disease to be a mutation in the ELANE gene. This is the first unambiguous multigenerational family with autosomal dominant inheritance of SCN, and the inheritance of the disease in this family proves that the ELANE mutation in this family is not a result of a pre-leukaemic disorder but the cause of the congenital neutropenia. Between the affected family members, heterogeneity exists in severity of the disease, which ranged from mild to severe. This may indicate the presence of modifying factors, but we were unable to identify these.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Briars GL, Parry HF, Ansari BM. Dominantly inherited severe congenital neutropenia. J Infect. 1996;33:123–126. doi: 10.1016/S0163-4453(96)93081-9. [DOI] [PubMed] [Google Scholar]
- 2.Reimann HA, de Berardinis CT. Periodic (cyclic) neutropenia, an entity. A collection of sixteen cases. Blood. 1949;4:1109–1116. [PubMed] [Google Scholar]
- 3.Zuelzer WW. Myelokathexis: a new form of chronic granulocytopenia. New Eng J Med. 1964;270:699–704. doi: 10.1056/NEJM196404022701402. [DOI] [PubMed] [Google Scholar]
- 4.Shwachman H, Diamond LK, Oski FA, Khaw KT. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr. 1964;65:645–663. doi: 10.1016/S0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 5.Dong F, Hoefsloot LH, Schelen AM, Broeders CA, Meijer Y, Veerman AJ, Touw IP, Lowenberg B. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci USA. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 7.Tidow N, Pilz C, Teichmann B, Muller-Brechlin A, Germeshausen M, Kasper B, Rauprich P, Sykora KW, Welte K. Clinical relevance of point mutations in the cytoplasmic domain of the granulocyte colony-stimulating factor receptor gene in patients with severe congenital neutropenia. Blood. 1997;89:2369–2375. [PubMed] [Google Scholar]
- 8.Dong F, Dale DC, Bonilla MA, Freedman M, Fasth A, Neijens HJ, Palmblad J, Briars GL, Carlsson G, Veerman AJ, Welte K, Lowenberg B, Touw IP. Mutations in the granulocyte colony-stimulating factor receptor gene in patients with severe congenital neutropenia. Leukemia. 1997;11:120–125. doi: 10.1038/sj.leu.2400537. [DOI] [PubMed] [Google Scholar]
- 9.Ward AC, van Aesch YM, Gits J, Schelen AM, de Koning JP, van Leeuwen D, Freedman MH, Touw IP. Novel point mutation in the extracellular domain of the granulocyte colony-stimulating factor (G-CSF) receptor in a case of severe congenital neutropenia hyporesponsive to G-CSF treatment. J Exp Med. 1999;190:497–507. doi: 10.1084/jem.190.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande RV, Lalezari P, Pergolizzi RG, Moore MA. Structural abnormalities in the G-CSF receptor in severe congenital neutropenia. J Hematother Stem Cell Res. 1999;8:411–420. doi: 10.1089/152581699320171. [DOI] [PubMed] [Google Scholar]
- 11.Tschan CA, Pilz C, Zeidler C, Welte K, Germeshausen M. Time course of increasing numbers of mutations in the granulocyte colony-stimulating factor receptor gene in a patient with congenital neutropenia who developed leukemia. Blood. 2001;97:1882–1884. doi: 10.1182/blood.V97.6.1882. [DOI] [PubMed] [Google Scholar]
- 12.Germeshausen M, Ballmaier M, Schulze H, Welte K, Flohr T, Beiske K, Storm-Mathisen I, Abrahamsen TG. Granulocyte colony-stimulating factor receptor mutations in a patient with acute lymphoblastic leukemia secondary to severe congenital neutropenia. Blood. 2001;97:829–830. doi: 10.1182/blood.V97.3.829. [DOI] [PubMed] [Google Scholar]
