Abstract
Repeated DNA in heterochromatin presents enormous difficulties for whole-genome sequencing; hence, sequence organization in a significant portion of the genomes of multicellular organisms is relatively unknown. Two sequenced BACs now allow us to compare telomeric retrotransposon arrays from Drosophila melanogaster telomeres with an array of telomeric retrotransposons that transposed into the centromeric region of the Y chromosome >13 MYA, providing a unique opportunity to compare the structural evolution of this retrotransposon in two contexts. We find that these retrotransposon arrays, both heterochromatic, are maintained quite differently, resulting in sequence organizations that apparently reflect different roles in the two chromosomal environments. The telomere array has grown only by transposition of new elements to the chromosome end; the centromeric array instead has grown by repeated amplifications of segments of the original telomere array. Many elements in the telomere have been variably 5′-truncated apparently by gradual erosion and irregular deletions of the chromosome end; however, a significant fraction (4 and possibly 5 or 6 of 15 elements examined) remain complete and capable of further retrotransposition. In contrast, each element in the centromere region has lost ≥40% of its sequence by internal, rather than terminal, deletions, and no element retains a significant part of the original coding region. Thus the centromeric array has been restructured to resemble the highly repetitive satellite sequences typical of centromeres in multicellular organisms, whereas, over a similar or longer time period, the telomere array has maintained its ability to provide retrotransposons competent to extend telomere ends.
THE wealth of genome sequences now available has revealed much about genome organization and how this organization has evolved. These sequences have greatly extended our understanding of the ways in which transposable elements have added to and shaped eukaryotic genomes (Slotkin and Martienssen 2007; Bourque 2009; Cordaux and Batzer 2009). For most eukaryotes, however, a significant portion of the genome is still poorly understood. This portion is the heterochromatin, which makes up about one-fifth of the human genome and a one-third of the Drosophila genome (Hoskins et al. 2007). Heterochromatin is very rich in transposable element sequences, and it would be especially interesting to understand how these sequences are related to the properties that distinguish heterochromatin from euchromatin. However, the large number of highly repeated and rapidly evolving DNA sequences in heterochromatin presents problems for accurately assembling sequences long enough to give a complete picture of the organization of these elements and their possible roles in specific heterochromatic regions such as centromeres and telomeres.
We are studying the three non-LTR retrotransposons that maintain the length of Drosophila telomeres HeT-A, TART, and TAHRE. These elements transpose by means of a poly(A)+ RNA that is reverse-transcribed directly onto the end of the chromosome (Pardue et al. 2005). Successive transpositions form long arrays of head-to-tail repeats. These repeats are analogous to the repeats that telomerase adds in most other organisms except that the Drosophila repeats are copies of retrotransposons, which are three orders of magnitude longer than the repeats added by telomerase. Recently, we analyzed a BAC and a finished scaffold of equal quality containing sequence from two Drosophila melanogaster telomeres. These sequences gave us our first overview of both the organization of transposable elements within a telomere (George et al. 2006) and, now, the mechanisms by which these telomeres are maintained.
Although the telomeric retrotransposons appear capable of transposing only onto chromosome ends—either natural telomeres or broken chromosomes—HeT-A DNA was found to hybridize to the centromere region of the D. melanogaster Y chromosome (Agudo et al. 1999) and later shown to colocalize with antibody to centromere-specific histone on the Y chromosomes of other members of the melanogaster species subgroup (Berloco et al. 2005). Thus, HeT-A-related sequences appear to have been maintained at the centromere of the Y for >13 MY, even though the structure of the chromosome has diverged so that the Y is now metacentric in some species and telocentric in others. Mendez-Lago et al. (2009) have recently reported sequence of a BAC from this D. melanogaster Y centromere cluster.
The assembled sequences of the telomeric BAC and the centromeric BAC give the first opportunity to examine arrays long enough to allow us to analyze the organization, maintenance, and evolution of these retrotransposon arrays in two different heterochromatic environments. Comparison of the telomeric and centromeric sequences reveals that HeT-A arrays in telomeric heterochromatin are maintained very differently from those in centromeric heterochromatin; each is structured in ways that appear to be compatible with their different roles at the telomere and the centromere. It is frequently said that transposable elements have done much to shape eukaryotic genomes; in these cases, the genome is shaping the elements.
MATERIALS AND METHODS
Sequences analyzed:
The BAC-containing telomere sequence from the 4R telomere is AC010841. The XL telomere sequence is CP000372, a scaffold made by directed finishing of the sequence including the most distal gene on XL, CG17636, and extending some 20 kb into the telomere array. The BAC-containing sequence from the Y centromere region is BACR26J21 (Mendez-Lago et al. 2009). The canonical HeT-A element is 23Zn-1 (U06920, bp 1015–7097); For Drosophila canonical sequences, see http://flybase.org/static_pages/downloads/FB2010_09/transposons/transposon_sequence_set.embl.txt.gz.
Definition of sequence included in HeT-A/TART telomere arrays:
We define HeT-A/TART telomere arrays as the sequence on the chromosome end that is distal to the most distal HeT-A or TART element disrupted by insertion of a nontelomere transposable element. The disrupted element marks the end of the transition zone, the proximal end of the telomere where nontelomere elements have invaded HeT-A and TART sequences. HeT-A Tags are recognizable 3′-most sequences of HeT-A that are transferred to the 5′ end of a downstream element in the course of HeT-A transcription. They can form long “strings” on the 5′ end of complete elements; each complete element in these arrays carries a Tag string. A fifth Tag string lies just 5′ of the transition zone on 4R where its parent element has apparently been invaded by a 1360 transposon.
