Abstract
The kinetochore is a dynamic multiprotein complex assembled at the centromere in mitosis. Exactly how the structure of the kinetochore changes during mitosis and how its individual components contribute to chromosome segregation is largely unknown. Here we have focused on the contribution of the Mis12 complex to kinetochore assembly and function throughout mitosis in Drosophila. We show that despite the sequential kinetochore recruitment of Mis12 complex subunits Mis12 and Nsl1, the complex acts as a single functional unit. mis12 and nsl1 mutants show strikingly similar developmental and mitotic defects in which chromosomes are able to congress at metaphase, but their anaphase movement is strongly affected. While kinetochore association of Ndc80 absolutely depends on both Mis12 and Nsl1, BubR1 localization shows only partial dependency. In the presence of residual centromeric BubR1 the checkpoint still responds to microtubule depolymerization but is significantly weaker. These observations point to a complexity of the checkpoint response that may reflect subpopulations of BubR1 associated with residual kinetochore components, the core centromere, or elsewhere in the cell. Importantly our results indicate that core structural elements of the inner plate of the kinetochore have a greater contribution to faithful chromosome segregation in anaphase than in earlier stages of mitosis.
ACCURATE segregation of sister chromatids in mitosis is crucial for viability and normal fate of daughter cells. Missegregation leads to aneuploidy, a phenomenon associated with developmental defects and cancer (Kops et al. 2005). Accurate segregation of chromosomes requires function of kinetochores, multiprotein complexes assembled on the outer sides of centromeres on each sister chromatid (Allshire and Karpen 2008; Welburn and Cheeseman 2008). The molecular composition and functions of the kinetochore change during mitosis. Kinetochore assembly begins with the accumulation of the Mis12 complex on centromeres in late G2 (Goshima et al. 2003; Kline et al. 2006). This is joined by Spc105 and the Ndc80 complex to form the stable “core kinetochore” on mitotic entry (Cheeseman and Desai 2008). The core kinetochore provides a platform for kinetochore-associated proteins recruited after nuclear envelope breakdown (NEBD) (Santaguida and Musacchio 2009). After NEBD, microtubules emanating from the two opposite poles of the spindle form transient connections with kinetochores (Vorozhko et al. 2008). Once sister kinetochores have made attachments to opposite poles, the bioriented chromosomes are able to congress to the equator of the mitotic spindle in prometaphase. Proper chromosome biorientation and congression is monitored by the spindle assembly checkpoint (SAC), which recognizes unattached kinetochores (Welburn and Cheeseman 2008) and reduced inter- and/or intrakinetochore tension (Elowe et al. 2007; Maresca and Salmon 2009; Uchida et al. 2009), and responds by delaying anaphase promoting complex/cyclosome (APC/C) activation. Erroneous microtubule (MT) kinetochore connections are recognized, disassembled, and corrected through an Aurora B-mediated mechanism (Ruchaud et al. 2007). Once alignment has been achieved at metaphase, securely bound k-fiber MTs enable the core kinetochores to sustain two anaphase functions: to maintain stable connections between segregating chromosomes and the k-fiber MTs and to provide docking sites for proteins that contribute to MT plus-end dynamics and for motor proteins required for chromosome movement (Cheeseman et al. 2006; Dong et al. 2007).
The Mis12 complex is an inner subcomplex of the core kinetochore, which in the majority of organisms comprises four subunits, Mis12, Dsn1, Nnf1, and Nsl1. In cultured human cells, localization of these subunits to the kinetochore was found to be interdependent (Kline et al. 2006). Partial depletion of any of the components by RNAi led to reduced Ndc80 complex and Blinkin/Spc105 at the kinetochore and similar defects in chromosome biorientation and segregation (Kline et al. 2006). Cenp-E and BubR1 kinetochore localization were also reduced and SAC activity was compromised. In contrast, depletion of the Dsn1 homolog of Caenorhabditis elegans was found to give a more severe, “kinetochore null,” phenotype than depletion of the other subunits: chromosomes failed to align to the metaphase plate and failure of chromosome segregation was associated with premature spindle elongation and mislocalization of all other kinetochore components (Desai et al. 2003). Depletion of the Mis12, Nnf1, and Nsl1 orthologs led to much less severe chromosome segregation and spindle elongation defects and reduced but did not prevent recruitment of other core kinetochore components (Cheeseman et al. 2004). The differences in phenotypes after depletion of Mis12 complex subunits may reflect incomplete knockdown, but may be also interpreted as real differences between function of individual proteins in different species. Recent high-resolution light microscopy studies (Schittenhelm et al. 2007; Joglekar et al. 2009; Wan et al. 2009) and low-resolution structural studies on yeast and human Mis12 complexes (Maskell et al. 2010; Petrovic et al. 2010) suggest linear, sequential arrangement of subunits in the complex. However, together these data do not currently provide a consistent view about the position or the function of individual Mis12 complex subunits in the architecture of the kinetochore.
