Abstract
Background
Improved outcome measures in systemic sclerosis are critical to finding active therapeutics for this disease. The modified Rodnan skin score (MRSS) is the current standard for evaluation of skin disease in systemic sclerosis, but is not commonly employed in the clinical setting in part because it requires specialized training to perform accurately and consistently between different physicians. We have investigated whether skin gene expression might serve as a more objective, surrogate outcome measure to supplement skin score evaluations.
Methods
Skin RNAs from a group of patients with diffuse cutaneous systemic sclerosis were studied for expression levels of known transforming growth factor-beta (TGFβ) and interferon (IFN)-regulated genes. These levels were correlated with the MRSS using multiple regression analyses to obtain best-fit models.
Results
Skin expression of TGFβ-regulated genes, cartilage oligomeric matrix protein (COMP) and thrombospondin-1 (THS1) correlated moderately well with the MRSS, but the addition of other TGFβ-regulated genes, failed to significantly improve best-fit models. IFN-regulated genes were also found to correlate with the MRSS and the addition of interferon-induced 44 (IFI44) and sialoadhesin (SIG1) to COMP and THS1 in multiple regression analyses significantly improved best-fit modelsachieving an R2= 0.89. These results were validated using an independent group of skin biopsies. Longitudinal scores using this four-gene biomarker indicated that it detects change over time corresponding to changes in the MRSS.
Conclusions
We describe a four-gene predictor of the MRSS and validate its performance. This objective measure of skin disease could provide a strong surrogate outcome measure for patient care and for clinical trials.
Biomarkers are increasingly recognized as important supplements to clinical evaluations and are particularly important to consider in rheumatologic diseases, in which clinical disease can be hard to define accurately and quantitatively. Such measures can be especially useful in clinical trials, in which changes in disease status must be assessed over time. Thus, increased accuracy of outcome measures provides better discrimination of change, resulting in the potential to decrease subject enrollment or improve study power. Surrogate outcome measures are already components of commonly used measures of rheumatoid arthritis and systemc lupus erythematosus disease activity(1–3). The difficulty in carrying out large studies on SSc patients makes better outcome measure particularly important.
The modified Rodnan skin score (MRSS) has become the standard primary outcome in most recent studies of therapeutics targeting skin disease for diffuse cutaneous systemic sclerosis (SSc). Inter and intra observer variability is relatively low(4) and it has been adjudicated as a valid measure by OMERACT (Outcome Measures in Rheumatologic Clinical Trials) consensus (5, 6). Despite the utility of the MRSS, it has limitations. Consistent evaluation of the MRSS requires careful training of scorers(7) andinterobserver scoring variability is significantly higher than intraobserver variability(8), requiring availability of consistent scorer(s) at clinical study sites and potentially adding variability to scoring in multicenter studies.
A variety of changes in cytokines and collagen metabolites have been found in sera and/or urine from patients with SSc. SSc patients have on average increased levels of endothelin-1, interleukins-4, -6,-10, -12, -13, -17, and MCP-1, but these have not been shown to correlate with clinical markers adequately to be used as surrogate outcome measures (9–11). Collagen metabolites, serum amino-terminal procollagens type I and III(12, 13), andurinary desmosine and isodesmosine (14) are increased in SSc patients. However, these markers alsocorrelate only modestly with the MRSS(13, 15). Thus, no biomarker has been generally embraced for use in SSc.
Biomarkers also have the potential to reveal insights into underlying disease pathogenesis. Biochemical changes that correlate with changes in clinical measures of disease activity are more likely involved in pathogenesis. Several cytokines have been implicated in disease pathogenesis in SSc patients. Transforming growth factor beta (TGFβ) is the most potent known pro-fibrotic cytokine, stimulating the development of myofibroblasts, a profibrotic cell type(16). We have previously shown that the degree of infiltration of skin with myofibroblasts and expression of the TGFβ-regulated gene, cartilage oligomeric protein (COMP)correlate highly with the mRSS(17, 18). These data further implicate TGFβ in disease pathogenesis and indicate that a skin biopsy at a “landmark” site on the mid-forearm provides information on disease status over all lesional skin as defined by the MRSS. We have also shown that some patients with SSc show increased expression of IFN-regulated genes in peripheral blood mononuclear cells (PBMCs)(19), suggesting that type I IFNs might also play a role in SSc pathogenesis as has been implicated in systemic lupus erythematosus and other autoimmune diseases.
We report here studies on skin expression of a variety of TGFβ- and IFN-regulated genes. These studies provide strong evidence that specific genes within each of these groups provide complementary information, permitting the development of a highly informative surrogate outcome measure for skin disease in patients with SSc.
MATERIAL AND METHODS
Study subjects
All study subjects met criteria for dcSSC with proximal skin disease as defined previously (20). The study was conducted under a protocol approved by the Boston University Medical Center Institutional Review Boardand all subjects gave written informed consent. 3 mm punch skin biopsies were performed over the dorsal mid-forearm (lesional skin), or shoulder or back (non-lesional skin) and placed immediately into RNAlater (Qiagen). Samples in RNAlater were stored at −20°C until preparation of RNA, within 6 months of the biopsy. The MRSS was evaluated on each patient on the day of the biopsy.
