Abstract
Adaptation in haploid organisms has been extensively modeled but little tested. Using a microvirid bacteriophage (ID11), we conducted serial passage adaptations at two bottleneck sizes (104 and 106), followed by fitness assays and whole-genome sequencing of 631 individual isolates. Extensive genetic variation was observed including 22 beneficial, several nearly neutral, and several deleterious mutations. In the three large bottleneck lines, up to eight different haplotypes were observed in samples of 23 genomes from the final time point. The small bottleneck lines were less diverse. The small bottleneck lines appeared to operate near the transition between isolated selective sweeps and conditions of complex dynamics (e.g., clonal interference). The large bottleneck lines exhibited extensive interference and less stochasticity, with multiple beneficial mutations establishing on a variety of backgrounds. Several leapfrog events occurred. The distribution of first-step adaptive mutations differed significantly from the distribution of second-steps, and a surprisingly large number of second-step beneficial mutations were observed on a highly fit first-step background. Furthermore, few first-step mutations appeared as second-steps and second-steps had substantially smaller selection coefficients. Collectively, the results indicate that the fitness landscape falls between the extremes of smooth and fully uncorrelated, violating the assumptions of many current mutational landscape models.
THE question of how populations adapt to changes in their environment has long been a central one in evolutionary biology. While Fisher (1930) and Wright (1932) proposed the foundational models of adaptation, the discoveries linking DNA sequence to amino acid sequence to functional protein changed the way that biologists conceived of adaptation. In the mutational landscape framework proposed by Maynard Smith (1970) and further developed especially by Gillespie (1984, 1991) and Orr (2000, 2002, 2005), organisms occupy points in discrete sequence space, where sequences are either DNA or protein. Spatially adjacent sequences differ from each other by one mutational change. Each sequence has an associated fitness yielding a fitness surface. An altered environment changes the fitness surface and shifts the peak away from the wild type. In a nonrecombinant haploid system, as is our focus here, mutations on the parental background(s) allow the population to explore neighboring regions of sequence space and thereby climb a fitness peak.
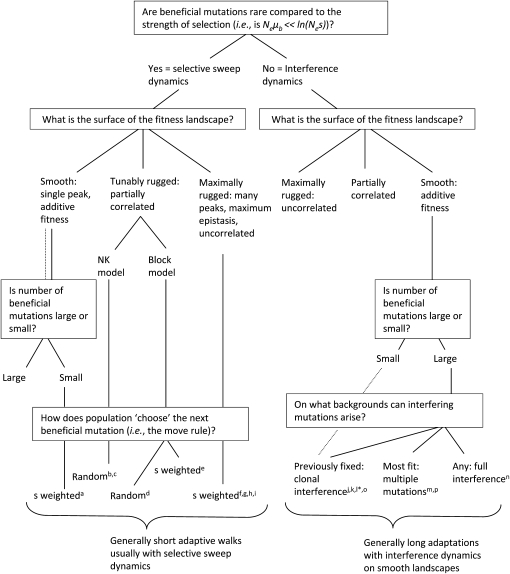
A conceptual map showing how the assumptions of many of the mutational landscape models relate one to another is presented in Figure 1. Our objective here is to first describe several of these assumptions in the Introduction and then examine their validity using real data from experimental evolution. We caution from the outset, however, that neither our survey of models nor our list of important modeling questions is exhaustive. We do not, for example, address extensions of Fisher's geometrical model, the effect of deleterious mutations, recombination, or a continually changing environment to the adaptive process, and we generally omit models that delve into these realms.
Figure 1.—
Relationships among many mutational landscape models based on their assumptions. Superscripts indicate where models are published: a, Orr 2000; b, Kauffman 1993; c, Macken and Perelson 1989; d, Perelson and Macken 1995; e, Orr 2006; f, Gillespie 1991; g, Orr 2002; h, Rokyta et al. 2006a; i, Joyce et al. 2008; j, Gerrish and Lenski 1998; k, Wilke 2004; l, Kim and Orr 2005; m, Desai and Fisher 2007; n, Park and Krug 2007; °, Campos and de Oliveira 2004; p, Brunet et al. 2008. *The dotted line tracks the model of (l) Kim and Orr (2005), which assumes a small number of beneficial mutations on a smooth landscape and, therefore, a bounded walk. It thus does not match the “long adaptation” description at the bottom. The models of Jain and Krug (2007), Wahl and Krakauer (2000), and Hegreness et al. (2006) are not shown because they can occupy more than one region of the conceptual map.
A central issue in all models of adaptation involves how frequently beneficial mutations arise in the population. When rare, the population will be fixed for one background for an extended time, “waiting” for a beneficial mutation to arise. Upon arising, it sweeps to fixation rapidly (relative to the waiting time). We refer to this situation as selective sweep dynamics, and the conditions that produce it are known as strong selection, weak mutation (SSWM) (Gillespie 1984, 1991). Selective sweep dynamics depend on the probability of fixation of each of the possible beneficial mutations. Some models have assumed that the population fixes a random beneficial mutation (e.g., the NK models in Figure 1), but Gillespie (1984) showed that mutations should be chosen with probability equal to their selection coefficient divided by the sum of all beneficial selection coefficients (Figure 1, bottom left). The weight dictates the probability that a mutation survives drift. This assumes, however, that all mutations are equally likely to arise. Rokyta et al. (2005) showed that data from a G4-like DNA virus do not follow this move rule when naively applied, but that a good fit is obtained once differences in mutation rates (i.e., transitions vs. transversions) are incorporated.
When mutations are not rare, for example in large populations, a beneficial mutation that is increasing in frequency will not fix before one or more secondary beneficial mutations arise and begin competing with it. We refer to this as interference dynamics. Different models make differing assumptions about the backgrounds upon which the interfering mutations arise (Figure 1, bottom right). The clonal interference model of Gerrish and Lenski (1998) assumes that secondary beneficial mutations occur only on what was originally the fixed (i.e., wild-type) background and that no second-steps will arise before one of the first-steps fixes. By contrast, the multiple mutations model (Desai and Fisher 2007; Desai et al. 2007; Brunet et al. 2008) assumes that secondary beneficial mutations may arise on any background. Because in this model all mutations are assumed to have the same effect, beneficial mutations on backgrounds of the same fitness do not compete with each other. Thus, the mutations most influential for adaptation are those occurring on the most-fit background. Park and Krug (2007) develop a full interference model where beneficial mutations may occur on any background present in the population but do compete with each other. Hegreness et al. (2006) begin with a similar assumption of full interference dynamics. However, their strategy is to simplify by showing that much of the evolutionary dynamics from this complex model can be approximated by an equivalent model where mutations arise at an effective mutation rate and each has a fixed, effective, selection coefficient.
Models general enough to encompass both selective sweep and interference dynamics include those of Wahl and Krakauer (2000) and Jain and Krug (2007). These models also allow for dynamics at higher mutation incidence where all one-step mutations are produced in the population in one generation, and double mutants, triple mutants, etc., may also arise. This generally yields more deterministic dynamics as the population searches sequence space more systematically (Quer et al. 1996).
Another important component of modeling adaptation is the fitness landscape's surface. At one theoretical extreme lies a smooth, additive, single-peak landscape and at the other, a highly rugged one with many local peaks. Additive landscapes occur when there is no epistasis such that a fixed number of beneficial mutations are available and each has a fixed effect on fitness irrespective of background. (Note that this is not the same as a smooth landscape in Fisher's geometric model; there it is phenotype and not genotype that maps smoothly onto fitness.) Adaptation is then simply the process of accumulating all the beneficial mutations. With the exception of Wahl and Krakauer (2000) and Jain and Krug (2007), models that allow interference dynamics assume fitness landscapes are additive (Figure 1, right side). These interference-on-additive-landscape models also generally assume that the number of beneficial mutations is large, Kim and Orr (2005) being an exception. The adaptive process consequently extends a long time, achieves a steady-state property, and makes it meaningful to focus on the rate of evolution.
At the opposite extreme are maximally rugged landscapes. These assume that the fitness at every sequence is an independent draw from a single probability distribution. There is no correlation in fitness for sequences that are similar to each other. Hence, these landscapes are often called uncorrelated. A biological interpretation is that epistasis is pervasive, with every site affecting every other site in a complex manner. Knowing the fitness of a particular mutation on background A tells you nothing about its fitness on background B—even if background B is just one step away from A. On uncorrelated landscapes there are many local peaks, randomly scattered across the landscape, and no starting sequence is far from one. As a population climbs a local peak, the number of beneficial one-step neighbors is expected to shrink rapidly (Kauffman 1993; Orr 2002; Rokyta et al. 2006a). As long as mutations are restricted to one step at a time, adaptations on such landscapes tend to be short with expected values <5 (Orr 2002; Rokyta et al. 2006a). Most of the models that assume selective sweep dynamics also assume uncorrelated landscapes (Figure 1, left side).
Intermediate landscapes allowing positive correlation in fitness between similar sequences have also been modeled. One example includes the NK model (Macken and Perelson 1989; Kauffman 1993), where N represents the total number of nucleotide or amino acid sites and K is related to the number of epistatically interacting sites. When K = 0, an additive landscape results. When K = N − 1, a maximally rugged landscape is produced. Intermediate values of K yield intermediate landscapes. The block model (Perelson and Macken 1995; Orr 2006) takes a different approach: the sequence is partitioned into blocks (e.g., the domains of a protein). Sites within a block interact in an uncorrelated, maximally epistatic manner, while different blocks interact in a purely additive way. As the number of blocks moves from small to large, landscapes shift from rugged to smooth. While these illustrate two simple ways to model landscapes of intermediate ruggedness, an innumerable array of such landscapes is conceivable. Maximally smooth and maximally rugged landscapes provide convenient extremes, but it is important to realize that the possibilities between them are vastly more complex than a one-dimensional continuum would imply.
Another central feature of most models of adaptation is a distribution of beneficial mutation effects. Important exceptions to this are the models of Hegreness et al. (2006) and Desai and Fisher (2007) where all beneficial mutations have the same fixed effect. Among models where effects come from a distribution, most assume that beneficial effects are random draws from an exponential distribution. To understand why, note that although the wild type is not most fit (the environment has changed), it should have high fitness relative to all the possible sequences. The beneficial mutations represent the small fraction of other sequences with fitnesses above wild type and, therefore, compose the upper tail of the parent fitness distribution. Extreme value theory posits that if the parent distribution comes from one of the familiar tailed distributions like the normal or the gamma (or formally, from the Gumbell domain), then the uppermost tail will be exponentially distributed (Gillespie 1991; Orr 2003). It is important to note that if the parent distribution is not in the Gumbell domain, for example if it is a bounded distribution like the uniform, then the uppermost tail is not exponential. The generalized Pareto distribution (GPD) is a family of distributions that encompasses the extreme tails of a broad range of distributions. Joyce et al. (2008) examined the properties of adaptive walks with selective sweep dynamics on uncorrelated landscapes where beneficial effects are GPD distributed.
