Abstract
Purine nucleotides are structural components of the genetic material, function as phosphate donors, participate in cellular signaling, are cofactors in enzymatic reactions, and constitute the main carriers of cellular energy. Thus, imbalances in A/G nucleotide biosynthesis affect nearly the whole cellular metabolism and must be tightly regulated. We have identified a substitution mutation (G388D) that reduces the activity of the GMP synthase Gua1 in budding yeast and the total G-nucleotide pool, leading to precipitous reductions in the GDP/GTP ratio and ATP level in vivo. gua1–G388D strongly reduces the rate of growth, impairs general protein synthesis, and derepresses translation of GCN4 mRNA, encoding a transcriptional activator of diverse amino acid biosynthetic enzymes. Although processing of pre-tRNAiMet and other tRNA precursors, and the aminoacylation of tRNAiMet are also strongly impaired in gua1–G388D cells, tRNAiMet-containing complexes with the macromolecular composition of the eIF2·tRNAiMet.GTP complex (TC) and the multifactor complex (MFC) required for translation initiation accumulate ∼10-fold in gua1–G388D cells and, to a lesser extent, in wild-type (WT) cells treated with 6-azauracil (6AU). Consistently, addition of an external supply of guanine reverts all the phenotypes of gua1–G388D cells, but not those of gua1–G388D Δhpt1 mutants unable to refill the internal GMP pool through the salvage pathway. These and other findings suggest that a defect in guanine nucleotide biosynthesis evokes a reduction in the rate of general protein synthesis by impairing multiple steps of the process, disrupts the gene-specific reinitiation mechanism for translation of GCN4 mRNA and has far-reaching effects in cell biology and metabolism.
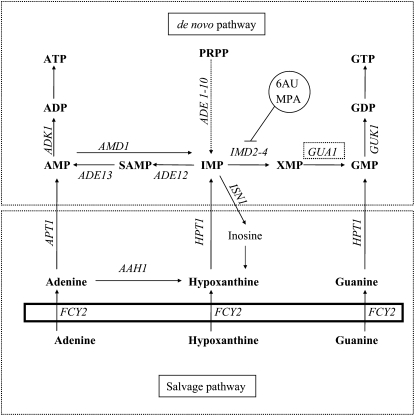
IMBALANCE in purine nucleotide biosynthesis affects nearly the whole of cellular metabolism and, hence, this process must be tightly regulated depending on nutrient availability and the conditions of growth. In the yeast Saccharomyces cerevisiae, regulation of purine nucleotide biosynthesis occurs at the enzymatic and genetic levels (Daignan-Fornier and Fink 1992; Rolfes 2006) and is finely coordinated with phosphate consumption (Pinson et al. 2009). The GMP is synthesized from IMP through a de novo synthesis pathway in a two-step reaction (Figure 1). First, IMP is converted into XMP by the IMP dehydrogenase (Impdh) (Hyle et al. 2003) and second, the GMP-synthase Gua1 mediates the amination of XMP to GMP (Dujardin et al. 1994). Transcription of the IMD genes is strongly repressed by guanine, but activated by treatment with the Impdh inhibitors 6AU or mycophenolate (MPA) (Exinger and Lacroute 1992; Bremer et al. 2009). Partial guanine deprivation results in a drastic reduction of the internal GTP pool in a yeast gua1 mutant, inducing meiosis and sporulation in homothallic strains (Varma et al. 1985) and leads to concomitant reductions of the methionine and S-adenosyl-methionine (SAM) pools (Freese et al. 1984). Recently the effects on the yeast transcriptome of a marked depletion of guanine nucleotides have been reported (Saint-Marc et al. 2009). Remarkably, 71 genes are affected under conditions that lead to GDP/GTP shortage, most of which are involved in amino acid metabolism and regulated by Gcn4 (Natarajan et al. 2001).
Figure 1.—
Pathways of purine nucleotide biosynthesis in Saccharomyces cerevisiae. Cytosolic reactions are shown over a rectangle representing the plasmatic membrane. The de novo pathway to synthesize IMP from PRPP involves 10 enzymatic reactions that are catalyzed by the products of the ADE 1–10 genes. The gene products of ADE12 and ADE13 catalyzed the production of AMP from IMP, and the products of IMD2/3/4 and GUA1 the synthesis of GMP from IMP. The salvage pathway involves three reactions that produce IMP, GMP, and AMP directly from their nonphosphorylated precursors, and adenine is converted to hypoxanthine by the Aah1 enzyme. The reaction products are indicated in bold font: ADP, adenosine-5′-diphosphate; AMP, adenosine-5′-monophosphate; ATP, adenosine-5′-triphosphate; GDP, guanosine-5′-diphosphate; GMP, guanosine-5′-monophosphate; GTP, guanosine-5′-triphosphate; IMP, inosine-5′-monophosphate; PRPP, 5-phosphoribosyl-1-pyrophosphate; SAMP, S-adenosine-5′-monophosphate; XMP, xanthosine-5′-monophosphate. Genes indicated in italics encode the following enzymatic activities: ADE12, adenylosuccinate synthetase; ADE13, adenylosuccinate lyase; ADK1, AMP kinase; AMD1, AMP deaminase; APT1, adenine phosphoribosyltransferase; FCY2, purine cytosine permease; GUK1, GMP kinase; HPT1, hypoxanthine-guanine phosphoribosyltransferase; and IMD2, IMD3, and IMD4, IMP dehydrogenase. GUA1, GMP synthetase.
In addition to the de novo synthesis, the salvage pathway for GMP production is physiologically important because it allows cells to reutilize nucleosides and nucleotides produced by degradation of nucleic acids and permits the interconversion of adenine with guanine-containing nucleotides (Figure 1). Deletion of HPT1 impairs guanine uptake from the medium and leads to deregulation of the purine de novo pathway (Guetsova et al. 1997). Strikingly, accumulation of guanine nucleotides in deregulated hpt1 mutants has deleterious consequences for yeast cells, indicating that an overabundance of GMP derivatives can be as detrimental as starvation (Breton et al. 2008).