RNA preparation and RT-PCR
Tissues were transferred into 600 µl of RLT buffer (Qiagen, Valencia, CA), minced and disrupted using a Polytron homogenizer. RNAs were purified from RLT buffer supernatants using the RNeasy total RNA kit (Quiagen, Valencia, CA). cDNAs were synthesized from 0.05-0.01 µg of total RNA using Superscript II RNase H− reverse transcriptase and random primers (Invitrogen Life Technologies, Rockville, MD). All RT-PCR was carried out using a Prism 7700 Sequence Detector and primers (see supplemental methods) as recommended by the supplier (Applied BioSystems). Expression was normalized to 18S rRNA expression(human 18S TaqMan primer set) assayed in the same samples. Changes in the relative expression of each gene were calculated using ΔΔCt formula, choosing a healthy donor sample as the control(21). Fold change in all samples was normalized to this single healthy skin, so that fold changes in other healthy skin as well as SSc samples were compared to this control.
Primers (synthesized by Integrated DNA Technologies) for quantitative real-time polymerase chain reaction (RT-PCR) were designed using Primer Express software (Applied BioSystems): COMP forward: 5` AGC ACC GGC CCC AAG T 3`; reverse: 5` GGT TGT GCC AAG ACC ACG TT 3`; PAI1 forward: 5` AGC TCA TCA GCC ACT GGA AAG 3`; reverse: 5` GGA GGA CTT GGG CAG AAC CA 3`, THS1 forward: 5` CAC AGT TCC TGA TGG AGA ATG C 3`; reverse: CAT GGA GAC CAG CCA TCG T 3`; CTGF forward, 5` TGT GTG ACG AGC CCA AGG A 3`; reverse: 5` TCT GGG CCA AAC GTG TCTT C 3`; COL4 forward: 5`GCA AAT GTG ACT GCC ATG GA3`, reverse: 5` GAA ACC CAA TGA CAC CTT GTA ACC 3`. COMP, PAI1 CTGF, COL4 and THS1 mRNA expression was measured using SYBR Green. For these amplifications Sybr Green target genes and Taqman control amplifications were run in separate wells on the same PCR plate. To assure the specificity of the COMP, PAI-1 CTGF, COL4 and THS1 primer sets, amplicons generated from PCR reactions were analyzed for specific melting temperatures. For IFI44, MX11, OAS2 and SIG1 TaqMan primers and probes were used (Applied Biosystems) with target and control reactions run on separate wells of the same PCR plate.
Statistical analyses
Statistical significance for differences between SSc and control mRNA gene expression was determined by the Mann-Whitney test using SPSS (SPSS Inc., Chicago, IL). Multiple regression analyses and best-fit modeling were performed using Minitab 15 Statistical Software (Minitab Inc., State College, PA). Further statistical analyses for gene expression correlations with the MRSS and graphing were carried out using Excel (Microsoft, Seattle, WA). The biomarker skin score was calculated using the 4-gene best fit equation: mRSS = 1.49 + 0.20(COMP) + 1.19(THS1) + 0.267(SIG1)+ 1.59(IFI44), where the value for each gene is the fold-change by RT-PCR normalized to 18S.
Validating the biomarker
To validate the predictor, RT-PCR was completed simultaneously on 12 test samples RNA from skin not previously analyzed, as well as on 7 samples that had been amplified previously and that were part of the original predictor (internal controls). These internal controls were used to create a regression equation for each gene, comparing the previous and new PCR expression of these controls. Test gene expression levels were normalized using these regression equations, and then used to calculate the four-gene biomarker using the equation above.
RESULTS
Expression of TGFβ-regulated genes in SSc skin correlates with the mRSS
We have previously shown that COMP, a gene regulated by TGFβ, and myofibroblasts, a fibroblast phenotype associated with TGFβ stimulation, each correlate with the mRSS(17, 18). To clarify whether expression of other TGFβ-regulated genes might allow us to identify a biomarker that correlates with skin score better than these measures, we tested mRNA expression in the skin of diffuse cutaneous SSc patients (see demographics and disease duration in Table I-supplemental) of a series of other genes known from our previous studies to be particularly responsive to TGFβ(22, 23): connective tissue growth factor (CTGF, also known as CCN2), plasminogen activator inhibitor-1 (PAI1), thrombospondin-1 (THS1) and type IV, alpha 1 collagen (COL4).