A last and nuanced point about models of uncorrelated landscapes is that they generally assume the fitness effects of beneficial mutations in subsequent steps come from the same distribution that produces first-steps (Orr 2002; Joyce et al. 2008). More precisely, subsequent steps are drawn from an ever-winnowing, rescaled upper tail of the parent distribution. When the upper tail is assumed exponential, then rescaling is irrelevant since a rescaled exponential is the same exponential. When the upper tail is GPD but not exponential, the distribution changes only by a scale parameter in a specified way (Beisel et al. 2007).
The purpose of this research is to examine several of these major modeling assumptions in the context of experimental evolution of microbial populations. We use a G4-like phage adapting to laboratory conditions in a flask-passage design. Using full genome sequencing of individual plaques, we track the appearance of mutations and their change in frequency over passages. We estimate mutant fitness in the same flask-passage conditions used for selection. From these data, we address a number of questions: What type of dynamics characterizes our experimental populations and do the dynamics change as we increase population size? What distribution do the beneficial mutations come from? Is the distribution the same across different backgrounds? What type of surface is the fitness landscape?
MATERIALS AND METHODS
Strains:
Research was conducted using the bacteriophage ID11, a member of family Microviridae described by Rokyta et al. (2006b). ID11 is a single-stranded DNA icosahedral virus with a genome 5577 bases long encoding 11 genes. ID11 was also used by Rokyta et al. (2005) in a similar experiment to study the fitness distribution of first-step mutations. The bacterial host used throughout was Escherichia coli C.
Passage experiment:
Adaptations were conducted using a passage design modeled after Rokyta et al. (2002, 2005). Six replicate adaptations were conducted: three at a 104 (small) and three at a 106 (large) bottleneck size. A preliminary large bottleneck line was also conducted prior to the other six to fine-tune techniques. Experiments were carried out in phage-LB+Ca media (a modified Luria–Bertani broth containing 10 g tryptone, 5 g yeast extract, 10 g NaCl per liter supplemented with CaCl2 to 2 mm). All replicate lines were initiated from the same ancestral stock population that was derived from a single wild-type plaque grown at 37° and suspended in 1 ml of sterile phage-LB+Ca. Each line was carried out for 20 flask-passage increments. Each increment of each line involved the following stages: (1) titer previous flask phage population on plates; (2) add 10 ml sterile phage-LB+Ca to a 125-ml Erlenmeyer flask capped with a loose inverted plastic cup; (3) shake flask in a water bath (water depth = 2.5 cm above liquid level in flask) at 200 rpm at 37° for 5 min; (4) add 45 μl of 100× E. coli C freshly thawed freezer stock into a shaking flask and allow to grow for 1 hr (45 μl of cell stock was previously calibrated to yield ∼3 × 108 cells/ml after 1 hr growth); (5) add ∼104 (small line) or 106 (large line) phage to flask on the basis of titer from step 1; (6) allow phage to grow for 40 min (0.67 hr); (7) kill cells by adding 200 μl chloroform to a 2-ml sample from the flask; (8) titer the population at beginning and end of growth to estimate population growth rate (i.e., fitness, w) expressed as doublings per hour based on the exponential relationship N0.67 = N02w0.67; and (9) freeze a portion of the sample in 20% glycerol at −80° for archived storage, and use the other portion to passage to the next flask.
Sampling and sequencing:
After 20 passages were completed, freezer stocks were used to sample individuals from the populations at time points (i.e., passage numbers) 3, 5, 10, 15, and 20. For each replicate line at each of these time points, the stock was plated and 16 (time-point 3) or 32 (all other time points) individual plaques were suspended in 50 μl of sterile phage-LB+Ca. A sample of each was frozen in 20% glycerol at −80° for future use in fitness assays. Another sample of each was used to sequence the entire genome at single coverage through a contract with Sequetech Corporation (Mountain View, CA). Genomes were assembled and SNPs identified using the application SeqMan Pro (version 7.2.2) within Lasergene software (DNASTAR, Madison, WI). After one round of gap filling was conducted, sequences were accepted only if either the entire genome was obtained or if all of the following conditions were satisfied: (i) >85% coverage, (ii) any observed SNPs were also observed in other samples from the same line, and (iii) coverage included all sites known to be polymorphic. To improve accuracy, all assembled genomes were independently analyzed by two people. Under these criteria, 631 genomes were accepted into the data set. Complete coverage was obtained for 569 (90%) of these. Among the remaining 10%, mean coverage was 96%. A whole-genome population sequence was also obtained for the preliminary large bottleneck line from time-point 18.
Fitness estimates:
Fitness was estimated for the wild type and all first- and second-step mutants. We also estimated fitness for all one-step mutations observed by Rokyta et al. (2005), but not by us. First-step mutants were assayed 5 times each in two batches while second-steps were assayed 10 times each, also in two batches. By batch we mean a group of assays done during 1 wk from a common set of isolates. The wild type was included in all batches as a control. In the second-step assays, the first-step background was also included.
Fitness assays were virtually identical to a single round of passaging (steps 1–8 in Passage experiment above, including use of the growth equation shown there) except for the source and number of phage added. As a source, frozen, sequenced isolates were plated and picked and plaques were suspended in 1 ml sterile phage-LB+Ca for 1–3 days to allow titers to stabilize. These were titered and a target of 2000 phage was added at step 4 (instead of 104 or 106). A small preliminary study on sources of variation in estimating fitness revealed that isolate age can have a significant effect. Consequently, all assays were done on isolates between 2 and 7 days of age.
Statistical analysis:
To estimate mutant fitness and control for other factors (batch and assay order), fitness data were analyzed using the general linear model in R (R Development Core Team 2009). Models of increasing complexity were considered beginning with observed fitness as a function of mutation identity alone and then adding (i) batch and (ii) within-batch assay order. The best model was selected using Akaike's information criterion (AIC) (Akaike 1974).
To test the uncorrelated landscape's inherent assumption that the fitness effects of second-steps are drawn from the same distribution as those of first-steps (see Introduction), we used the GPD framework developed by Beisel et al. (2007). We emphasize that our test regards the distribution of fitness effects and not fixed fitness effects (Rozen et al. 2002). Our data are appropriate for such a test because they come from repeated sampling of the same pool of beneficial mutations where the identity of each mutation is known by whole-genome sequencing and is counted in the data set only once. One complexity with these data is that, even with repeated sampling, small-effect mutations may be missed because they are more likely to be lost to drift or competition before detection. Like Beisel et al. (2007), we correct for this inherent bias by shifting the first-step data to the smallest mutation significantly more fit than the wild type (i.e., defining its fitness effect as zero). By contrast, a data set appropriate for studying fixed effects would include every observation and, ideally, the same mutations would never appear twice (since repeated sampling from a continuous distribution, as the model assumes, should yield repeat observations with probability zero).
The GPD describes the upper tails of most distributions using a shape parameter, κ, and a scale parameter, τ. Beisel et al. (2007) tested if fitness effects are exponentially distributed using a likelihood-ratio test to compare the κ = 0 GPD (exponential) to the best-fit GPD. Similarly, we compute the likelihood ratio of a null model where all the data come from one GPD (with parameters κ, τ) to an alternative model where first- and second-step effects come from separate GPD distributions (κ1, τ1, κ2, τ2). Uncertainty in the true fitness of mutants due to assay noise is accounted for using importance sampling (Ripley 1987). We ask where the observed likelihood ratio (Λobs) is located in the distribution of Λ when the null model is true. The mathematical details of our likelihood ratio test are presented in appendix a.
Note that in the likelihood-ratio test on the combined data set, we cannot shift the second-step data like we do the first-steps. While shifting first-steps merely truncates the null distribution in a different spot, shifting second-steps creates a discontinuity in the null distribution that makes calculating the likelihood problematic. There remains, however, a sampling bias against second-step mutations of small effect. This bias should be less influential than it might seem since it affects both the null and the alternative models in the same way, and its effect should largely cancel in the ratio of their likelihoods. To further guard against possible effects of the sampling bias, we designed a second test that is based on the distance between the largest second-step mutation and its first-step background (the largest first-step). This distance does not depend on observing the small-effect second-step mutations. In the test, we simulate the distribution of this distance under the null using parametric bootstrap and compare the observed distance to this distribution. Details are provided in appendix b.
Conditional on rejecting the null hypothesis, we then treat the first- and second-step mutations as separate data sets. This allows us to shift both data sets to correct for sampling bias against small effect. For each data set, we calculate confidence intervals on κ as well as test the hypothesis that κ < 0 (i.e., that the GPD is truncated) vs. κ ≥ 0 with parametric bootstrap. Simulation under known parameter values revealed that, as is common with estimating boundary parameters, our estimator is biased. Our methods for correcting for this bias, estimating parameters, and testing for truncated GPDs is covered in appendix c.
An uncorrelated landscape also implies that the number of beneficial mutations on increasingly fit backgrounds should diminish rapidly. We test this assumption by estimating the number of beneficial mutations available on a high-fitness first-step background and then comparing it to the expectation. Formally, if a first-step background has rank r among first-steps, then the number of beneficial mutations available on this background will follow the negative binomial with parameters r and 0.5 and have an expected value of r (Rokyta et al. 2006a). To understand why this is, note that with the fixation of a first-step beneficial mutation, the background has changed, and a new set of mutational neighbors becomes accessible. In the uncorrelated landscape model where genetic proximity is independent of fitness, the new set is simply a fresh draw of the same size from the same distribution as the original set of mutational neighbors. The only difference is that the threshold defining what is beneficial has increased. Gumbel and Schelling (1950) showed that, for large sample sizes and independent of the distribution, the number of values in the new sample exceeding the rth-ranked value from a previous sample is negative binomial.
Assuming a random transition is equally likely to be beneficial as a random transversion, then this argument should also hold for transitions. That is, if background B has rank rB among the first-step transitions (where B itself could be a transition or not), the number of second-step beneficial transitions accessible from background B should be negative binomial (rB, 0.5). We employ this second argument since we have only a reasonable sample size for transitions. To be conservative, we use the observed number of beneficial transitions (tobserved) as a lower bound on the true number and calculate the negative binomial probability of observing tobserved on a background of rank rB. Substantial doubt is cast on the uncorrelated landscape if this probability is sufficiently small.
