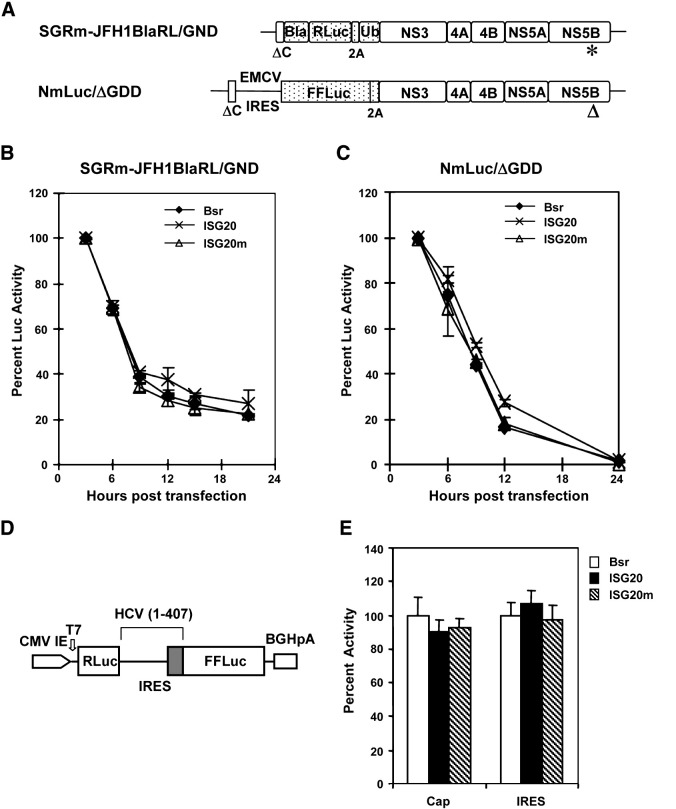
Fig. 3.
ISG20 does not accelerate the degradation of transfected HCV RNAs, nor does it inhibit HCV IRES-directed translation. (A) Schematic representation of the replication-incompetent, luciferase-encoding, subgenomic HCV RNAs derived from JFH1 (SGRm-JFH1BlaRL/GND) and HCV-N (NmLuc/ΔGDD) sequences. SGRm-JFH1BlaRL/GND is identical to SGRm-JFH1BlaRL (Fig. 2A) except for the aa mutation in the GDD motif (to GND) in NS5B (indicated by “*”). In NmLuc/ΔGDD, the GDD motif in NS5B is deleted (indicated by “Δ”). Translation of Firefly luciferase (FFLuc), FMDA 2A autoprotease and the downstream NS3 to NS5B polyprotein is directed by EMCV IRES. (B) and (C) 7.5-Bsr, 7.5-ISG20 and 7.5-ISG20m cells were transfected with in vitro transcribed SGRm-JFH1BlaRL/GND (B) or NmLuc/ΔGDD (C) RNAs. At indicated time points post transfection, cells were lysed and subjected to Renilla (B) or Firefly (C) luciferase activity assays. HCV RNA degradation is reflected in the percent decrease in luciferase activity (normalized to that at 3 h post-transfection, when luciferase activity results from translation of initial input HCV RNA). (D) Schematic showing the organization of the bicistronic reporter construct, pRL-HL. CMV-IE: CMV immediate early promoter; BGHpA: bovine growth hormone polyadenylation signal. (E) 7.5-Bsr, 7.5-ISG20, and 7.5-ISG20m cells in triplicate wells were transfected with pRL-HL for 24 h and then lysed for dual luciferase assay determining the translation efficiencies directed by 5′cap (RLuc) and HCV IRES (FFLuc), respectively. Results are normalized to the luciferase activity from transfected Bsr cells (100%).