Abstract
Behavior is influenced by an organism's genes and environment, including its interactions with same or opposite sex individuals. Drosophila melanogaster perform innate, yet socially modifiable, courtship behaviors that are sex specific and require rapid integration and response to multiple sensory cues. Furthermore, males must recognize and distinguish other males from female courtship objects. It is likely that perception, integration, and response to sex-specific cues is partially mediated by changes in gene expression. Reasoning that social interactions with members of either sex would impact gene expression, we compared expression profiles in heads of males that courted females, males that interacted with other males, or males that did not interact with another fly. Expression of 281 loci changes when males interact with females, whereas 505 changes occur in response to male–male interactions. Of these genes, 265 are responsive to encounters with either sex and 240 respond specifically to male–male interactions. Interestingly, 16 genes change expression only when a male courts a female, suggesting that these changes are a specific response to male–female courtship interactions. We supported our hypothesis that socially-responsive genes can function in behavior by showing that egghead (egh) expression, which increases during social interactions, is required for robust male-to-female courtship. We predict that analyzing additional socially-responsive genes will give us insight into genes and neural signaling pathways that influence reproductive and other behavioral interactions.
BEHAVIORS are complex processes resulting from an organism's ability to integrate sensory cues into physiological and motor outputs. Adding to the complexity of this process are the effects from the organism's genetics and environment, including social interactions, on behavior, brain morphology, and gene expression (Siegel and Hall 1979; Levine et al. 2002; Shen et al. 2004; Stewart and McLean 2004; Burmeister et al. 2005; Kozorovitskiy et al. 2006; Yurkovic et al. 2006; Carney 2007; Technau 2007; Ellis and Carney 2009).
It is possible to use microarray technology to assess changes in mRNA expression occurring during or in response to behavioral interactions to gain insight into corresponding physiological changes. Several studies, particularly in songbirds, bees, and fruit flies, have examined transcript-level changes in freely behaving animals. In songbirds, 33 genes are regulated by singing behavior, including loci involved in signal transduction and synaptic signaling (Wada et al. 2006), and a variety of social environments and stimuli impact honeybee brain gene expression (Grozinger et al. 2003; Whitfield et al. 2003, 2006; Sen Sarma et al. 2009). Similarly, male Drosophila melanogaster show rapid changes in transcript levels due to social interactions with females (Carney 2007; Ellis and Carney 2009). However, we do not know if these are specific responses to females or more general responses to interacting with a second individual. Although the signaling cascades mediating changes in mRNA levels due to behavior and social interactions are unclear, by studying these changes we can clarify the intracellular processes affecting nervous system function, physiology, and behavior. An advantage of such studies in Drosophila is that mutant strategies can be employed to characterize behavioral requirements for responsive loci.
The courtship behaviors of male Drosophila are influenced by genetics (reviewed in Billeter et al. 2002) and social interactions (Ewing 1983; reviewed in Greenspan and Ferveur 2000; Mehren et al. 2004). The somatic sex-determination pathway regulates these behaviors (reviewed in Cline 2005; Shirangi and McKeown 2007) and sexually dimorphic development, including that of the nervous system (Finley et al. 1997; Kimura et al. 2005; Manoli et al. 2005; Stockinger et al. 2005; Rideout et al. 2007; Sanders and Arbeitman 2008; Mellert et al. 2010; Rideout et al. 2010; reviewed in Billeter et al. 2006). Although target loci of the transcriptional regulatory members of this pathway are known (Burtis et al. 1991; Cann et al. 2000; Kopp et al. 2000; Dauwalder et al. 2002; Fujii and Amrein 2002; Drapeau et al. 2003; Arbeitman et al. 2004; Goldman and Arbeitman 2007; Lazareva et al. 2007; Fujii et al. 2008; Dalton et al. 2009), few have clearly defined functions in behavior and neural development. Several Drosophila microarray studies were key to identifying most of these downstream targets (Arbeitman et al. 2004; Goldman and Arbeitman 2007; Dalton et al. 2009), but the strategies used do not allow us to distinguish target genes that affect development of the nervous system from those that impact physiology and behavior post development.
During courtship or other social interactions, males are exposed to sensory information that must be rapidly interpreted to create the appropriate behavioral response (e.g., to continue courtship directed toward that fly or to seek a new mate). In males, interacting with a second individual causes rapid expression-level changes detectable in whole animals (Carney 2007; Ellis and Carney 2009). These rapid responses are likely mediated by signaling in the nervous system, sensory organs, and other tissues that affect neural physiology. Our expression analysis approach has the advantage of using wild-type animals performing behaviors to identify adult-expressed gene products that are impacted by behavior, including target genes of the somatic sex-determination hierarchy.
Since our earlier studies did not address the possibility that some of the loci that respond to male–female interactions might be generally “socially-responsive” genes rather than specifically “courtship-responsive” genes, we examine this possibility in our study by examining gene expression changes occurring in the male head (rather than in the whole body) during interactions with either females or males. We also expanded on our earlier studies by showing that socially-responsive loci can function in behavior. Our data indicate that social interactions cause expression changes in loci expressed in neuronal tissue as well as in non-neuronal adipose tissue that may modulate neural signaling and behavior.
MATERIALS AND METHODS
Microarray analysis:
We used an isogenized wild-type Canton-S (CS) strain and handled flies similarly to Carney (2007) except that the females' genitals were electrically cauterized to prevent mating (non-mateable females). Twenty or fewer virgin isogenic CS males were aged collectively for 3 days, and ≤20 virgin isogenic CS females were aged collectively for 3 days. On day 4, males were aspirated into individual vials, and females had their genitals cauterized by passing a 4-mA current over two fine tungsten wires on the external genitalia of the female to prevent mating. Females recovered for 1 day in a new vial. All flies were kept on a 12-hr light/dark cycle at 25°, and we performed all procedures within 2 hr of lights on to control for circadian effects on gene expression and behavior.
Analysis of courtship behavior on day 5 included equally dividing males into three groups: (1) courting male, (2) male–male, and (3) control. For the courting male treatment, one cauterized female was aspirated into a male's vial. For the male–male group, a second male was aspirated into the vial of a single male. Control males were treated in the same way except that a second fly was not transferred during the aspiration process. Courtship or male–male exposure lasted for 20 min. In courting-male treatments, the presence of courtship was assessed at 1-min intervals. Only males that courted a female for at least 70% of the observation time were collected for analysis. During this time, brief male–male interactions (lasting only a few seconds) were observed. We did not detect locomotor differences among males in the three treatment groups (two-tailed t-test, P > 0.05). After 20 min, males were removed from the vials, quick-frozen in liquid nitrogen, and stored at −80° for future RNA extraction.
We separated heads from the rest of the bodies by vortexing quick-frozen flies. For each treatment, 20 male heads were randomly assigned to 1 of 10 groups, giving us 10 RNA preparations for each of the courting-male, male–male, or control treatments. Total head RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) following standard protocols. The University of Kentucky MicroArray Core Facility labeled and hybridized 5 RNA preparations each from courting-male, male–male, and control heads (15 individual samples) to Affymetrix Drosophila 2.0 Genome Arrays following standard Affymetrix (Santa Clara, CA) protocols. Therefore, animals for all three treatment groups were collected and analyzed at the same time, and all 15 microarray hybridizations were carried out concurrently.