RESULTS
Number and identity of mutations:
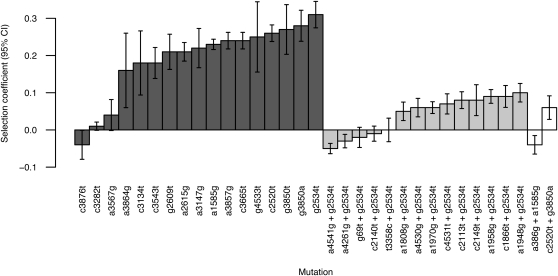
The total number of first-step mutations observed in the six passage lines is 14 (Table 1). Mutations are herein identified by nucleotide position and base change relative to the published wild-type sequence (GenBank accession AY751298). Two first-step mutations (a3875g and c3876t) have estimated fitness less than the wild type. The sites are near the origin of replication and may represent mutational hotspots. Removing these two and combining the rest with mutations from Rokyta et al. (2005), who employed the same experimental conditions, 16 first-step beneficial mutations are observed.
TABLE 1.
Mutations observed during phage adaptation
| Mutation type | Mutation position in genome (nucleotide substitution) | Background | Amino acid positiona | Amino acid change | Fitnessb | SE (95% C.I.) | Selection coefficient | No. lines observed (this study, Rokyta et al. 2005)c |
|---|---|---|---|---|---|---|---|---|
| — | Wild type | — | — | — | 14.3 | 0.17 (14.0–14.6) | 0 | — |
| First step | 3875 (a → G) | Wild type | F425 | T → A | 11.0 | 0.27 (10.2–11.8) | −0.23 | 1 (1, 0) |
| 3876 (c → T) | Wild type | F425 | T → I | 13.7 | 0.20 (13.1–14.3) | −0.04 | 2 (2, 0) | |
| 3282 (c → T) | Wild type | F227 | S → F | 14.5 | 0.05 (14.4–14.6) | 0.01 | 1 (1, 0) | |
| 3567 (a → G) | Wild type | F322 | N → S | 14.8 | 0.30 (14.0–15.6) | 0.04 | 2 (1, 1) | |
| 3864 (a → G) | Wild type | F421 | D → G | 16.6 | 0.51 (15.2–18.0) | 0.16 | 1 (0, 1) | |
| 3134 (c → T) | Wild type | F178 | R → C | 16.8 | 0.44 (15.6–18.0) | 0.18 | 1 (1, 0) | |
| 3543 (c → T) | Wild type | F314 | A → V | 16.9 | 0.21 (16.3–17.5) | 0.18 | 1 (0, 1) | |
| 2609 (g → T) | Wild type | F3 | V → F | 17.2 | 0.25 (16.5–17.9) | 0.21 | 1 (0, 1) | |
| 2615 (a → G) | Wild type | F5 | T → A | 17.3 | 0.13 (16.9–17.7) | 0.21 | 1 (1, 0) | |
| 3147 (a → G) | Wild type | F182 | N → S | 17.4 | 0.27 (16.7–18.1) | 0.22 | 1 (6, 0) | |
| 1585 (a → G) | Wild type | A509 | Silent | 17.6 | 0.07 (17.4–17.8) | 0.23 | 1 (1, 0) | |
| A*296 | Silent | |||||||
| B104 | T → A | |||||||
| 3857 (a → G) | Wild type | F419 | T → A | 17.6 | 0.12 (17.3–17.9) | 0.24 | 1 (0, 1) | |
| 3665 (c → T) | Wild type | F355 | P → S | 17.6 | 0.11 (17.3–17.9) | 0.24 | 6 (5, 5) | |
| 4533 (g → T) | Wild type | G172 | V → F | 17.9 | 0.48 (16.6–19.2) | 0.25 | 1 (1, 0) | |
| 2520 (c → T) | Wild type | J15 | A → V | 17.9 | 0.12 (17.6–18.2) | 0.26 | 7 (6, 6) | |
| 3850 (g → T) | Wild type | F416 | M → I | 18.1 | 0.35 (17.1–19.1) | 0.27 | 2 (1, 1) | |
| 3850 (g → A) | Wild type | F416 | M → I | 18.2 | 0.22 (17.6–18.8) | 0.28 | 4 (3, 2) | |
| 2534 (g → T) | Wild type | J20 | V → L | 18.7 | 0.25 (18.2–19.2) | 0.31 | 2 (6, 1) | |
| Second step | 4541 (a → G) | 2534 (g → T) | G174 | Silent | 17.8 | 0.12 (17.5–18.1) | −0.05 | 1 (1, —) |
| 4261 (a → G) | 2534 (g → T) | G81 | N → S | 18.1 | 0.14 (17.8–18.4) | −0.03 | 1 (1, —) | |
| 69 (g → T) | 2534 (g → T) | A4 | T → I | 18.3 | 0.22 (17.8–18.8) | −0.02 | 1 (1, —) | |
| 2140 (c → T) | 2534 (g → T) | D55 | Silent | 18.5 | 0.17 (18.1–18.9) | −0.01 | 1 (1, —) | |
| 3358 (t → C) | 2534 (g → T) | F252 | Silent | 18.7 | 0.27 (18.1–19.3) | 0.00 | 1 (1, —) | |
| 1808 (a → G) | 2534 (g → T) | K57 | Stop → W | 19.6 | 0.20 (19.1–20.1) | 0.05 | 1 (1, —) | |
| C30 | D → G | |||||||
| 4530 (a → G) | 2534 (g → T) | G171 | T → A | 19.8 | 0.20 (19.3–20.3) | 0.06 | 2 (2, —) | |
| 1970 (a → G) | 2534 (g → T) | C84 | N → S | 19.8 | 0.13 (19.5–20.1) | 0.06 | 1 (1, —) | |
| 4531 (c → T) | 2534 (g → T) | G171 | T → I | 19.9 | 0.23 (19.4–20.4) | 0.07 | 2 (2, —) | |
| 2113 (c → T) | 2534 (g → T) | D46 | Silent | 20.1 | 0.18 (19.7–20.5) | 0.08 | 1 (1, —) | |
| 2149 (c → T) | 2534 (g → T) | D58 | Silent | 20.2 | 0.34 (19.4–21.0) | 0.08 | 2 (2, —) | |
| 1958 (a → G) | 2534 (g → T) | C80 | N → S | 20.4 | 0.15 (20.1–20.7) | 0.09 | 2 (2, —) | |
| 1866 (c → T) | 2534 (g → T) | C49 | Silent | 20.4 | 0.24 (19.9–20.9) | 0.09 | 1 (1, —) | |
| 1948 (a → G) | 2534 (g → T) | C77 | T → A | 20.6 | 0.20 (20.1–21.1) | 0.10 | 1 (1, —) | |
| 386 (a → G) | 1585 (a → G) | A110 | I → V | 16.9 | 0.19 (16.5–17.3) | −0.04 | 1 (1, —) | |
| 2520 (c → T) | 3850 (g → A) | J15 | A → V | 19.3 | 0.25 (18.7–19.9) | 0.06 | 1 (1, —) | |
| Third step | 3010 (a → G) | 2534 (g → T) + 1970 (a → G) | F136 | Silent | NA | NA | NA | 1 (1, —) |
Mutations observed in passage experiments on ID11 are identified by nucleotide substitution and sorted by fitness. Also included are mutations observed by Rokyta et al. (2005) under the same adaptive conditions. NA, not assayed. Note that some genes in ID11 overlap; mutations 1585 and 1808 affect multiple genes.
The amino acid position gives the gene name followed by the residue number.
Fitness is reported as log2 increase in phage per hour.
“—” indicates not applicable because Rokyta et al. (2005) did not progress past one-step mutations.
In our experiment, four of these mutations—c2520t, g2534t, g3665t, and a3147g—were observed by time-point 5 in all, or all but one, line. Several of these four may have been present in the ancestral stock at low frequencies. The starting stock used in all lines was obtained from a single plaque that was grown from the non-lab-adapted wild-type phage exposed to selective lab conditions (e.g., 37°). With plaques growing from one phage to 108−109 phage and mutations occurring at an estimated rate of 1/300 replications (Drake et al. 1998), we expect many mutations to arise. None will be present at high frequency, but adaptive mutations could increase in the plaque to modest frequency. With inocula sizes of ≥104, the frequency of such adaptive mutations could still have been quite low (≥0.0003) and have had ≥95% probability of making it into all lines (on the basis of binomial probabilities). Most importantly for interpreting our results, there are good reasons to believe that g2534t was present in the ancestral stock. As a transversion, g2534t should be rare. Indeed, it arose only once in 20 opportunities for Rokyta et al. (2005) and yet it was observed here in every line. To independently determine if it was present in the starting stock, we sequenced the end point from the preliminary line seeded with the same ancestral stock, several weeks before the other adaptations were conducted. That population-level sequence also contained g2534t.
The fortuitous consequence of using a single plaque grown under selective conditions was that the high-fitness mutant g2534t became abundant in every line. As a result, g2534t was the background upon which 13 of the observed 15 second-step mutations occurred (Table 1). Four of these (a4541g, a4261g, g69t, and c2140t) may not be beneficial given their lack of persistence (Figure 2) and low fitness estimates (Table 1). Only one of the second-step mutations (g3850a + c2520t) involves two mutations that have been observed individually as beneficial as first-steps. One third-step mutation was observed.
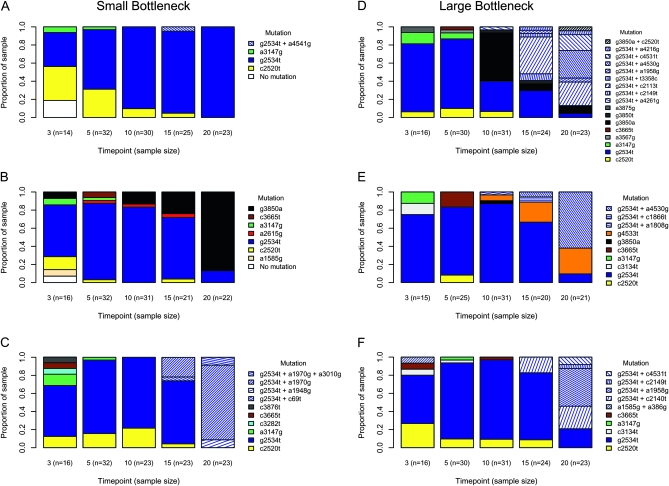
Figure 2.—
Observed mutation frequencies across time points in replicate adaptations. To the left (A–C) are three small bottleneck (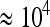 ) replicates; to the right (D–F) are three large bottleneck (
) replicates; to the right (D–F) are three large bottleneck ( ) replicates. Sample sizes (number of sequenced isolates) are shown on the horizontal axis. Note that first-step mutations are shown as solid colors, and second- and third-steps are hatched with hatch coloring corresponding to their background.
) replicates. Sample sizes (number of sequenced isolates) are shown on the horizontal axis. Note that first-step mutations are shown as solid colors, and second- and third-steps are hatched with hatch coloring corresponding to their background.