Drosophila has one mis12, one nsl1 gene, and two nnf1 genes that encode isoforms with different developmental expression patterns (Schittenhelm et al. 2007). Despite extensive studies, a Dsn1 ortholog has not been identified (Meraldi et al. 2006; Przewloka et al. 2007, 2009; Schittenhelm et al. 2007). In contrast to the uniform recruitment of Mis12 complex members in human cells, EGFP-tagged Mis12 and Nnf1a were found to localize to centromeres of cultured cells in interphase and thus in advance of the mitotic localization of Nsl1 and Nnf1b. In accord with this finding, kinetochore recruitment of Drosophila Mis12 was independent of Nsl1, whereas Nsl1 recruitment required Mis12. Knockdown of several centromeric (CID, CENP-C), Mis12 complex (Mis12, Nsl1), or Ndc80 complex (Nuf2, Ndc80, Spc25/Mitch) proteins led to similar phenotypes in cultured cells: chromosome congression was impaired, anaphases showed chromosome missegregation and bridging, and spindles became markedly elongated (Przewloka et al. 2007).
Here we wished to clarify whether the apparent differences in organization and recruitment of Mis12 complex components between Drosophila tissue culture cells and other metazoans had any significance in the whole organism. To this end, we have investigated the phenotypes of mutants for two representative genes of the Mis12 complex, mis12 and nsl1. We confirm that their gene products are differentially localized to the kinetochore in the embryonic cell cycles. Nevertheless, mis12 and nsl1 mutants show strikingly similar developmental and mitotic defects in flies. Interestingly, chromosome congression can occur following depletion of these molecules in the mutants. Residual Mis12 and Nsl1 molecules on kinetochores are sufficient to perform the function required for the chromosome congression. However, having a functional kinetochore and the correct stoichiometry of Mis12 complex subunits appear to be essential for correct anaphase chromosome movement. Despite kinetochore disruption, we found residual BubR1 still present in centromeric regions and the spindle assembly checkpoint, although weakened, remained functional.
MATERIALS AND METHODS
cDNAs and constructs:
Full-length cDNA clones for Mis12 (RE19545), Nuf2 (SD05495), Ndc80 (LD33040), and Nsl1 (RE03006) were ordered from the Drosophila Genomics Resource Centre. Constructs used in this study were generated by the Gateway Cloning System (Invitrogen). Entry clones for C- and N-terminal protein fusions were generated for all of the cDNAs listed above. Constructs for bacterial protein expression and for transgenic flies were made by LR reactions between the entry clones and the following destination vectors: pDEST42 (for expression in Escherichia coli with C-terminal 6xHis fusion, Invitrogen), pKM596 (for expression in E. coli with N-terminal maltose binding protein (MBP) fusion; Fox et al. 2003), pDEST12 and pDEST13 (for UASp promoter-driven expression of N-terminal and C-terminal EGFP fusions in transgenic flies, constructed by Frederik Wirtz-Peitz in our laboratory).
Fly stocks:
The Drosophila mutant stocks nsl1P{lacW}G0237 (Peter et al. 2002), mis12PBac{WH}f03756 (Thibault et al. 2004), and the Df(3L)BSC27, Df(3L)BSC224 deficiencies were obtained from the Bloomington Stock Center. Stocks for Df(1)52 and Dp(1:Y)Bs- v+ y+ (Meller and Rattner 2002) were kindly provided by Victoria Meller. We used GFP balancer chromosomes to distinguish between homozygous or hemizygous mutants from sibling progeny derived from heterozygous parents. For embryonic stages 9–12, we used the balancer chromosomes FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5 or TM3, P{GAL4-Kr.C}DC2, P{UAS-GFP.S65T}DC10, Sb1 (Casso et al. 2000). In late embryos and in larval stages we used the FM7i, P{Act-GFP}JMR3 or TM3, P{w[+mC]=Act-GFP}JMR2, Ser balancer chromosomes (Ferrandon et al. 1998).
To investigate the function of mis12 and nsl1 in mitotic chromosome segregation in living animals, mutant alleles were combined with a second chromosome bearing the P{His2AvD-EGFP.C} insertion (Crest et al. 2007). To investigate the cal1 phenotype in flies cal1PBac{PB}cal1c03789 and Df(3R)Exel6176 were combined.
Transgenic strains expressing kinetochore proteins fused to EGFP were generated by standard P-element–mediated germ line transformation and by selection on the basis of the use of the mini-white dominant marker gene. Functionality of Mis12∷EGFP and Nsl1∷EGFP transgenes were confirmed by rescue experiments. Features of the transgenes are summarized in supporting information, Table S1. To express fusion proteins in the early embryo for the purpose of live imaging, we used the α-tubulin4–GAL4–VP16 driver (Duffy 2002). Single copies of the driver and the UASp transgenes were combined in female flies by standard crosses. The animals were raised on standard food and kept on at 25°.
Cell culture and RNAi experiments:
The Dmel-2 cell line (Invitrogen) was cultured in Drosophila Express Five SFM medium (Invitrogen) supplemented with Pen/Strep and l-glutamine at 25° according to standard protocols. dsRNAs for mis12, nsl1, and ndc80 were synthesized and RNAi treatments were carried out as described previously (Przewloka et al. 2007).
Antibodies and immunofluorescence microscopy:
Polyclonal antibodies were generated in rabbits against bacterially expressed Ndc80–6xHis, Nsl1–6xHis, MBP–Mis12, and against the mixture of two Mis12 peptides: DFNSLAYDQKFFNF and CKLKQFWNQVPLQIKKETD; IgGs were purified from the sera. The anti-MBP–Mis12 serum was preabsorbed to MBP before using it in Western blot experiments.