Protein synthesis requires great amounts of cellular GTP, mainly during the elongation phase. However, the initiation process is the most tightly regulated step and consumes two GTP molecules. The translation initiation factor 2 (eIF2) bound to GTP recruits the methionyl initiator tRNA (tRNAiMet) in a ternary complex (TC) and delivers it to the 40S ribosome. This process also involves the formation of an intermediate multi factor complex (MFC) by the addition of other eIFs (eIF1, eIF3, and eIF5) that can exist unbound to 40S in the yeast cells (Asano et al. 2000). After the recognition of the initiation AUG codon, GTP hydrolysis promotes tRNAiMet release and the eIF2·GDP complex dissociation from the ribosome prior to 60S ribosomal subunit joining, which requires the hydrolysis of another GTP molecule bound to eIF5B (Lee et al. 2002). Next, eIF2B mediates the recycling of eIF2·GDP to eIF2·GTP, increasing by ∼10-fold the affinity of eIF2 for Met–tRNAiMet and stimulating the beginning of a new round of initiation (Kapp and Lorsch 2004). This process is inhibited by the phosphorylation of eIF2α in serine 51, a key event in translational regulation under starvation and stress in higher eukaryotes (Pestova et al. 2007). In yeast, the protein kinase Gcn2 phosphorylates eIF2α in response to single amino acid starvation, leading to a general inhibition of translation initiation due to impaired eIF2B function and the attendant reduced rates of TC formation (Hinnebusch 1985; Rolfes and Hinnebusch 1993). By contrast, small defects in TC formation or its recruitment to the 40S ribosome that do not appreciably affect global protein synthesis specifically promotes the translation of the mRNA encoding transcription factor Gcn4 (Dever et al. 1995). Gcn4 activates the expression of many amino acid biosynthetic genes when cells are starved for a single amino acid, a response known as general amino acid control (GAAC). More recently Gcn4 was shown to be required for full induction of at least 539 genes under histidine starvation conditions (∼1/10 of the yeast genome), including 77 amino acid biosynthetic genes (Natarajan et al. 2001).
GCN4 translation is regulated through a mechanism of reinitiation that depends on the presence of four short upstream open reading frames (uORFs −1 to −4) in the GCN4 mRNA leader and on the action of positive (Gcn) and negative (Gcd) trans-acting factors (Hinnebusch and Fink 1983; Harashima and Hinnebusch 1986). Under amino acid replete conditions, ∼50% of the ribosomes that translate uORF1 remain bound to the leader and are able to resume scanning and reacquire the TC in time to reinitiate translation at the inhibitory uORFs −3 and −4 downstream, which elicits high rates of dissociation of 40S ribosomal subunits from the GCN4 mRNA leader and attendant low levels of Gcn4 synthesis. However, low rates of TC formation under amino acid starvation conditions reduce the likelihood that ribosomes reacquire the TC in time to reinitiate at uORFs −3 and −4, favoring reinitiation at the GCN4 AUG instead. The resulting increase in Gcn4 levels allows the transcriptional activation of amino acid biosynthetic genes under its control.
Mutations in GCD genes (Harashima and Hinnebusch 1986; Niederberger et al. 1986; Cuesta et al. 1998; Calvo et al. 1999) or in the recently identified GCD17/RPL33A gene (Martin-Marcos et al. 2007), lead to constitutive derepression of GCN4 translation independent of the positive regulators Gcn2 and Gcn3 and of amino acid availability. In this article, we report the identification of a gua1–G388D mutation in a GMP synthase that provokes constitutive derepression of GCN4 translation (Gcd− phenotype), unconditional slow growth (Slg−), thermosensitivity at 37° (Tsm−) and altered polysome profiles. In addition, gua1–G388D cells exhibit defects in mature ribosome levels, processing and accumulation of pre-tRNA and reductions of mature tRNA species, including tRNAiMet, a component of the TC. Surprisingly, whereas the total guanine nucleotide level is reduced, and GDP is undetectable, GTP is elevated in gua1–G388D vs. WT cells. Moreover, we obtained evidence that TC and MFC accumulate to high levels in the gua1–G388D mutant. We propose that imbalance in the guanine nucleotide pool evokes a reduction in the rate of protein synthesis by impairing multiple steps in the process, including one or more reactions involved in the reinitiation mechanism governing GCN4 translation.
MATERIALS AND METHODS
Plasmids:
Cloning of GCD18 and gcd18-1 alleles:
Plasmid pDI2 bearing a ∼17-kb DNA fragment of yeast chromosome XIII was isolated from a yeast genomic library in YCp50 (Rose et al. 1987). Subclones of the genomic insert in pDI2 were constructed in the shuttle-vector pRS316 to define the boundaries of GCD18. Plasmid pDI3 bears a 4619-bp HindIII–EcoRV genomic fragment from pDI2 cloned into the unique SmaI site of pRS316. The 4.6-kb insert contains the GUA1 ORF flanked by 614-bp belonging to the 5′ region of the SKY1 ORF and 1456 bp belonging to the 3′ end of the TRS130 ORFs. A 6649-bp BamHI–HindIII fragment from pDI3 was cloned into pRS426 to produce the high-copy number plasmid pDI12. The gcd18-1 mutation (a transition from G to A at nucleotide 1163) was identified by independent PCR amplifications of the corresponding mutant allele from genomic DNAs of strains H166 and Hm458 (Table 1) using as primers oligonucleotides 1 and 2 (Table 2), and followed by automatic sequencing of the amplification products on the two DNA strands. The gua1–G388D mutant allele was cloned as a 2498-bp fragment into pGEM-T Easy (Promega) to produce pgcd18. A 2516-kb EcoRI fragment from pgcd18 was cloned into the EcoRI site of pRS316 yielding pDI9.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| H96 | MATα gcn2-101 gcn3-101 his1-29 ura3-52 (HIS4∷lacZ ura3-52) | Harashima and Hinnebusch (1986) |
| H117 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ URA3) | Harashima and Hinnebusch (1986) |
| H166 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ URA3) gua1-G388D | Harashima and Hinnebusch (1986) |
| Hm455 | MATα gcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ ura3-52) gua1-G388D | This study |
| Hm458 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ ura3-52) gua1-G388D | This study |
| Hm500 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 leu2∷hisG-URA3-hisG (HIS4∷lacZ ura3) gua1-G388D | This study |
| Hm510 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 leu2∷hisG (HIS4∷lacZ ura3-52) gua1∷hisG | This study |
| Hm512 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 leu2∷hisG-URA3-hisG (HIS4∷lacZ ura3) GUA1 | This study |
| Hm534 | MATα gcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ ura3-52) hpt1∷Kan-MX4 gua1-G388D | This study |
| Hm535 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4∷lacZ URA3) hpt1∷Kan-MX4 | This study |
| H2894 (KAY35) | MATagcn2Δ, ura3-52, leu2-3, -122, trp1-Δ63, Δtif5∷HisG p (TIF5-FL, LEU2) | Asano et al. (1999) |
| Hm520 | MATagcn2Δ, ura3-52, leu2-3, -122, trp1-Δ63, Δtif5∷HisG p (TIF5-FL, LEU2) GUA1 | This study |
| Hm521 | MATagcn2Δ, ura3-52, leu2-3, -122, trp1-Δ63, Δtif5∷HisG p (TIF5-FL, LEU2) gua1-G388D | This study |
| H2888 (KAY25) | MATα Δgcn2∷hisG, ura3-52, leu2-3, -122, ino1, Δsui3 (HIS4-lacZ ura3-52) YCpSUI3(SUI3-FL, LEU2) | Asano et al. (1999) |
| Hm522 | MATα Δgcn2∷hisG, ura3-53, leu2-3,-122, ino1, Δsui3 YCpSUI3(SUI3-FL,LEU2) GUA1 | This study |
| Hm523 | MATα Δgcn2∷hisG, ura3-53, leu2-3, -122, ino1, Δsui3 YCpSUI3(SUI3-FL, LEU2) gua1-G388D | This study |
| F35 | MATα, ura3-52, ino1, can1, (HIS4∷lacZ URA3) | Lucchini et al. (1984) |
| MY10 | MATa, ura3-52, leu2-3, leu2-112, gcn4∷LEU2 | Ramirez et al. (1992) |
| H4 | MATa, ura3-52, leu2-3, leu2-112 | Harashima et al. (1987) |
TABLE 2.