The most interior Tags in long strings can become too decayed to be recognized and are indistinguishable from the 5′ UTR of the element to which they are attached; by default, we include these decayed Tags in the 5′ UTR. An exhaustive search for Tags was made during final annotation of the 4R and XL sequences after it had been recognized just how important their role might be in telomere maintenance. Using lowered stringency Blastn, we searched for terminal fragments of HeT-A's well-conserved 30-nucleotide 3′ terminus in short overlapping regions of the 5′ end of each HeT-A and the 3′ end of its upstream neighbor. (Only searches of short sequences identified all Tags.) Our search protocol ended the search at the last clearly defined Tag found, even if another could be seen within the nearby 5′ UTR. This procedure was adopted because short TA(n) or TTA(n) (n < 5) abound within the AT-rich 5′ UTR. Also, estimating conservatively, because it is likely that they are the result of replication slippage long after they moved into the interior of the telomere, we omit all but the first of several consecutive short sequences that make up the proximal end of the Tag string on XL. Not unexpectedly, Tag length is not distributed normally. In this article, we find it most descriptive to report mean lengths and their 95% confidence intervals. For most calculations, we omit elements truncated by cloning at the end of 4R and XL.
End erosion and terminal deletion data processing:
Analysis was performed using Excel and JMP 8 (SAS Institute, Cary, NC).
Dot-plot analysis:
Dot plots comparing the canonical HeT-A with the sequences in the BACs were compared by the National Center for Biotechnology Information (NCBI) BLAST (bl2seq) using the default parameters for somewhat similar sequences (blastn). This procedure gives adequate alignment of the canonical sequence with sequences of all known HeT-A subfamilies. More stringent BLAST parameters show less sequence in the alignment.
RESULTS AND DISCUSSION
HeT-A elements in telomeric heterochromatin
Sequences analyzed:
Our comparison is based on a telomeric BAC from 4R, plus sequence from directed finishing of a scaffold from the telomere of chromosome XL, reported previously, but with some revisions in annotation. Both the 4R and the XL sequences begin in their assembled chromosome and extend into the telomere, thus showing the precise relationship between these telomere arrays and the rest of the genome (George et al. 2006). Neither sequence extends to the distal end of the telomere, but together they give nearly 100 kb of telomeric HeT-A/TART array (76 kb from 4R and 20 kb from XL). Importantly, both include the most proximal elements of the array. Because telomere elements are added sequentially, the most proximal elements must be the oldest; thus, these arrays present the most accurate history available of D. melanogaster telomere maintenance.
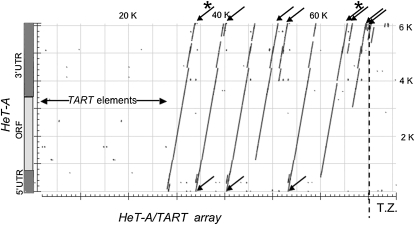
Neither the 4R nor XL sequences have nontelomeric elements in the telomeric array except for a small transition zone at the proximal edge of the array where there are some fragments of nontelomeric elements. We do not include these transition zones in our discussion of telomere arrays. Figure 1 gives an overview of the organization of HeT-A sequences in the telomere array and transition zone of 4R. The sequence of this segment of the BAC is compared to the sequence of the canonical HeT-A, 23Zn-1, which is diagrammed on the x-axis of the dot plot. Although none of the elements in the array belong to the same subfamily as the canonical element, all show linear similarity except for small gaps or repetitions in the 3′ UTR. Each complete or partial HeT-A in the array is intact at the 3′ end. Essentially all sequence loss has been at the 5′ end. Complete elements match the entire 5′ end of the canonical HeT-A. Two elements that have mildly truncated 5′ UTRs are grouped with partial elements because we do not know whether they contain 5′ UTR sequence required for transposition competence.
Figure 1.—
Analysis of HeT-A-related sequences in the 4R telomere array. The most distal 76 kb of sequence on the 4R telomere is compared with the sequence of the canonical 6-kb complete HeT-A (23Zn-1) by the least stringent NCBI BLAST algorithm (blastn) to ensure alignment with the several subfamilies of HeT-A elements in this array. Sequence of the 76 kb of sequence assembled from the telomere on this chromosome end is shown on the x-axis with the distal end on the left. The HeT-A sequence is diagrammed on the y-axis, showing the 5′ UTR, ORF, and 3′ UTR. The dotted line near the right end defines the 6.2-kb transition zone (T.Z.) where elements from the euchromatic and subtelomeric regions have invaded the telomeric array. For clarity, only HeT-A sequences are shown on the plot. All HeT-A elements, complete and partial, touch the top line because they have complete 3′ ends. Arrows above the top line mark the eight partial HeT-As. Asterisks indicate partial elements with less than full-length 5′ UTRs; these two elements may still be replicatively competent. Arrows below the bottom line mark the three complete HeT-As. The empty space on the left is occupied by two complete and two partial TARTs. A very short partial TART lies just after the most proximal full-length HeT-A. Empty space in the T.Z. is occupied by fragments of nontelomeric elements and also by TART fragments.
HeT-A elements are much more abundant than TART elements in all D. melanogaster genomes that we have analyzed. Consistent with this, there are very few TART elements in the 4R array and none in the shorter XL sequence. Because we are making comparisons with a segment of the centromere array that consists entirely of HeT-A, we have not included TART data in the following analyses and discussion. Empty space in Figure 1 contains complete or partial TARTs. The third (very rare) telomeric retrotransposon, TAHRE, is also omitted from discussion because it is not found in any of the arrays considered here.
Each telomere array is a chronological record of events at the end of the chromosome:
The studies reported here analyze the 4R and XL data to investigate sequence management within an intact Drosophila telomere. These sequences present a unique opportunity: the BAC and the sparser, but still useful, finished scaffold data are the first long sequences that give the exact order of elements in the telomere and their relationship to the rest of the genome. This relationship is informative because HeT-A, TART, and TAHRE transpose only onto the ends of chromosomes so that the order of elements in an array reflects the order of transposition onto the chromosome, with the oldest elements at the proximal end. Rearrangements and imprecise recombination are ruled out by the large-scale integrity of all the elements present; the full-length elements evidence replicative capability and even the partial elements are simply 5′-truncated with no evidence of sequence rearrangement or decay.