For immunofluorescence imaging cultured cells were fixed and stained as described before (Bettencourt-Dias et al. 2004). Blocking and staining with primary and secondary antibodies was carried out as described previously (Sullivan et al. 2000). Briefly, embryos of the requested age were collected, dechorionated by incubation in 50% bleach for 2 min, rinsed in water, and immobilized on coverslips. Immobilized embryos were covered by a drop of fixation buffer. The vitelline membranes were permeabilized mechanically by a glass needle at the ends of the embryos. The fixative was allowed to diffuse into the embryos for 15 min. Fixed embryos were manually removed from the vitelline membrane, rinsed twice in PBS, and once in PBT. Primary rabbit antibodies were used in 1:500 dilutions and were against: Mis12 peptide, Nsl1, Ndc80, and Ser10-Phospho-histone H3 (Abcam) and BubR1 (generated in our laboratory). A mouse monoclonal α-tubulin DM1A (Sigma) was used at a 1:1000 dilution. Chicken anti-CID antibody (generated in our laboratory) was used at a 1:3000 dilution. For Westerns anti-MBP–Mis12 and anti-Ndc80 antibodies were used at 1:2000 dilutions.
Drug treatment of embryos:
mis12, nsl1 mutant, and wild-type stage 14 embryos were dechorionated using 50% domestic bleach. The vitelline membrane of the embryos was permeabilized by brief octane treatment (Su et al. 1999). Subsequently embryos were incubated for 1 hr in Drosophila Express Five SFM medium (Invitrogen) supplied with 25 μm colcemid or with equal volume of DMSO. After drug treatment embryos were fixed and processed for immunofluorescence as described above.
Live imaging:
To characterize mitotic defects in mis12 and nsl1 mutants, eggs were collected from fertilized Drosophila females having the P{His2AvD-EGFP.C} insertion on the second chromosome and nsl1P{lacW}G0237/FM7i, ftz-lacZ or mis12PBac{WH}f03756/TM3, ftz-lacZ genotype and aged for 180–210 min. Partner males had the P{His2AvD-EGFP.C} insertion on the second chromosome and Y/FM7i, ftz-lacZ or Df(3L)BSC224/TM3, ftz-lacZ chromosome combinations, respectively. Staining for lacZ activity enabled fixed mutant and wild-type embryos to be distinguished after live imaging. Embryos were dechorionated, aligned, and immobilized on coverslips. After covering embryos with halocarbon oil, spinning disc confocal laser microscopy was carried out on an inverted Zeiss Axiovert 200 microscope equipped with a Perkin Elmer UltraVIEW RS system. A Zeiss ×63/1.4 oil immersion objective was used for time-lapse imaging of GFP fluorescence signals. GFP signals in the embryonic cortex were filmed through mitoses 14–16. Z-stacks of 25 images (stack step 0.5 μm) were acquired every 20 sec. Experiments were carried out at 24° in a temperature-controlled room.
RESULTS AND DISCUSSION
Drosophila Mis12 complex proteins, Mis12 and Nsl1, differ in the dynamics of their recruitment to centromeres:
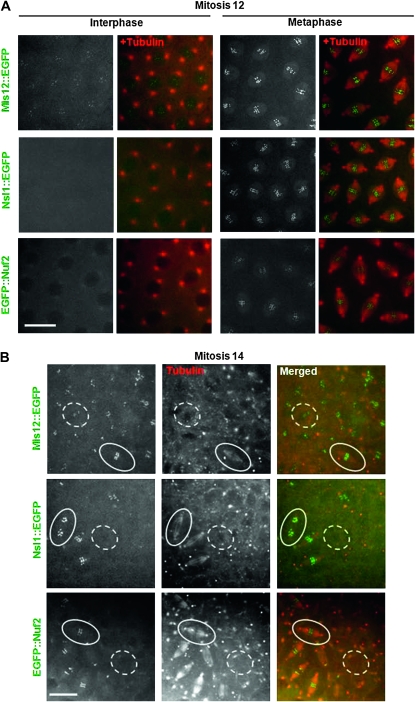
We wished to determine whether the differences in timing of recruitment of EGFP-tagged Mis12 or Nsl1 (Przewloka et al. 2007) or their endogenous counterparts (Figure S1 and Figure S2) to the kinetochore in cultured Dmel-2 cells were also seen in cells in the intact organism. We therefore constructed transgenic lines of flies expressing EGFP-tagged Mis12 or Nsl1 to follow the dynamics of recruitment of these proteins to the kinetochore. We found that in both the nuclear division cycles of the syncytium (Figure 1A, File S1, File S3) or in the mitotic domains of the cellularized embryo (Figure 1B, File S2, File S4), EGFP–Mis12, but not EGFP–Nsl1, was present at the centromere in interphase. The signal intensity of EGFP–Mis12 at the centromere was, however, weaker in interphase than in mitosis when it appeared to be recruited to the kinetochore immediately before nuclear envelope breakdown. In contrast, Nsl1 was first recruited to the kinetochore in prophase. In this aspect the timing of Nsl1 recruitment was more similar to the Ndc80 complex subunit Nuf2 than to Mis12 (Figure 1, File S5, File S6). Thus in Drosophila, association of a population of Mis12 with the centromere in interphase appears to anticipate the assembly of the Mis12 complex on mitotic entry. This is quite different from the weak localization of all members of the Mis12 complex to the centromere during interphase and the wave of their recruitment just before mitosis in human cells (Goshima et al. 2003; Kline et al. 2006; McAinsh et al. 2006; Hemmerich et al. 2008). This could reflect either differences in organization of the Mis12 complex between insects and mammals or the increase in centromere component complexity in mammals (reviewed in Przewloka and Glover 2009).