We extracted expression values from the microarrays using five algorithms: GeneChip Operating Software (MAS 5.0, Affymetrix), Gene Spring (Agilent, Santa Clara, CA), PM, and PM-MM (dChip; Li and Wong 2001), and GCRMA (R Development Core Team). For paired-data analysis comparing courting male and control treatments, we conducted a Bayesian t-test (CyberT; Baldi and Long 2001) and false-discovery rate analyses (q < 0.05, Storey and Tibshirani 2003), requiring that P < 0.001 in at least three of five algorithms. For a combined analysis of the three data sets, we used the SAS Mixed procedure (SAS Institute, Cary, NC) and identified significantly up- and downregulated socially-responsive genes (P < 0.05 in at least three of five algorithms).
Courtship-responsive genes are those for which expression in courting-male heads differs from that in control and male–male heads (control = male–male expression). Male–male-responsive genes are those that differ only in male–male interactions (control = courting male expression). Other genes that respond to interactions with both sexes were placed in the general category of socially-responsive genes.
Quantitative PCR:
We validated the microarray results by quantitative PCR (qPCR) analysis on the 5 control and 5 courting male RNA preparations not used for microarray hybridization. cDNA was synthesized from poly+A purified (Oligotex mRNA mini kit, Qiagen) RNA using the SuperScript First-Strand Synthesis System (Invitrogen).
Since few of the socially-responsive loci identified from the paired analysis (courting male compared to control) had known or predicted functions in behavior, primers were designed for a randomly chosen set of six upregulated (CG9377, CG10621, egh, HLHmβ, Lsp2, sug) and three downregulated (CG31181, Rim, Sh) candidate genes. We chose a range of genes with adult expression predicted to be enriched in the brain (CG9377, Rim, Sh), fat body (Lsp2, sug), or both tissues [CG10621, CG31181, HLHmβ, egh (egh expression is very low in fat)] (Chintapalli et al. 2007). Genes with low predicted transcript levels in the head were not tested (Chintapalli et al. 2007). Of these selected genes, only egh and Sh had previously described reproductive behavioral roles in females and males, respectively. To control for amplification specificity, primer pairs were designed across introns when possible. No template controls as well as controls with template but without reverse transcriptase were included.
Using the ABI7500 and its default parameters (Applied Biosystems), each template was run in triplicate, using 2 μl of a 1:4 cDNA dilution and the SYBR Green PCR Mastermix (Applied Biosystems). We used dissociation curve analysis to determine primer-specific amplification and the relative standard curve method (Applied Biosystems) to determine transcript levels. Normalization to rp49 levels generated relative transcript abundance values for control or courting-male samples. The relative fold change for each gene was measured as the ratio of courting-male relative abundance to control-male relative abundance, and significance was determined by a two-tailed t-test. Upregulation of egh and HLHmβ and downregulation of CG31181 were confirmed by secondary qPCR analysis.
A regression analysis of microarray mean expression fold changes compared to independent qPCR fold changes indicated a significant positive correlation between results obtained by both methods (r = 0.68, N = 9, P = 0.006).
In situ hybridization:
We performed in situ hybridization for a subset of socially-responsive genes using cDNA clones for CG9377 (GH08193), CG10621 (RE64786), cwo (LD15411), egh (GH01085), and sug (LD36528). Antisense and sense probes were made from the above clones using the digoxigenin (DIG)-labeling kit's standard protocol (Roche, Nutley, NJ). Probes were hydrolyzed into 200-bp fragments and hybridized to dissected male tissues (brains, heads, or abdominal carcasses) as previously described (Arbeitman et al. 2004).
To confirm that fit expression increased in courting males compared to control males, we generated antisense and sense probes directed against fit using the RH40291 clone. Control and courting-male heads were cryosectioned and incubated with fit probes as described above. We only detected signal using antisense probes.
Courtship behavior analysis:
Flies were maintained on a 12-hr light/dark cycle at 25°, except when noted otherwise. The Bloomington Stock Center supplied P-element insertion mutants (eghEP804, eghEY03917). Both insertions are located within the first egh exon and reduce egh expression to barely detectable levels (supporting information, File S1 and Figure S1). For both X-linked P-element insertions, we crossed P-element females to isogenic CS males, and we crossed P-element males to isogenic CS females, generating experimental and control males, respectively, in genetically similar backgrounds. For behavioral analysis, P-element and control males were aged at 25° in individual vials for 4–5 days and CS virgin females (≤20) were aged collectively for 3–5 days. All courtship tests with egh mutants were performed in dim red-light conditions because mutations in egh affect photoreceptor pathfinding (Fan et al. 2005) and therefore likely impact eye function. In red-light conditions, fly courtship relies more heavily upon sensory systems other than the eye.
We analyzed courtship behavior under red light at 22°. A male was aspirated into a mating chamber (diameter = 1 cm) and a virgin CS female was introduced 2 min later. The pair was video recorded for 10 min. The courtship index (CI)—i.e., the time performing courtship divided by the total observation time—was calculated. CI values were arcsine-transformed, and two-tailed t-test comparisons between mutants and controls were calculated to determine significance (P < 0.05).
To reduce egh specifically in the adult nervous system, we utilized two egh RNA interference (RNAi) alleles, eghv45160 and eghv45161, from the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al. 2007). We used in situ hybridization to verify reduced egh expression upon activation of each RNAi allele (Figure S1).
We targeted egh reduction pan-neuronally with elavc155-Gal4 (Lin and Goodman 1994) and more specifically with apmd544-Gal4 (Calleja et al. 1996), which is expressed in ap-expressing neurons in larval and adult nervous systems (Figure S2). We increased the efficiency of the RNAi process by adding one copy of UAS-Dicer-2 (VDRC). To reduce egh specifically in adults, the RNAi alleles were under the control of UAS-tubulin-Gal80ts (reviewed in McGuire et al. 2004). Crosses were maintained at the permissive temperature. Control males had UAS-Dicer-2 and UAS-tubulin-Gal80ts and either the RNAi allele or the Gal4 driver. We collected virgin males and stored them in individual vials at either 20° or 29°. The courtship objects, CS virgin females, were collected and stored collectively at 25°. Behavioral analysis was conducted under red light at the aforementioned temperatures. We used ANOVA and Tukey's post-hoc analysis to determine significant changes in CI due to temperature and genotype.
To restore egh expression, we crossed a genomic rescue construct (eghP2) to eghEY03917 and compared CIs of eghEY03917; eghP2 males to eghEY03917 males. To narrow down which cells require egh expression for proper courtship behavior, we utilized the rescue construct, UAS-eghHA (Soller et al. 2006). We crossed UAS-eghHA to the apmd544-Gal4 driver in the eghEY03917 background. eghEY03917 males with either component of the Gal4/UAS system served as controls. Both rescue experiments were carried out at 22° under red light.
RESULTS
Changes in male gene expression during social interactions with females or males:
Within 5 min of male-to-female social interactions, whole-animal transcript profiles are altered in courting males, and there is a differential response to conspecific compared to heterospecific females (Carney 2007; Ellis and Carney 2009). Next, we focused solely on male-head gene expression in response to courtship since the head contains the brain as well as other tissues and sensory organs that impact behavioral and physiological responses to sensory inputs. We extended the courtship interaction period to 20 min to ensure a robust response and used Affymetrix Drosophila 2.0 Genome Arrays to examine ∼18,500 transcripts for expression-level changes in males performing courtship toward non-mateable females (referred to as “courting males”) compared to males that were not given a female courtship object (“control males”) (see materials and methods).