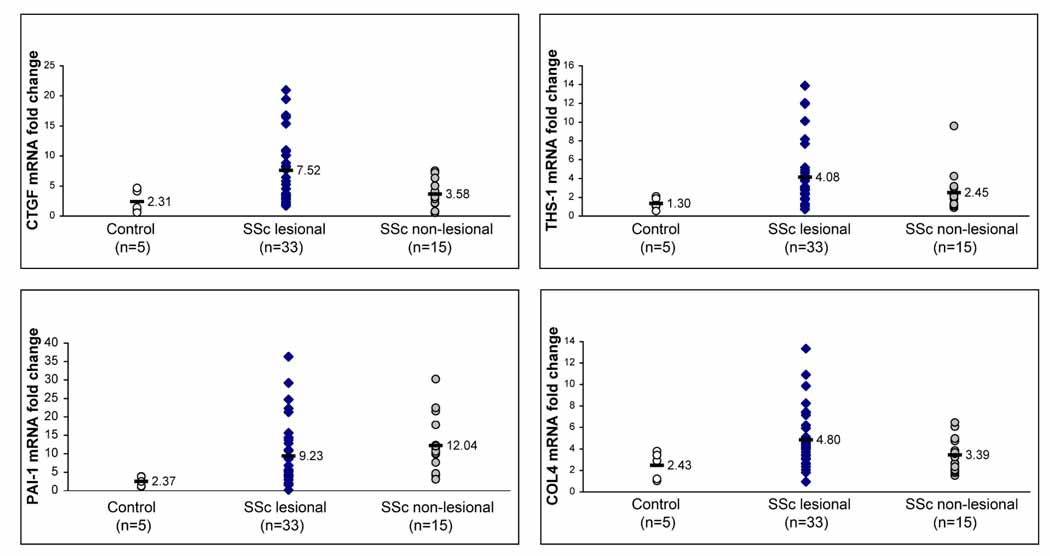
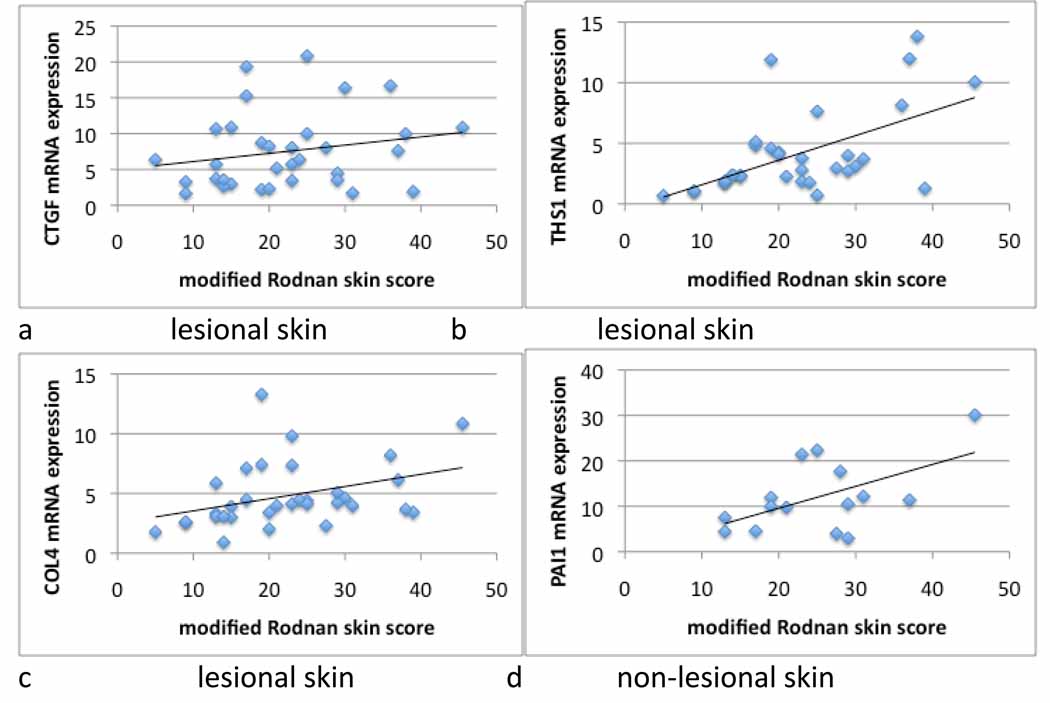
As we have shown previously with COMP, average expression for each of these genes was higher in both lesional and non-lesional SSc skin compared to healthy control skin, although in non-lesional skin only the increase in PAI1 expression reached statistical significance (Fig. 1). All of the genes except PAI1 showed higher average expression levels in lesional than non-lesional skin. When the expression of each of these genes in lesional skin was analyzed by linear regression, several were found to correlate significantly with the MRSS. As we have shown previously, COMP levels correlate moderately well with the MRSS (Table I and(18)). We found that THS1 and COL4 also correlated moderately well with the mRSS (R2=0.32 and 0.13, respectively), suggesting that TGFβ-regulated gene expression is, more generally, a good marker for the extent of skin disease in patients with SSc (Fig, 2, panels b and c). However, expression of PAI1 and CTGF in lesional skin correlated very weakly with the mRSS (Table II, R2=0.05 for both and Fig. 2a (CTGF) and Fig. 1 - supplemental (PAI1)).
Figure 1. Expression of TGFβ-regulated genes in skin from patients with dcSSc.
mRNA expression of TGFβ-regulated genes, CTGF, THS1, COL4 and PAI1, in lesional (33 biopsies) and non-lesional (15 biopsies) skin from dcSSc patients and healthy controls (5 biopsies). Fold-change shown on the graph is normalized to mRNA expression by one of the healthy controls. The average fold-change compared to this control reference sample of CTGF, THS1, PAI1 and COL4 in lesional dcSSc skin (7.52, 4.08, 9.23 and 4.80, respectively) compared to the average fold-change in control, healthy skin compared to the control reference sample (2.31, 1.30, 2.37 and 2.43, respectively) was increased for CTGF (3.25-fold increase, p=0.014), THS1 (3.14-fold increase, p=0.012), COL4 (3.89-fold increase, p=0.028) and PAI1 (1.97-fold increase, p=0.016). Only PAI1 showed statistically significant increased expression in non-lesional skin compared to healthy control skin (5.08-fold increase, p<0.001).
TABLE I.