Dynamics:
The dynamics of mutation frequencies across passages differ substantially among the six passage lines (Figure 2). While models of adaptation assume that the population begins as fixed for the wild type, our populations were probably polymorphic at founding. This somewhat complicates categorizing dynamics of each line. Beginning with the small bottleneck lines, the dynamics range from being sweep-like to showing full interference. In line A (Figure 2A), the only mutations observed are first-steps (except the double mutant a4541g, which is probably not beneficial), and fixation has occurred by passage 20. In the sense that multiple haplotypes are competing in line A, the frequency dynamics are obviously akin to clonal interference. But in the sense that only putative founder mutations are observed and no new beneficial mutations appear after the first sample, the mutational dynamics are selective sweep-like. The implication is if the population began as monomorphic, a selective sweep might have occurred. Line B shows a similar pattern of only first-step mutations arising and a trend toward fixation. But in line B there are several unique mutations in addition to the founder set, making it more likely that some mutations arose on the wild-type background after passaging began. This is consistent with the clonal interference model. Up until time-point 10, line C shows patterns similar to those of lines A and B. However, before any mutation fixes, several second-steps and then a third-step arise. With more than one mutation arising on the g2534t background and then a third-step mutation arising before any of the second-steps fix, line C is consistent with full interference dynamics.
The three large passage lines (D–F) display extensive interference dynamics and, again, the pattern generally favors full interference over strictly defined clonal interference. Inconsistent with the clonal interference model, we observe a case in line D where second-step mutations arise on two different first-step backgrounds (on g2534t and on g3850a) rather than waiting for one background to fix. The multiple-mutations model is harder to evaluate. While the assumption of a fixed fitness effect is obviously violated (Table 1), the model's real claim is not that a fixed effect is literally true, but rather that long-term adaptation can be approximated with a fixed effect equal to an average among variable effects. A less obvious test of the multiple-mutations model is its assumption that the suite of next-step beneficial mutations in the population should arise on currently established most-fit background(s). This assumption is generally supported, but not without exception. In three of the four lines where beneficial second-step mutations arise (C–F), they arise on the most-fit g2534t background (C, E, and F). In line D, however, the frequency changes suggest that g3850a is the most-fit background and line B echoes this ranking; yet the second-step beneficial mutations that come to dominate by time-point 20 are on the g2534t background.
In their development of a model of clonal interference, Gerrish and Lenski (1998) defined a leapfrog event as one in which a mutant type rises to >50% frequency, but is then superseded by another mutation from which it differs by two mutational changes (the most possible when only one-step mutations are permitted). In passage line B we observe this effect when g2534t is replaced by g3850a as the dominant type between time-points 15 and 20. The same event occurs in passage line D between time-points 5 and 10. In line D between time-points 10 and 15 we observe a more dramatic leapfrog event where g3850a is superseded by a suite of second-step mutations on the g2534t background—mutations that differ from it by three mutational steps.
Fitness landscapes and distribution of fitness effects:
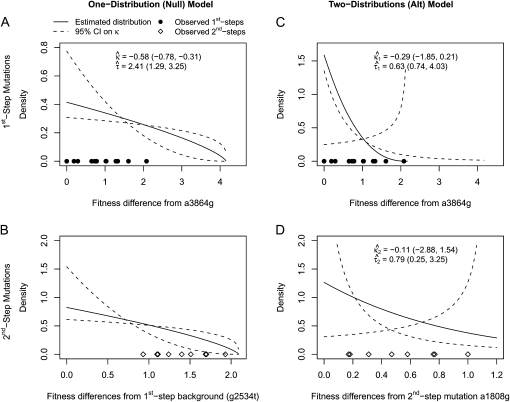
We tested whether the fitness values of first- and second-step mutations come from the same distribution. In doing so, we first tested our fitness assay data for sources of systematic error. The general linear model indicates that batch had a significant effect on fitness estimates (ΔAIC = −57.0), but that assay order within batch was not significant (ΔAIC = 2.0). To reduce bias in analyzing distributions, first-step fitness data were shifted so mutant a3864g had fitness of zero. The likelihood-ratio test for one- vs. two-fitness-distributions models (appendix a) favors the two-distribution model P = 0.03. The test for distance between largest first-step and largest second-step similarly rejects the null model (P = 0.02). The bias-corrected estimates for κ under the two-distribution model (with 95% C.I.'s) are κ1 = −0.29 (−1.85, 0.21) and κ2 = −0.11 (−2.88, 1.54). These distributions, along with the best-fit null and the fitness data, are presented in Figure 3. When the hypothesis that the fitness distributions are truncated (κ < 0 vs. κ ≥ 0) is tested for the first-step data using bias-corrected bootstrapping, the nontruncated distributions cannot be rejected (P = 0.14). For second-steps, where sample noise is high compared to effect size, there is little information regarding truncated vs. nontruncated distributions (P = 0.61). Using the Beisel et al. (2007) test of the similar exponential hypothesis (κ < 0 vs. κ = 0) returns the same qualitative results: P = 0.08 and P = 0.20.
Figure 3.—
Observed fitness effects and fitted GPD distributions under the one-distribution (null) and two-distributions (alternative) models. Point estimates and 95% confidence intervals are given in insets and presented visually as solid and dashed lines, respectively. Note that confidence bounds are calculated on κ only (see materials and methods). The bounds on τ are defined on the basis of upper and lower bounds on κ to maintain a scale consistent with the point estimates. (A) First-step mutations and fitted null model. (B) Second-step mutations and fitted null model. Parameter estimates are the same as in A. (C) First-step mutations and fitted alternative model. (D) Second-step mutations and fitted alternative model. Note changes in scale on both axes between sections.
Nine beneficial transitions are observed on the g2534t background (Table 1). To test whether this number is consistent with the uncorrelated model expectation of a negative binomial distribution, we rank g2534t among the first-step transitions (see materials and methods for explanation). By fitness assay, g2534t ranks first among all observed mutations, including transitions (Table 1). In the frequency data (Figure 2), the only transition that may rank above it is g3850a. This suggests that g2534t ranks first or second, but, clearly, we have not observed all first-step beneficial transitions. Would its rank drop if we could observe all such mutations? As we argue below, the large bottleneck lines were large enough to generate most beneficial transitions every few passages. If there were any very high fitness transitions, they should have established and been observed. It is therefore likely that g2534t genuinely ranks among the top two transitions. Since the expected number of beneficial mutations under the negative binomial model is equal to the rank (Rokyta et al. 2006a), on a background of rank 1, 2, or 3, we expect there to be one, two, or three total beneficial transitions. In fact, we observed nine (Table 1). The P-values associated with observing nine on a background of rank 1, 2, or 3 are 0.001, 0.006, and 0.02. Thus, the data do not support the assumption of the fully uncorrelated landscape.
DISCUSSION
Frequency of beneficial mutations:
The dynamics of adaptation depend strongly on how often beneficial mutations occur. At one extreme, beneficial mutations are rare and selective sweeps dominate. As the supply of mutations rises, say because population size increases, a secondary beneficial mutation will tend to establish before the first one fixes, but which secondary mutation and when it does so remain stochastic. As the incidence of mutations further increases, the population produces more than one beneficial mutation each generation, then many, then most, and then all such mutations. With larger populations becoming increasingly saturated with the available beneficial mutations, the set of best mutations will establish and compete, and adaptation becomes more deterministic (Wahl and Krakauer 2000; Jain and Krug 2007). This inevitable competition between these highly fit mutations also means that the rate of adaptation will tend to plateau with increasing mutation supply (De Visser et al. 1999). One of our objectives in passaging at two substantially different bottleneck sizes (104 and 106) was to observe and perhaps demarcate where these transitions in dynamics occur.
The 104 bottleneck size appears to put the population in the transition between selective sweep dynamics and highly stochastic interference dynamics. As explained in materials and methods, the source population was very likely polymorphic and included the highly fit mutation g2534t. Therefore, the appearance of multiple first-step mutations in all three lines is not illuminating. More telling is the appearance of second-step mutations in the third line (interference dynamics), but their absence in the other two small lines (selective sweep-like dynamics; Figure 2C vs. 2A and 2B). Note that g4541t in line A is probably not beneficial on the basis of the fitness data from Table 1 as well its failure to persist. Numerous beneficial mutations on the g2534t background exist (Table 1) and g2534t was the majority background in all three small lines for most of the passages. So the opportunity for second-steps to arise in lines A and B clearly existed. This begs the question: Around what population size did the transition between selective sweeps and interference dynamics occur?
To convert the population size data under periodic bottlenecks to effective sizes (Ne), we use the equation Ne = NbGr from Wahl and Gerrish (2001), where Nb is the transfer size, G is the number of generations between passages, and r is the per generation exponential growth rate. In all three small bottleneck lines, Ne was ∼5 × 104. This suggests that in similar biological systems, populations will tend to adapt under sweep dynamics when Ne < 104 and under interference dynamics when Ne > 105.
How similar must other biological systems be for this generalization to hold? Sweep dynamics will prevail when the time it takes for a beneficial mutation to arise and escape the vagaries of drift (i.e., to become established) is longer than the time for an established mutation to fix. The expected number of generations to establish and fix is known from population genetics theory: testablish = 1/Neμbs and tfix ≈ [ln(Nes)]/s, respectively where s is the selection coefficient and μb is the beneficial mutation rate (Gillespie 1991; Desai and Fisher 2007). Sweep conditions will prevail when Neμb ≪ ln(Nes) (Desai and Fisher 2007). Hence, the similarity of other systems to ours depends not just on Ne, but also on s and μb.
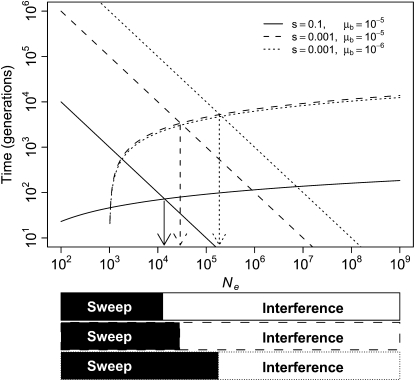
The s-values observed here (e.g., 0.05–0.1 for second-steps) are relatively large (Table 4). However, the size of s has relatively little influence on which dynamics prevail. Mathematically, this is because s appears only in the log term of the inequality above. The magnitude of s instead dictates the timescale upon which the process plays out with large coefficients leading to both much shorter establishment and much shorter fixation times as illustrated in Figure 5.
Figure 5.—
Transition from sweep to interference dynamics across Ne. Descending, straight lines are expected times for a beneficial mutation to establish and ascending, curved lines are expected times to fixation for an established mutation {based on testablish = 1/Neμbs and tfix ≈ [ln(Nes)]/s}. Arrows indicate Ne where approximate transitions in dynamics occur as emphasized by solid and open bands below the horizontal axis.