Bayesian CyberT analysis comparing expression values from heads of courting males to those from controls identified 35 loci with altered expression due to male–female interactions (see materials and methods). Sixteen transcripts were upregulated (Table 1) and 19 were downregulated (Table 2) after 20 min of courtship. These changes are not likely due to locomotor differences since courting and control males have similar activity levels during the assay period (two-tailed t-test, P > 0.05). The small number of loci with altered expression is consistent with results from other behavioral studies (e.g., Lawniczak and Begun 2004; Mack et al. 2006; Wada et al. 2006) and is partially a consequence of our extremely conservative criteria for identifying responsive genes (see materials and methods).
TABLE 1.
Candidate genes upregulated after 20 min of courtship
| Gene identifier | Gene name | Average fold change | GO molecular function | GO biological process |
|---|---|---|---|---|
| CG1897 | Drop (Dr) | 1.46 | Sequence-specific DNA binding | Nervous system development |
| CG3850 | sugarbabe (sug) | 1.6 | Transcription activator activity | Regulation of transcription |
| CG6494 | hairy (h) | 1.52 | Transcription repressor activity | Nervous system development |
| CG6806 | Larval serum protein 2 (Lsp2) | 1.34 | Nutrient reservoir activity | Transport |
| CG9377 | 1.7 | Serine-type endopeptidase activity | Proteolysis | |
| CG9659 | egghead (egh) | 1.36 | β-1,4-mannosyltransferase activity | Axon guidance and oogenesis |
| CG10142 | Ance-5 | 1.34 | Metallopeptidase activity | Proteolysis |
| CG10621 | 1.44 | Homocysteine S-methyltransferase activity | Unknown | |
| CG10812 | drosomycin-5 (dro5) | 1.34 | Unknown | Defense response to fungus |
| CG14489 | olf186-M | 1.26 | Unknown | Unknown |
| CG14548 | E(spl) region transcript mβHLHmβ | 1.58 | Transcription repressor activity | Nervous system development |
| CG14688 | 1.28 | Unknown | Unknown | |
| CG17100 | clockwork orange (cwo) | 1.45 | Transcription repressor activity | Regulation of circadian rhythm |
| CG17820 | female-specific independent of transformer (fit) | 1.38 | Unknown | Unknown |
| CG18477 | 1.52 | Serine-type endopeptidase activity | Proteolysis | |
| CG42370 | 1.30 | Metalloendo-peptidase activity | Proteolysis |
Comparing control male heads to courting male heads revealed that 16 genes are significantly (P < 0.001) upregulated in male heads after 20 min of courtship.
TABLE 2.
Candidate genes downregulated after 20 min of courtship
| Gene identifier | Gene name | Average fold change | GO molecular function | GO biological process |
|---|---|---|---|---|
| CG1522 | cacophony (cac) | −3.08 | Voltage-gated calcium channel activity | Courtship behavior |
| CG2217 | −1.36 | Unknown | Unknown | |
| CG3738 | Cyclin-dependent kinase subunit 30A (Cks30A) | −1.36 | Cyclin-dependent protein kinase regulator activity | Cyclin catabolic process |
| CG4269 | −3.92 | Unknown | Unknown | |
| CG9266 | −1.28 | Unknown | Unknown | |
| CG9983 | hnRNA-binding protein 1 (Hrb98DE) | −1.24 | Nucleic acid binding | Unknown |
| CG10077 | −1.3 | ATP-dependent helicase activity | Unknown | |
| CG10851 | B52 | −1.3 | Nucleic acid binding | Regulation of nuclear mRNA splicing via spliceosome |
| CG12295 | straightjacket (stj) | −1.3 | Voltage-gated calcium channel activity | Synaptic vesicle fusion to presynaptic membrane |
| CG12348 | Shaker (Sh) | −1.32 | Voltage-gated cation channel activity | Regulation of synaptic activity and courtship behavior |
| CG12449 | Glutamine: fructose-6-phosphate aminotrans-ferase 1 (Gfat) | −1.38 | Glutamine-fructose-6-phosphate transaminase activity | Carbohydrate biosynthetic process |
| CG12478 | bruno-3 (bru-3) | −1.26 | RNA binding | Negative regulation of translation |
| CG31181 | −1.36 | Unknown | Unknown | |
| CG31182 | −1.44 | Unknown | Unknown | |
| CG33197 | muscleblind (mbl) | −1.44 | Zinc ion binding | Muscle development |
| CG33547 | Rim | −1.44 | Small GTPase regulator activity | Regulation of exocytosis |
| CG42492 | −1.34 | Unknown | Unknown | |
| CG42670 | pasilla (ps) | −1.34 | Unknown | Unknown |
| CG42698 | pou domain motif 3 (pdm3) | −1.38 | Unknown | Unknown |
Average fold changes, molecular functions, and biological processes are shown for 19 genes that are significantly (P < 0.001) downregulated in male heads after 20 min of courtship.
We performed a second analysis of our data that included a third comparison to gene expression changes occurring as a consequence of male–male interactions. This strategy allowed us to distinguish loci whose expression changes due to interactions specifically with females from loci that change due to interaction with another individual of either sex (see materials and methods). A total of 505 genes responded to male–male interactions, while 281 genes responded to male–female interactions. Most expression changes that occur due to male–female interactions also occur as a consequence of male–male interactions (Table S1). The list of 265 genes in Table S1 includes 24 genes present in the original comparison between courting-male and control heads. We also identified 240 genes whose expression changes specifically in response to paired male interactions (Table S2).
Sixteen genes were responsive to male–female interactions but not to male–male interactions and therefore appear to be true courtship-responsive loci (Table 3). Five of these 16 loci [Drop, sugarbabe (sug), hairy, olf186-M, HLHmβ] also were present on the list (Table 1) from the paired comparison of courting males to control males. Six genes with strong statistical support in the initial comparison of courting males and control males [egh, Lsp2, clockwork orange (cwo), cacophony (cac), CG2217, CG4629] were not present in the new list of genes that responded to encounters with both sexes. However, cwo is on the list specific to male–male interactions (Table S2), suggesting that it may be socially-responsive. It is possible that expression changes in the remaining five genes from the paired comparison are also a specific response to courtship rather than a general response to social interactions. In support of this argument, cac functions in production of male courtship song. We refer to the broad group of genes identified here as “socially-responsive genes” and refer to specific subcategories (e.g., “courtship-responsive,” “male–male-responsive”) as appropriate.
TABLE 3.
Courtship-responsive genes
| Average fold change in courting male heads compared to: |
|||||
|---|---|---|---|---|---|
| Gene identifier | Gene name | Male–male heads | Control male heads | GO molecular function | GO biological process |
| CG1416 | 1.12 | 1.12 | ATPase activator activity | Response to stress | |
| CG1897 | Drop (Dr) | 1.35 | 1.44 | Sequence-specific DNA binding | Nervous system development |
| CG3850 | sugarbabe (sug) | 1.3 | 1.61 | Transcription activator activity | Regulation of transcription |
| CG6461 | 1.1 | 1.1 | Transferase activity | Unknown | |
| CG6494 | hairy (h) | 1.63 | 1.52 | Transcription repressor activity | Nervous system development |
| CG11877 | 1.11 | 1.12 | Unknown | Unknown | |
| CG13116 | 1.21 | 1.25 | Unknown | Unknown | |
| CG14489 | olf186-M | 1.18 | 1.27 | Unknown | Unknown |
| CG14548 | E(spl) region transcript mβ (HLHmβ) | 1.39 | 1.58 | Transcription repressor activity | Nervous system development |
| CG30445 | Tyrosine decarboxyl-ase 1 | 1.22 | 1.24 | Tyrosine decarboxylase activity | Unknown |
| CG31137 | twin | 1.28 | 1 | Transcription regulator activity | Nuclear-transcribed mRNA poly(A) tail shortening |
| CG4962 | CG4962 | −1.28 | −1.20 | Unknown | Unknown |
| CG14210 | CG14210 | −1.23 | −1.02 | Protein binding | Unknown |
| CG6081 | Cyp28d2 | −1.11 | −1.18 | Oxidoreductase activity | Oxidation reduction |
| CG32491 | modifier of mdg4 [mod(mdg4)] | −1.09 | −1.16 | Transcription factor activity | Induction of apoptosis |
| CG18525 | Serine protease inhibitor 5 (Spn5) | −1.14 | −1.11 | Serine-type endopeptidase inhibitor activity | Unknown |
Eleven upregulated and 5 downregulated genes are courtship-responsive when comparing all three treatment groups (control, courting, and male-exposed) by mixed ANOVA and Tukey's post-hoc analyses (P < 0.05).