Figure 1.—
Localization of EGFP-tagged Mis12 and Ndc80 complex proteins in living transgenic embryos. (A) In syncytial blastoderm, a subpopulation of Mis12∷EGFP is constantly present on centromeres. In contrast, Nsl1 does not localize to interphase centromeres and EGFP∷Nuf2 is excluded from interphase nuclei. In mitosis, all the three fusion proteins localize identically to kinetochores. (B) Following cellularization, these three proteins show similar subcellular localization to syncytial blastoderm: Mis12 is present throughout the cell cycle but increases in levels at the kinetochore during mitosis; Nsl1 and Nuf2 only associate with chromosomes at kinetochores during mitosis. Microtubules were detected by injected rhodamine-conjugated tubulin. Examples of single mitotic cells are surrounded by intact lines; single interphase cells are surrounded by dotted lines. Bars, 10 μm.
mis12 and nsl1 mutants display embryonic lethality:
To gain insight into the in vivo functions of these two Mis12 complex subunits in Drosophila we have characterized mutant alleles of their genes. The mis12PBac{WH}f03756 allele has a transposon insertion in the first intron of mis12 and was lethal when homozygous or when placed over two different deficiency chromosomes, Df(3L)BSC27 or Df(3L)BSC224, that each uncover only a narrow cytological region on the third chromosome. A total of 95% of mis12PBac{WH}f03756 homozygotes and 98% of mis12PBac{WH}f03756/Df(3L)BSC224 individuals died during embryogenesis, with the hatched mutant larvae dying uniformly at first larval instar (L1). The nsl1P{lacW}G0237 allele has a P transposon inserted into the second exon of nsl1 (after codon 87) and was lethal in homozygous and in nsl1P{lacW}G0237/Y hemizygous combinations and when placed against a 50-kb deficiency, Df(1)52, which removes the entire nsl1 gene (Meller and Rattner 2002). Additionally Dp(1:Y)Bs v+ y+ (a translocation including the nsl1 genomic region; Meller and Rattner 2002) completely rescued the lethality of the nsl1P{lacW}G0237/Df(1)52 allele combination. We found that 65% of nsl1P{lacW}G0237/− individuals died as embryos and 35% reached the L1 larval stage. The hatched nsl1 mutant larvae were strongly retarded in movement and all of them died in the first instar (n = 400). Together, these data indicate that the mis12PBac{WH}f03756 and nsl1P{lacW}G0237 alleles are strong hypomorphic or potential null alleles of mis12 and nsl1. This was confirmed by our failure to detect Mis12 or Nsl1 proteins either by Western blotting or immunostaining in the late mitotic cycles of embryonic development (Figure S3).
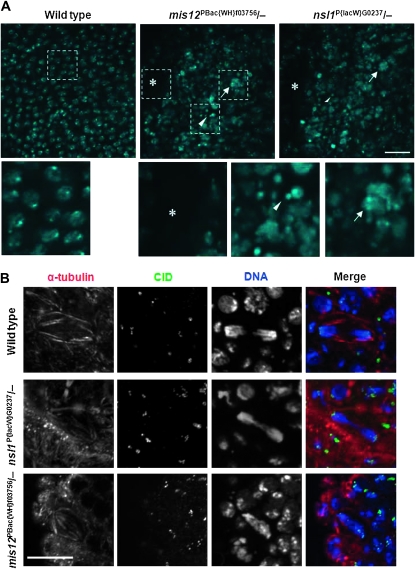
Neither nsl1 nor mis12 mutant embryos displayed any major gross defects in their morphological patterning at late stages (Bowne's stages 15–17; data not shown). However they did have body areas with lower cell densities in which cells displayed a high number of micronuclei and/or presumptive polyploid nuclei, indicative of accumulated defects in chromosome segregation from previous mitotic divisions (Figure 2A). In fixed mutant embryos there were no detectable mitotic defects up to mitosis 14, indicating that their maternal proteins can drive mitosis until this stage. Throughout cell division cycle 15 and 16, the size and shape of mitotic spindles in fixed mis12 and nsl1 mutant embryos did not differ significantly from wild type (Figure S4). We could detect no signs of any chromosome condensation or congressional defects in the fixed (tubulin-, CID-, and DNA-stained) embryo preparations in mitoses 15 and 16 (data not shown). On the other hand, there were a high number of chromosome segregation defects in anaphase and telophase (Figure 2B), indicating a requirement of mis12 and nsl1 for the proper segregation of mitotic chromosomes. At a stage where only a few CNS restricted cells of wild-type embryos perform one last mitosis before hatching, both the mis12 and nsl1 mutants had an increased number of cells in mitosis (see also below). Many of these cells were dispersed throughout the whole body, suggesting they represented mitotically delayed cells from earlier cycles.
Figure 2.—
Cellularized embryos of mis12 and nsl1 mutants show similar mitotic defects. (A) Distribution and morphology of nuclei (stained with DAPI to reveal DNA) in mis12 and nsl1 mutant embryos. Areas showing a lower nuclear density (*), high numbers of micronuclei (arrowheads), or large, presumably polyploid nuclei (arrows) are characteristic for both mutants. (B) Wild-type, mis12, and nsl1 mutant embryos stained to reveal α-tubulin (red), CID (green), or DNA (blue). Lagging chromosomes and bridges were seen in anaphase cells of the mis12 and nsl1 mutants in mitosis 15 and 16. Bars, 10 μm.