Correlations between modified Rodnan skin score and skin gene expression (R2(p-value))
| TGFβ-regulated genes | |||||
|---|---|---|---|---|---|
| COMP | CTGF | THS1 | PAI1 | COL4 | |
| Lesional skin (n=33) | 0.41 (0.00004) | 0.05 (0.24) | 0.32 (0.0006) | 0.05 (0.22) | 0.13 (0.038) |
| Non-lesional skin (n=15) | 0.0003 (0.95) | 0.13 (0.18) | 0.0002 (0.96) | 0.30 (0.034) | 0.12 (0.15) |
| Interferon-regulated genes | |||||
| MX1 | OAS2 | IFI44 | SIG1 | ||
| Lesional skin (n=18) | 0.30 (0.18) | 0.33 (0.013) | 0.41 (0.004) | 0.17 (0.068) | |
Figure 2. Linear regression in skin from patients with dcSSc.
mRNA expression in lesional (CTGF, THS1 and COL4) or non-lesional (PAI1) skin related to the mRSS in subjects with dcSSc (see also Table II).
To test the possibility that gene expression in non-lesional skin might better reflect overall skin involvement, we also tested the correlation of COMP, THS1, CTGF or PAI1 mRNA expression in non-lesional skin with the MRSS. Notably PAI1, which correlated weakly with the mRSS in lesional skin, correlated relatively highly with the mRSS in non-lesional skin (Table I and Fig. 2d, R2 = 0.30 p=0.034). CTGF and COL4 expression in non-lesional skin correlated weakly with the mRSS, and COMP and THS1 expression in non-lesional skin showed no correlation with the mRSS (Table I and Fig. 1-supplemental).
Inclusion of expression levels of multiple TGFβ-regulated genes improves prediction models
We postulated that multiple markers of TGFβ activity might enhance the correlation of gene expression and the mRSS and thus tested different models of COMP, CTGF, THS1, COL4 and PAI1 gene expression using multiple regression analysis. In an initial model, all 5 genes and cross products were tested, yielding a best-fit equation with R2 of 0.82 (data not shown). However, the p-values for each of the individual terms in this equation were greater than 0.2 for most of the terms, making it unlikely that such an equation would correlate with future skin scores with this level of accuracy.
To provide a more conservative estimate of the how accurately TGFβ-regulated gene expression might be used to predict mRSS, the best fit calculation was re-done using only the parameter genes from the initial analysis with p<0.2 and gene cross-products with p<0.1. This left only the constant, COMP, THS1, CTGF, and COMP/THS and COL4/THS1 cross products. The best fit linear equation using these values provided an R2 = 0.621, but several more of the parameters now showed p>0.2. Thus, again, parameters were eliminated showing p>0.1, leaving only COMP (p<0.001), THS1 (p=0.004), the COMP/THS1 cross product (p=0.073) and the constant (p=0.066). The best fit recalculated using these parameters provided an R2 of 0.602.
Several other variations were tested. Leaving out the COMP/THS1 cross product (using only COMP and THS1 expression) decreased the R2 to 0.55. Using COMP, THS1 and COMP/THS1 cross product in a power-law fit also slightly increased the quality of the fit (R2=0.616). However, all of the power (squared) parameters in this analysis showed p>0.4. In summary, using these five TGFβ-regulated genes, it appeared that the best model for predicting the mRSS using gene expression from lesional skin, included only COMP, THS1 and the COMP/THS1 cross product, giving a moderately high R2= 0.602.
Interferon-regulated genes correlate with the mRSS
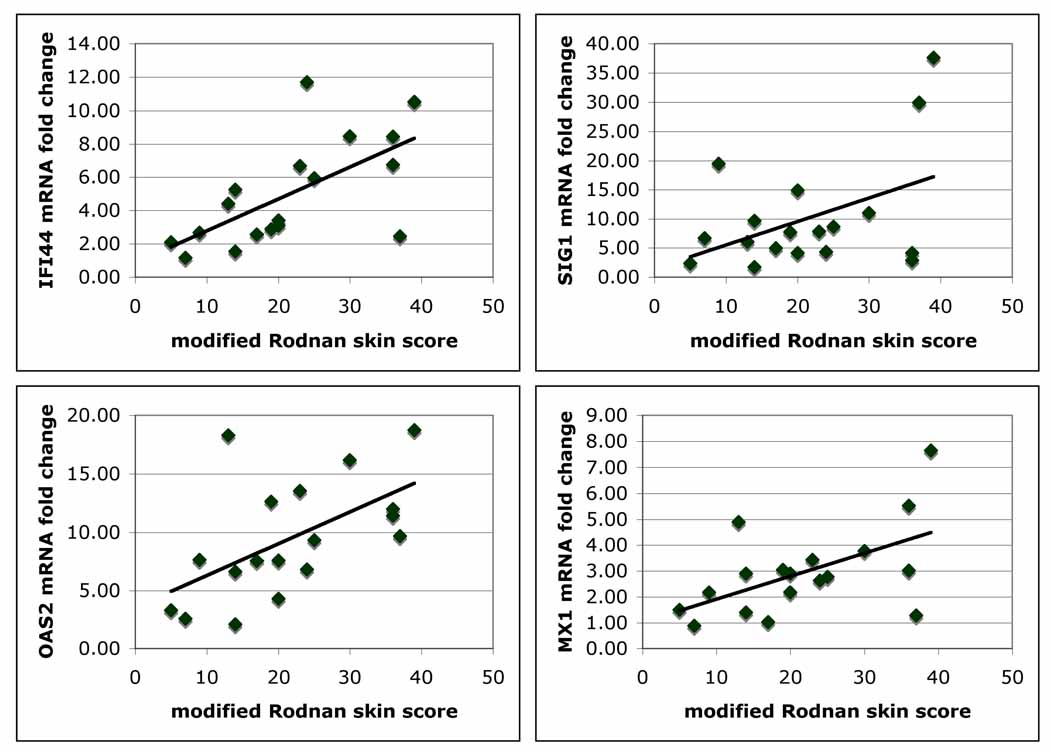