qPCR validation of microarray results:
To verify our microarray results, we used qPCR to analyze transcript levels from a subset of socially-responsive genes from Tables 1 and 2. We compared expression in control and courting-male head RNA preparations not used for microarray hybridization. The six upregulated and three downregulated socially-responsive genes tested showed the expected trends in expression (Table 4; see also materials and methods) with fold changes comparable to those from the microarrays. Four of the genes, including the courtship-responsive loci sug and HLHmβ, showed statistically significant changes in courting males compared to controls. Since all nine genes showed the expected trend by qPCR, this is strong support for the validity of the microarray data. Increasing the sample sizes would likely increase the statistical support.
TABLE 4.
qPCR confirmation of the microarray results
| Gene identifier | Gene symbol | Average relative expression level in control male heads ±SEM | Average relative expression level in courting male heads ±SEM | Relative qPCR fold change ±SEM | Relative array fold change ±SEM |
|---|---|---|---|---|---|
| CG9377 | 0.07 ± 0.01 | 0.09 ± 0.02 | 1.25 ± 0.25 | 1.7 ± 0.07 | |
| CG10621 | 1.95 ± 0.46 | 2.42 ± 0.63 | 1.31 ± 0.28 | 1.44 ± 0.03 | |
| CG9659 | egh | 3.2 ± 1.07 | 6.62 ± 1.72 | 2.04 ± 0.42 | 1.36 ± 0.02 |
| CG14548 | HLHmβ | 0.65 ± 0.16 | 1.92 ± 0.62 | 3.13 ± 0.82* | 1.58 ± 0.05 |
| CG6806 | Lsp2 | 0.57 ± 0.17 | 1.5 ± 0.72 | 2.79 ± 1.09 | 1.34 ± 0.03 |
| CG3850 | sug | 3.95 ± 1.2 | 9.31 ± 2.58 | 2.36 ± 0.58* | 1.6 ± 0.05 |
| CG31181 | 0.6 ± 0.2 | 0.15 ± 0.11 | −2.7 ± 0.09* | −1.36 ± 0.03 | |
| CG33547 | Rim | 0.4 ± 0.09 | 0.03 ± 0.004 | −12.5 ± 0.006 | −1.44 ± 0.06 |
| CG12348 | Sh | 0.39 ± 0.09 | 0.12 ± 0.04 | −3.13 ± 0.09* | −1.32 ± 0.04 |
Average relative expression of nine genes was assessed by qPCR. An asterisk indicates a significant (P < 0.05) difference in the average relative expression level in control male heads compared to courting-male heads as determined by qPCR. SEM, standard error of the mean.
Socially-responsive genes are expressed in the brain and other head tissues:
Because we assayed head tissue, identified loci may be expressed in the brain, sensory structures, the fat body, or a combination of these tissues. Expression of many socially-responsive genes is enriched in the head relative to the brain, indicating higher expression in tissues outside of the brain (Chintapalli et al. 2007). Although some socially-responsive genes are enriched in the eye, others are enriched in head tissues other than the brain or eye, including the adipose tissue lining the brain (Table S3).
Two socially-responsive genes, Lsp2 and female-specific independent of transformer (fit), are expressed in fat surrounding the brain in both sexes (Benes et al. 1990; Fujii and Amrein 2002). fit was named for its high level of expression in females compared to males and because its expression is regulated by the somatic sex-determination hierarchy gene Sex-lethal (Fujii and Amrein 2002).
In an earlier whole-male microarray analysis, we detected a statistically significant increase in fit transcripts in courting males; this increase was validated by qPCR (Carney 2007). fit also increases in 20-min courting-male heads (Table 1) and is responsive to same-sex interactions as well (Table S1). Since we had not examined the specific tissue in which this increased expression occurs, we examined fit's response to 20-min courtship in sectioned male heads. Although expression is low in virgin males, fit levels increased in response to male–female interactions (Figure 1). This increase was detected in the fat body, an adipose tissue previously implicated in modulation of courtship behavior (reviewed in Dauwalder 2008). In situ hybridization confirmed that other socially-responsive genes are expressed in the male fat body (CG10621, sug), the male brain (CG9377, egh), or both tissues (cwo) (Figures 2 and 3).
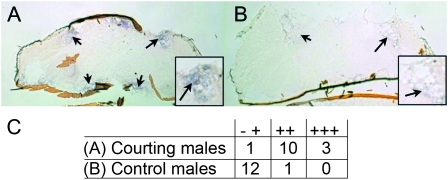
Figure 1.—
Courting males show increased fit expression in the fat body. A DIG-labeled fit RNA antisense probe was made from the RH40291 cDNA clone. In situ hybridization was performed on cryosectioned male heads and confirmed that fit transcript levels are upregulated in the adipose tissue (arrows) of courting males (A) compared to control males (B). A qualitative assessment of signal intensity in both treatment groups is presented in C.
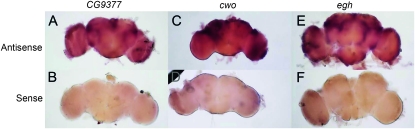
Figure 2.—
Socially-responsive genes CG9377, cwo, and egh are expressed in the male brain. Antisense (A, C, and E) or sense (B, D, and F) RNA probes were designed for cDNA clones for CG9377 (A and B), cwo (C and D), and egh (E and F). In situ hybridization to whole-mounted male CS tissue reveals that courtship-responsive genes are expressed in male brains. Dark pink staining in A, C, and E indicates expression due to hybridization of antisense probes.
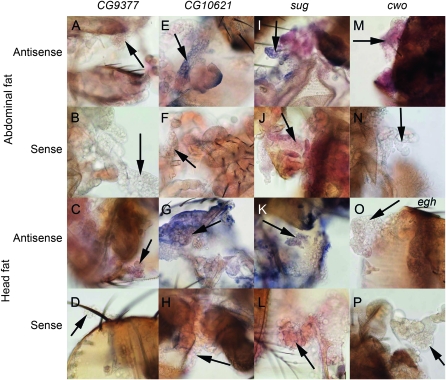
Figure 3.—
Socially-responsive genes CG10621, sug, and cwo are expressed in male adipose tissue. Antisense (A, C, E, G, I, K, M, and O) or sense (B, D, F, H, J, L, N, and P) RNA probes were designed to cDNA clones for CG9377 (A–D), CG10621 (E–H), sug (I–L), cwo (M and N), and egh (O and P). In situ hybridization to whole-mounted male CS tissue shows candidate gene expression in the fat body tissue (arrows) on abdominal (A, B, E, F, I, J, M–P) or head (C, D, G, H, K, and L) cuticle. Expression is indicated by light (cwo) or dark purple (CG10621, sug).
egghead is required in the adult male brain for robust courtship:
We hypothesized that genes with altered expression patterns due to male–female interactions likely modulate courtship behavior either by regulating the performance of courtship steps or by making the male a more efficient courter by increasing the efficiency of stimulus processing. This increased efficiency could affect the current courtship interaction or, more likely, subsequent courtship encounters. We predicted that we could identify behavioral functions for these loci by testing mutations in the genes for effects on male courtship behavior.