The more important factor influencing dynamics is μb, about which relatively little is known. For our system, we can get an order of magnitude estimate of μb by again noting that second-step mutations established in only one of the three small bottleneck lines despite the opportunity. Since these lines represent ∼60 phage generations, the time to establishment must be very roughly on the order of 60 generations. Assuming that s = 0.075 (the midpoint of observed values) and that Ne = 4 × 104 (∼Ne of the highly fit g2534t background upon which most beneficial mutations could have occurred), we solve the equation testablish = 1/Neμbs for μb to get μb ≈ 5 × 10−6. The following cross-check of this estimate suggests this value is probably low, but not unreasonable. When this rate is divided by an estimate of the per-genome mutation rate of 1.0 × 10−2 from the closely related φX174 virus (Raney et al. 2004; Cuevas et al. 2009), it suggests that ∼1 in every 2000 mutations is beneficial. However, we observed 9 beneficial transitions on the g2534t background and a simple mark–recapture model (Miller et al. 2005) estimates that 18 exist. There are 5577 transitions possible from this background, and 18/5577 = 0.003, suggesting that ∼1 in 300 mutations is beneficial. In other words, μb is probably >5 × 10−6. Nonetheless, our approximation of μb is relatively similar to a recent estimate in E. coli of ∼10−5, or 1/150 mutations (Perfeito et al. 2007). Drake et al. (1998) showed that the per-genome mutation rate is on the same order of magnitude across most microbes. It is conceivable that μb will also be in the vicinity of 10−5 across many microbes.
Figure 5 plots the expected times to establishment and fixation on the basis of the above equations, emphasizing the transition from sweep to interference dynamics. If μb is indeed near 10−5, then the transition from sweep to interference will occur between 104 and 105 across a wide range of selection coefficients. The dotted line in Figure 5 illustrates that if μb is decreased an order of magnitude to 10−6, the effect is to increase the Ne where transition occurs approximately an order of magnitude. Lynch (2006) estimated long-term Ne in prokaryotes, nonparasitic unicellular eukaryotes, invertebrates, and land plants on the orders of 108, 107, 106, and 106, respectively. Pathogens may have lower long-term Ne due to their host dependence. While these estimates come from a dramatically different temporal scale than our research, they indicate that most microbes spend most of the time at effective sizes where interference dynamics are expected to dominate.
Given these arguments, it is not surprising that in the three large bottleneck lines where the mean Ne ≈ 107, all lines exhibit interference dynamics. More interesting is that the large lines may be in another transition zone, one where mutation supply is great enough to begin pushing the population down a more deterministic path. By passage 20, each large line has a different set of second-step mutations, giving the impression that the process is highly stochastic. However, in all three large lines, secondary mutations affecting residue 171 in gene G (which encodes the major spike protein) appear to be emerging or have emerged as the most fit haplotype. Specifically, in line E, mutation a4530g causes the T171A amino acid substitution in gene G and is clearly dominant. In line D, three mutations divide most of the frequency at time-point 20: c2113t, a4530g, and c4531t. However, c2113t has declined in frequency since passage 15, suggesting it has lower fitness. Note that this rank order is inferred from changes in frequency and not fitness assay estimates (Table 1) as confidence intervals from the fitness assays overlap substantially (Figure 4). While mutation c4531t affects the same residue as a4530g, the substitution is T171I. Further note that in line D the increase of a4530g and c4531t also comes at the expense of a1958g and c2149t. This is consequential for understanding line F. Here, four mutations are observed in the last time point: c2140t, a1958g, c2149t, and c4531t. Mutation c2140t is little changed from passage 15, suggesting it is not most fit. The other three types are first observed at passage 20. If the rank order deduced from line D above is correct, then c4531t affecting residue 171 in gene G would be the most fit among them.
Figure 4.—
Selection coefficients and their 95% confidence intervals for all mutations in this study. Boxes with dark shading show first-step mutations on the wild-type background. Boxes with light shading show second-step mutations on the g2534t background. Open boxes show second-step mutations on another first-step background. The first-step deleterious mutation a3875g is omitted for purposes of scale. Fitness was not measured on the third-step mutation a3010g.
The convergence of the large bottleneck lines along similar adaptive paths is consistent with the argument that they lie in the transition zone of waning stochasticity. However, Ne ≈ 107 is not nearly large enough to produce all beneficial mutations every generation. Other beneficial mutations on the g2534t background (besides a4530g and c4531t) did establish in all three lines. In general, 5–10 passages (15–30 generations) passed before mutations affecting residue G171 appeared. This implies that when Ne ≪ 107, a population is far less likely to obtain the most beneficial mutation. Consistent with this, the only small bottleneck line to proceed past first-step mutations (C) has committed to a totally different second-step on the g2534t background, either a1970g or a1948g.
Genetic background on which beneficial mutations arise:
Besides how often they occur, the background on which beneficial mutations may arise will dictate adaptive dynamics. Several interference models have examined this topic in some detail (Figure 1, right side) and made differing assumptions, yet virtually all assume an additive fitness landscape. Ironically, while differing assumptions about the backgrounds on which mutations occur lead to different dynamics and rates of adaptation, the ultimate outcome is unaffected—the same pool of mutations will eventually fix regardless of order. Background is much more important on uncorrelated landscapes because the background a mutation arises upon will uniquely define the adaptive trajectory. The issue also bears on if and how populations can cross fitness valleys in the adaptive landscape. Several authors have argued that weakly beneficial, neutral, or even mildly deleterious mutations may establish and have transient existences in the population (Iwasa et al. 2004; Weinreich and Chao 2005; Jain and Krug 2007). If a beneficial mutation occurs on such a background in a rugged or semirugged landscape, it may spawn a mutation that is more fit than any existing type and thereby steer adaptation. Evolution of the influenza virus in humans via neutral networks may be a real world example of this type of dynamic (Koelle et al. 2006).
Our data indicate that beneficial mutations can arise on any background present in the population. To illustrate, we compare our data to several of the aforementioned models. Consistent with the clonal interference model of Gerrish and Lenski (1998), a multitude of first-step mutations are observed on the wild-type background. An important conclusion of clonal interference models is that the rate of fixation is slowed compared to selective sweep dynamics. This slowdown is implied in portions of our data. Using the estimated fitness from Table 1 and imagining that g2534t alone is competing with the wild type, we would deterministically expect a single g2534t mutation to be virtually fixed in 6–8 passages (depending on the bottleneck size). In no lines did g2534t reach fixation by passage 10, presumably because its ascent was dampened by the presence of other first-step mutations that were substantially more fit than the wild type.
However, the data do not conform to the clonal interference assumption that multiple first-steps will compete and one will fix before any second-steps arise. In the four lines where second-steps appear (Figure 2, C–F), they establish before any first-step reaches fixation. The multiple-mutations model of Desai and Fisher (2007) assumes that one or more beneficial mutations of equal fitness will increase in frequency and, before fixing, spawn one or more second-step beneficial mutations. While less-fit backgrounds may give rise to mutations, it is only the mutations on the most-fit background(s) that will yield the new most-fit haplotype(s). The data generally support this assumption, though not universally. In all lines where second-steps arise (C–F), they do so before first-steps fix. In lines C, E, and F, the second-steps appear on the high-fitness g2534t background. However, in the large bottleneck line D, frequency changes indicate that g3850a is probably outcompeting g2534t at time-point 10 (Figure 2). Small line B further supports this rank order. But before g3850a fixes, g2534t spawns several second-step mutations that are more fit than g3850a and by time-point 20 have come to predominance. Mutation g2534t is, thus, “rescued” from its decline by producing second-step mutations.
Interestingly, time-point 20 in line D contains a second-step mutation on the waning g3850a background (g3850a + c2520t). This double mutant may or may not have changed the fate of g3850a had the experiment continued, but like the g2534t rescue event earlier in line D, it illustrates that the full interference models are likely correct: beneficial mutations may establish on any background present in the population and thereby affect dynamics. In a recent flask-passage adaptation experiment similar to ours, but using an RNA bacteriophage, Betancourt (2009) also observed instances of both clonal interference and more complex interference dynamics, further supporting the full interference model.
Leapfrog events:
An interesting case of mutations on a suboptimal background affecting dynamics comes from what Gerrish and Lenski (1998) called the leapfrog event. Here, a mutant that is temporarily the most abundant in the population is supplanted by another genotype from which it differs by more than one mutational change. By facilitating a more rapid genetic shift at the population level than is possible under selective sweeps, leapfrog events may be biologically significant when the population is under selective pressure from another evolving entity like a competitor or a host immune system. Three instances of leapfrog events are observed in our data: line B time-points 15–20, line D time-points 5–10, and again in line D at time-points 10–20 (Figure 2; see results for details). In the last case, the event involves a change of three mutations. The data suggest leapfrog events may be common, and because beneficial mutations may arise on a diversity of backgrounds, leaps may involve multiple mutational steps.
Distribution of beneficial effects:
Most models of adaptation assume fitness effects of beneficial mutations are distributed exponentially (see Introduction). The empirical evidence for this has been mixed (Sanjuán et al. 2004; Rokyta et al. 2005, 2008; Betancourt and Bollback 2006; Kassen and Bataillon 2006; Eyre-Walker and Keightley 2007). Rokyta et al. (2008) analyzed data from two phage systems—including the one studied here—and rejected the exponential in favor of a truncated distribution. That analysis of ID11 was based on nine beneficial first-step mutations. We (re)assayed fitness for those nine mutations plus an additional seven. Our analysis of first-step mutations favors a truncated distribution (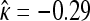 , Figure 3), but the exponential distribution cannot be rejected (P = 0.14 or P = 0.08, depending on which test is used). Analysis of second-steps yields extremely wide confidence intervals on κ (Figure 3), indicating that our assay noise is too large to make meaningful inferences about the shape of the second-step distribution.
, Figure 3), but the exponential distribution cannot be rejected (P = 0.14 or P = 0.08, depending on which test is used). Analysis of second-steps yields extremely wide confidence intervals on κ (Figure 3), indicating that our assay noise is too large to make meaningful inferences about the shape of the second-step distribution.
Another common assumption in models of adaptation is that the distribution from which fitness effects come remains unchanged as adaptation proceeds. For purely additive landscapes this is literally true, while for uncorrelated landscapes in a static environment, the assumption is more precisely that the distribution at each step is always the rescaled upper tail of the same distribution. We return to the topic of additive landscapes in the next subsection and here address the fixed distribution assumption under uncorrelated landscapes. A qualitative pattern that arises from this assumption is that the size of fitness effects will decrease as fitness increases along a walk. This pattern has been observed previously in experimental evolution (Holder and Bull 2001; Silander et al. 2007; Betancourt 2009). Our data show the same pattern, although in greater detail for only the first two steps: second-steps have both smaller absolute fitness effects (Figure 3) and smaller selection coefficients than first-steps (Figure 4).