Oligonucleotides used in this study
| Oligo number | Sequence |
|---|---|
| 1 | 5′ gcgcaagcttcagaaagctgtatgcagacg 3′ |
| 2 | 5′ gcgcggatccctcttcaacctcaaaagggg 3′ |
| 3 | 5′ gcgcggatccgtgtcactgacccagaaaag 3′ |
| 4 | 5′ tgctccaggggaggttcgaactctcgacc 3′ |
| 5 | 5′ tcggtttcgatcccgaggacatcaagggttatga 3′ |
| 6 | 5′ gcgcctcgagctttgtcgccccgaacaatc 3′ |
| 7 | 5′ cgcggaattctaccaaatgacccagggaag 3′ |
| 8 | 5′ gcgcgaattcgcaggagaaatgtaagtagg 3′ |
| 9 | 5′ cgcggcggccgcattgctacgaaatcacagcc 3′ |
| 10 | 5′ tgctcgaggtggggttgaacccacgacgg 3′ |
| 11 | 5′ cagttgatcggacgggaaac 3′ |
| 12 | 5′ tgcgttcttcatcgatgcgagaacc 3′ |
| 13 | _5′ a_g_g_t_a_t_t_c_c_a_a_a_a_a_t_t_c_c_c_t_ _3′ |
Construction of a gua1 null allele:
A 1.3-kb fragment containing sequences belonging to the 5′-upstream region of GUA1 was amplified by PCR using genomic DNA of H117 as template and primers 6 and 7 (Table 2). The 1.3-kb PCR product was cloned at the XhoI/EcoRI sites of pRS316 to obtain pDI31. A 0.9-kb fragment containing sequences belonging to the 3′-downstream region of GUA1 was amplified by PCR using DNA of H117 as template and primers 9 and 10 (Table 2), and cloned at the EcorI/NotI sites of pDI31 to obtain pDI32. Finally, the hisG–URA3–hisG cassette (Alani et al.1987), cloned at the EcoRI site of pDI32, in between the 5′ (1.3-kb) and 3′ (0.9-kb) flanking sequences of GUA1, yielded pDI33. The 5.5-kb null allele gua1∷hisG–URA3–hisG was excised from pDI33 by digestion with KpnI. This allele was subcloned at the unique EcoRI site of pRS425 to obtain pDI34.
Other vectors and plasmids used in this work:
The pRS vectors were previously described (Sikorski and Hieter 1989). Plasmids p180, p226, and p227 contain distinct GCN4–LacZ fusions (Mueller and Hinnebusch 1986). Two fragments of 890-bp containing sequences belonging to the 3′-end coding regions of wild-type GUA1 and of the gua1–G388D allele were amplified by PCR using as template genomic DNA from strains H117 and H166, respectively, and primers 1 and 3 (Table 2). The two fragments were independently cloned into the integrative pRS306 vector, producing, respectively, plasmids pDI30 and pDI22. The GCN2 gene was excised from p585 as a 9.1-kb PstI/SalI fragment (Wek et al. 1990) and cloned at the same restriction sites of pRS314 to produce pDI29.
Yeast strain construction:
The S. cerevisiae strains used in this study are listed in Table 1. The mutant strains Hm455 and Hm458 (ura3-52) were selected from the offspring of a cross between H96 (GUA1ura3-52) and H166 (gua1–G388D, URA3). Mutant Hm500 is an isogenic leucine auxotroph of Hm458 that was obtained by interrupting the LEU2 gene with a leu2∷hisG–URA3–hisG cassette from plasmid PNKY85 (Alani et al. 1987). The WT strain Hm512 is an isogenic GUA1 derivative of Hm500 that was obtained by the replacement of a 1.2-kb BglII–BstEII fragment internal to gua1–G388D with the equivalent WT fragment cloned from a GUA1 strain (H96). The WT transformant was selected in SD medium at 37° and the correct replacement verified by PCR amplification of the resulting GUA1 allele followed by automatic sequencing. Hm510, containing a chromosomal deletion of GUA1 (Δgua1) was generated by integration the gua1∷hisG–URA3–hisG cassette into the Hm500 strain followed by the selection of the uracile auxotroph clones in 5-FOA medium. Chromosomal deletions of HPT1 (Δhpt1) were generated by replacing a 820-bp BglII internal fragment of HPT1 with a KanMx4 cassette (Wach et al. 1994) in strains Hm458 and H117, obtaining isogenic strains Hm534 (gua1–G388D Δhpt1∷kanMX4) and Hm535 (GUA1 Δhpt1∷kanMX4). Isogenic Hm520 (GUA1TIF5–FL) and Hm521 (gua1–G388D TIF5–FL) strains bearing a tagged TIF5–FLAG allele were constructed by transforming strain KAY35 (Asano et al. 1999) with integrating plasmids pDI30 or pDI22, respectively, previously linearized with Tth111I, followed by selection of a Ura− derivative in 5-FOA plates (Boeke et al. 1984). The same strategy was used to generate from strain KAY25 bearing a plasmid-borne SUI3–FLAG-tagged allele (Asano et al. 1999) a pair of isogenic GUA1 and gua1–G388D strains, Hm522 (GUA1SUI3–FL) and Hm523 (gua1–G388D SUI3–FL).
Media:
Yeast were grown in rich YPD medium or in standard-dextrose SD medium supplemented as required. Starvation for histidine with 3AT was performed as described (Harashima and Hinnebusch 1986). 5-FOA plates were prepared as described (Boeke et al. 1984).
Biochemical techniques:
Assay of HIS4–lacZ and GCN4–lacZ fusions:
β-Galactosidase assays were conducted as previously described (Burke and Kwast 2000) on ∼1 OD600 of cells grown in SD medium that contained only the required supplements. For repressing conditions, cultures were harvested in midlogarithmic phase after 8 hr of growth. For derepressing conditions, cells were grown for 3 hr under repressing conditions and then for 6 hr after 3AT was added to 10 mm. The values shown in Figure 2, D and F are the averages of three independent determinations. β-Galactosidase activities are expressed in β-galactosidase units at OD420 (nmoles of o-nitrophenyl-β-d-galactopyranoside cleaved per minute per ∼100 μg of protein). For 6AU treatment, cells were grown to early logarithmic phase (DO600 ∼0.2) in SD medium and then incubated for 12 hr after 6AU was added to 100 μg/ml, or in 6AU absence (final OD600 ∼0.8).