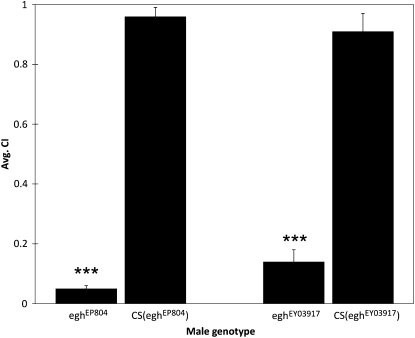
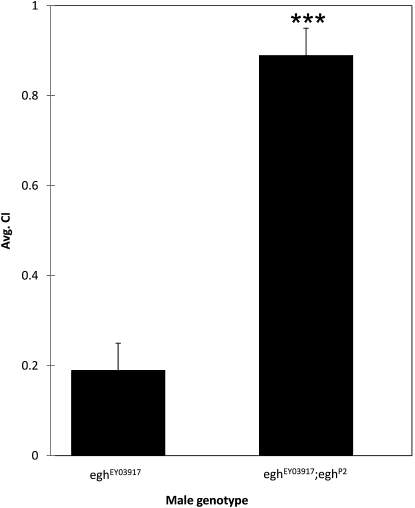
Therefore, we tested P-element insertions or VDRC strains targeting some of the male–female-responsive genes from Tables 1 and 2 for effects on male courtship activity (measured as the CI). For several of the alleles, we observed weak phenotypic effects on behavior. However, mutations in egh had strong effects on male–female courtship, so we focused our current downstream analysis on this locus. Males with either of two independent insertions in egh (eghEP804 and eghEY03917) performed all standard courtship behaviors but had significantly reduced CI values compared to genetically similar controls (Figure 4, two-tailed t-test, P < 0.001). We did not observe male–male courtship or aggressive interactions in groups of aged mutant males that were placed together in vials and observed over a 2-week period. Therefore, reduced egh expression led to an overall reduction in time spent courting a female but did not appear to affect male–male interactions. Reintroduction of a genomic copy of egh in the eghEY03917 background restored male-to-female courtship activity to wild-type levels (Figure 5, P <0.001), verifying that the courtship phenotype is due to disruption of the egh locus.
Figure 4.—
egh is required for robust male courtship behavior. Under red light, males with either X-linked egh insertion (eghEP804 or eghEY03917) showed significant (***P < 0.001) decreases in CI values compared to control males in a similar genetic background [CS(eghEP804) or CS(eghEY03917)] under similar conditions. Error bars reflect the SEM. N = 10 males for each genotype.
Figure 5.—
egh expression rescues male courtship behavior. Restoring egh expression in egh-expressing cells (eghP2) in the eghEY03917 mutant background significantly (***P < 0.001) rescued the courtship defect in eghEY03917 mutant males. N = 10 males for both genotypes.
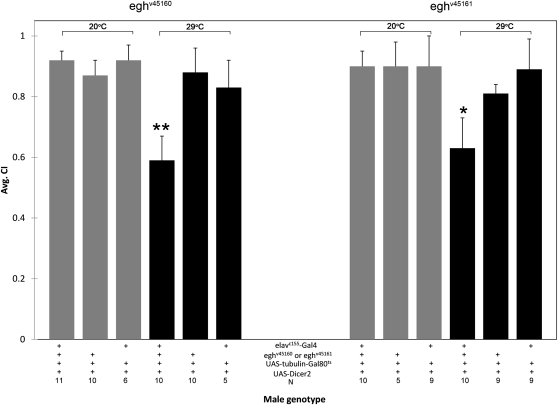
We selectively reduced egh in the adult nervous system with UAS-egh-RNAi under the control of UAS-tubulin-Gal80ts and neural-expressed elavc155-Gal4. This adult-specific decrease in egh resulted in significantly reduced CI values for experimental males at the restrictive temperature (29°) compared to all controls (Figure 6).
Figure 6.—
Male courtship requires egh expression in the adult nervous system. Expressing UAS-egh-RNAi alleles, eghv45160 or eghv45161, in the adult nervous system using elavc155-Gal4, UAS-Dicer-2, and UAS-tubulin-Gal80ts at the restrictive temperature (29°, black bars) significantly (**P < 0.01; *P < 0.05) reduced male courtship activity compared to 29° controls lacking elavc155-Gal4 or UAS-egh-RNAi or compared to males at the permissive temperature (20°, gray bars).
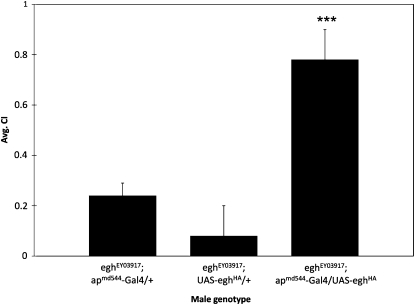
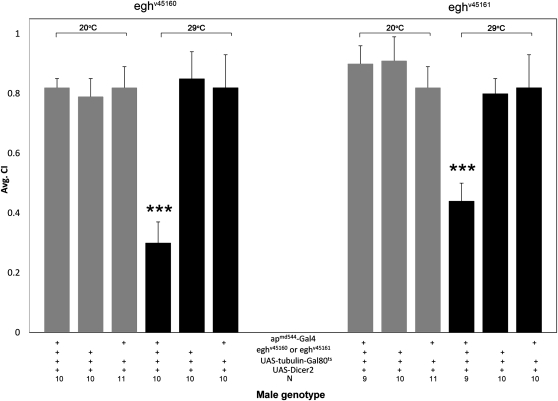
Larval egh expression is required in ap-expressing ventral nerve cord neurons for the female Sex-peptide response during adulthood (Soller et al. 2006). We asked whether this same circuit functioned in male reproductive behavior. Expressing egh (via UAS-eghHA) under control of apmd544-Gal4 in eghEY03917 mutant males was sufficient to restore male courtship behavior (Figure 7, P < 0.001), indicating that Ap neurons modulate reproductive behaviors in both sexes. Although Soller et al. (2006) attributed modulation of the Sex-peptide response to developmental expression, apmd544-Gal4 expresses in the male and female adult nervous system (Figure S2). Therefore, we expressed egh-RNAi via apmd544-Gal4 to specifically reduce egh expression in adult males (Figure 8). Targeted egh reduction significantly decreased courtship activity (P < 0.001), confirming that egh is needed in Ap neurons during adulthood for proper courtship behavior.
Figure 7.—
egh expression in ap-expressing neurons restores male courtship behavior. Narrowing egh expression to ap neurons by expressing UAS-eghHA under the control of apmd544-Gal4 in the eghEY03917 background significantly (***P < 0.001) restored male courtship activity compared to control eghEY03917 males lacking either component of the Gal4/UAS system. Ten males of each genotype were tested.