The sequence at the 5′ end of each internal element is a record of the sequence that remained on a terminal retrotransposon when that element was capped by a newly transposing element and thus demoted to an internal position in the telomere. Comparing that capped sequence with the 5′ end of its presumed RNA template gives an estimate of the amount of sequence lost while that element served as the end of the chromosome. (This is a maximum estimate because some sequence could have been lost in transposition.)
The only measurements of the rate of end erosion and the addition of new elements on Drosophila chromosomes have come from broken chromosomes that have lost all telomeric and subtelomeric DNA. Such chromosomes shortened gradually by ∼70 nucleotides per fly generation (Levis 1989; Biessmann et al. 1990; Mikhailovsky et al. 1999), calculated to represent an average loss of 2–3 nt per cell generation (Biessmann et al. 1990). The rate at which a broken end is healed by addition of HeT-A varied from <2 × 10−5 to 2 × 10−3, depending on the background genotype (Kahn et al. 2000). Thus, broken chromosome ends show a fairly regular slow loss of end sequence accompanied by infrequent addition of large retrotransposons; HeT-A is ∼6 kb; TART and TAHRE are 10–12 kb.
The 4R and XL sequences give us the first opportunity to analyze the turnover of sequence on established telomeres. For this analysis we first discuss two indicators of sequence loss in the telomere arrays: (1) the Tags (see 5′ Tags reveal slow sequence erosion of the telomere end) of nonessential sequence on the 5′ end of each HeT-A RNA and (2) the distribution of complete and 5′ truncated elements in the telomere array.
These data fall into two classes: (1) qualitative observations that describe the nature of the processes governing telomere maintenance and renewal (these observations strongly constrain models based on numerical analysis) and (2) quantitative statistical analyses that help in distinguishing and judging competing conclusions. As discussed below, we conclude that maintenance of established telomeres involves at least three processes acting in concert to maintain relatively stable conditions: relatively short-range terminal erosion, long-range terminal deletion, and irregular transpositions.
5′ Tags reveal slow sequence erosion of the telomere end:
Tags are short sequences added to the 5′ end of HeT-A RNA by HeT-A's unusual promoter that initiates transcription within the 3′ UTR of the upstream element (Danilevskaya et al. 1997; Traverse et al. 2010). The resulting Tag of upstream sequence becomes a de facto extension of the 5′ UTR and is reverse-transcribed with the rest of the RNA when the element transposes, providing expendable sequence to buffer the loss of essential 5′ sequence from chromosome end erosion. Part or all of a Tag can be eroded while it forms the end of the telomere (Figure 2A). When a new element transposes to cap the chromosome end, erosion of the capped Tag is halted, leaving a partial Tag. Repeated transcription of an element will add a new Tag to any already on its 5′ end. Thus, an element can have a 5′ string of variably truncated Tags—evidence that it has transposed multiple times (Figure 2B). Tags are the hallmark of complete HeT-As, which carry a string of them. We note that there is evidence to suggest that erosion can at times continue into the 5′ UTR (see Distribution of 5′-truncated elements in the telomere array provides evidence of sporadic terminal deletions).
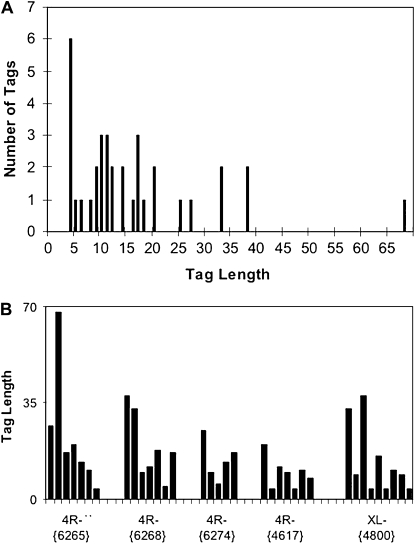
Figure 2.—
HeT Tags in telomere arrays on 4R and XL. Tag length includes oligoA. (A) Histogram of Tag length. The 35 individual Tags in this study are grouped by size. (B) Organization of Tags into strings. Within each string, individual Tags are ordered from distal to proximal with distal on the left. Each cluster is labeled by the FlyBase identifier of its associated HeT-A element (see chromosomes X and 4 at http://flybase.org/cgi-bin/gbrowse/dmel/). Cluster 4R-{4617} lies at the border of the transition zone without an associated complete HeT-A but contributes so much statistically typical data that it is included for Tag analysis. It is identified by the immediately 5′ partial HeT-A element, HeT-A{4617}, which is the first element in the transition zone.
Tags are the best indicators of nucleotide loss on the end of an established telomere. There is only now a statistically robust sample of Tags and, because Tags are born in discrete short lengths, their end erosion is directly measurable. (It is frequently convenient to differentiate between a Tag's 3′-oligoA sequence, its “Tag-tail,” and its more 5′ sequence, the “bare Tag”). The initial length of each bare Tag is determined by its transcription start site, which can be 93, 62, or 31 nt from the 3′ end of the element serving as a promoter; the 3′-oligoA of the promoting element then forms the tail of this new Tag (Traverse et al. 2010). Thus the longest Tag should have a 93-nt “bare Tag” plus its tail; the Tags in our arrays are all much shorter than this (Figure 2A).
If end erosion is regular and averages ∼70 nt per fly generation, as found for broken ends, most Tags should last <1 generation, but complete elements typically have several Tags in various states of erosion (Figure 2B). Strings of partially eroded Tags indicate that these expendable sequences provide enough protection to allow a significant number of elements to survive with intact 5′ ends.