Figure 2.—
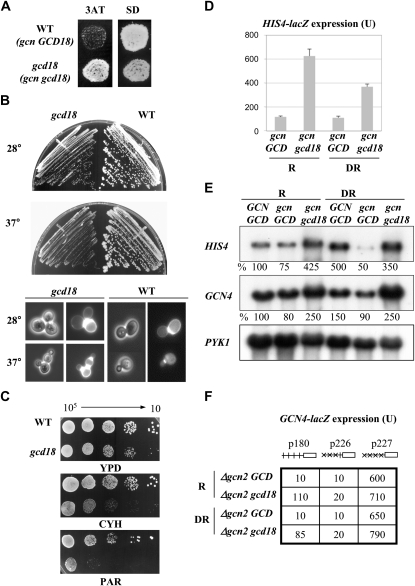
Gcd− (3ATR), slow growth (Slg−), cell morphology defects, and antibiotic sensitivity phenotypes of the H166 mutant (gcd18-1). (A) 3ATR phenotype of H166 (gcn2-101 gcn3-101 gcd18-1) relative to the 3ATs of the isogenic wild-type strain H117 (gcn2-101 gcn3-101 GCD18). Isolated colonies of each strain were grown on minimally supplemented SD plates for 2 days and replica printed to plates containing 10 mm 3AT and to new SD plates, which were incubated at 28° for 3 days. (B) Slow growth and aberrant cell morphologies of the gcd18 mutant. Cells of H117 and of the H166 mutant were streaked for single colonies on yeast extract-peptone dextrose medium plates (YPD) that were incubated at 28° or 37° for 3 to 7 days (top). Cells were grown for 12 hr in liquid YPD at 28° or 37° fixed with 4% formaldehyde and stained with Calcofluor White (Ufano et al. 2004). Pictures were taken under a phase-contrast microscope using visible or an UV filter (×40). (C) Increased sensitivity of gcd18 mutants to compounds that inhibit mRNA translation. Serial dilutions (105–10) of cells from the same strains as above growing exponentially were plotted on YPD plates and on YPD containing 25 ng/ml of cycloheximide (CYH) or 500 μg/ml of paromomycin (PAR). (D and E) The gcd18-1 mutation increases expression of the HIS4 gene. (D) Cells of isogenic strains H117 (gcn GCD18) and H166 (gcn gcd18-1) were grown under repressing (R), amino acid replete conditions (SD medium), and under derepressing (DR) conditions of histidine starvation induced by 3-aminotriazole (10 mm 3AT) during 6 hr at 28°. The β-galactosidase activity synthesized from an integrated HIS4–lacZ allele was measured in the corresponding WCE (materials and methods). Results are the average of three independent determinations. (E) Total RNA was extracted from strains F35 (GCN GCD), H117 (gcn GCD18), and H166 (gcn gcd18) grown as indicated above, and 10 μg analyzed by Northern using radiolabeled probes specific to visualize HIS4, GCN4, and PYK1 mRNAs (materials and methods). The hybridization signals were quantified with a phosphorimager and values, normalized relative to PYK1 mRNA, are given in percentages below each panel relative to the corresponding values in F35, which were set to 100%. (F) The gcd18-1G388D mutation leads to constitutive derepression of GCN4–lacZ independently of the positive Gcn factors. GCN4–lacZ fusions were introduced into isogenic strains Hm520 (Δgcn2 GCD18) and Hm521(Δgcn2 gcd18-1) on low-copy number plasmids p180, p226, and p227. The four uORFs in the leader sequence of p180 are shown as small vertical lanes, and point mutations that remove the AUG codons of uORFs 1–3 (p226) or 1–4 (p227) are shown as Xs. β-Galactosidase activity was measured in WCE of cells grown to midlogarithmic phase under nonstarvation, repressing (R) conditions or derepressing (DR) conditions of histidine starvation induced by 3AT. Values are average of results obtained in three independent determinations with two independent transformants.
Northern analysis:
Yeast total RNAs to quantify the steady-state levels of several mRNA transcripts were purified and analyzed by Northern exactly as described previously (Hinnebusch 1985). A 3.1-kb PCR amplification product containing HIS4 sequences was used as radiolabeled probe for the HIS4 mRNA; a 6.7-kb HindIII fragment containing the entire pyruvate kinase-coding sequence (PYK1) was used as the probe for PYK1 mRNA; a 1.24-kb BglII/BstEII fragment internal to the GUA1 gene was used as probe for the GUA1 mRNA; and a 0.45-kb KpnI–MluI fragment internal to the GCN4 gene was used as probe for the GCN4 mRNA.
Yeast total RNAs to quantify the steady-state levels of pre-tRNA, tRNA and small 5S and 5.8S rRNAs were purified by the phenol-acid method (Schmitt et al. 1990) and RNAs were separated by electrophoresis on a 10% polyacrylamide, pH 8.0, 8 m urea gels. Aminoacylated tRNAs were prepared under acidic conditions (0.3 m NaOAc, pH 4.5, and 10 mm EDTA) via glass bead lysis (Sarkar et al. 1999) and RNAs were separated by electrophoresis on a 10% polyacrylamide, pH 4.5, 8 m urea gel. Cells were grown in liquid SD medium at 28° to OD600 ∼1 (t = 0) and then transferred for 3–8 hr to 37°. Samples containing 15 μg of total RNA were electrophoresed in polyacrylamide/urea gels, electroblotted to positively charged nylon membranes (Roche) and immobilized by UV cross-linking with an UV Stratalinker 2400 (Stratagene). The blots were sequentially hybridized with oligonucleotides 5′ end labeled with [γ-32P] ATP (3000 Ci/mmol), and direct quantification of the corresponding hybridization signals was performed by phosphorimage analysis, using the MacBas v2.5 software and a BAS-1500 PhosphorImager. Oligonucleotides 4, 5, and 10–13 (Table 2) were used as probes for  ,
, 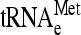 ,
, 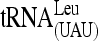 , 5S rRNA, 5.8S rRNA, and U4 RNA species, respectively.
, 5S rRNA, 5.8S rRNA, and U4 RNA species, respectively.
Western analysis:
These analyses were carried out exactly as described (Nielsen et al. 2004). Briefly, immunoprecipitates and whole cell extracts (WCEs) were boiled in SDS-loading buffer for 10 min and separated by SDS–PAGE and transferred to a PVDF membrane (Roche). Antibodies against Nip1, Prt1 Sui3, and eiF5 (Nielsen et al. 2004) and monoclonal anti-Flag M2-(HRP) antibody (Sigma) were used for protein detection.