However, the fixed-distribution assumption makes a more precise prediction than simply that fitness effects should decrease. It specifies by how much they should decrease. We tested this more specific prediction by asking whether the first- and second-step fitness effects fit a one-distribution model well compared with a two-distribution model. The results of the likelihood-ratio test indicate that our second-steps do not come from the upper tail of the same distribution that generated the first-steps (P = 0.03, Figure 3). Given the large uncertainty in κ, this cannot be the result of the first- and second-steps having incompatible shapes. Instead, the result is driven by second-steps being too large to be explained under the null model. Recall that for bounded GPD distributions, first-steps are drawn from the full distribution and second-steps are drawn from the region of the distribution above their first-step background—hence the specific prediction about how much fitness effects should decrease. The one-distribution (null) model does not fit the data well because there is a big gap between the largest first-step mutation and the estimated boundary (Figure 3A). We confirmed this explanation using a second test that asks whether the difference between the largest first-step and the largest second-step (20.6 − 18.7 = 1.9; Table 1) is compatible with the null model (appendix b). The rarity of a difference this large (P = 0.020) again indicates that the one-distribution model can be rejected. This suggests an unpleasant reality (from a modeling perspective) that the distribution of fitness effects may change with the background.
Landscape ruggedness:
Mutational landscape models have tended to rely heavily on fitness landscapes at the theoretical extremes: maximally rugged (uncorrelated effects) or maximally smooth (additive effects). We now critique our data from the two ends of this spectrum, asking whether they are consistent with either one. As we saw in the previous subsection, the likelihood-ratio test casts doubt on the one-distribution assumption and, by extension, the uncorrelated landscape. Another expectation of an uncorrelated landscape is that the number of beneficial mutations (i.e., exceedences) should be small for high-ranking backgrounds. Nine beneficial second-step transitions are observed on the high-fitness, high-ranking g2534t background. Ignoring the fact that the true number must be larger than this, nine is too many compared to the negative binomial expectation of two to four beneficial transitions (P < 0.02). Thus, the data do not support an uncorrelated landscape.
Nor are the data consistent with a smooth fitness landscape. One expectation for such landscapes is that mutations beneficial as first-steps will also be beneficial as second-steps and have consistent fitness effects (when measured as selection coefficients). Among the 11 second-step beneficial mutations, only one involves the pairing of known first-step beneficial mutations (g3850a + c2520t). In this case, the observed fitness of the second-step c2520t on the g3850a background (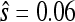 ) is substantially smaller than its effect on the wild-type background (
) is substantially smaller than its effect on the wild-type background (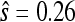 ). Rather than additive, this observation is consistent with a model of diminishing returns epistatis (De Visser et al. 1999; Bull et al. 2000) where a mutation's fitness effect decreases as the background fitness increases.
). Rather than additive, this observation is consistent with a model of diminishing returns epistatis (De Visser et al. 1999; Bull et al. 2000) where a mutation's fitness effect decreases as the background fitness increases.
The data from background g2534t (the only well-sampled first-step background on which to evaluate second-step mutations) also disfavors an additive landscape. On the g2534t background, zero of nine observed second-steps (all transitions) were observed beneficial first-steps. While there are nine first-step mutations with values >0.15, all observed second-steps have values ≤0.1 (Figure 4; Table 1). If s-values do not change with background as additivity dictates, the first-step mutations should have been selectively favored and some should have appeared as second-steps. Whether the first-step mutations on the g2534t background generally have negative fitness effects (sign epistasis; Weinreich et al. 2005), or are simply less than additive (magnitude epistasis), is unknown.
Thus the landscape is likely neither totally additive nor fully uncorrelated. The truth, of course, must lie in between and the question is where. Not only is the answer important for understanding the dynamics of adaptation, but also it bears on the long-standing debate about the evolutionary advantages of sex. Much interest in interference dynamics in asexuals owes to how interference slows the rate at which beneficial mutations fix in the population (Muller 1932; Gerrish and Lenski 1998; Orr 2000; Wilke 2004; Kim and Orr 2005). In this framework, all beneficial mutations that are eventually outcompeted are “wasted” and interference is viewed as hindering adaptation. Sex circumvents the problem by allowing the competing beneficial mutations to be combined in the same genome. Yet a central premise of this argument is that beneficial mutations are additive or at least semiadditive (i.e., magnitude epistasis). If mutations instead show extensive sign epistasis (i.e., the landscape is moderately rugged), then the tendency for sex to combine individual beneficial mutations is of no advantage since they tend not to be beneficial in combination (Jain and Krug 2007). Note, however, that this argument does not extend beyond individual mutations. Sex may be important because it can also recombine genomic units that compose more extensive variation (e.g., regions with multiple substitutions). This can produce combinations of mutations that are both highly beneficial and, if the landscape is rugged, not available through stepwise adaptation (Watson et al. 2006).
Regardless of the advantages of sexual reproduction, we suggest that for asexuals, interference dynamics may not be detrimental as generally is assumed. Interference may instead be a creative evolutionary force because it allows the population to explore more sequence space than is possible under selective sweep dynamics (Jain and Krug 2007). On rugged landscapes, this amounts to exploring more evolutionary trajectories before one is chosen by selection.
Conclusion:
Our research has several implications about adaptive evolution and attempts to model it. Except when populations are quite small (≤104), selective sweep dynamics are unlikely for many microbial populations. Larger populations experience extensive interference dynamics where multiple beneficial mutations may arise on any background, although they are more likely to arise on the most fit backgrounds. As population size and the amount of interference increase, the particular mutations that establish and increase in frequency become less stochastic. We find evidence that the distribution of fitness effects changes between two backgrounds. This is not consistent with a fully uncorrelated landscape. Also inconsistent is the large number of beneficial mutations on a high-fitness first-step background. Nor do the data support a smooth, fully additive landscape. The truth likely lies somewhere in between. The data from our system suggest that the most reasonable modeling assumptions involve interference dynamics on partially correlated landscapes where mutations may occur on any background present in the population. While these findings may not be surprising from a biological perspective, an examination of Figure 1 illustrates that models conforming to them do not currently exist. Alternatively, some existing models may be robust to violations of these assumptions, but with a few important exceptions (e.g., Hegreness et al. 2006) this has not been well demonstrated.
Acknowledgments
We thank J. J. Bull and Darin Rokyta for helpful comments on this manuscript. This work was supported by grants from the National Institutes of Health: 1R01GM076040 to P.J., with analytical resources provided by 3P20 RR16448 and 2P20 RR16454.
APPENDIX A: LIKELIHOOD-RATIO TEST FOR THE CHANGE OF FITNESS EFFECTS DISTRIBUTION ON DIFFERENT BACKGROUNDS
Fitness effects are determined by averaging the results of replicate fitness assays. The variance associated with these assays demonstrates that measurement error contributes significantly to the overall pattern of variation. Measurement error must be accounted for when attempting to distinguish between competing hypotheses regarding the distribution of fitness effects. Kassen and Bataillon (2006) as well as Beisel et al. (2007) recognized this fact when considering the fitness distribution of adaptive mutations in the first step of an adaptive walk. However, accounting for measurement error when two steps of adaptation are considered turns out to be a much more delicate problem with many more technical issues. There are two reasons for this. First, our data demonstrate that second-step mutations tend to have much smaller effects than first-steps, yet measurement error is on the same order of magnitude for the fitness effects of both first- and second-step mutations (Table 1, Figure 4). So relative measurement error associated with second-steps is quite high. The second problem involves boundary issues associated with the GPD. Since the data tend to favor truncated distributions and since the truncation boundaries are important parameters in the model, standard likelihood approaches are problematic.
Distribution of the fitness effect of a single mutant:
To simplify the presentation we begin by developing an approximation to the distribution of the average fitness effects of a single adaptive mutation. All of the more complex formulas needed to calculate the likelihood-ratio statistic will be variants of this basic distribution.
Let x be the fitness effect of a mutant relative to a particular wild type. Following Beisel et al. (2007) we assume that x follows the GPD, which is given as
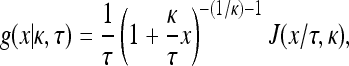 |
(A1) |
where J(z, κ) = I{κ < 0}I{0 ≤ z < −1/κ} + I{κ > 0}I{z > 0}. I represents an indicator function that takes the value 1 when the condition within parentheses is met and 0 otherwise. We evaluate Equation A1 at κ = 0 by taking the limit as κ goes to zero and noting
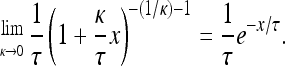 |
Let Y1, Y2,…, Ym be the replicate observed fitness values for a mutant with (unobserved) fitness x. Using the normal distribution to account for measurement error, we assume that conditional on x, Y1, Y2,…, Ym is a random sample from a normal distribution with mean x and variance σ2. Let 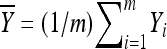 and
and 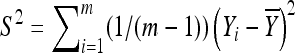 be the sample mean and variance, respectively. It follows from basic statistical theory that (given x)
be the sample mean and variance, respectively. It follows from basic statistical theory that (given x) 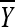 and S2 are sufficient statistics and
and S2 are sufficient statistics and 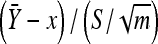 follows the t-distribution with m − 1 d.f. Given the sufficiency of the statistics
follows the t-distribution with m − 1 d.f. Given the sufficiency of the statistics 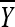 and S2 and the need to eliminate the nuisance parameter σ2, we replace the conditional likelihood of Y1, Y2,…, Ym given x with
and S2 and the need to eliminate the nuisance parameter σ2, we replace the conditional likelihood of Y1, Y2,…, Ym given x with
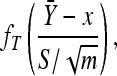 |
where 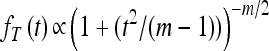 is the standard t-distribution.
is the standard t-distribution.