Tag properties:
On average, our Tags are surprisingly short (Figure 2A). Their median length, including the oligoA tail, is 11 nt, mean of 14.0 with the 95% confidence interval (C.I.) of the mean 10.7–17.3 nt. Furthermore, the very shortest Tags are overrepresented (18% have the sequence TAAA but contain <5% of the total Tag sequence), suggesting that the rate of sequence loss is reduced as Tags are eroded toward their oligoA tail. There is also one very long Tag (68 nt) that is a distinct outlier; the next longest is 38 nt. The paucity of long Tags may be evidence that the two distant transcription starts, −93 and −62, are very rarely used. Alternatively, the longest Tags may be subject to more severe erosion; in fact, there may be an accumulation at the transition to bare lengths ∼31 nt. The evidence for this conjecture is too weak to assert with confidence.
We note that the narrow limit on Tags per string (five to nine) and Tag string length (69–161 nt) indicates that erosion is regulated to balance transposition because we find neither intact HeT-As without Tags nor strings that have grown without limit, as they would if not effectively pruned.
HeT-A oligoA tails (mean 8.3 nt, 95% C.I. 4.6–12.1 nt) are very short compared to TART oligoA tails (18.3 nt, range 13–23 nt). The facts that HeT-A oligoA tails give rise to Tags and that TART does not utilize Tags to protect its 5′ end suggest that HeT-A oligoA length is one adaptation to help control the overall length of HeT-A Tag strings. With TART-like tails, the average Tag and Tag string were more than twice as long,
These analyses show that erosion of the telomere end is more complex than the relatively regular loss described by studies of broken chromosomes. It also shows that many, perhaps all, new transpositions occur before the terminal Tag has been completely eroded. In the maintenance of the extreme ends of chromosomes, there are at least two stochastic processes at work: relatively regular erosion of Tags and a mechanism that protects the very shortest ones. However, it is important to recognize that Tag erosion is nevertheless a relatively closely regulated process.
Distribution of 5′-truncated elements in the telomere array provides evidence of sporadic terminal deletions:
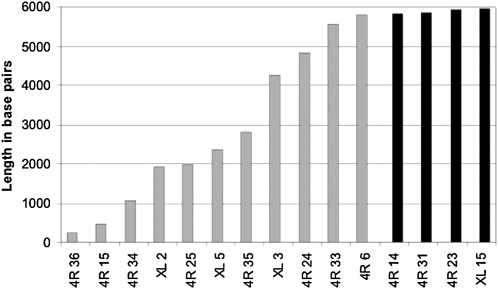
Within experimental uncertainties, the rate of terminal erosion measured from Tag lengths approximates that measured on broken chromosomes. In contrast, sequence loss from 5′-truncated elements is on a much larger scale. There are 11 truncated HeT-As in these arrays. All contain the 3′-most 150 nt (which are almost completely conserved among HeT-A subfamilies), but they differ significantly in the amount of sequence lost from the 5′ end (Figure 3). Their lengths are broadly distributed between 5892 and 241 bp, showing no correlation with position in the array. The longest two of these HeT-As have partial 5′ UTRs, the next longest three contain partial ORFs, and all others have only 3′ UTR sequence. All have enough 3′ UTR to provide promoter activity for a downstream neighbor, although the 274-bp element would provide only weak activity.
Figure 3.—
Complete and 5′-truncated HeT-As in telomere arrays on 4R and XL rank ordered by length. Elements are named by their position (distal-to-proximal) in their telomere array. Shaded bars, 5-truncated elements; solid bars, intact elements (with Tag clusters removed).
If the sequence loss on a telomere occurs at the gradual rate measured for broken ends, and the 11 truncated elements here were unprotected by 5′ capping Tags, then they would have resided on the end of the telomere for periods ranging from one fly generation (the longest truncated element) to 81 fly generations (the shortest element). Because the Tag analysis shows that elements frequently remain on the extreme end of the chromosome for less than one fly generation it seems unlikely that many of the more truncated elements result from gradual end erosion. Instead, we favor the idea that truncation can result from terminal deletion. These terminal deletions may occur at many places within the array, leading to occasional rebuilding of all or part of the array.
Sequence loss from the longest truncated element (asterisk in Figure 1) falls within the range of sequence loss measured by Tag erosion, suggesting that it was produced by the same erosional process, rather than by terminal deletion. With much lower probability, the same might be true of the second longest (asterisk in Figure 1). It is also possible that one or both of these elements are transposition-competent since each retains part of its 5′ UTR. (Unlike the typical non-LTR element, HeT-A does not have its promoter in the 5′ UTR; thus these two elements have not lost essential promoter sequence.) However, 5′ UTR sequence might also have other functions, such as directing second-strand synthesis during transposition. Until more is known about activities of the 5′ UTR we cannot know whether either of these elements is competent to produce new HeT-A transpositions. Therefore, we do not include them as complete elements.
Although Drosophila telomeres have telomere-specific retrotransposons rather than telomerase, their telomeres appear to be functionally analogous to telomeres in other organisms. The use of occasional terminal deletions to maintain telomere length is another point of similarity with other organisms. The first evidence that terminal deletion is used to regulate telomere length came from studies of terminal rapid deletion in budding yeast (Li and Lustig 1996; Bucholc et al. 2001; Lustig 2003). More recently, mammalian telomeres have been shown to utilize a similar mechanism (Wang et al. 2004; Pickett et al. 2009). These examples show that loss of long segments of telomeres can be part of the regulation of telomere length. We suggest that similar rapid losses are also utilized by Drosophila telomeres, although the mechanism of the deletion may be different. Terminal deletions in mammals are most likely due to homologous recombination between their short, identical telomere repeats. It is less likely that such recombination is a major cause of deletion in the more complex repeats of Drosophila telomeres.
It is possible that some HeT-A elements become truncated during the process of transposition (Traverse et al. 2010). However, we suggest that most truncated elements in the arrays result from terminal deletion. Evidence of repeated loss of complete telomere arrays, discussed below, suggests that large terminal deletions are not uncommon, especially if we consider that any terminal deletions not reaching into subtelomere regions would almost certainly have escaped detection.