Immunoprecipitation assays:
Co-immunoprecipitation assays of Sui3–FL and eIF5–FL proteins were performed using anti-FLAG antibodies as described (Asano et al. 1999, 2000). Briefly, cells were cultured in 100 ml of the appropriate medium, harvested at an OD600 of ∼0.8, washed, concentrated in 0.6 ml of buffer A (20 mm Tris, pH 7.5; 100 mm KCl; 5 mm MgCl2; 0.1 mm EDTA; 7 mm β-mercaptoethanol, 5 mm NaF; 1 mm phenylmethylsulfonyl fluoride, PMSF) with protease inhibitors and broken with glass beads by three 15-sec pulses in a Fastprep, with 30 sec of cooling between pulses. Lysates were clarified by 10 min of centrifugation at 16,000 × g at 4° and used as WCEs. Approximately 0.5 mg of each WCE was incubated with 15 μl of FLAG affinity resin (Sigma) at 4° for 4 hr. After washing the samples four times with 1 ml of buffer A both the immunoprecipitates and ∼4% of WCEs were resolved by SDS–PAGE electrophoresis and analyzed by Western as indicated above. For tRNA analysis, the immunoprecipitates were resuspended in water, total RNA purified by organic extraction and precipitated with ethanol (adding 50 ng of total wheat tRNA as carrier). Samples of total RNA were denatured in 15 μl of buffer D (1 m Glyoxal, 10 mm NaH2PO4, 50% DMSO) for 1 hr at 50° and analyzed by slot blot using radiolabeled oligonucleotides specific for tRNAiMet (4) and tRNAeMet (5) (Table 2).
Polysome analysis:
Polysomes analysis by sucrose gradient centrifugation was done basically as previously described (Foiani et al. 1991). Cells growing in SD at 28° were harvested at OD600 ∼1 and cycloheximide was added at a final concentration of 100 μg/ml. WCEs were obtained as described (Foiani et al. 1991), and resolved onto 7–50% gradients, which were scanned at OD254. For ribosomal subunit quantification, low-Mg2+ sucrose gradients and WCEs were prepared in the absence of cycloheximide. Translation elongation rates were evaluated by polysome analysis of WCE obtained after incubate the cells in glucose-free SD media, as previously described (Ashe et al. 2000). Briefly, cells growing in SD at 28° were harvested at OD600 ∼0.5 and resuspended in 200 ml of medium either with or without glucose as carbon source. After a specific time (1–5 min in the majority of experiments), the culture was added to cold 200-ml centrifuge bottles containing 2 ml of 10 mg/ml cycloheximide. WCEs were obtained and polysome analysis performed as indicated above.
Analysis of purine nucleosides and nucleotides by HPLC:
WCEs were essentially prepared as described by Gonzalez et al. (1997), a reliable and reproducible method that combines a rapid quenching using cold methanol to arrest cellular activity with a boiling buffered ethanol step to extract major metabolites of the intermediary metabolism. Briefly, cells growing in SD at 28° were harvested at OD600 ∼0.3 from cultures, and 2 × 107 cells (∼7 ml) were mixed immediately with 25 ml 60% (v/v) methanol/10 mm Hepes, pH 7.1 solution at −40° in 50 ml Nalgene tubes. The tubes were then centrifuged at 3000 × g for 5 min in a Beckman centrifuge at −20°. The extraction of metabolites according to Gonzalez et al. (1997) consisted of adding 3 ml of boiling buffered ethanol (BE: 75% ethanol in 10 mm Hepes, pH 7.1) to the cell pellet and incubation of the suspension for 3 min at 80°. The ethanol solution was eliminated using a rotavapor apparatus. The residue was resuspended in 120 μl of sterile water. After eliminating the insoluble particles by centrifugation at 5000 × g, 10 min at 4°, purine nucleotides on the supernatant was determined by high-pressure liquid chromatography, (HPLC). Samples of 5 μl were mixed with 45 μl of mobile phase (20 mm sodium pyrophosphate buffered at pH 5.79 with phosphoric acid (Breton et al. 2008) and injected on a C18 reverse-phase column (Gemini-NX 110A; 5 μm; 250 mm × 4.6 mm), obtained from Phenomex (Torrance, CA). Separation of purine nucleotides was performed at a constant flow rate (1 ml/min) of mobile phase. The eluent was monitored at 260 nm and purine nucleotides identified by UV absorption spectrum with a Waters 996 photodiode array detector (Waters, Milford, MA) in accordance with absorption spectra of purified purine nucleotide standards (Sigma, St. Louis). The amount of the nucleotides in the samples was determined from the areas of the corresponding peaks, using the absorption coefficients obtained from standard curves with the Millenium32 quantification software (Waters).
RESULTS
The gcd18-1 mutation elicits constitutive derepression of GCN4 translation:
All gcn mutations confer sensitivity to 3-aminotriazole (3ATS), impairing derepression of GCN4 and of histidine biosynthetic genes regulated by Gcn4 (Hinnebusch and Fink 1983). The Gcd− mutant H166 was originally isolated as a spontaneous 3ATR revertant of the Gcn− (3ATS) phenotype of H117 (gcn2-101 gcn3-101) (Harashima and Hinnebusch 1986) (Figure 2A), carries a monogenic gcd mutation that is recessive in heterozygous gcd/GCD diploids (Cuesta et al. 1998) and, as demonstrated below, defines a new complementation group of Gcd− mutants, designated as gcd18. The 3ATR phenotype conferred by gcd18-1 suggests that it derepresses GCN4 translation. In addition, at 28° mutant H166 exhibits a severe slow-growth phenotype (Slg−) on rich YPD medium (Figure 2B, top), and on minimally supplemented SD medium (Figure 4A), and is unable to form colonies at 37°. The doubling time of mutant H166 in liquid SD at 28° (∼7.5 hr) was threefold greater than that of the isogenic wild-type strain H117 (∼2.2 hr). Furthermore, ∼20% of the mutant cells grown at 28° had two buds and ∼80% formed chains after 12 hr incubation at 37° in liquid YPD. Staining with calcofluor white revealed a defect in mother–daughter cell separation (Figure 2B, bottom). The Gcd− and Slg− phenotypes, and the increased sensitivity to drugs that affect protein synthesis of gcd18 mutants (Figure 2C), all suggest that Gcd18 has an essential function in the initiation of protein synthesis, as with all other known Gcd factors.
Figure 4.—
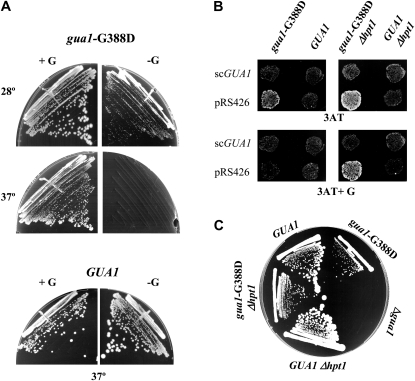
Guanine reverts the pleiotropic phenotype of the gua1–G388D mutant. (A) Reversion of the Slg− phenotype of gua1–G388D by guanine. Strains Hm458 (gua1–G388D) and H117 (GUA1) were streaked for single colonies on minimally supplemented SD plates (right), and on SD plates containing 130 μm guanine (+G, left) that were incubated for 3 days at the indicated temperatures. (B) Reversion of the Gcd phenotype by guanine requires the integrity of the salvage pathway. Strains H117 (GUA1) and Hm535 (GUA1 Δhpt1), and transformants of Hm458 (gua1–G388D) and Hm534 (gua1–G388D Δhpt1) with scGUA1 (pDI3) or with empty vector (pRS426), were replica printed to 10 mm 3AT plates (top) and to 3AT plates containing 130 μm guanine (+G, bottom) that were incubated for 4 days at 28°. (C) Growth phenotypes of the five strains used to obtain WCE for HPLC assays (Table 3 and materials and methods). Cells of the five strains with the relevant genotypes indicated were streaked for single colonies on minimally supplemented SD plates and incubated for 8 days at 25°.