- 13.Sinha S, Zhu SS, Romero G, Corey SJ. Deletional mutation of the external domain of the human granulocyte colony-stimulating factor receptor in a patient with severe chronic neutropenia refractory to granulocyte colony-stimulating factor. J Pediatr Hematol Oncol. 2003;25:791–796. doi: 10.1097/00043426-200310000-00010. [DOI] [PubMed] [Google Scholar]
- 14.Druhan LJ, Ai J, Massullo P, Kindwall-Keller T, Ranalli MA, Avalos BR. Novel mechanism of G-CSF refractoriness in patients with severe congenital neutropenia. Blood. 2005;105:584–591. doi: 10.1182/blood-2004-07-2613. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama T, et al. A novel mutation in the juxtamembrane intracellular sequence of the granulocyte colony-stimulating factor (G-CSF) receptor gene in a patient with severe congenital neutropenia augments GCSF proliferation activity but not through the MAP kinase cascade. Int J Hematol. 2005;82:28–34. doi: 10.1532/IJH97.05010. [DOI] [PubMed] [Google Scholar]
- 16.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 17.Link DC, Kunter G, Kasai Y, Zhao Y, Miner T, McLellan MD, Ries RE, Kapur D, Nagarajan R, Dale DC, Bolyard AA, Boxer LA, Welte K, Zeidler C, Donadieu J, Bellanne-Chantelot C, Vardiman JW, Caligiuri MA, Bloomfield CD, DiPersio JF, Tomasson MH, Graubert TA, Westervelt P, Watson M, Shannon W, Baty J, Mardis ER, Wilson RK, Ley TJ. Distinct patterns of mutations occurring in de novo AML versus AML arising in the setting of severe congenital neutropenia. Blood. 2009;110:1648–1655. doi: 10.1182/blood-2007-03-081216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, Papayannopoulou T, Grimes HL, Horwitz M. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochberg JC, et al. Mosaic tetraploidy and transient GFI1 mutation in a patient with severe chronic neutropenia. Pediatr Blood Cancer. 2008;50:630–632. doi: 10.1002/pbc.21094. [DOI] [PubMed] [Google Scholar]
- 20.Dale DC, Person RE, Bolyard AA, Aprikyan AG, Bos C, Bonilla MA, Boxer LA, Kannourakis G, Zeidler C, Welte K, Benson KF, Horwitz M. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. [PubMed] [Google Scholar]
- 21.Li FQ, Horwitz M. Characterization of mutant neutrophil elastase in severe congenital neutropenia. J Biol Chem. 2001;276:14230–14241. doi: 10.1074/jbc.M010279200. [DOI] [PubMed] [Google Scholar]
- 22.Ancliff PJ, Gale RE, Liesner R, Hann IM, Linch DC. Mutations in the ELA2 gene encoding neutrophil elastase are present in most patients with sporadic severe congenital neutropenia but only in some patients with the familial form of the disease. Blood. 2001;98:2645–2650. doi: 10.1182/blood.V98.9.2645. [DOI] [PubMed] [Google Scholar]
- 23.Ancliff PJ, Gale RE, Watts MJ, Liesner R, Hann IM, Strobel S, Linch DC. Paternal mosaicism proves the pathogenic nature of mutations in neutrophil elastase in severe congenital neutropenia. Blood. 2002;100:707–709. doi: 10.1182/blood-2002-01-0060. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi H, Kobayashi M, Nakamura K, Konishi N, Miyagawa S, Sato T, Toyoda H, Komada Y, Kojima S, Todoroki Y, Ueda K, Katoh O. Dysregulation of transcriptions in primary granule constituents during myeloid proliferation and differentiation in patients with severe congenital neutropenia. J Leukoc Biol. 2003;73:225–234. doi: 10.1189/jlb.0902427. [DOI] [PubMed] [Google Scholar]
- 25.Bellanne-Chantelot C, Clauin S, Leblanc T, Cassinat B, Rodrigues-Lima F, Beaufils S, Vaury C, Barkaoui M, Fenneteau O, Maier-Redelsperger M, Chomienne C, Donadieu J. Mutations in the ELA2 gene correlate with more severe expression of neutropenia: a study of 81 patients from the French Neutropenia Register. Blood. 2004;103:4119–4125. doi: 10.1182/blood-2003-10-3518. [DOI] [PubMed] [Google Scholar]