Figure 8.—
Adult expression of egh in ap-expressing neurons is necessary for robust courtship behavior. Expressing UAS-egh-RNAi alleles, eghv45160 or eghv45161, in Ap neurons during adulthood using apmd544-Gal4, UAS-Dicer-2, and UAS-tubulin-Gal80ts at the restrictive (29°, black bars) temperature significantly (***P < 0.001) reduced male courtship activity compared to controls lacking the Gal4 or UAS-egh-RNAi component or compared to males at the permissive temperature (20°, gray bars).
DISCUSSION
Social interactions alter male gene expression:
Drosophila perform stereotypical sex-specific courtship behaviors that are influenced by genetics, including the somatic sex-determination pathway, and by environmental cues, including social interactions. Previous studies have shown that male–female social interactions cause rapid (within 5 min) changes in whole-male transcript abundance (Carney 2007). In this study we focused on male-head tissue and found that 521 genes are socially-responsive in a 20-min interaction period. Expression of 281 genes changes during male–female interactions, while 505 genes are affected by male–male interactions. At least 16 of these loci are specifically courtship responsive (Table 3). Similarly to genes identified in array studies on songbirds and honeybees responding to behavioral cues (Grozinger et al. 2003; Whitfield et al. 2003, 2006; Wada et al. 2006; Sen Sarma et al. 2009), the 16 Drosophila courtship-responsive genes include several loci that regulate gene expression and neural development and signaling, but their specific relationship to behavior is not clear. These loci may control gene cascades important for subsequent courtships, such as those that fine-tune neural connections due to courtship and mating experience. An additional set of five genes identified only in a paired comparison of courting and control males (Lsp2, egh, cac, CG2217, and CG4269) may also be courtship-responsive since their transcript levels were not affected by male–male interactions. If not specifically courtship responsive, they are likely to be generally socially-responsive or to have behavioral functions. In support of this hypothesis, we showed that egh expression is important for robust male–female courtship.
Interestingly, a much larger group of genes is responsive to interactions with both sexes (265 genes, Table S1) or is male–male responsive (240 genes, Table S2). Therefore, social interactions have an impact on gene expression patterns that depends on the sex of the interacting individuals. This result is not surprising since social experience affects a variety of behaviors and morphological phenotypes in flies and other animals (Siegel and Hall 1979; Levine et al. 2002; Shen et al. 2004; Stewart and McLean 2004; Burmeister et al. 2005; Kozorovitskiy et al. 2006; Yurkovic et al. 2006; Technau 2007). Some of the expression changes identified in our study may underlie observed effects of social interactions on circadian behavior and pheromone profiles (Levine et al. 2002; Kent et al. 2008; Krupp et al. 2008).
The large number of male–male responsive genes was surprising. However, male–male interactions such as those involved in aggressive encounters may have greater effects on gene expression than male–female interactions. Males of many Drosophila species, including D. melanogaster, compete for mates and territories, and aggressive behavior is correlated with mating success (Dow and von Schilcher 1975); both factors correlated with genotype (Cabral et al. 2008). Social experience with other males reduces aggressive behavior during competition for territories, and experienced males are more likely to regain territories (Hoffmann 1990). Although our male–male assays were performed under conditions under which there is predicted to be little male–male competition (e.g., no food source or female), it is likely that sensory processing and gene expression was affected by the brief encounters between individuals. Observed changes in gene expression due to male–male interactions may contribute to the phenotypic plasticity in behaviors important for obtaining territories, food sources, and mates. Further investigation is required to understand fully the importance of the large number of changes that occur due to general social interactions or specifically in response to male–male interactions.
Socially-responsive genes and the sex-determination hierarchy:
We predicted that some socially-responsive loci would function as downstream targets of the somatic sex-determination pathway that regulates male courtship behavior. Three transcription factors—Fruitless (Fru), Doublesex (Dsx), and Dissatisfaction (Dsf)—are important regulatory components of this pathway. One courtship-responsive gene, CG13116, is negatively regulated by the female-specific Doublesex (Dsx) protein, and one upregulated male–male-responsive gene, CG16713, is downstream of transformer (tra) (Goldman and Arbeitman 2007). Four upregulated socially-responsive genes are regulated by the sex-determination pathway. fit is regulated by tra; CG9377 is downstream of fru; and CG9837 and CG8539 are regulated by dsx (Goldman and Arbeitman 2007). The surprisingly small number of socially-responsive genes that are known sex-determination hierarchy targets may indicate that our lists include many target genes that could not be detected by the strategies used previously to identify output genes of the hierarchy. For example, genes from our study may function downstream of dsf, which is expressed in both males and females; transcriptional targets of dsf are not known. Another possibility is that the hierarchy does not regulate expression of many socially-responsive genes, indicating an alternative regulation.
Gene expression in the male brain:
Since brain gene expression has a clear function in behavior, we expected some socially-responsive genes to be expressed in the brain. In situ hybridization showed that CG9377 and egh were expressed in the male brain but were not detected in adipose tissue (Figures 2 and 3). Two downregulated, socially-responsive genes function in courtship behavior and are expressed in the brain. cac encodes a calcium voltage-gated channel needed for courtship song production (reviewed in Greenspan and Ferveur 2000; Billeter et al. 2002); Shaker (Sh) encodes a potassium channel that functions in olfactory memory and learning (reviewed in Greenspan and Ferveur 2000). Other socially-responsive genes (e.g., Drop, egh, hairy, and Sh) regulate nervous system development and function (Giniger et al. 1994; Heng and Tan 2003; Zhong and Wu 2004; Fan et al. 2005; Ueda and Wu 2006; Urbach et al. 2006) and may modulate adult neural signaling and behavior.
Changes in brain gene-expression patterns due to social interactions are likely a result of signaling pathways, including G-protein-coupled receptor signaling, functioning within the brain to mediate the perception and integration of sensory cues. Such signaling pathways may coordinate motor output pathways necessary for courtship and relay information to the brain to establish a male brain that is more readily perceptive to courtship cues than a naive male brain.
Gene expression in male adipose tissue:
Signals mediating social cues are not likely restricted to the brain, however. Adipose tissue, or the fat body, surrounding the brain and in the thoracic and abdominal cavities is a secretory tissue (reviewed in Schlegel and Stainier 2007) that could influence neuronal signaling or transmit signals to other reproductively important tissues. Indeed, there is growing evidence that fat body-expressed genes modulate reproductive behaviors (reviewed in Dauwalder 2008).
fit and Lsp2 are expressed in the female and male fat body (Figure 1) (Benes et al. 1990; Fujii and Amrein 2002), and in situ hybridization confirmed fat body expression of three additional socially-responsive genes (CG10621, cwo, and sug) (Figure 3). cwo is also expressed in the male brain, but we did not detect CG10621 or sug transcripts in the male brain. Many socially-responsive genes are enriched in head tissue, including fat body but not including the brain (Figures 2 and 3; Table S3). This suggests that the circuitry responding to and governing social interactions such as courtship likely is modulated by both neuronal and non-neuronal signals. The response to social interactions involves complex and specific changes that may mediate various downstream effects, including neural plasticity.
egghead and courtship behavior:
To determine if mutations in candidate genes affected courtship behavior, we measured CI values in various mutants. Our analysis showed that a specific locus, egh, is needed for robust male courtship behavior (Figure 4). egh encodes a 1,4-mannosyltransferase that regulates glycosphingolipid biosynthesis (Wandall et al. 2003), affects Drosophila neural development and behavior, and is required in Ap neurons for female Sex-peptide response post mating (Soller et al. 2006).