Under amino acid starvation conditions, Gcn4 activates transcription of a large number of amino acid biosynthetic genes, including the HIS4 gene (Natarajan et al. 2001). Gcd− mutants exhibit constitutive high levels of HIS4 transcription under conditions of amino acid sufficiency owing to constitutive derepression of GCN4 expression. We found that gcd18-1 elicits high levels of β-galactosidase activity from a HIS4–lacZ reporter under both nonstarvation conditions of growth in minimal medium (SD) and under histidine starvation imposed by 3AT (Figure 2D). The steady-state levels of the authentic HIS4 mRNA were approximately sevenfold higher in the gcn gcd18 mutant than in the isogenic gcn GCD18 strain under both conditions (Figure 2E). Whereas levels of GCN4 mRNA increased by approximately threefold in the mutant, levels of HIS4–mRNA increased by a larger factor, consistent with translational derepression of GCN4 mRNA in gcd18 cells (Figure 2E).
To obtain direct evidence that gcd18-1 derepresses translation of GCN4 we first measured levels of β-galactosidase expressed from a GCN4–lacZ fusion on plasmid p180 in isogenic strains Hm520 (Δgcn2GCD18) and Hm521 (Δgcn2gcd18-1). This fusion contains the wild-type GCN4 mRNA leader with the four uORFs and thus, exhibits efficient translational regulation of GCN4 expression (Mueller and Hinnebusch 1986). As expected, low-level expression of GCN4–lacZ from p180 was observed in the Δgcn2GCD18 strain under nonstarvation (R) or histidine starvation conditions (DR), because the Δgcn2 allele present in this strain impairs derepression of GCN4 translation (Gcn− phenotype) (Figure 2F). Levels of β-galactosidase were 8- to 10-fold greater in Δgcn2gcd18-1 than in Δgcn2GCD18 cells under R and DR conditions, showing that gcd18-1 produces a strong, constitutive derepression of GCN4–lacZ expression. These data indicate that Gcd18 is required for repression of GCN4 expression under conditions of amino acid sufficiency.
Derepression of GCN4 translation under amino acid starvation requires uORF1 whereas efficient repression requires the presence of the negative elements uORFs −3 or −4. We found that gcd18-1 had little effect on expression of a GCN4–lacZ construct lacking all four uORFs (p227), whereas the presence of uORF4 alone impairs derepression of GCN4–lacZ expression to nearly the same extent as in GCD18 cells, under repressing and derepressing conditions (p226) (Figure 2F). Together, these data indicate that Gcd18 is a novel repressor of GCN4 translation under conditions of amino acid sufficiency.
Cloning of GCD18 and the gcd18-1 mutant allele:
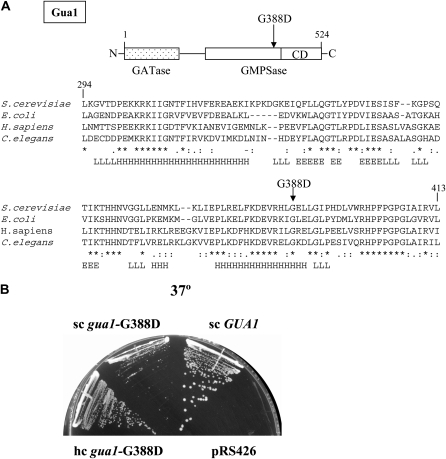
The wild-type allele of GCD18 was cloned in plasmid pDI2 from a yeast genomic library (materials and methods) by complementing the recessive Slg− phenotype at 28° of the gcd18-1 ura3-52 mutant Hm458 (whose complete genotype is listed in Table 1). Analysis of subclones of the genomic insert contained in pDI2 identified the YMR217W/GUA1 open reading frame as the gene responsible for the complementation. To distinguish between complementation by GUA1 and dosage-dependent suppression of gcd18-1, an 895-bp fragment of the pDI2 insert corresponding to the 3′ end of the GUA1 gene (http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=GUA1) was subcloned into a nonreplicating URA3 plasmid (pDI30) and shown to direct plasmid integration to the GUA1 locus in Hm458 (data not shown). As the integration event restored wild-type growth to Hm458, this analysis also verified by marker rescue that GUA1 is the wild-type allele of GCD18, and mapped the gcd18-1 mutation to the 695-bp sequences at the 3′ end of the gua1-1 ORF. We determined that gcd18-1 is a single-point mutation (materials and methods), consisting of a transition from G to A at nucleotide 1163 of the GUA1 open reading frame, replacing a glycine codon (GGU) with an aspartic acid codon (GAU) at amino acid 388 of the Gua1 protein (Figure 3A). Henceforth, gcd18-1 is referred to as gua1–G388D.
Figure 3.—
Evidence that the gua1–G388D enzyme is partially functional. (A) Schematic representation of the two functional domains of Gua1, glutamine-amido-transferase (GATase) and GMP-synthetase (GMPSase), and location of the G388/D mutation relative to the GMPSase catalytic domain (CD). Alignment of 119 amino acid residues of the Gua1 primary sequence with those of Escherichia coli, Caenorhabditis elegans, and human GMP synthetases, and secondary-structure predictions for Gua1 (H, α-helix; L, loop; E, β-sheet). The G388/D mutation is indicated in boldface type. (B) Transformants of Hm458 (gua1–G388D) carrying empty vector (pRS426), low-copy number plasmids bearing GUA1 (pDI3) and (pDI9), and a high-copy number plasmid bearing (pDI10), were streaked for single colonies on minimally supplemented SD medium and incubated at 37° for 3 days.
Gua1 is a highly conserved protein of 524 amino acids with orthologs in bacteria and eukaryotes. The gua1–G388D mutation maps to the GMPase domain of Gua1, defined by homology with the equivalent enzymes from other organisms, replacing the highly conserved glycine located 43 residues upstream of the catalytic center of this enzyme (amino acid residues 431–523) (Figure 3A). Bioinformatic predictions suggest that the G388D mutation maps to a α-helix near to a turn and would not drastically modify the secondary structure of the Gua1 protein (http://www.predictprotein.org).