Although Drosophila telomeres may not undergo terminal deletion by homologous recombination, there is evidence that they do undergo loss of complete telomere arrays and then rebuild by addition of telomere retrotransposons. The evidence is perhaps more convincing because some of it is a by-product of investigations directed, not at chromosome ends, but at P-element expression. P-elements frequently insert in subtelomeric sequences, and three inserts on the tips of X (Marin et al. 2000), 3R (Sheen and Levis 1994), and 2L (Golubovsky et al. 2001) were found to have terminal deletions that removed the telomere array and part of the P-element inserted in telomere-associated sequences. In most cases, it appears that the P-element did not cause the deletion. Further evidence of long terminal deletions comes from studies of subtelomeric regions (Walter et al. 1995; Kern and Begun 2008). These regions have high levels of gene presence/absence polymorphism not seen in the adjacent euchromatin. At least some of this structural polymorphism has been shown to be due to terminal deletions that have been healed by transposition of HeT-A (Kern and Begun 2008). Early studies of lethal (2) giant larvae, near the 2L tip (Walter et al. 1995), and a recent, more extensive study of the tip of 3L (Kern and Begun 2008) have revealed deletions similar to those found in the P-element studies mentioned above.
Telomere arrays contain an unexpected number of complete elements with coding regions that show no signs of sequence decay:
If terminal sequence loss and new transpositions were relatively regular continuous processes, it might be expected that each element in the array would undergo approximately the same amount of 5′-truncation. As reported earlier (George et al. 2006), this is not what the arrays show. Complete HeT-A and TART elements are overrepresented in the 4R and XL telomere arrays. (This includes two complete TARTs making up 24.6 kb of the 4R array. For simplicity they are not shown in Figure 1 and will not be considered here.) The 4R telomere sequence has three complete HeT-A elements, and the most proximal element in the XL array is also complete. None of these elements shows evidence of sequence decay. Each has a complete 5′ UTR with an associated cluster of Tags, indicating that it has transposed several times.
HeT-A elements have a single ORF. It encodes a Gag protein involved in localization to telomere regions and apparently is required for transposition (Rashkova et al. 2002, 2003; Pardue et al. 2005). Complete coding regions in the HeT-A elements of the 4R and XL arrays identify four HeT-A subfamilies whose gag genes range from 2766 to 2856 nt (George et al. 2006). Most of the difference is in a length polymorphic region near the N terminus of the protein. None of these polymorphisms interrupt the reading frame, and all subfamilies share conserved sequences involved in specific interactions with other telomeric Gags (Fuller et al. 2010).
Even the three truncated HeT-A gag genes in these arrays show no degradation except loss of 5′ sequence. and although some gag sequences lie in the most proximal, and therefore the oldest, part of the 4R and XL telomeres, they show no sign of sequence decay, even though they should no longer be under selection for function. This suggests that these arrays turn over more frequently than other chromosomal regions.
This study suggests that at least three processes may be operating in telomere arrays—slow erosion, terminal deletion, and irregular transposition:
Like Tag erosion, transposition cannot be purely stochastic. It must be regulated in concert with end erosion and relatively frequent terminal deletion to control telomere length in response to environmental cues, to preserve a reservoir of replicatively competent HeT-A elements, and to balance the fact that each transposition adds orders of magnitude more sequence than the loss measured from Tag erosion (HeT-A, 6 kb, or TART or TAHRE, 10–12 kb).
It appears that telomere retrotransposons have two major functions. They provide telomere-specific DNA, analogous to the telomere-specific repeats produced by telomerase. They also maintain a population of functional elements capable of adding to the transposon array. The first function can be fulfilled by truncated elements, but the second function requires that some elements escape 5′ truncation and sequence decay. Our studies show that the 5′ Tags could provide protection against gradual terminal erosion, at least for elements that do not remain in the terminal position very long. Terminal deletions could maintain a more regular telomere length in spite of the very long additions added by each transposition. Perhaps more importantly, they would remove decayed elements, allowing replacement by transposition-competent elements when the deleted telomere is regenerated by new transpositions.
We propose that together the processes of slow erosion, terminal deletion, and irregular transposition maintain an environment that forms telomeric heterochromatin and also assures a supply of new HeT-A elements competent for maintenance of chromosome length by telomere-specific transposition.
HeT-A elements in centromeric heterochromatin
Sequences analyzed:
The BAC sequenced by Mendez-Lago et al. (2009) contained part of a telomere array that apparently transposed into the centromere region of the Y chromosome >13 MYA (Berloco et al. 2005). This conserved localization suggests that the HeT-A cluster has some role at the centromere, possibly forming the kinetochore, affecting sister-chromatid cohesion, or maintaining the heterochromatic environment.
Mendez-Lago et al. (2009) suggest that the sequence initially consisted of nine telomere retrotransposons in a typical telomeric head-to-tail array. Five retrotransposons, four HeT-As, and one TART at the distal end on the telomere were all extremely 5′-truncated. The remainder of the founder array consisted of four complete HeT-A elements, numbers 6, 7, 8, and 9 in their notation (see Table 1). This founder sequence could have been either a Y chromosome telomere that moved to the interior by an inversion or a segment of telomere from another chromosome inserted into the Y, which has a record of accepting sequence from other chromosomes (Koerich et al. 2008).
TABLE 1.
Elements in the Y chromosomal HeT-A array
| Element | Length | Longest ORF |
|---|---|---|
| A (6) | 2139 | 510 |
| B (7, 6) | 1668 | 558 |
| C (7, 6) | 1887 | 558 |
| D (7) | 2854 | 447 |
| E (6) | 1884 | 558 |
| F (7) | 3178 | 318 |
| G (8) | 3640 | 528 |
| H (9) | 3192 | 381 |
| I (9, 8) | 2567 | 246 |
| J (9) | 3185 | 381 |
| K (9, 8) | 1609 | 246 |
Elements are designated by single letters in their order on the BAC. Numbers in parentheses following the letter show the ancestral elements identified by Mendez-Lago et al. (2009) as contributing sequence to that element. Any sequence from other transposable elements was removed before analysis of these elements. Length of the element is given in nucleotides including both 5′ tags and 3′ oligoA. The longest ORF shows the number of nucleotides currently in the longest open reading frame. Element K was truncated at the 3′ end by cloning. For comparison, the mean length of complete HeT-A elements in telomere arrays is 5927 ± 104 bp, and the mean length of intact HeT-A ORFs is 2821 ± 41 bp.