Since apmd544-Gal4 (a Gal4 insertion in ap) is expressed in the adult nervous system of both sexes (Figure S2), we examined whether this neural circuit also functioned in males to regulate courtship behavior. In eghEY03917 mutant males, egh expression in Ap neurons rescued the courtship defect (Figure 7). Decreased adult egh expression (via RNAi) in ap-expressing neurons also resulted in decreased courtship (Figure 8). Although Fru neuron expression of the EcR transcription factor is important for courtship behavior (Dalton et al. 2009), decreasing EcR in adult Ap neurons did not affect courtship (Figure S3 and Figure S4). Therefore, egh appears to have a specific behavioral function in Ap-expressing neurons. Ap is a transcription factor that regulates developmental as well as post-developmental neural gene expression (Benveniste et al. 1998). ap mutant males also have decreased levels of male-to-female courtship (Ringo et al. 1992). Given the similarity between the ap and egh mutant phenotypes and the requirement for egh expression in ap neurons for male courtship, the hypothesis that ap regulates egh expression should be tested in future experiments.
Differences in sex-specific behaviors may be due to dimorphisms in neural architecture, including the number or morphology of neurons, such as those present in the fruP1 circuit that modulates male courtship behavior (Kimura et al. 2005; Stockinger et al. 2005; Rideout et al. 2007; Clyne and Miesenböck 2008; Datta et al. 2008). The same circuit could be co-opted by each sex for different behaviors. We hypothesize that this is the case for the egh circuit. egh is required in both male and female Ap neurons but modulates sex-specific reproductive behaviors. This may occur because of changes in neural physiology resulting from the perception of sex-specific cues that trigger different signaling cascades between the sexes. However, it is possible that different subsets of Ap neurons regulate sex-specific behavior. The egh circuit important for male behavior does not appear to rely directly upon fru neurons since expressing eghRNAi in fru neurons did not cause the behavioral defects observed in egh mutant or apmd544-Gal4/eghRNAi males (data not shown). Therefore, egh neurons may interact indirectly with fru neurons to modulate reproductive behaviors.
Our study strengthens the growing body of work demonstrating that animals respond to social interactions by altering transcript abundance. By investigating the function of these socially-responsive loci, we can clarify the relationship between genetics and the intracellular processes governing behavior and physiology. Additional studies are needed to understand the relationship of courtship-responsive and other socially-responsive loci to the somatic sex-determination hierarchy or other pathways that regulate Drosophila reproductive behaviors.
Acknowledgments
We thank Donna Wall and Kuey-Chu Chen at the University of Kentucky MicroArray Core Facility for microarray processing, Bruce Ellis for creating the electrical probes used for cauterization, Matthias Soller for providing eghP2 and UAS-eghHA, and the Vienna Drosophila RNAi Center for providing the egh-RNAi and UAS-Dicer2 alleles. We thank two anonymous reviewers for comments on the manuscript. Texas A&M University provided funding to G.E.C.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.122754/DC1.
The microarray data from this study are available through the Gene Expression Omnibus database via accession no. GSE24167.
References
- Arbeitman, M. N., A. A. Fleming, M. L. Siegal, B. H. Null and B. S. Baker, 2004. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131 2007–2021. [DOI] [PubMed] [Google Scholar]
- Baldi, P., and A. D. Long, 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17 509–519. [DOI] [PubMed] [Google Scholar]
- Benes, H., R. G. Edmondson, P. Fink, J. Kejzlarová-Lepesant, J. A. Lepesant et al., 1990. Adult expression of the Drosophila Lsp-2 gene. Dev. Biol. 142 138–146. [DOI] [PubMed] [Google Scholar]
- Benveniste, R. J., S. Thor, J. B. Thomas and P. H. Taghert, 1998. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development 125 4757–4765. [DOI] [PubMed] [Google Scholar]
- Billeter, J.-C., S. F. Goodwin and K. M. O'Dell, 2002. Genes mediating sex-specific behaviors in Drosophila. Adv. Genet. 47 87–116. [DOI] [PubMed] [Google Scholar]
- Billeter, J.-C., A. Villella, J. B. Allendorfer, A. J. Dornan, M. Richardson et al., 2006. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr. Biol. 16 1063–1076. [DOI] [PubMed] [Google Scholar]
- Burmeister, S. S., E. D. Jarvis and R. D. Fernald, 2005. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 3 1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis, K. C., K. T. Coschigano, B. S. Baker and P. C. Wensink, 1991. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk-protein gene enhancer. EMBO J. 10 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral, L. G., B. R. Foley and S. V. Nuzhdin, 2008. Does sex trade with violence among genotypes in Drosophila melanogaster? PLoS One 3 e1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja, M., E. Morena, S. Pelaz and G. Morata, 1996. Visualization of gene expression in living adult Drosophila. Science 274 252–255. [DOI] [PubMed] [Google Scholar]
- Cann, M. J., E. Chung and L. R. Levin, 2000. A new family of adenylyl cyclase genes in the male germline of Drosophila melanogaster. Dev. Genes Evol. 210 200–206. [DOI] [PubMed] [Google Scholar]
- Carney, G. E., 2007. A rapid genome-wide response to Drosophila melanogaster social interactions. BMC Genomics 8 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli, V. R., J. Wang and J. A. Dow, 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39 715–720. [DOI] [PubMed] [Google Scholar]
- Cline, T. W., 2005. Reflections on a path to sexual commitment. Genetics 169 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne, J. D., and G. Miesenböck, 2008. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell 133 354–363. [DOI] [PubMed] [Google Scholar]
- Dalton, J. E., M. S. Lebo, L. E. Sanders, F. Sun and M. N. Arbeitman, 2009. Ecdysone receptor acts in fruitless-expressing neurons to mediate Drosophila courtship behaviors. Curr. Biol. 19 1447–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S. R., M. L. Vasconcelos, V. Ruta, S. Luo, A. Wong et al., 2008. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452 473–477. [DOI] [PubMed] [Google Scholar]
- Dauwalder, B., 2008. Systems behavior: of male courtship, the nervous system and beyond in Drosophila. Curr. Genomics 9 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder, B., S. Tsujimoto, J. Moss and W. Mattox, 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Dow, M. A., and F. von Schilcher, 1975. Aggression and mating success in Drosophila melanogaster. Nature 254 511–512. [DOI] [PubMed] [Google Scholar]
- Drapeau, M. D., A. Radovic, P. J. Wittkopp and A. D. Long, 2003. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 55 53–72. [DOI] [PubMed] [Google Scholar]
- Ellis, L. L., and G. E. Carney, 2009. Drosophila melanogaster males respond differently at the behavioral and genome-wide levels to Drosophila melanogaster and Drosophila simulans. J. Evol. Biol. 22 2183–2191. [DOI] [PubMed] [Google Scholar]
- Ewing, A. W., 1983. Functional aspects of Drosophila courtship. Biol. Rev. 58 275–292. [Google Scholar]
- Fan, Y., M. Soller, S. Flister, M. Hollmann, M. Müller et al., 2005. The egghead gene is required for compartmentalization in Drosophila optic lobe development. Dev. Biol. 287 61–73. [DOI] [PubMed] [Google Scholar]
- Finley, K. D., B. J. Taylor, M. Milstein and M. McKeown, 1997. dissatisfaction, a gene involved in sex-specific behavior and neural development of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S., and H. Amrein, 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S., A. Toyama and H. Amrein, 2008. A male-specific fatty acid ω-hydroxylase, SXE1, is necessary for efficient male mating in Drosophila melanogaster. Genetics 180 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger, E., K. Tiejte, L. Y. Jan and Y. N. Jan, 1994. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120 1385–1398. [DOI] [PubMed] [Google Scholar]