However, since x is unknown, to calculate the likelihood, we must average. This leads to the following likelihood equation:
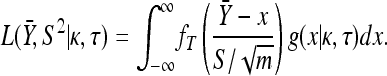 |
(A2) |
Importance sampling:
To calculate the integral in Equation A2 we use a method called importance sampling (Ripley 1987). To average over all possible x values we sample X1, X2,…, XB from a distribution Q(x) of our choosing and then approximate  by the following weighted average:
by the following weighted average:
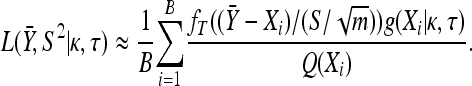 |
(A3) |
The validity of the above approximation follows from the law of large numbers. The efficiency of the above approximation depends critically on the choice of Q(x). The best choice for Q(x) would be the conditional distribution of x given 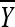 and S2. However, this distribution would depend on κ and τ and would actually require a priori knowledge of the integral in Equation A2. The purpose of importance sampling is to choose Q(x) close to the distribution of x given
and S2. However, this distribution would depend on κ and τ and would actually require a priori knowledge of the integral in Equation A2. The purpose of importance sampling is to choose Q(x) close to the distribution of x given 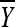 and S2. Recall that conditional on x,
and S2. Recall that conditional on x, 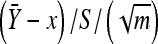 follows the t-distribution. This observation motivates the use of the following distribution for Q(x). Here we assume
follows the t-distribution. This observation motivates the use of the following distribution for Q(x). Here we assume 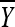 and S2 are fixed and define
and S2 are fixed and define
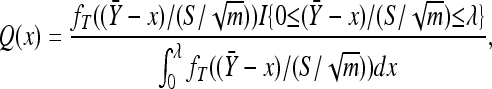 |
(A4) |
where λ = −τ/κ for κ < 0 and we assume that λ = ∞ when κ ≥ 0. Thus we use a truncated t-distribution for our importance sampling distribution Q(x) where we essentially reversed the roles of x and 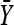 . This ensures that we efficiently sample unobserved Xi near the observed data
. This ensures that we efficiently sample unobserved Xi near the observed data 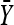 , but we must pay attention to the boundary conditions. It is easy to calculate the integral in the denominator of Equation A4 by using the standard t-distribution. For notational convenience we define
, but we must pay attention to the boundary conditions. It is easy to calculate the integral in the denominator of Equation A4 by using the standard t-distribution. For notational convenience we define
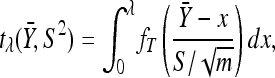 |
where T follows the standard t-distribution with m − 1 d.f. Substituting Equation A4 into A3,
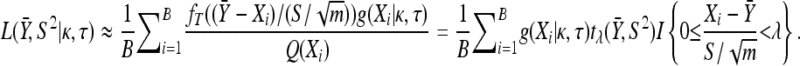 |
Likelihoods and likelihood ratios:
Formally, let 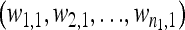 and
and 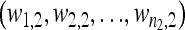 be the fitness values of the n1 and n2 observed first- and second-step mutations, respectively, listed from most fit to the least fit (i.e., according to rank order notation). Assume that the background for the second-step beneficial mutations is the most fit first-step background (i.e., is the background for second-steps). This assumption can be easily relaxed, but it holds true for the data we analyze. To reduce observational bias against small-effect mutations, we follow Beisel et al. (2007) and right-shift the data by redefining the smallest beneficial first-step mutation significantly more fit than wild type as zero. We shift two mutations instead of one because our smallest-effect first-step has fitness near the wild type (
be the fitness values of the n1 and n2 observed first- and second-step mutations, respectively, listed from most fit to the least fit (i.e., according to rank order notation). Assume that the background for the second-step beneficial mutations is the most fit first-step background (i.e., is the background for second-steps). This assumption can be easily relaxed, but it holds true for the data we analyze. To reduce observational bias against small-effect mutations, we follow Beisel et al. (2007) and right-shift the data by redefining the smallest beneficial first-step mutation significantly more fit than wild type as zero. We shift two mutations instead of one because our smallest-effect first-step has fitness near the wild type (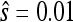 ; Table 1) and thus the one mutation shift would do little to protect against sampling bias. In vector form, the first- and second-step data are then defined as fitness differences above the second-smallest effect mutation:
; Table 1) and thus the one mutation shift would do little to protect against sampling bias. In vector form, the first- and second-step data are then defined as fitness differences above the second-smallest effect mutation:
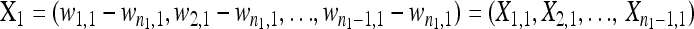 |
and
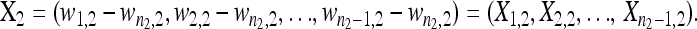 |
The null hypothesis states that 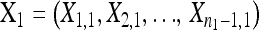 is a random sample drawn from a GPD (κ, τ) representing the fitness effects distribution of first-step mutations. Conditional on the largest-effect first-step mutation,
is a random sample drawn from a GPD (κ, τ) representing the fitness effects distribution of first-step mutations. Conditional on the largest-effect first-step mutation, 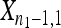 , the second-step fitnesses denoted by
, the second-step fitnesses denoted by 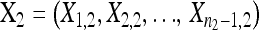 represent a random sample from a GPD (κ, τ*), where
represent a random sample from a GPD (κ, τ*), where 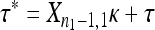 .
.
The alternative hypothesis states that 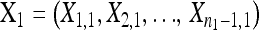 is a random sample from a GPD (κ1, τ1) representing the fitness effects distribution of first-step mutations. The second-step fitnesses, denoted by
is a random sample from a GPD (κ1, τ1) representing the fitness effects distribution of first-step mutations. The second-step fitnesses, denoted by 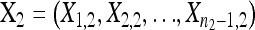 , represent a random sample from a GPD (κ2, τ2).
, represent a random sample from a GPD (κ2, τ2).
However, since X1 and X2 are not directly observable, we define
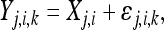 |
where ɛj,i,k is normally distributed with mean 0 and variance σj,i. Therefore, Yj,i,k is the observed fitness for the ith step (i = 1, 2), the jth mutant (j = 1, 2,…, ni − 1), and the kth replicate fitness assay (k = 1,…, mj). At each step (i) and for each mutant (j) we calculate the average (over replicates) 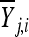 and variance
and variance 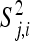 . In vector form,
. In vector form,
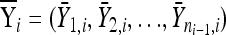 |
and
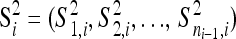 |
for each step i = 1, 2.
Then the likelihood of the data under the null hypothesis is given by
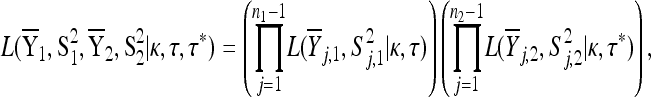 |
(A5) |
where 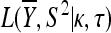 is given by Equation A2 and here
is given by Equation A2 and here 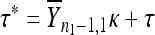 .
.
The likelihood equation under the alternative model is given by
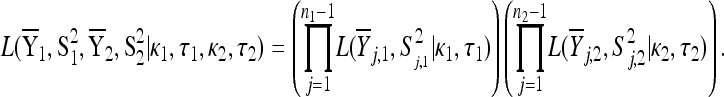 |
(A6) |
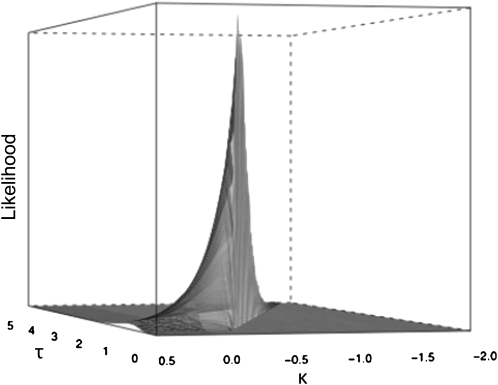
We obtain the maximum-likelihood estimates (MLEs) under both models by dividing the parameter space into discretized values and searching for the peak. The importance sampling size is set at 25 observations per mutation throughout. In practice, finding the MLEs turns out to be difficult because the likelihood surface is characterized by a ridge running diagonally across parameter values that is extremely narrow relative to the feasible resolution of the grid (Figure A1). The importance sampling furthers the problem by adding a slight stochasticity to where this ridge lies exactly. The strategy we use to map the likelihood surface is to begin with a value of λ (the boundary) slightly above the largest observation and calculate likelihoods across a discretized set of τ's and κ's that yield this λ (recall λ = −τ/κ). We then shift λ downward in small increments until the likelihood drops. In fact, the drop is cliff-like to nearly zero with decreasing λ (Figure A1). This is followed by shifting λ upward in small increments until the ridge top is crossed and peak likelihoods decline substantially. To avoid bias against unbounded distributions (which lack λ), we also search in a grid-like manner for incrementally increasing values of κ > 0 across a broad range of τ's. Whereas our initial grid-like search of parameter space yielded inconsistent results, this strategy of searching along the diagonal yielded highly repeatable MLEs.
Figure a1.
Figure A1.—Likelihood surface for first-step mutations under the alternative model. Note the severity of the high-likelihood ridge cutting diagonally across the surface (i.e., for a fixed λ). Also note the cliff-like drop as λ decreases (i.e., with decreasing τ and κ). The point of drop-off is defined by the largest observed fitness value.
To calculate the likelihood-ratio statistic, we begin by calculating the MLEs under the null. Denote the MLEs under the null as 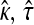 and
and 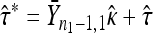 . To fit the alternative model to the data, we calculate the four MLEs under the alternative denoted by
. To fit the alternative model to the data, we calculate the four MLEs under the alternative denoted by 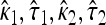 . Denote the likelihood-ratio test statistic by
. Denote the likelihood-ratio test statistic by
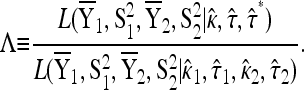 |
We calculate the distribution of Λ under the null hypothesis using the parametric bootstrap approach with 200 replicates, and from this distribution we calculate a P-value. Parametric bootstraps involve (1) resampling mutations from the estimated distributions, (2) resampling replicate fitness assays for each mutation with sample noise drawn from a normal with mean zero and variance equal to the average variance observed across fitness assays (0.77), and (3) averaging replicate assays for each mutant to obtain a data set for analysis.
APPENDIX B: DISTANCE BETWEEN LARGEST SECOND- AND FIRST-STEP TESTS
The observed distance between the largest second-step mutation and its background, the largest first-step, does not depend on how many small-effect second-step mutations were unobserved. Consequently, this distance provides a robust summary statistic to test the null hypothesis that both first- and second-steps come from the same distribution. The test is based on the distribution of the distance statistic under the null model and entails the following basic steps: (1) estimate τ and κ under the null model; (2) simulate 1000 data sets under the null; (3) for each data set, calculate the distance between the largest second-step and its background; and (4) compile distances into a distribution and define the P-value as the proportion of these equal to or greater than the observed distance. Note that in step 1, first-step data are shifted to the smallest first-step mutation significantly more fit than the wild type while second-steps are not shifted because such a shift is not feasible under the null (see materials and methods). In step 2, we are conservative by simulating under the upper bound on κ, rather than  , which shifts the simulated distances to larger values. For τ in the step 2 simulations, we use the value that yields the same λ produced by point estimates on τ and κ (on the basis of λ = −τ/κ). Finally, we note that simulations showed the test result (the P-value) was not sensitive to our choice of κ or τ as long as the effect sizes and the sampling noise remain on the same scale (i.e., λ is not changed appreciably).
, which shifts the simulated distances to larger values. For τ in the step 2 simulations, we use the value that yields the same λ produced by point estimates on τ and κ (on the basis of λ = −τ/κ). Finally, we note that simulations showed the test result (the P-value) was not sensitive to our choice of κ or τ as long as the effect sizes and the sampling noise remain on the same scale (i.e., λ is not changed appreciably).