The gua1–G338D mutation severely impairs GMP synthesis:
Unlike other GCD genes, GUA1 is nonessential because the redundant salvage pathway synthesizes GMP when an external supply of guanine is available (Figure 1). Because the GMP-synthetase activity of Gua1 is required for the synthesis de novo of GMP, null mutations in GUA1 confer auxotrophy for guanine (Gardner and Woods 1979). Consistently, addition of 130 μm guanine to minimally supplemented SD medium allows gua1–G388D mutants to grow almost identically to an isogenic GUA1 strain at 28° and to grow at a reduced rate at 37° (Figure 4A), and it fully suppresses the Gcd− (3-ATR) phenotype of gcn gua1–G388D cells (Figure 4B, columns 1 and 2). The recovery of the Gcd+ (3-ATS) phenotype depends on the salvage pathway and therefore, the Gcd− phenotype of a gcn gua1–G388D Δhpt1 mutant was not corrected by guanine (Figure 4B, columns 3 and 4)—nor was the Slg−—and Δgua1 and Δhpt1 mutations are synthetically lethal (data not shown). This last result implies that the gua1–G388D enzyme provides enough GMPase activity to permit biosynthesis of guanine at a level sufficient for viability in the absence of the salvage pathway. Consistent with this, the growth phenotype of the gua1–G388D mutant was partially complemented by extra copies of the mutant gua1–G388D allele on low- (pDI9) or high-copy number (pDI10) plasmids (Figure 3B).
The findings above suggest that the constitutive derepression of GCN4 translation in this mutant results from low levels of guanine nucleotides. To test this conclusion further, we asked whether a pharmacological reduction in guanine nucleotide pools is sufficient to derepress GCN4 translation. To this end, transformants of Δgcn2GUA1 cells harboring the WT GCN4–lacZ reporter with all four uORFs (p180) or the reporter lacking uORFs (p227) were treated for 12 hr with 100 μg/ml 6AU, a strong inhibitor of the IMP dehydrogenase activity (Figure 1) (Exinger and Lacroute 1992). The results showed that treatment with 6AU evoked a ninefold increase in β-galactosidase expression from the WT reporter, but only a twofold increase from the reporter lacking uORFs (data not shown). These findings indicate that reducing GTP/guanine nucleotide levels is sufficient to derepress GCN4 translation.
To provide direct evidence that gua1–G388D reduces levels of guanine nucleotides, we analyzed by HPLC the levels of purine nucleosides and nucleotides present in WCEs of gua1–G388D and WT cells. The technique used allowed us to quantify free nucleotides and those bound to cell components by salt bridges and hydrogen bonds, but not by covalent bonds. WCEs were prepared from cells grown in liquid SD medium at 28° to early logarithmic phase (A600 of ∼0.3) in the absence of guanine. The growth phenotypes of the isogenic strains employed for these experiments are shown in Figure 4C, and their doubling times are given in Table 3. As shown in Table 3, the total guanine nucleotide pool (GTP + GDP + GMP) is ∼40% lower in the gua1–G388D and gua1–G388D Δhpt1 cells than in the isogenic GUA1 cells. Whereas GDP was below the level of detection (ND), the GMP pool was either reduced by only ∼30% (HPT1 cells) or unchanged (Δhpt1 cells), and the levels of GTP were actually 40–60% higher in the gua1–G388D mutants vs. the cognate GUA1 strains. As discussed further below, the unexpected increase in the GTP pool on guanine starvation of gua1–G388D cells might result from a strong decrease in the metabolic rate and attendant reduced turnover of activated G proteins, leading to essentially complete conversion of GDP to GTP. An additional indication of reduced GMP synthetase activity in gua1–G388D and gua1–G388D Δhpt1 cells is the ∼1.5- to twofold accumulation of XMP (Table 3), the precursor of GMP in the de novo pathway (Figure 1). Presumably the synthesis of XMP from IMP by the Impdh enzymes proceeds at normal rates, but conversion of XMP to GMP is impaired in gua1–G388D cells. Both gua1–G388D and gua1–G388D Δhpt1 cells also display a dramatic accumulation of inosine concomitant with ∼25 to 40% reductions of the IMP pool, respectively (Table 3), suggesting that most of the IMP expected to accumulate in these cells is broken down to inosine (Itoh et al. 2003). The ATP pool was reduced by ∼40% in gua1–G388D HPT1 cells relative to the isogenic GUA1HPT1 strain; however, this was not observed in isogenic Δhpt1 cells (Table 3).
TABLE 3.
Intracellular concentration of purine nucleotides in several gua1 mutants
| Doubling time (hr)a |
Intracellular concentration (mm)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | Relevant genotype | GTP | GDP | GMP | XMP | ATP | IMP | INO | |
| Hm500 | gcn2 gcn3 gua1–G388D | 7.5 | 1.7 ± 0.08 | ND | 1.2 ± 0.08 | 5.0 ± 0.12 | 2.5 ± 0.23 | 6.7 ± 0.54 | 1.8 ± 0.20 |
| Hm512 | gcn2 gcn3 GUA1 | 2.3 | 1.0 ± 0.01 | 2.0 ± 0.47 | 1.7 ± 0.10 | 3.3 ± 0.30 | 4.2 ± 0.31 | 8.7 ± 0.89 | ND |
| Hm510 | gcn2 gcn3 Δgua1 +G (24 hr) | 1.5 | 3.3 ± 0.05 | 3.0 ± 0.92 | 2.8 ± 0.60 | 2.8 ± 0.17 | 1.8 ± 0.11 | 8.3 ± 0.77 | ND |
| −G (15 hr) | — | 4.2 ± 0.19 | ND | ND | 14.3 ± 1.48 | 4.0 ± 0.93 | 9.0 ± 0.92 | 11.7 ± 1.44 | |
| Hm534 | gcn2 gcn3 gua1–G388D Δhpt1 | 6.5 | 1.0 ± 0.15 | ND | 1.3 ± 0.07 | 4.2 ± 0.12 | 1.5 ± 0.20 | 4.7 ± 0.53 | 4.8 ± 0.90 |
| Hm535 | gcn2 gcn3 Δhpt1 | 2.4 | 0.7 ± 0.08 | 2.0 ± 0.98 | 1.3 ± 0.02 | 2.2 ± 0.30 | 1.5 ± 0.08 | 7.5 ± 0.82 | ND |
ND, compound not detected.
Colony growth phenotypes are shown in Figure 4.
Concentrations of purine nucleotides and inosine nucleoside, INO, were calculated using the values of 1 × 107 cells per OD600 and 0.6 ml intracellular water volume per OD600 (Freese et al. 1984). The results are expressed as the average ± standard deviation of four independent experiments.
In Δgua1 cells grown for 15 hr without external guanine (−G)—during which time they double three to four times—both GDP and GMP dropped to undetectable levels, but as observed in the gua1–G388D cells grown in minimal medium, the GTP levels exceeded those present in isogenic GUA1 cells (Table 3). As above, we suggest that most of the guanine nucleotides generated by the salvage pathway in Δgua1 cells prior to the shift to −G medium accumulate as GTP because the hydrolysis of GTP by cellular G proteins is arrested by a shutdown in metabolism occurring after several doublings in guanine starvation conditions. We also observed accumulation of XMP and inosine in the Δgua1cells in −G medium, but to a degree exceeding that seen in the gua1–G388D cells in the same medium (Table 3). Addition of external guanine (130 μm, +G) to the Δgua1 cells restores the GMP and GDP intracellular pools, and maintains the GTP pool, at levels even higher than those seen in GUA1 cells grown without guanine (Table 3). Addition of guanine also eliminates the accumulation of XMP and inosine in the Δgua1 cells (Table 3), which can be attributed to repression of IMD genes by the GDP or GTP produced by the salvage pathway (Escobar-Henriques and Daignan-Fornier 2001; Kuehner and Brow 2008).