The proposed nine-element founder sequence that moved into the Y would have been ∼30 kb long. In the Y chromosome, it has grown into a large sequence cluster: the BAC contains 159 kb of Y chromosome sequence, and the HeT-A cluster is truncated by cloning on both ends of the BAC. Part of the growth of the cluster is due to the insertion of members from seven families of nontelomeric transposable elements. Nevertheless, the majority of the expansion has come from amplification of regions within the original array. Mendez-Lago et al. (2009) propose that this amplification came about by a series of events involving different sections of the founder sequence.
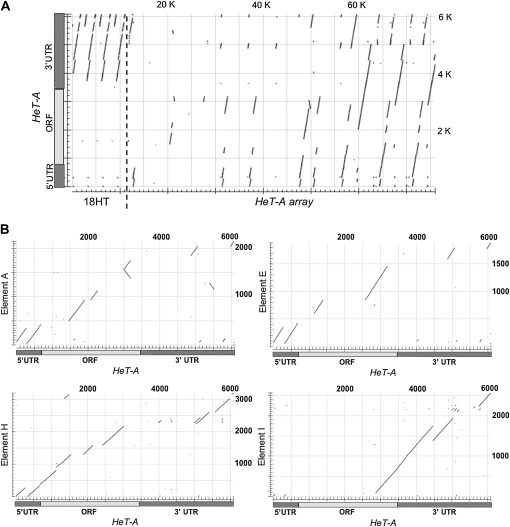
The amplifications divided the telomere sequence into two different kinds of arrays (Figure 4A). The severely truncated elements formed a 3.1-kb repeat that has been amplified to make up >100 kb of relatively homogeneous simple sequence repeats of the type classified as satellite DNA. This section is named the 18HT satellite.
Figure 4.—
(A) Analysis of HeT-A-related sequences in the HeT-A array in the centromeric region of the Y chromosome. Sequence of 76 kb of the cluster in the sequenced BAC is compared with the sequence of the canonical 6-kb complete HeT-A exactly as the 4R telomere sequence was compared in Figure 1. The HeT-A sequence is diagrammed on the y-axis, showing the 5′ UTR, ORF, and 3′ UTR. The dotted line near the left end of the plot marks the division between the sequence amplified in the 18HT satellite (first few repeats shown on the left) and the sequence of the ancestral complete HeT-A elements (on the right). These HeT-As gave rise to 11 elements, but the 3′end of the last element was cut off by cloning. Eleven 5′ ends can be detected as small fragments reaching the bottom line and the 10, 3′ ends as small fragments reaching the top line. These fragments are more easily seen in the higher magnification of B. Gaps in the elements indicate significant loss of sequence. The horizontal gaps contain sequence of nontelomeric transposable elements. (B) Comparisons of representative elements from the centromeric HeT-A array with the canonical HeT-A. Dot plots of individual elements were made as in A except that sequence of invading transposable elements was removed from each element. Note that here the HeT-A element is on the x-axis, and the centromere elements are on the y-axis. For description and order of elements, see Table 1. Element A now has three transposable elements inserted. Their sequence was removed before the dot plot, but some unidentified sequence remains. This element shows more sequence rearrangement than the others. There are two inversions (lines of opposite slant), and one of the inverted fragments is moved out of the collinear position. Element E was derived by amplification of A (Mendez-Lago et al. 2009), but both elements have undergone changes since the amplification. They now have 68% identity. Element H gave rise to J. These elements now have 99.7% identity. Element I gave rise to K, the element that was 3′-truncated by cloning. These elements have 98.9% identity where they overlap and are representative of the set. All have segments identical to the extreme 5′ and 3′ ends of HeT-A (lines touching the lower left and upper right corners) but some are difficult to see at this magnification.
The four complete HeT-A elements underwent a series of head-to-tail amplifications of different regions within their array to give 10 elements and an 11th element truncated by the cloning procedure. These elements, with the transposable elements inserted in them, now make up a complex set of repeats that stretches over >60 kb of the BAC. We refer to this region as the HeT-A array and designate the current set of elements as A–K (see Table 1 for their ancestral derivation as proposed by Mendez-Lago et al. 2009).
During their time on the Y chromosome, these HeT-A sequences have been conserved very differently from the HeT-As in telomere arrays. An overview of sequence from the centromeric BAC (Figure 4) shows that HeT-A sequences are much more fragmented than they are in the telomeric BAC (compare Figure 4A with Figure 1A).
Amplification events are not seen in telomere arrays:
In telomere arrays, each element is uniquely defined by combinations of subfamily sequence, 5′ truncation, presence or absence of Tag sequences, and length of the 3′ oligoA tail. These characters allowed Mendez-Lago et al. (2009) to identify elements amplified in the centromeric sequence. Similar amplifications have not been detected in telomere regions. Thus the amplification events provide the first evidence that the HeT-A array is being maintained differently in its centromeric position.
Full-length HeT-A elements in the centromeric array have undergone extensive internal deletions:
The 10 centromeric elements derived from ancestral intact elements 6–9 have undergone extensive sequence changes. Our analysis of this sequence (Figure 4B) shows that each of these centromeric elements has lost ∼40% or more of its sequence, and none would encode the Gag protein thought to be necessary for HeT-A transposition. In the original report (Mendez-Lago et al. 2009), these elements are described as decayed; however, our comparisons with HeT-A suggest that the changes are perhaps more accurately described as restructuring rather than decaying because they are not entirely random.