We have shown that peripheral blood mononuclear cells from SSc patients show increased interferon (IFN)-regulated gene expression(19). We therefore reasoned that these genes might also be upregulated in the skin of SSc patients. Because better correlations were found with TGFβ-regulated genes tested in lesional compared to non-lesional skin and ideally a biopsy of a single site would be much more practical as a biomarker, we focused our further studies and analyses on gene expression in lesional skin. We found that expression in the skin of several IFN-regulated genes we had found previously upregulated in SSc PBMCs correlated moderately well and significantly with the mRSS (see Table I and Fig. 3): MX1 (R2=0.30, p=0.018), OAS2 (R2=0.33 p=0.013), and IFI44 (R2=0.41, p=0.004). Siglec1 (SIG1, also known as sialoadhesin) expression, which we have shown is highly upregulated on circulating SSc monocytes and skin macrophages(19), correlated more weakly with the mRSS (R2=0.17, p=0.068).Other genes tested included IFN-regulated genes (IFITM1, CCL2 (MCP-1) and CXCL9), and other genes potentially important in immune regulation in skin, some of which have been seen increased in skin in a previous micorarray study of skin (24): TLR7, CD8B1, SOCS3 IGLJ3 (Ig lambda joining region 3), ADAM19, TNFSR12, TLR3, SPHK1 and IL3RA (CD123). These genes showed weak or no correlations with the MRSS (data not shown).
Figure 3. Expression of IFN-regulated genes in skin from patients with dcSSc.
Linear regression of mRNA expression of IFN-regulated genes from lesional skin of patients with dcSSc with the mRSS: IFI44 (R2=0.41, p=0.004), SIG1 (R2=0.17, p=0.068), OAS2 (R2=0.33 p=0.013) and MX1 (R2=0.30, p=0.018).
Expression of TGFβ- and IFN-regulated genes together correlate strongly with the MRSS
To test whether IFN-regulated gene expression provides additional information for designing a predictor for SSc skin disease, we correlated expression levels of IFN-regulated genes: OAS2, SIG1, IFI44 and MX1, and the TGFβ-regulated genes shown to be important in the predictor above: COMP and THS1, with the mRSS. We first included all 6 of these genes in the multiple regression analysis, giving a best-fitR2=0.896. Although, the p-values for COMP, THS1, SIG1 and IFI44 were 0.11, 0.02, 0.06 and 0.003, respectively, the p-values for MX1 and OAS2 were 0.4 and 0.46, respectively, indicating that these latter two genes were not contributing significantly (data not shown). Examining all possible five-gene fits gave similar results with a clear dichotomy between COMP, THS1, SIG1 and IFI44, in most cases giving p-values less than 0.05, and MX1 and OAS2, in most cases giving p-values greater than 0.4. Thus, although all the five-gene fits produced high R2 values between 0.762 and 0.896, the p-values suggested that MX1 and OAS2 were not likely contributing significantly to the predictor.
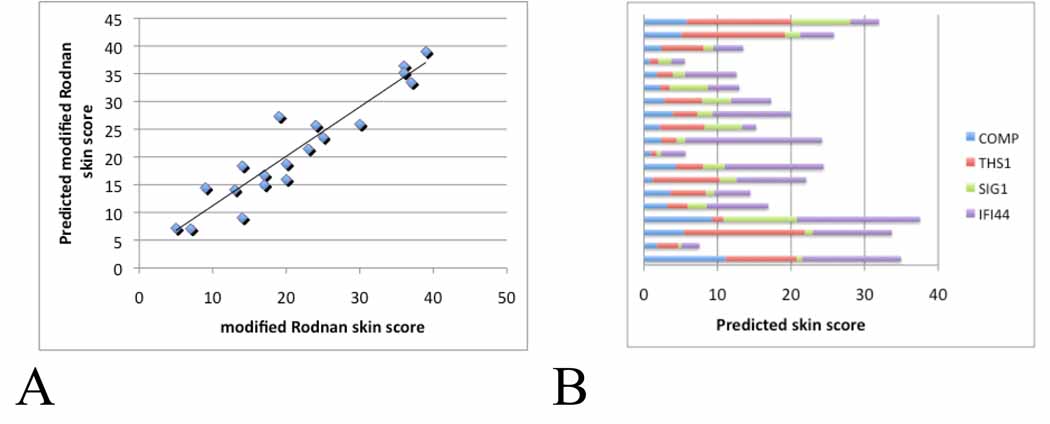
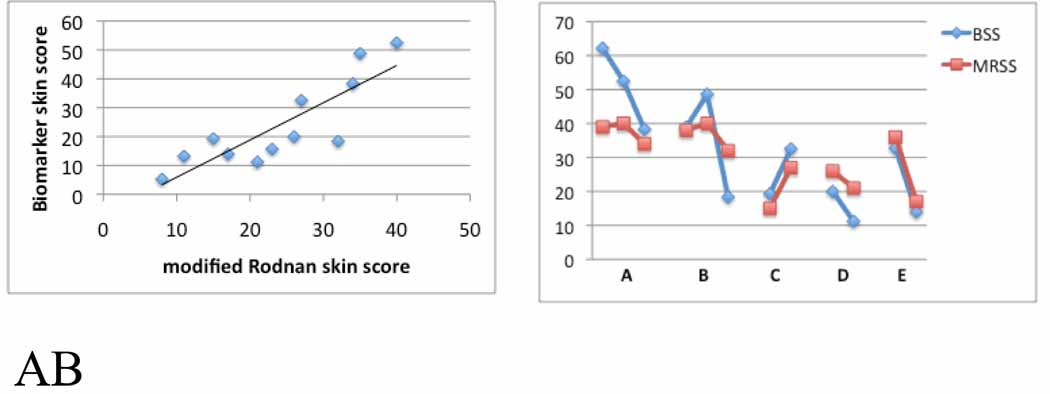
Consistent with this assessment, three- and four-gene regression analyses best-fits that included all or three of the genes COMP, THS1, SIG1 and IFI44 showed correlations with the mRSS that were nearly as high as those of the five- and six-gene regression analyses best-fits (adta not shown). Including all four of these genes showed a very high correlation with the MRSS, with R2=0.89 (Fig. 4A). The relative weights given to the gene expression in the four-gene best-fit equation shows the variability between samples and highlights the requirements for this complimentary set of genes in achieving this accurate predictor of the skin score (Fig.4B).