The developmental consequences of Mis12 or Nsl1 depletions are not only very similar to each other but also to those in mutants of centromeric components cid (Blower et al. 2006), cenp-c (Heeger et al. 2005), and cal1 (Erhardt et al. 2008 and see below) or the core kinetochore component Spc105 (Schittenhelm et al. 2009). In contrast, hypomorphic and null mutants for Ndc80 complex subunit, mitch (Williams et al. 2007) and nuf2 (our unpublished data) die only later in late larval stages. This could either reflect prolonged maternal contributions of Ndc80 complex components or a greater ability to tolerate the absence of the Ndc80 complex than the Mis12 complex from the kinetochore in embryonic cycles. Further experimentation will be required to distinguish between these two possibilities. The uniformity of spindle length between mis12 and nsl1 mutant and wild-type cells contrasts with the dramatic increase in spindle length upon knockdown of these genes in cultured cells. This may either represent different properties of the spindles per se between these different cell types or physical constraints upon cell size within tissues of the organism that are irrelevant for cultured cells. The depletion of other mitotic regulators, the Greatwall kinase for example, also leads to spindle elongation in tissue culture cells (Bettencourt-Dias et al. 2004) but not in the larval CNS (Yu et al. 2004; Archambault et al. 2007).
mis12 and nsl1 mutants show prometaphase delay and segregation defects:
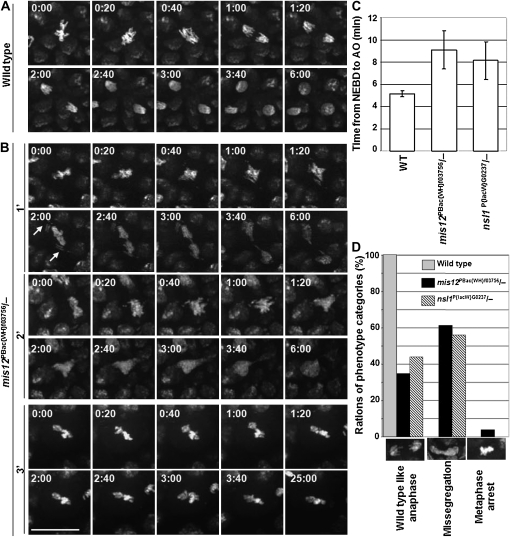
To understand more of the nature of the mitotic defects in mis12 and nsl1 hemizygous embryos, we carried out time-lapse imaging studies of mitoses 15 and 16 in which chromosome segregation defects lead to formation of aneuploid daughter cells (Figure 3, File S7, File S8, File S9, File S10, and File S11). We observed similar mitotic defects in both the mis12 and nsl1 mutants and classified them into three broad categories. In the first category were cells that showed a pronounced delay in anaphase onset (AO) (Figure 3, B and C). Nevertheless, chromosomes did congress to give a metaphase-like plate in this group and segregation then appeared to take place normally at anaphase. The second, and major, category comprised cells that also showed chromosome segregation defects (Figure 3, B and C). In the example of such a cell given in Figure 3B, the major chromosomes have remained at the equator of the cell where they have eventually undergone decondensation upon mitotic exit. In contrast, the tiny fourth chromosomes segregate to the poles. Indeed, we consistently observed cells in which smaller chromosomes (fourth and sex chromosomes) arriving at the spindle poles sooner than the larger ones (second and third). We found chromosome decondensation not accelerated or delayed in these cells. The chromosomes seemed to decondense with the same timing as in wild-type cells (compare Figure 3A and 1′ and 2′ from 3B). Since many of sister chromatids in the mutants were not fully detached as they started to decondense, the entangling of decondensing chromosomes most probably strongly contributed to the formation of telophase bridges.
Figure 3.—
Progression of mitosis 15 in living mis12 and nsl1 mutant embryos. (A) Congression and segregation of His2AvD∷EGFP-marked chromosomes in wild-type embryos at stage 9. (B) Three examples of congression and segregation of His2AvD∷EGFP-marked chromosomes in mis12 mutant embryos. In the majority of mutant cells, the breadth of the congressed metaphase plate did not differ from wild-type cells. Segregation defects were more severe for the larger chromosomes (second and third). The smaller chromosomes (fourth and sex chromosomes) were less effected (series 1′ time point 2:00). (C) Prolonged time between NEBD and AO is a characteristic of both mutants. This time interval was measured in 8 nuclei from two wild-type embryos and for 12 or 15 nuclei from three embryos of mis12 and nsl1 mutants. Error bars represent ±1 SD. (D) Categories of chromosome segregation phenotypes in wild-type, mis12, and nsl1 mutant embryos. N = 80 nuclei from three embryos for wild type; 150 nuclei from six embryos for mis12 mutants; and 150 nuclei from five embryos for nsl1 mutants.
The third category comprised cells in which mitotic chromosomes congressed to the equator of the cell and then remained there in a metaphase-like arrest (Figure 3, B and D).