APPENDIX C: BIAS-CORRECTED PARAMETER ESTIMATION
The LRT described in appendix a and applied to our data favors the alternative, two-distribution model (see results). We, therefore, wish to obtain estimates of κ and τ and confidence intervals around them under this model. Since first- and second-steps are independent under the alternative model, parameter estimation can be undertaken for one data set at a time. Analysis of simulated data sets with known parameters revealed that the MLEs from appendix a are biased. The nature of the bias is to favor more negative κ-values and the truncated distributions these produce. In essence, the likelihood is usually maximized by imposing a boundary (λ) just above the largest observation even when the true distribution has a tail beyond this, leading to a bias. Truncated distributions where the truncation point is a parameter in the model, referred to in the statistical literature as “range-dependent models,” often show this kind of bias. We need to correct for this bias and do so as follows. For a fixed value of κ, simulate 100 data sets on the basis of sampling from the GPD the same number of mutations as the real data and then generate observations equal in number to the real data, by adding normally distributed sampling error (μ = 0,  from the real data set). For the value of κ under consideration, fix τ so that the expected value of the resulting GPD (τ/(1 − κ)) will be equal to the observed sample average. This ensures that the magnitude of simulated fitness effects relative to the sampling error will remain consistent with the real data set across values of κ. For each of the 100 data sets, estimate κ by maximum likelihood as described in appendix a. Calculate the mean MLE across these 100 data sets as an approximation of
from the real data set). For the value of κ under consideration, fix τ so that the expected value of the resulting GPD (τ/(1 − κ)) will be equal to the observed sample average. This ensures that the magnitude of simulated fitness effects relative to the sampling error will remain consistent with the real data set across values of κ. For each of the 100 data sets, estimate κ by maximum likelihood as described in appendix a. Calculate the mean MLE across these 100 data sets as an approximation of  . Repeat this across a range of simulated κ-values (0, −0.25, −0.50, −0.75, −1.00, −1.25). We then plot
. Repeat this across a range of simulated κ-values (0, −0.25, −0.50, −0.75, −1.00, −1.25). We then plot 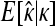 as a function of true κ and fit it to a simple function. This yields an approximation to the relationship
as a function of true κ and fit it to a simple function. This yields an approximation to the relationship 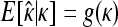 . Replacing
. Replacing 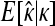 with
with 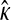 and solving for κ provides a bias-corrected estimate,
and solving for κ provides a bias-corrected estimate, 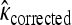 . The bias-correction functions, g(κ), along with the simulated data points they are based upon, are shown in Figure A2.
. The bias-correction functions, g(κ), along with the simulated data points they are based upon, are shown in Figure A2.
Figure a2.
Figure A2.—Bias correction in κ estimation. Data are simulated under known κ (horizontal axis) and used to estimate 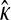 . Averaged over many data sets, this approximates
. Averaged over many data sets, this approximates 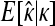 (vertical axis). Fitting these data points to simple functions and solving for κ provides a bias correction. As a comparison, the dashed line represents an unbiased estimator.
(vertical axis). Fitting these data points to simple functions and solving for κ provides a bias correction. As a comparison, the dashed line represents an unbiased estimator.
We apply this bias correction to the first- and second-step data sets separately. In both cases, the data are shifted to the smallest mutation substantially more fit than the background. To calculate approximate confidence intervals on the first- and second-step 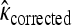 estimates, we again treat them separately and simulate 100 data sets under the bias-corrected estimates,
estimates, we again treat them separately and simulate 100 data sets under the bias-corrected estimates, 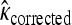 . For each data set, we obtain MLEs and apply the bias correction. The 100
. For each data set, we obtain MLEs and apply the bias correction. The 100 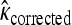 values are sorted, and those ranked second and 97th are used to form the confidence interval.
values are sorted, and those ranked second and 97th are used to form the confidence interval.
References
- Akaike, H., 1974. New look at statistical-model identification. IEEE Trans. Automat. Contr. Ac19 716–723. [Google Scholar]
- Beisel, C. J., D. R. Rokyta, H. A. Wichman and P. Joyce, 2007. Testing the extreme value domain of attraction for distributions of beneficial fitness effects. Genetics 176 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., 2009. Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage MS2. Genetics 181 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., and J. P. Bollback, 2006. Fitness effects of beneficial mutations: the mutational landscape model in experimental evolution. Curr. Opin. Genet. Dev. 16 618–623. [DOI] [PubMed] [Google Scholar]
- Brunet, E., I. M. Rouzine and C. O. Wilke, 2008. The stochastic edge in adaptive evolution. Genetics 179 603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J., M. R. Badgett and H. A. Wichman, 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17 942–950. [DOI] [PubMed] [Google Scholar]
- Campos, P. R., and V. M. de Oliveira, 2004. Mutational effects on the clonal interference phenomenon. Evolution 58 932–937. [DOI] [PubMed] [Google Scholar]
- Cuevas, J. M., P. Domingo-Calap, M. Pereira-Gómez and R. Sanjuán, 2009. Experimental evolution and population genetics of RNA viruses. Open Evol. J. 3 9–16. [Google Scholar]
- Desai, M. M., and D. S. Fisher, 2007. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics 176 1759–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, M. M., D. S. Fisher and A. W. Murray, 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser, J. A. G. M., C. W. Zeyl, P. J. Gerrish, J. L. Blanchard and R. E. Lenski, 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283 404–406. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., and P. D. Keightley, 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8 610–618. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Gerrish, P. J., and R. E. Lenski, 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103 127–144. [PubMed] [Google Scholar]
- Gillespie, J. H., 1984. Molecular evolution over the mutational landscape. Evolution 38 1116. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford.
- Gumbel, E. J., and H. V. Schelling, 1950. The distribution of the number of exceedances. Ann. Math. Stat. 21 247–262. [Google Scholar]
- Hegreness, M., N. Shoresh, D. Hartl and R. Kishony, 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311 1615–1617. [DOI] [PubMed] [Google Scholar]
- Holder, K. K., and J. J. Bull, 2001. Profiles of adaptation in two similar viruses. Genetics 159 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa, Y., F. Michor and M. A. Nowak, 2004. Stochastic tunnels in evolutionary dynamics. Genetics 166 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, K., and J. Krug, 2007. Deterministic and stochastic regimes of asexual evolution on rugged fitness landscapes. Genetics 175 1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, P., D. R. Rokyta, C. J. Beisel and H. A. Orr, 2008. A general extreme value theory model for the adaptation of DNA sequences under strong selection and weak mutation. Genetics 180 1627–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen, R., and T. Bataillon, 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38 484–488. [DOI] [PubMed] [Google Scholar]
- Kauffman, S. A., 1993. The Origins of Order. Oxford University Press, Oxford.
- Kim, Y., and H. A. Orr, 2005. Adaptation in sexuals vs. asexuals: clonal interference and the Fisher–Muller model. Genetics 171 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle, K., S. Cobey, B. Grenfell and M. Pascual, 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314 1898–1903. [DOI] [PubMed] [Google Scholar]
- Lynch, M., 2006. The origins of eukaryotic gene structure. Mol. Biol. Evol. 23 450–468. [DOI] [PubMed] [Google Scholar]
- Macken, C. A., and A. S. Perelson, 1989. Protein evolution on rugged landscapes. Proc. Natl. Acad. Sci. USA 86 6191–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. R., P. Joyce and L. P. Waits, 2005. A new method for estimating the size of small populations from genetic mark-recapture data. Mol. Ecol. 14 1991–2005. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Some genetic aspects of sex. Am. Nat. 8 118–132. [Google Scholar]
- Orr, H. A., 2000. The rate of adaptation in asexuals. Genetics 155 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2002. The population genetics of adaptation: the adaptation of DNA sequences. Evolution 56 1317–1330. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6 119–127. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2006. The population genetics of adaptation on correlated fitness landscapes: the block model. Evolution 60 1113–1124. [PubMed] [Google Scholar]
- Park, S. C., and J. Krug, 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104 18135–18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson, A. S., and C. A. Macken, 1995. Protein evolution on partially correlated landscapes. Proc. Natl. Acad. Sci. USA 92 9657–9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito, L., L. Fernandes, C. Mota and I. Gordo, 2007. Adaptive mutations in bacteria: High rate and small effects. Science 317 813–815. [DOI] [PubMed] [Google Scholar]
- Quer, J., R. Huerta, I. S. Novella, L. Tsimring, E. Domingo et al., 1996. Reproducible nonlinear population dynamics and critical points during replicative competitions of RNA virus quasispecies. J. Mol. Biol. 264 465–471. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2009. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Raney, J. L., R. R. Delongchamp and C. R. Valentine, 2004. Spontaneous mutant frequency and mutation spectrum for gene A of Phi X174 grown in E-coli. Environ. Mol. Mutagen. 44 119–127. [DOI] [PubMed] [Google Scholar]
- Ripley, B. D., 1987. Stochastic Simulation. Wiley, New York.
- Rokyta, D., M. R. Badgett, I. J. Molineux and J. J. Bull, 2002. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol. Biol. Evol. 19 230–238. [DOI] [PubMed] [Google Scholar]
- Rokyta, D. R., P. Joyce, S. B. Caudle and H. A. Wichman, 2005. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat. Genet. 37 441–444. [DOI] [PubMed] [Google Scholar]
- Rokyta, D. R., C. J. Beisel and P. Joyce, 2006. a Properties of adaptive walks on uncorrelated landscapes under strong selection and weak mutation. J. Theor. Biol. 243 114–120. [DOI] [PubMed] [Google Scholar]
- Rokyta, D. R., C. L. Burch, S. B. Caudle and H. A. Wichman, 2006. b Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 188 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta, D. R., C. J. Beisel, P. Joyce, M. T. Ferris, C. L. Burch et al., 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, D. E., J. A. G. M. de Visser and P. J. Gerrish, 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12 1040–1045. [DOI] [PubMed] [Google Scholar]
- Sanjuán, R., A. Moya and S. F. Elena, 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander, O. K., O. Tenaillon and L. Chao, 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5 e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. M., 1970. Natural selection and the concept of a protein space. Nature 225 563–564. [DOI] [PubMed] [Google Scholar]
- Wahl, L. M., and P. J. Gerrish, 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55 2606–2610. [DOI] [PubMed] [Google Scholar]
- Wahl, L. M., and D. C. Krakauer, 2000. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics 156 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. A., D. M. Weinreich and J. Wakeley, 2006. Effects of intra-gene fitness interactions on the benefit of sexual recombination. Biochem. Soc. Trans. 34 560–561. [DOI] [PubMed] [Google Scholar]
- Weinreich, D. M., and L. Chao, 2005. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution 59 1175–1182. [PubMed] [Google Scholar]
- Weinreich, D. M., R. A. Watson and L. Chao, 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59 1165–1174. [PubMed] [Google Scholar]
- Wilke, C. O., 2004. The speed of adaptation in large asexual populations. Genetics 167 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1932. The rules of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of the Sixth International Congress of Genetics, pp. 356–366. New York Brooklyn Botanic Garden, Brooklyn, NY.