Figure 4. Multiple linear regression of four-gene biomarker with the MRSS in patients with dcSSc.
Panel A: Best-fit model of skin gene expression of COMP, THS1, IFI44 and SIG1 with the MRSS. Panel B: The contribution of expression of each gene, COMP, THS1, IFI44 and SIG1, to the biomarker predicted skin score. The contribution of each gene to the overall skin score was calculated by multiplying the PCR fold-change value for each gene by the constant associated with that value in the best-fit regression equation. For the 4-gene biomarker this equation takes the form: mRSS = 1.49 + 0.200(COMP) + 1.19(THS1) + 0.267(SIG1)+ 1.59(IFI44). Each bar represents the biomarker predicted skin score of one patient.
Validating the biomarker
To further validate the utility of this biomarker the regression equation derived from the initial analysis was utilized on a new dataset. Gene expression was measured from 12 additional, lesional skin biopsies, separate from the ones used to generate the predictor. Comparing predicted skin scores with clinical MRSS confirmed a strong correlation (Fig. 5A, R2=0.73).
Figure 5. Validation of the four-gene biomarker for the skin score.
Panel A: 4-gene biomarker prediction of the skin score using skin biopsies (n=12), independent from those used to develop the biomarker. The biomarker skin score was calculated from normalized mRNA expression of COMP, THS1, IFI44 and SIG1, using the best-fit equation determined by multiple linear regression shown in Fig. 3. Panel B: Comparison of changes in the MRSS (red lines) with changes in the four-gene biomarker (BSS, blue lines) over time in 5 patients with dcSSc. Patients A and B, and C, D and E show biomarker and MRSS, respectively, at baseline, 6 and 12 months, or at baseline and 6 months.
Some of the skin samples in the final series of 12 test and 7 normalization samples tested, included biopsies taken from the same patients at approximately 6-month intervals. The 4-gene biomarker was compared with change in the MRSS to further indicate the utility of the biomarker in longitudinal studies. The four-gene biomarker in all 5 patients in whom such data was available paralleled the change in skin score. This included both patients showing improvement and progression in skin score (pts. C, D and E; Fig. 5B). In some cases, the biomarker appeared to exaggerate (pt. A) or presage (pt. B) changes detected by the MRSS.
DISCUSSION
We identify a four-gene biomarker that is highly predictive of the MRSS in patients with diffuse cutaneous SSc. We propose that inclusion of this biomarker in clinical assessments and clinical trials of SSc skin disease would provide a valuable surrogate outcome measure. The biomarker changes dynamically in parallel with the MRSS, but does not require any specialized training and provides an objective measure of change over time. We anticipate that ongoing development of this four-gene biomarker will make this measure widely available for future evaluations of SSc skin disease.
The observed differences of TGFβ-regulated gene expression and the correlation of these values with the MRSS are not surprising in light of our previous data showing that the MRSS correlates with dermal myofibroblast infiltration and COMP expression(17, 18). However, differences in the correlation between lesional and non-lesional skin are of note. Some gene expression correlated with the MRSS much more highly in lesional than non-lesional skin (COMP and THS1), whereas other gene expression (CTGF and PAI1) correlated much more highly in non-lesional than lesional skin. This is consistent with our previous observations that myofibroblast skin infiltration correlates highly with the MRSS in lesional skin(17) but poorly in non-lesional skin (R. Lafyatis, unpublished observation). These data suggest either some temporal change in TGFβ-regulated gene expression over time as skin progresses from lesoinal to non-lesional, or that other different, unknown factors serve to control TGFβ-regulated gene expression at lesional and non-lesional sites.