The ability of chromosomes to congress in the great majority of mis12 and nsl1 mutant cells indicates that full kinetochore function is not required and that redundant mechanisms are being used for this process. Of these, chromosome movement utilizing chromokinesin function seems likely to be most significant (Afshar et al. 1995). Although several outer kinetochore components, supposedly missing from kinetochores of mis12 and nsl1 mutants (e.g., Ndc80, see below) have been shown to be required for chromosome congression in human cells, this process can occur in the absence of k-fibers (Cai et al. 2009). The fact that chromosomes congress in the mis12 and nsl1 mutants may either reflect sufficiency of alternative mechanisms for this process in Drosophila compared to vertebrates (see Kops et al. 2010 for review) or the incomplete depletion of Mis12 and Nsl12 in mutant embryos. Perhaps even a low amount of residual Mis12 and Nsl1 on kinetochores is sufficient to perform the function required for the chromosome congression. However, a functional kinetochore and the correct stoichiometry of Mis12 complex subunits appear to be essential for correct anaphase chromosome movement. The uniform rate of chromosome movement in a normal anaphase may reflect the equivalent density of kinetochore fiber microtubules that has been observed by electron microscopy for all chromosomes (Maiato et al. 2006). It may be that the ability of some chromosomes to move poleward at anaphase in the mis12 and nsl1 mutants may reflect some residual ability of microtubules to connect to kinetochore remnants, which could be sufficient to move the smaller, but not the larger, chromosomes.
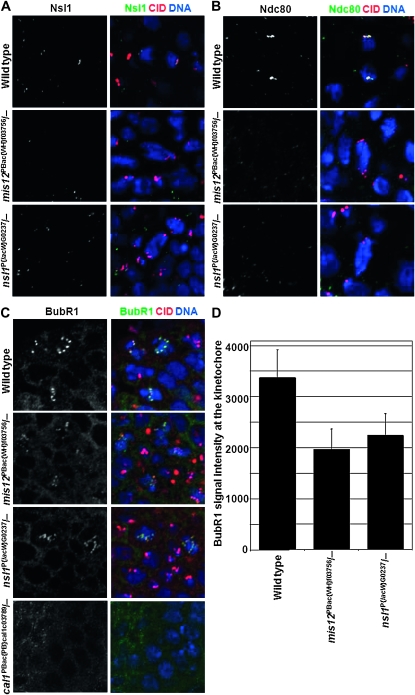
mis12 and nsl1 mutants fail to recruit Ndc80 complex but still have BubR1 at centromeres:
To uncover the extent to which kinetochore organization was disrupted in mis12 and nsl1 mutants, we examined the localization of the Ndc80 protein, a key component of the Ndc80 complex that links the Mis12 complex to kinetochore fiber MTs. In addition to providing the attachment site for kinetochore microtubules, the Ndc80 complex is also crucial for recruitment of certain of the SAC proteins including the RZZ complex and Mad2 (Martin-Lluesma et al. 2002; DeLuca et al. 2003; Lin et al. 2006). We were unable to detect Ndc80 at kinetochores in both mis12 and nsl1 mutant stage-14 embryos (Figure 4). This accords with the high levels of chromosome segregation defects observed. In contrast, the centromeric localization of BubR1 was only partially diminished in mis12 or nsl1 mutant cells (Figure 4, C and D) and is similar to spc105 mutants, as previously reported (Schittenhelm et al. 2009).
Figure 4.—
Recruitment dependencies of Nsl1 (A), Ndc80 (B), and BubR1(C and D) in mis12 and nsl1 mutant stage 14 embryos. Green staining indicates the Nsl1, Ndc80, and BubR1 proteins relative to CID (red) and DNA (blue). Localization of BubR1 in cal1 mutant is also shown for comparison in C. Error bars represent ±1 SD.
When we examined cal1 mutant embryos, in which the centromere does not form, as shown by the absence of CID (Cal1 protein is involved in the CENP-A/CID loading onto centromeric chromatin; Erhardt et al. 2008), we were unable to detect BubR1 on chromosomes (Figure 4C). In human cells the kinetochore localization of BubR1 seems to be dependent on Spc105 (Kiyomitsu et al. 2007) and such an interaction is also possible in Drosophila. Our studies of mis12, nsl1, and cal1 mutants as well as studies of spc105 and cid mutants published by other groups (Blower et al. 2006; Schittenhelm et al. 2009) suggest that BubR1 may have at least two different binding sites: one on the centromere, the other one on the kinetochore. Indeed two different localizations of BubR1, at inner and outer kinetochore, have in fact been previously reported (Jablonski et al. 1998; reviewed in Huang and Yen 2009). We are at present unable to account for the mechanism of the kinetochore independent centromeric localization of BubR1. Additionally, it is still necessary to explain how BubR1 localization is restricted in space to the centromeric region of chromosomes and in time to the prophase-to-metaphase interval. The fact that BubR1 is not detectable at the centromeres of cid (Blower et al. 2006) and cal1 mutants suggests that these two proteins might be essential for BubR1 localization at the centromere. BubR1 was also recently described to be bound to DNA tethers that have been observed to form between centric and acentric chromosome fragments of broken chromosome arms (Royou et al. 2010) and at unprotected telomeres (Musaro et al. 2008). Understanding BubR1's partners on DNA tethers, uncapped telomeres, centromeres, and kinetochores is an important challenge requiring further investigation.