The four dot plots (Figure 4B) comparing individual centromeric elements to canonical HeT-A give a representative view of the changes in this array. In contrast to the telomere elements where loss is always from the 5′ end, each centromere element has several large internal deletions scattered through its sequence. There has been little rearrangement of the remaining sequence, most of which is collinear with the canonical HeT-A and has relatively few inversions and rearrangements. Surprisingly, the only regions that are conserved in every centromere element are the extreme 5′ and 3′ ends.
Comparisons of these elements show that many deletions are shared with siblings derived from the same amplification. Thus these 10 elements and the partial element have become a complex array of repeats. Because amplification events apparently involved more than a single unit, higher orders of repeat arise, as can be seen in the full-length view in Figure 4A. A more specific example of a higher-order repeat, elements H, I, J, and K, is shown in Figure 4B. H and J are nearly identical while the alternating elements I and K are also nearly identical but quite different from their flanking elements (H and J).
As a result of both sequence loss and nucleotide changes, these HeT-A elements have lost much of their protein-coding capacity. The longest open reading frames in these elements range from 246 to 558 nt while the shortest complete HeT-A gag gene is 2766 nt.
The centromeric HeT-A array contains several nontelomeric transposable elements:
At the telomere, nontelomeric elements are found only in small transition zones at the junction between the telomere array and the rest of the chromosome; elements in the transition zones are only fragments. In contrast, the centromeric BAC contains nontelomeric elements from seven different families, three in the 18HT sequences and four in the HeT-A array. In the HeT-A array, one element, the non-LTR retrotransposon F, arrived early in the expansion of the sequence and was included in two of the amplification events. More recent arrivals, mdg1, diver, and 1731, are present as single copies.
The LTR retrotransposon 1731 is especially interesting. It is completely intact and its two LTR sequences are identical. This element is a member of the 1731 subfamily in which the gag and pol reading frames are fused to produce an ORF of 3852 nt (Kalmykova et al. 1999). This entire reading frame is open and has 100% nucleotide identity with other elements in the database. Therefore, this element is potentially active. Other studies have suggested that there is at least one active 1731 on the Y chromosome. The 1731 fused Gag-Pol protein is expressed in testis (Kalmykova et al. 2004), where Y chromosomes are genetically active. Also, polytenized 1731 Y sequence has been found in salivary glands (Junakovic et al. 2003). Thus the 1731 in the centromere array may be involved in these activities.
The centromere array has been shaped into clusters resembling the repeated sequences found in centromeric heterochromatin:
As discussed above, the maintenance of HeT-A at the Y centromere has been dramatically different from the maintenance of HeT-A at telomeres. As a result, the ancestral telomere has given rise to two different types of clustered repeats: the more uniform 18HT satellite and a complex set of repeats derived from amplifications of several regions of internally deleted elements. Both types of repeats are head-to-tail arrays, rather than the palindromes that are abundant in protein-coding regions on human and chimpanzee Y chromosomes (Hughes et al. 2010). Both types of repeats are also similar to repeated sequences that characterize the heterochromatic centromere regions in chromosomes of multicellular organisms (Sullivan et al. 2001; Sun et al. 2003; Schueler and Sullivan 2006; Pertile et al. 2009).
Because centromeres in multicellular organisms are determined epigenetically (Sullivan et al. 2001; Malik and Henikoff 2009), it is not possible to identify centromeres by sequence alone. Nevertheless, this Y cluster is similar in size, repeated sequence structure, and presence of transposable elements to the only functionally characterized centromere in Drosophila, the X chromosome centromere (Sun et al. 2003). The similarities between the Y cluster and the X centromere do not extend to the level of the nucleotide sequence. Because the Y chromosome is the one chromosome that does not pair normally with its homolog, it is not surprising that the Y does not share centromeric sequences with the homolog. Y-specific centromere sequences could well be either a cause or an effect of this nonpairing behavior. [The mouse Y centromere is a known example of Y-specific centromere sequences (Pertile et al. 2009).]
There are reports that some characteristics of replication and/or repair in heterochromatin differ from those in euchromatin (Anderson et al. 2008; Peng and Karpen 2008). We suggest that the structure of these HeT-A-derived clusters might in part be determined by mechanisms preferentially used in pericentric regions, in addition to being driven by selection for function. For example, repeated amplification of portions of this sequence is an effective way of providing the rapid evolution that has been noted for centromere regions (Henikoff and Malik 2002).
The strong conservation of the 3′-most end of HeT-A in both 18HT and the more complex repeats in the Y centromeric complex may also be driven by function. There is growing evidence that RNA transcripts may be important for the formation of heterochromatin and for some aspects of centromere function (Savitsky et al. 2006; Slotkin and Martienssen 2007; Lee 2009; Malik and Henikoff 2009). Start sites for both the sense and the antisense transcripts of HeT-A lie in the conserved 3′ region; sense-strand start sites are found 93, 62, and 31 nt upstream of the 3′oligoA (Danilevskaya et al. 1997; Maxwell et al. 2006), while multiple antisense starts have been found 220 to 190 nt from the oligoA (Shpiz et al. 2009). Thus it is possible that the 3′ region is conserved to direct transcription from this sequence. Northern blots show several transcripts <1 kb in length with sequence homology to the 3′ region (Danilevskaya et al. 1999). Some of these RNAs are found only in males and might be products of the Y centromere. However, we have also found 3′ fragments of HeT-A in several intergenic regions on the Y, so the origin of the transcripts will require further study.
Acknowledgments
We thank Madeleine Crosby, Harvard University, for aid and advice while performing final annotation of the 4R and XL sequences. This work was supported by National Institutes of Health grant GM50315 to M.-L. P.