The complementary contribution of TGFβ- and IFN-regulated genes to the four-gene biomarker is consistent with the observation that both fibrosis (regulated by TGFβ) and inflammation (regulated by IFN) play a role in SSc skin disease. Further, the observation that TGFβ- and IFN-regulated genes correlate closely with the MRSS implicates these cytokines in SSc dermal pathogenesis. However, expression of these genes can be regulated by other factors. For examples, IL-13, CTGF/CCN2 and endothelin-1 can regulate genesalso regulated by TGFβ(25–27). Thus, although our results suggest that TGFβ mediates fibrosis in SSc skin, these or other mediators might be contributing to expression of the TGFβ-regulated genes. IFN-regulated gene expression in autoimmune rheumatic diseases has been most often linked to type I IFNs, particularly in patients with the related disease systemic lupus erythematosus(28). However, both type I (primarily α and β) and type II (primarily γ) IFNs can stimulate very similar series of genes(29). Thus, further study will be required to clarify the type and source of IFN regulating these genes in the skin of SSc patients. This is particularly important, as both type I and type II IFNs have been shown to block collagen induction by TGFβ(30, 31).In addition TGFβ has been shown to upregulate IFNβ transcription through an interaction between smad3 with IRF-7 (32). Thus it appears that TGFβ upregulation of IFN might normally act as a homeostatic control on collagen production.
Most of the patients in this study had relatively early diffuse cutaneous SSc. Although not designed for utilization on patients with limited cutaneous SSc, in five patient biopsies tested the four-gene biomarker predicted low skin scores consistent with the low skin scores from the patients biopsied (data not shown). Although we have previously shown that peripheral blood mononuclear cells from some SSc patients express increased levels of IFN-regulated genes, we have not seen a correlation between expression of IFN-regulated genes by peripheral blood mononuclear cells and skin score ((19) and data not shown).
Other methods to assess skin involvement have been examined, particularly durometry, which shows a high interobserver intraclass correlation coefficient(33, 34) and a moderate correlation with the MRSS. We suggest that such supplemental clinical measure(s) may also provide valuable information that could be combined with the four-gene biomarker to provide more consistent and accurate assessments of change in skin disease over time as a better clinical outcome measure for future therapeutic trials for SSc.
Supplementary Material
Acknowledgments
This study was supported by NIH:NIAMS grant U01AR055063.
REFERENCES
- 1.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35(6):630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 2.Yee CS, Farewell V, Isenberg DA, Prabu A, Sokoll K, Teh LS, et al. Revised British Isles Lupus Assessment Group 2004 index: a reliable tool for assessment of systemic lupus erythematosus activity. Arthritis Rheum. 2006;54(10):3300–3305. doi: 10.1002/art.22162. [DOI] [PubMed] [Google Scholar]
- 3.Felson DT, Anderson JJ, Boers M, Bombardier C, Furst D, Goldsmith C, et al. American College of Rheumatology. Preliminary definition of improvement in rheumatoid arthritis. Arthritis Rheum. 1995;38(6):727–735. doi: 10.1002/art.1780380602. [DOI] [PubMed] [Google Scholar]
- 4.Clements P, Lachenbruch P, Siebold J, White B, Weiner S, Martin R, et al. Inter and intra observer variability of total skin thickness score (modified Rodnan TSS) in systemic sclerosis. J Rheumatol. 1995;22(7):1281–1285. [PubMed] [Google Scholar]
- 5.Furst D, Khanna D, Matucci-Cerinic M, Clements P, Steen V, Pope J, et al. Systemic sclerosis - continuing progress in developing clinical measures of response. J Rheumatol. 2007;34(5):1194–1200. [PubMed] [Google Scholar]
- 6.Furst DE. Outcome measures in rheumatologic clinical trials and systemic sclerosis. Rheumatology (Oxford) 2008;47 Suppl 5:v29–v30. doi: 10.1093/rheumatology/ken269. [DOI] [PubMed] [Google Scholar]
- 7.Czirjak L, Nagy Z, Aringer M, Riemekasten G, Matucci-Cerinic M, Furst DE. The EUSTAR model for teaching and implementing the modified Rodnan skin score in systemic sclerosis. Ann Rheum Dis. 2007;66(7):966–969. doi: 10.1136/ard.2006.066530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope JE, Baron M, Bellamy N, Campbell J, Carette S, Chalmers I, et al. Variability of skin scores and clinical measurements in scleroderma. J Rheumatol. 1995;22(7):1271–1276. [PubMed] [Google Scholar]
- 9.Scala E, Pallotta S, Frezzolini A, Abeni D, Barbieri C, Sampogna F, et al. Cytokine and chemokine levels in systemic sclerosis: relationship with cutaneous and internal organ involvement. Clin Exp Immunol. 2004;138(3):540–546. doi: 10.1111/j.1365-2249.2004.02642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato S, Hasegawa M, Takehara K. Serum levels of interleukin-6 and interleukin-10 correlate with total skin thickness score in patients with systemic sclerosis. J Dermatol Sci. 2001;27(2):140–146. doi: 10.1016/s0923-1811(01)00128-1. [DOI] [PubMed] [Google Scholar]