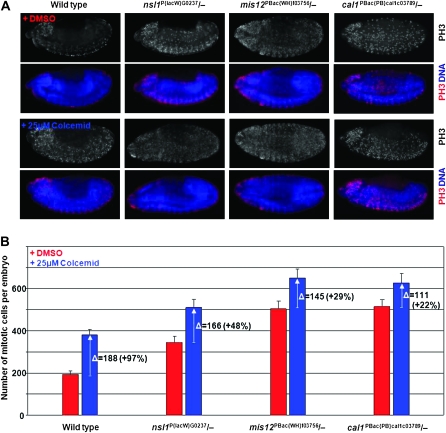
The extended time between NEBD and AO seen in mis12 and nsl1 mutants (Figure 3C) suggests that the SAC is able to induce mitotic delay in response to aberrant kinetochore function. If indeed the checkpoint were functional, then the checkpoint response might be enhanced following disruption of microtubules. To test this, we treated both wild-type and mutant embryos for 1 hr with colcemid at stage 14 (Figure S5) and determined the effect upon the mitotic index (Figure 5). In both wild-type and mutant embryos the number of mitotic cells increased significantly after MT disruption (Figure 5), indicating the presence of an active SAC. However the increase in mis12 and nsl1 mutant embryos was less than in wild-type, suggesting the SAC might be weakened as a result of reduced BubR1 levels at kinetochores. Furthermore in cal1 and cid mutants that fail to assemble the centromere and are unable to recruit detectable amounts of BubR1 (Figure 4C; Blower et al. 2006), the mitotic index was less prominent, but still significantly increased after drug treatment (Figure 5; Blower et al. 2006). Together, these data indicate that the SAC is able to work through pathways that do not require BubR1 to be robustly located at the centromere. Irrespective of whether BuR1 is present at the centromere in reduced amounts, following disruption of the Mis12 complex, or absent, in cal1 and CID mutants, an increased number of cells arrest in mitosis following destabilization of microtubules.
Figure 5.—
SAC response to microtubule depolymerization in stage 14 embryos. (A) Distribution of mitotic cells in fixed embryos stained to reveal DNA (blue) and Ser 10-phosphohistone H3 (red; PH3) after treatment for 1 hr with DMSO or colcemid. (B) Numbers of mitoses per embryo in wild-type, mis12, nsl1, and cal1 mutant stage 14 embryos following treatment of embryos of the indicated genotypes with DMSO or colcemid. Ten embryos scored for each measurement. Error bars represent ±1 SD. Number of mitotic cells significantly increased in the presence of colcemid for all genotype categories (two-tailed independent t-test, P < 0.001).
The facts that in mis12 and nsl1 mutants the SAC response to MT depolymerization is reduced and that this correlates with reduction of BubR1 at the centromere, suggest that both centromeric and the kinetochore associated populations of BubR1 contribute to the robustness of the SAC. It has previously been described that the SAC was unaffected in the absence of centromere and kinetochore formation in cid null mutants (Blower et al. 2006). Our finding of a SAC response to MT depolymerization in cal1 mutants, where CID is also lost from centromeres, is in accord with this earlier report. However, we also observe a weakening of SAC activity in the cal1 mutants, suggesting the possibility of a CID-independent involvement of Cal1 in SAC activity. This adds even greater complexity to the previous conclusion that the SAC could function independently of the association of its component molecules with the centromere (Blower et al. 2006). In addition to its role in the SAC, BubR1 has been proposed to have additional functions to establish proper connections between the kinetochore and microtubules and also as part of the basal timer of the interval between prophase and anaphase onset (Rahmani et al. 2009). Our data cannot address such roles, although we might speculate that a kinetochore-associated subpopulation of BubR1 might participate more in the k-fiber MT capturing function.
In conclusion, we have demonstrated that despite the sequential recruitment of its subunits to the kinetochore, the Drosophila Mis12 complex acts as a single functional unit in mitosis. mis12 and nsl1 mutants show strikingly similar developmental and mitotic defects in flies and confirm the necessity of the intact Mis12 complex in multiple kinetochore functions, of which mediating anaphase chromosome movement is of principal importance for these particular cell types. Finally, our findings reveal multiple levels of association of BubR1 with chromosomes that are likely to affect the multiple functions of this checkpoint protein.
Acknowledgments
We thank members of our laboratory for helpful discussions. This work was supported by a project grant from the Biotechnology and Biological Sciences Research Council.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.119628/DC1.
References
- Afshar, K., N. R. Barton, R. S. Hawley and L. S. Goldstein, 1995. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell 81 129–138. [DOI] [PubMed] [Google Scholar]
- Allshire, R. C., and G. H. Karpen, 2008. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 9 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault, V., X. Zhao, H. White-Cooper, A. T. Carpenter and D. M. Glover, 2007. Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 3 e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., R. Giet, R. Sinka, A. Mazumdar, W. G. Lock et al., 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432 980–987. [DOI] [PubMed] [Google Scholar]
- Blower, M. D., T. Daigle, T. Kaufman and G. H. Karpen, 2006. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, S., C. B. O'Connell, A. Khodjakov and C. E. Walczak, 2009. Chromosome congression in the absence of kinetochore fibres. Nat. Cell Biol. 11 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso, D., F. Ramirez-Weber and T. B. Kornberg, 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91 451–454. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., and A. Desai, 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9 33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., S. Niessen, S. Anderson, F. Hyndman, J. R. Yates, 3rd et al., 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., J. S. Chappie, E. M. Wilson-Kubalek and A. Desai, 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127 983–997. [DOI] [PubMed] [Google Scholar]
- Crest, J., N. Oxnard, J. Y. Ji and G. Schubiger, 2007. Onset of the DNA replication checkpoint in the early Drosophila embryo. Genetics 175 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J. G., B. J. Howell, J. C. Canman, J. M. Hickey, G. Fang et al., 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13 2103–2109. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Hyman et al., 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., K. J. Vanden Beldt, X. Meng, A. Khodjakov and B. F. McEwen, 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, J. B., 2002. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34 1–15. [DOI] [PubMed] [Google Scholar]