References
- Agudo, M., A. Losada, J. P. Abad, S. Pimpinelli, P. Ripoll et al., 1999. Centromeres from telomeres? The centromeric region of the Y chromosome of Drosophila melanogaster contains a tandem array of telomeric HeT-A- and TART-related sequences. Nucleic Acids Res. 27 3318–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. A., Y. S. Song and C. H. Langley, 2008. Molecular population genetics of Drosophila subtelomeric DNA. Genetics 178 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berloco, M., L. Fanti, F. Sheen, R. W. Levis and S. Pimpinelli, 2005. Heterochromatic distribution of HeT-A- and TART-like sequences in several Drosophila species. Cytogenet. Genome Res. 110 124–133. [DOI] [PubMed] [Google Scholar]
- Biessmann, H., S. B. Carter and J. M. Mason, 1990. Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. USA 87 1758–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., 2009. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19 607–612. [DOI] [PubMed] [Google Scholar]
- Bucholc, M., Y. Park and A. J. Lustig, 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux, R., and M. A. Batzer, 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O. N., I. R. Arkhipova, K. L. Traverse and M. L. Pardue, 1997. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell 88 647–655. [DOI] [PubMed] [Google Scholar]
- Danilevskaya, O. N., K. L. Traverse, N. C. Hogan, P. G. Debaryshe and M. L. Pardue, 1999. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol. Cell. Biol. 19 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, A. M., E. G. Cook, K. J. Kelley and M.-L. Pardue, 2010. Gag proteins of Drosophila telomeric retrotransposons: collaborative targeting to chromosome ends. Genetics 184 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, J. A., P. G. Debaryshe, K. L. Traverse, S. E. Celniker and M. L. Pardue, 2006. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 16 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky, M. D., A. Y. Konev, M. F. Walter, H. Biessmann and J. M. Mason, 2001. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics 158 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and H. S. Malik, 2002. Centromeres: selfish drivers. Nature 417 227. [DOI] [PubMed] [Google Scholar]
- Hoskins, R. A., J. W. Carlson, C. Kennedy, D. Acevedo, M. Evans-Holm et al., 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J. F., H. Skaletsky, T. Pyntikova, T. A. Graves, S. K. Van Daalen et al., 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463 536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junakovic, N., D. Fortunati, M. Berloco, L. Fanti and S. Pimpinelli, 2003. A subset of the elements of the 1731 retrotransposon family are preferentially located in regions of the Y chromosome that are polytenized in larval salivary glands of Drosophila melanogaster. Genetica 117 303–310. [DOI] [PubMed] [Google Scholar]
- Kahn, T., M. Savitsky and P. Georgiev, 2000. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol. Cell. Biol. 20 7634–7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova, A., C. Maisonhaute and V. Gvozdev, 1999. Retrotransposon 1731 in Drosophila melanogaster changes retrovirus-like expression strategy in host genome. Genetica 107 73–77. [PubMed] [Google Scholar]
- Kalmykova, A. I., D. A. Kwon, Y. M. Rozovsky, N. Hueber, P. Capy et al., 2004. Selective expansion of the newly evolved genomic variants of retrotransposon 1731 in the Drosophila genomes. Mol. Biol. Evol. 21 2281–2289. [DOI] [PubMed] [Google Scholar]
- Kern, A. D., and D. J. Begun, 2008. Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics 179 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich, L. B., X. Wang, A. G. Clark and A. B. Carvalho, 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. T., 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis, R. W., 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58 791–801. [DOI] [PubMed] [Google Scholar]
- Li, B., and A. J. Lustig, 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10 1310–1326. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J., 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4 916–923. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2009. Major evolutionary transitions in centromere complexity. Cell 138 1067–1082. [DOI] [PubMed] [Google Scholar]
- Marin, L., M. Lehmann, D. Nouaud, H. Izaabel, D. Anxolabehere et al., 2000. P-element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics 155 1841–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, P. H., J. M. Belote and R. W. Levis, 2006. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 34 5498–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lago, M., J. Wild, S. L. Whitehead, A. Tracey, B. De Pablos et al., 2009. Novel sequencing strategy for repetitive DNA in a Drosophila BAC clone reveals that the centromeric region of the Y chromosome evolved from a telomere. Nucleic Acids Res. 37 2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailovsky, S., T. Belenkaya and P. Georgiev, 1999. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma 108 114–120. [DOI] [PubMed] [Google Scholar]
- Pardue, M. L., S. Rashkova, E. Casacuberta, P. G. Debaryshe, J. A. George et al., 2005. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 13 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. C., and G. H. Karpen, 2008. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 18 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertile, M. D., A. N. Graham, K. H. Choo and P. Kalitsis, 2009. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 19 2202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, H. A., A. J. Cesare, R. L. Johnston, A. A. Neumann and R. R. Reddel, 2009. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova, S., S. E. Karam, R. Kellum and M. L. Pardue, 2002. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J. Cell Biol. 159 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova, S., A. Athanasiadis and M. L. Pardue, 2003. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J. Virol. 77 6376–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, M., D. Kwon, P. Georgiev, A. Kalmykova and V. Gvozdev, 2006. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler, M. G., and B. A. Sullivan, 2006. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 7 301–313. [DOI] [PubMed] [Google Scholar]
- Sheen, F. M., and R. W. Levis, 1994. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc. Natl. Acad. Sci. USA 91 12510–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz, S., D. Kwon, Y. Rozovsky and A. Kalmykova, 2009. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 37 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R. K., and R. Martienssen, 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8 272–285. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., M. D. Blower and G. H. Karpen, 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2 584–596. [DOI] [PubMed] [Google Scholar]
- Sun, X., H. D. Le, J. M. Wahlstrom and G. H. Karpen, 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse, K. L., J. A. George, P. G. Debaryshe and M. L. Pardue, 2010. Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons. Proc. Natl. Acad. Sci. USA 107 5064–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M. F., C. Jang, B. Kasravi, J. Donath, B. M. Mechler et al., 1995. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma 104 229–241. [DOI] [PubMed] [Google Scholar]
- Wang, R. C., A. Smogorzewska and T. De Lange, 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119 355–368. [DOI] [PubMed] [Google Scholar]