- 11.Vancheeswaran R, Magoulas T, Efrat G, Wheeler-Jones C, Olsen I, Penny R, et al. Circulating endothelin-1 levels in systemic sclerosis subsets--a marker of fibrosis or vascular dysfunction? J Rheumatol. 1994;21(10):1838–1844. [PubMed] [Google Scholar]
- 12.Lee YJ, Shin KC, Kang SW, Lee EB, Kim HA, Song YW. Type III procollagen N-terminal propeptide, soluble interleukin-2 receptor, and von Willebrand factor in systemic sclerosis. Clin Exp Rheumatol. 2001;19(1):69–74. [PubMed] [Google Scholar]
- 13.Scheja A, Wildt M, Wollheim FA, Akesson A, Saxne T. Circulating collagen metabolites in systemic sclerosis. Differences between limited and diffuse form and relationship with pulmonary involvement. Rheumatology (Oxford) 2000;39(10):1110–1113. doi: 10.1093/rheumatology/39.10.1110. [DOI] [PubMed] [Google Scholar]
- 14.Stone PJ, Korn JH, North H, Lally EV, Miller LC, Tucker LB, et al. Cross-linked elastin and collagen degradation products in the urine of patients with scleroderma. Arthritis Rheum. 1995;38(4):517–524. doi: 10.1002/art.1780380409. [DOI] [PubMed] [Google Scholar]
- 15.Denton CP, Merkel PA, Furst DE, Khanna D, Emery P, Hsu VM, et al. Recombinant human anti-transforming growth factor beta1 antibody therapy in systemic sclerosis: a multicenter, randomized, placebo-controlled phase I/II trial of CAT-192. Arthritis Rheum. 2007;56(1):323–333. doi: 10.1002/art.22289. [DOI] [PubMed] [Google Scholar]
- 16.Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122(1):103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kissin EY, Merkel PA, Lafyatis R. Myofibroblasts and hyalinized collagen as markers of skin disease in systemic sclerosis. Arthritis Rheum. 2006;54(11):3655–3660. doi: 10.1002/art.22186. [DOI] [PubMed] [Google Scholar]
- 18.Farina G, Lemaire R, Pancari P, Bayle J, Widom RL, Lafyatis RA. Cartilage-oligomeric-matrix protein expression in systemic sclerosis reveals heterogeneity of dermal fibroblast responses to transforming growth factor-{beta} Ann Rheum Dis. 2008 doi: 10.1136/ard.2007.086850. [DOI] [PubMed] [Google Scholar]
- 19.York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum. 2007;56(3):1010–1020. doi: 10.1002/art.22382. [DOI] [PubMed] [Google Scholar]
- 20.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15(2):202–205. [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Farina G, WR L, DS, HG, Korn JH. Gene expression profiling in scleroderma: Confirmation of results using real-time quantitative PCR. Arthritis & Rheumatism. 2002;46:S894. [Google Scholar]
- 23.Gardner H, Strehlow D, Bradley L, Widom R, Farina A, de Fougerolles A, et al. Global expression analysis of the fibroblast transcriptional response to TGF beta. Clin Exp Rheumatol. 2004;22(3 Suppl 33):S47–S57. [PubMed] [Google Scholar]
- 24.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3(7):e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu SW, Howat SL, Renzoni EA, Holmes A, Pearson JD, Dashwood MR, et al. Endothelin-1 induces expression of matrix-associated genes in lung fibroblasts through MEK/ERK. J Biol Chem. 2004;279(22):23098–23103. doi: 10.1074/jbc.M311430200. [DOI] [PubMed] [Google Scholar]
- 26.Gore-Hyer E, Shegogue D, Markiewicz M, Lo S, Hazen-Martin D, Greene EL, et al. TGF-beta and CTGF have overlapping and distinct fibrogenic effects on human renal cells. Am J Physiol Renal Physiol. 2002;283(4):F707–F716. doi: 10.1152/ajprenal.00007.2002. [DOI] [PubMed] [Google Scholar]
- 27.Kaviratne M, Hesse M, Leusink M, Cheever AW, Davies SJ, McKerrow JH, et al. IL-13 activates a mechanism of tissue fibrosis that is completely TGF-beta independent. J Immunol. 2004;173(6):4020–4029. doi: 10.4049/jimmunol.173.6.4020. [DOI] [PubMed] [Google Scholar]
- 28.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50(12):3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 29.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga J, Olsen A, Herhal J, Constantine G, Rosenbloom J, Jimenez SA. Interferon-gamma reverses the stimulation of collagen but not fibronectin gene expression by transforming growth factor-beta in normal human fibroblasts. Eur J Clin Invest. 1990;20(5):487–493. doi: 10.1111/j.1365-2362.1990.tb01890.x. [DOI] [PubMed] [Google Scholar]
- 31.Duncan MR, Berman B. Gamma interferon is the lymphokine and beta interferon the monokine responsible for inhibition of fibroblast collagen production and late but not early fibroblast proliferation. J Exp Med. 1985;162(2):516–527. doi: 10.1084/jem.162.2.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qing J, Liu C, Choy L, Wu RY, Pagano JS, Derynck R. Transforming growth factor beta/Smad3 signaling regulates IRF-7 function and transcriptional activation of the beta interferon promoter. Mol Cell Biol. 2004;24(3):1411–1425. doi: 10.1128/MCB.24.3.1411-1425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kissin EY, Schiller AM, Gelbard RB, Anderson JJ, Falanga V, Simms RW, et al. Durometry for the assessment of skin disease in systemic sclerosis. Arthritis Rheum. 2006;55(4):603–609. doi: 10.1002/art.22093. [DOI] [PubMed] [Google Scholar]
- 34.Merkel PA, Silliman NP, Denton CP, Furst DE, Khanna D, Emery P, et al. Validity, reliability, and feasibility of durometer measurements of scleroderma skin disease in a multicenter treatment trial. Arthritis Rheum. 2008;59(5):699–705. doi: 10.1002/art.23564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.