- Elowe, S., S. Hummer, A. Uldschmid, X. Li and E. A. Nigg, 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21 2205–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt, S., B. G. Mellone, C. M. Betts, W. Zhang, G. H. Karpen et al., 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., A. C. Jung, M. Criqui, B. Lemaitre, S. Uttenweiler-Joseph et al., 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, J. D., K. M. Routzahn, M. H. Bucher and D. S. Waugh, 2003. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 537 53–57. [DOI] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda and M. Yanagida, 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger, S., O. Leismann, R. Schittenhelm, O. Schraidt, S. Heidmann et al., 2005. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich, P., S. Weidtkamp-Peters, C. Hoischen, L. Schmiedeberg, I. Erliandri et al., 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., and T. J. Yen, 2009. BubR1 is an effector of multiple mitotic kinases that specifies kinetochore: microtubule attachments and checkpoint. Cell Cycle 8 1164–1167. [DOI] [PubMed] [Google Scholar]
- Jablonski, S. A., G. K. Chan, C. A. Cooke, W. C. Earnshaw, and T. J. Yen, 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107 386–396. [DOI] [PubMed] [Google Scholar]
- Joglekar, A. P., K. Bloom and E. D. Salmon, 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu, T., C. Obuse and M. Yanagida, 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13 663–676. [DOI] [PubMed] [Google Scholar]
- Kline, S. L., I. M. Cheeseman, T. Hori, T. Fukagawa and A. Desai, 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G. J., B. A. Weaver and D. W. Cleveland, 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5 773–785. [DOI] [PubMed] [Google Scholar]
- Kops, G. J., A. T. Saurin and P. Meraldi, 2010. Finding the middle ground: how kinetochores power chromosome congression. Cell. Mol. Life Sci. 67 2145–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. T., Y. Chen, G. Wu and W. H. Lee, 2006. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene 25 6901–6914. [DOI] [PubMed] [Google Scholar]
- Maiato, H., P. J. Hergert, S. Moutinho-Pereira, Y. Dong, K. J. Vandenbeldt et al., 2006. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma 115 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca, T. J., and E. D. Salmon, 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V. M. Stucke and E. A. Nigg, 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297 2267–2270. [DOI] [PubMed] [Google Scholar]
- Maskell, D. P., X. W. Hu and M. R. Singleton, 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh, A. D., P. Meraldi, V. M. Draviam, A. Toso and P. K. Sorger, 2006. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 25 4033–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., and B. P. Rattner, 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., A. D. McAinsh, E. Rheinbay and P. K. Sorger, 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro, M., L. Ciapponi, B. Fasulo, M. Gatti and G. Cenci, 2008. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat. Genet. 40 362–366. [DOI] [PubMed] [Google Scholar]
- Peter, A., P. Schottler, M. Werner, N. Beinert, G. Dowe et al., 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic, A., S. Pasqualato, P. Dube, V. Krenn, S. Santaguida et al., 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190 835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka, M. R., and D. M. Glover, 2009. The kinetochore and the centromere: a working long distance relationship. Annu. Rev. Genet. 43 439–465. [DOI] [PubMed] [Google Scholar]
- Przewloka, M. R., W. Zhang, P. Costa, V. Archambault, P. P. D'Avino et al., 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2 e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka, M. R., Z. Venkei and D. M. Glover, 2009. Searching for Drosophila Dsn1 kinetochore protein. Cell Cycle 8 1292–1293. [DOI] [PubMed] [Google Scholar]
- Rahmani, Z., M. E. Gagou, C. Lefebvre, D. Emre and R. E. Karess, 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou, A., M. E. Gagou, R. Karess and W. Sullivan, 2010. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell 140 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud, S., M. Carmena and W. C. Earnshaw, 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8 798–812. [DOI] [PubMed] [Google Scholar]
- Santaguida, S., and A. Musacchio, 2009. The life and miracles of kinetochores. EMBO J. 28 2511–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm, R. B., S. Heeger, F. Althoff, A. Walter, S. Heidmann et al., 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm, R. B., R. Chaleckis and C. F. Lehner, 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, T. T., S. D. Campbell and P. H. O'Farrell, 1999. Drosophila grapes/CHK1 mutants are defective in cyclin proteolysis and coordination of mitotic events. Curr. Biol. 9 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, W., Ashburner, M. and R. S. Hawley, 2000. Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36 283–287. [DOI] [PubMed] [Google Scholar]
- Uchida, K. S., K. Takagaki, K. Kumada, Y. Hirayama, T. Noda et al., 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko, V. V., M. J. Emanuele, M. J. Kallio, P. T. Stukenberg and G. J. Gorbsky, 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma 117 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, X., R. P. O'Quinn, H. L. Pierce, A. P. Joglekar, W. E. Gall et al., 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell 137 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, J. P., and I. M. Cheeseman, 2008. Toward a molecular structure of the eukaryotic kinetochore. Dev. Cell 15 645–655. [DOI] [PubMed] [Google Scholar]
- Williams, B., G. Leung, H. Maiato, A. Wong, Z. Li et al., 2007. Mitch a rapidly evolving component of the Ndc80 kinetochore complex required for correct chromosome segregation in Drosophila. J. Cell Sci. 120 3522–3533. [DOI] [PubMed] [Google Scholar]
- Yu, J., S. L. Fleming, B. Williams, E. V. Williams, Z. Li et al., 2004. Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J. Cell Biol. 164 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]