- 26.Sera Y, Kwaguchi H, Nakamura K, Sato T, Habara M, Okada S, Ishikawa N, Kojima S, Katoh O, Kobayashi M. A comparison of the defective granulopoiesis in childhood cyclic neutropenia and in severe congenital neutropenia. Haematologica. 2005;90:1032–1041. [PubMed] [Google Scholar]
- 27.Thomas M, Jayandharan G, Chandy M. Molecular screening of the neutrophil elastase gene in congenital neutropenia. Indian Pediatr. 2006;43:1081–1084. [PubMed] [Google Scholar]
- 28.Boxer LA, Stein S, Buckley D, Bolyard AA, Dale DC. Strong evidence for autosomal dominant inheritance of severe congenital neutropenia associated with ELA2 Mutations. J Pediatr. 2006;148:633–636. doi: 10.1016/j.jpeds.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Salipante SJ, Benson KF, Luty J, Hadavi V, Kariminejad R, Kariminejad MH, Rezaei N, Horwitz MS. Double de novo mutations of ELA2 in cyclic and severe congenital neutropenia. Hum Mutat. 2007;28:874–881. doi: 10.1002/humu.20529. [DOI] [PubMed] [Google Scholar]
- 30.Grenda DS, Murakami M, Ghatak J, Xia J, Boxer LA, Dale D, Dinauer MC, Link DC. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donini M, Fontana S, Savoldi G, Vermi W, Tassone L, Gentili F, Zenaro E, Ferrari D, Notarangelo LD, Porta F, Facchetti F, Notarangelo LD, Dusi S, Badolato R. G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood. 2007;109:4716–4723. doi: 10.1182/blood-2006-09-045427. [DOI] [PubMed] [Google Scholar]
- 32.Shiohara M, Shigemura T, Saito S, Tanaka M, Yanagisawa R, Sakashita K, Asada H, Ishii E, Koike K, Chin M, Kobayashi M, Koike K. Ela2 mutations and clinical manifestations in familial congenital neutropenia. J Pediatr Hematol Oncol. 2009;31:319–324. doi: 10.1097/MPH.0b013e3181984dbe. [DOI] [PubMed] [Google Scholar]
- 33.Lee ST, Yoon HS, Kim HJ, Lee JH, Park JH, Kim SH, Seo JJ, Im HJ. A novel mutation Ala57Val of the ELA2 gene in a Korean boy with severe congenital neutropenia. Ann Hematol. 2009;88:593–595. doi: 10.1007/s00277-008-0629-y. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg PS, Alter BP, Link DC, Stein S, Rodger E, Bolyard AA, Aprikyan AA, Bonilla MA, Dror Y, Kannourakis G, Newburger PE, Boxer LA, Dale DC. Neutrophil elastase mutations and risk of leukaemia in severe congenital neutropenia. Br J Haematol. 2008;140:210–213. doi: 10.1111/j.1365-2141.2007.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donadieu J, Leblanc T, Bader-Meunier B, Barkaoui M, Fenneteau O, Bertrand Y, Maier-Redelsperger M, Micheau M, Stephan JL, Phillipe N, Bordigoni P, Babin-Boilletot A, Bensaid P, Manel AM, Vilmer E, Thuret I, Blanche S, Gluckman E, Fischer A, Mechinaud F, Joly B, Lamy T, Hermine O, Cassinat B, Bellanne-Chantelot C, Chomienne C, French Severe Chronic Neutropenia Study Group Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90:45–53. [PubMed] [Google Scholar]
- 36.Matsushita H, Asai S, Komiya S, Inoue H, Yabe H, Miyachi H. A family of severe congenital neutropenia with −199c to a substitution in ELA2 promoter. Am J Hematol. 2006;81:985–986. doi: 10.1002/ajh.20637. [DOI] [PubMed] [Google Scholar]
- 37.Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Germeshausen M, Schulze H, Ballmaier M, Zeidler C, Welte K. Mutations in the gene encoding neutrophil elastase (ELA2) are not sufficient to cause the phenotype of congenital neutropenia. Br J Haematol. 2001;115:222–224. doi: 10.1046/j.1365-2141.2001.03069.x. [DOI] [PubMed] [Google Scholar]
- 39.Jeha S, Chan KW, Aprikyan AG, Hoots WK, Culbert S, Zietz H, Dale DC, Albitar M. Spontaneous remission of granulocyte colony-stimulating factor-associated leukemia in a child with severe congenital neutropenia. Blood. 2000;96:3647–3649. [PubMed] [Google Scholar]