- Goldman, T. D., and M. N. Arbeitman, 2007. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3 e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, R. J., and J.-F. Ferveur, 2000. Courtship in Drosophila. Annu. Rev. Genet. 34 205–232. [DOI] [PubMed] [Google Scholar]
- Grozinger, C. M., N. M. Sharabash, C. W. Whitfield and G. E. Robinson, 2003. Pheromone mediated gene expression in the honey bee brain. Proc. Natl. Acad. Sci. USA 100(Suppl. 2): 14519–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, J. I. T, and S. S. Tan, 2003. The role of class I HLH genes in neural development: Have they been overlooked? BioEssays 25 709–716. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A., 1990. The influences of age and experience with conspecifics on territorial behavior in Drosophila melanogaster. J. Insect Behav. 3 1–12. [Google Scholar]
- Kent, C., R. Azanchi, B. Smith, A. Formosa and J.D. Levine, 2008. Social context influences chemical communication in D. melanogaster males. Curr. Biol. 18 1384–1389. [DOI] [PubMed] [Google Scholar]
- Kimura, K., M. Ote, T. Tazawa and D. Yamamoto, 2005. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438 229–233. [DOI] [PubMed] [Google Scholar]
- Kopp, A., I. Duncan, D. Godt and S. B. Carroll, 2000. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408 553–559. [DOI] [PubMed] [Google Scholar]
- Kozorovitskiy, Y., M. Hughes, K. Lee and E. Gould, 2006. Fatherhood affects dendritic spines and vasopressin V1a receptors in the primate prefrontal cortex. Nat. Neurosci. 9 1094–1095. [DOI] [PubMed] [Google Scholar]
- Krupp, J. J., C. Kent, J.-C. Billeter, R. Azanchi, A. K.-C. So et al., 2008. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr. Biol. 18 1373–1383. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K. N., and D. J. Begun, 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47 900–919. [DOI] [PubMed] [Google Scholar]
- Lazareva, A. A., G. Roman, W. Mattox, P. E. Hardin and B. Dauwalder, 2007. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 3 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, J. D., P. Funes, H. B. Dowse and J. C. Hall, 2002. Resetting the circadian clock by social experience in Drosophila melanogaster. Science 298 2010–2012. [DOI] [PubMed] [Google Scholar]
- Li, C., and W. H. Wong, 2001. Model based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D. M., and C. S. Goodman, 1994. Ectopic and increased expression of fasciclin II alters motorneuron growth cone guidance. Neuron 13 507–523. [DOI] [PubMed] [Google Scholar]
- Mack, P. D., A. Kapelnikov, Y. Heifetz and M. Bender, 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli, D. S., M. Foss, A. Villella, B. J. Taylor, J. C. Hall et al., 2005. Male-specific fruitless specifies the neural substrates of Drosophila courtship behavior. Nature 436 395–400. [DOI] [PubMed] [Google Scholar]
- McGuire, S. E., Z. Mao and R. L. Davis, 2004. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 220 pl6. [DOI] [PubMed] [Google Scholar]
- Mehren, J. E., A. Ejima and L. C. Griffith, 2004. Unconventional sex: fresh approaches to courtship learning. Curr. Opin. Neurobiol. 14 745–750. [DOI] [PubMed] [Google Scholar]
- Mellert, D. J., J.-M. Knapp, D. Manoli, G. W. Meissner and B. S. Baker, 2010. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex, and the Roundabout receptors. Development 137 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2006. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (http://www.R-project.org).
- Rideout, E. J., J.-C. Billeter and S. F. Goodwin, 2007. The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr. Biol. 17 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout, E. J., A. J. Dornan, M. C. Neville, S. Eadie and S. F. Goodwin, 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo, J., R. Werczberger and D. Segal, 1992. Male sexual signaling is defective in mutants of the apterous gene of Drosophila melanogaster. Behav. Genet. 22 469–487. [DOI] [PubMed] [Google Scholar]
- Sanders, L. E., and M. N. Arbeitman, 2008. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel, A., and D. Y. R. Stainier, 2007. Lessons from “lower” organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 3 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Sarma, M., S. L. Rodriguez-Zas, F. Hong, S. Zhong and G. E. Robinson, 2009. Transcriptomic profiling of central nervous system regions in three species of honey bee during dance communication behavior. PLoS ONE 4 e6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. P., Y. Tsimberg, C. Salvadore and E. Meller, 2004. Activation of Erk and JNK MAPK pathways by acute swim stress in rat brain regions. BMC Neurosci. 5 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi, T. R., and M. McKeown, 2007. Sex in flies: What ‘body-mind’ dichotomy? Dev. Biol. 306 10–19. [DOI] [PubMed] [Google Scholar]
- Siegel, R. W., and J. C. Hall, 1979. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. USA 76 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller, M., I. U. Haussmann, M. Hollmann, Y. Choffat, K. White et al., 2006. Sex-peptide-regulated female sexual behavior requires a subset of ascending ventral nerve cord neurons. Curr. Biol. 16 1771–1782. [DOI] [PubMed] [Google Scholar]
- Stewart, B. A., and J. R. McLean, 2004. Population density regulates Drosophila synaptic morphology in a Fasciclin-II-dependent manner. J. Neurobiol. 61 392–399. [DOI] [PubMed] [Google Scholar]
- Stockinger, P., D. Kvitsiani, S. Rotkopf, L. Tirian and B. J. Dickson, 2005. Neural circuitry that governs Drosophila male courtship behavior. Cell 121 795–807. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau, G. M., 2007. Fiber number in the mushroom bodies of adult Drosophila melanogaster depends on age, sex and experience. J. Neurogenet. 21 183–196. [DOI] [PubMed] [Google Scholar]
- Ueda, A., and C. F. Wu, 2006. Distinct frequency-dependent regulation of nerve terminal excitability and synaptic transmission by IA and IK potassium channels revealed by Drosophila Shaker and Shab mutations. J. Neurosci. 26 6238–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach, R., D. Volland, J. Siebert and G. M. Technau, 2006. Segment-specific requirements for dorsoventral patterning genes during early brain development in Drosophila. Development 133 4315–4330. [DOI] [PubMed] [Google Scholar]
- Wada, K, J. T. Howard, P. McConnell, O. Whitney, T. Lints et al., 2006. A molecular and neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc. Natl. Acad. Sci. USA 103 15212–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall, H. H., J. W. Pedersen, C. Park, S. B. Levery, S. Pizette et al., 2003. Drosophila egghead encodes a beta 1,4-mannosyltransferase predicted to form the immediate precursor glycosphingolipid substrate for brainiac. J. Biol. Chem. 278 1411–1414. [DOI] [PubMed] [Google Scholar]
- Whitfield, C. W., A. M. Cziko and G. E. Robinson, 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302 296–299. [DOI] [PubMed] [Google Scholar]
- Whitfield, C. W., Y. Ben-Shahar, C. Brillet, I. Leoncini, D. Crauser et al., 2006. Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. USA 103 16068–16075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovic, A., O. Wang, A. C. Basu and E. A. Kravitz, 2006. Learning and memory associated with aggression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103 17519–17524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Y., and C. F. Wu, 2004. Neuronal activity and adenylyl cyclase in environment-dependent plasticity of axonal outgrowth in Drosophila. J. Neurosci. 24 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]