- 40.Hiltermann JTN, Lapperre TS, van Bree L, Steerenberg PA, Brahim JJ, Sont JK, Sterk PJ, Hiemstra PS, Stolk J. Ozone-induced inflammation assessed in sputum and bronchial lavage fluid from asthmatics: a new noninvasive tool in epidemiologic studies on air pollution and asthma. Free Radic Biol Med. 1999;27:1448–1454. doi: 10.1016/S0891-5849(99)00191-4. [DOI] [PubMed] [Google Scholar]
- 41.Kuijpers TW, Maianski NA, Tool ATJ, Smit GP, Rake JP, Roos D, Visser G. Apoptotic neutrophils in the circulation of patients with glycogen storage disease type 1b (GSD1b) Blood. 2003;101:5021–5024. doi: 10.1182/blood-2002-10-3128. [DOI] [PubMed] [Google Scholar]
- 42.Kuijpers TW, Maianski NA, Tool ATJ, Becker K, Plecko B, Valianpour F, Wanders RJA, Pereira R, Van Hove J, Verhoeven AJ, Roos D, Baas F, Barth PG. Neutrophils in Barth syndrome (BTHS) avidly bind annexin-V in the absence of apoptosis. Blood. 2004;103:3915–3923. doi: 10.1182/blood-2003-11-3940. [DOI] [PubMed] [Google Scholar]
- 43.Elsner J, Roesler J, Emmendorffer A, Zeidler C, Lohmann-Matthes M, Welte K. Altered function and surface marker expression of neutrophils induced by rhG-CSF treatment in severe congenital neutropenia. Eur J Haematol. 1992;48:10–19. doi: 10.1111/j.1600-0609.1992.tb01787.x. [DOI] [PubMed] [Google Scholar]
- 44.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson J, Carlsson G, Ramme KG, Hägglund H, Fadeel B, Nordenskjöld M, Henter JI, Palmblad J, Pütsep K, Andersson M. Low plasma levels of the protein pro-LL-37 as an early indication of severe disease in patients with chronic neutropenia. Br J Haematol. 2007;137:166–169. doi: 10.1111/j.1365-2141.2007.06530.x. [DOI] [PubMed] [Google Scholar]
- 46.Skokowa J, Fobiwe JP, Dan L, Thakur BK, Welte K. Neutrophil elastase is severely down-regulated in severe congenital neutropenia independent of ELA2 or HAX1 mutations but dependent on LEF-1. Blood. 2009;114:3044–3051. doi: 10.1182/blood-2008-11-188755. [DOI] [PubMed] [Google Scholar]
- 47.Andersson M, Karlsson J, Carlsson G, Putsep K. Expression of granule-associated proteins in neutrophils from patients with severe congenital neutropenia. Blood. 2007;110:2772–2773. doi: 10.1182/blood-2007-03-083063. [DOI] [PubMed] [Google Scholar]
- 48.Bonilla MA, Gillio AP, Ruggeiro M, Kernan NA, Brochstein JA, Abboud M, Fumagalli L, Vincent M, Gabrilove JL, Welte K, et al. Effects of recombinant human granulocyte colony-stimulating factor on neutropenia in patients with congenital agranulocytosis. N Engl J Med. 1989;320:1574–1580. doi: 10.1056/NEJM198906153202402. [DOI] [PubMed] [Google Scholar]
- 49.Rosenberg PS, Zeidler C, Bolyard AA, Alter BP, Bonilla MA, Boxer LA, Dror Y, Kinsey S, Link DC, Newburger PE, Shimamura A, Welte K, Dale DC. Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br J Haematol. 2010;150:196–199. doi: 10.1111/j.1365-2141.2010.08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonilla MA, Dale D, Zeidler C, Last L, Reiter A, Ruggeiro M, Davis M, Koci B, Hammond W, Gillio A, et al. Long-term safety of treatment with recombinant human granulocyte colony-stimulating factor (r-metHuG-CSF) in patients with severe congenital neutropenias. Br J Haematol. 1994;88:723–730. doi: 10.1111/j.1365-2141.1994.tb05110.x. [DOI] [PubMed] [Google Scholar]
- 51.Freedman MH, Bonilla MA, Fier C, Bolyard AA, Scarlata D, Boxer LA, Brown S, Cham B, Kannourakis G, Kinsey SE, Mori PG, Cottle T, Welte K, Dale DC. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96:429–436. [PubMed] [Google Scholar]