Evidence that the gua1–G388D mutation reduces general translation rates and impairs ribosomal subunit accumulation:
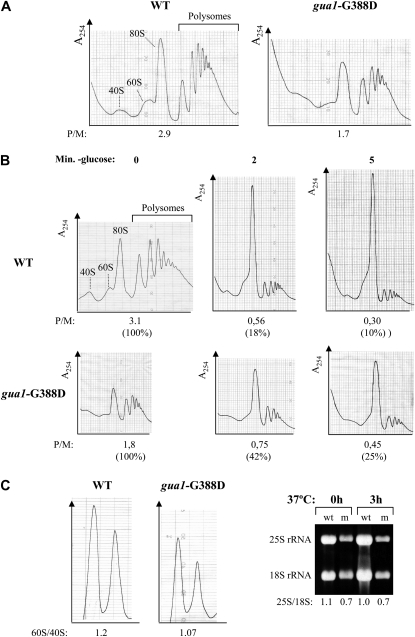
In addition to derepressing GCN4 translation, most gcd mutations also diminish the rates of general translation initiation, and gcd17-1 impairs ribosome biogenesis and subunit joining (Martin-Marcos et al. 2007). To determine whether gua1–G388D diminishes the rates of general translation initiation, we obtained total polysome profiles from gua1–G388D and WT strains by fractionating WCEs on sucrose gradients by velocity sedimentation (materials and methods). The extracts were obtained in the presence of the elongation inhibitor cycloheximide (CYH) to avoid continuous elongation and ribosomal runoff during extract preparation in vitro. We examined polysome profiles for the isogenic strains Hm500 (gua1–G388D) and Hm512 (GUA1) grown at 28° since gua1–G388D cells do not form colonies at 37° on SD (Figure 2B). As shown in Figure 5A, the gua1–G388D mutant had reduced amounts of 80S monosomes and polysomes. Importantly, the average size of the polysomes was smaller than that of the WT, averaging three to four ribosomes per polysome in gua1–G388D and six to seven ribosomes in WT cells, and quantification of the profiles revealed that the polysome to monosome ratio (P/M) was also lower in the mutant (P/M ratio of 1.7) than in the isogenic wild-type strain (P/M ratio of 2.9). Both of these alterations are indicative of a reduced rate of translation initiation.
Figure 5.—
Translational defects of gua1–G388D mutants. (A) Total polysome profiles of isogenic strains Hm500 (gua1–G388D) and Hm512 (GUA1) grown in liquid SD to midlogarithmic phase at 28° (OD600 ∼0.8). Cycloheximide was added at 100 μg/ml before harvesting cells, and whole-cell extracts (WCE) containing ribosomes and polyribosomes prepared in the presence of 10 mm Mg+2 and separated by velocity sedimentation on 7–50% sucrose gradients. Peaks representing free-ribosomal 40S and 60S subunits and 80S monosomes are indicated. The polysome to monosome ratio (P/M) was estimated, and the average of values from three independent determinations indicated below the A254 tracings. (B) Polysome profiles of the same strains as in A were obtained before and after subjecting cells to glucose starvation to inhibit translation initiation (Ashe et al. 2000). Aliquots were harvested of cells growing in liquid SD to midlogarithmic phase at 28° (0 min) and after being resuspended for 2 or 5 min in glucose-free SD medium and incubated at 28°. Cycloheximide was added at 100 μg/ml 5 min before harvesting cells and the rates of polysomal runoff estimated relative to the polysomal content of each strain before glucose starvation (0 min) that was set to 100%. The polysome to monosome ratio (P/M) was calculated and the average of values from two independent determinations indicated below the A254 tracings. (C) The same strains were cultured as described in A, but WCE were prepared in the absence of cycloheximide and Mg+2, and resolved by velocity sedimentation through 7–50% sucrose gradients. The mean ratios of total 60S/40S subunits determined from two replicate experiments are indicated below the A254 tracings (left). Total RNAs obtained from Hm512 (WT) and Hm500 (mutant, m) grown at 28° to midlogarithmic phase and after being transferred to 37° for 3 hr electrophoresed in a native agarose 1.1% gel. Electrophoretograms of the same RNA samples were obtained with an Agilent Technologies 2100 Bioanalyzer and the estimated 25S/18S ratios are indicated below (right).
To evaluate a possible translation elongation defect in the gua1–G388D mutant we analyzed total polysome profiles from gua1–G388D and WT cells that were starved for glucose for a few minutes just before obtaining the WCE, because glucose withdrawal from the medium produces an immediate inhibition of translation initiation (Ashe et al. 2000). This approach allows the detection of an effect on ribosomal transit without the added complication of de novo translation initiation (Shenton et al. 2006). WT and gua1–G388D cells were grown at 28° for 2 or 5 min after glucose withdrawal, cycloheximide was added for 5 min to fix the positions of elongating ribosomes, and the P/M ratios were quantified from polysome profiles. As shown in Figure 5B, gua1–G388D caused a slower rate of polysomal runoff under these conditions. Despite a reduced polysome content in gua1–G388D cells prior to glucose withdrawal, the amounts of polysomes were higher in the mutant vs. WT cells after glucose withdrawal. Because we obtained polysome profiles for the same cultures prior to glucose withdrawal, we could calculate the P/M ratios after glucose withdrawal as a percentage of the values observed with glucose present, as indicated in parenthesis beneath the P/M ratios of Figure 5B. Thus, the P/M ratio dropped approximately two- to threefold faster in WT than in gua1–G388D cells after glucose withdrawal, a phenotype that indicates an inhibition of translation elongation or termination in the mutant. A reduced rate of elongation alone is expected to increase the average size of the polysomes. However, as noted above, the average size of polysomes in glucose-replete medium was smaller in gua1–G388D than in WT cells (Figure 5, A and B, 0 min). We conclude that the decreased rate of elongation or termination is overpowered by an even stronger defect in translation initiation due to reduced rates of GMP synthesis in the gua1–G388D mutant. Consistent with this conclusion, the rate of incorporation of radioactive methionine into acid-insoluble material was reduced by ∼45% at 28° in gua1–G388D compared to isogenic GUA1 cells (Table 4).
TABLE 4.
Rates of protein synthesis in gua1–G388D
| Strain | Relevant genotype | l-[35S]methionine (cpm/OD600)a |
|---|---|---|
| H96 | gcn2 gcn3 | 15.540 (±52) |
| Hm458 | gcn2 gcn3 gua1–G388D | 7.400 (±95) |
Values are averages of results obtained from assays on two or three independent experiments (standard errors are shown inside parentheses).
It was recently reported that pharmacological depletion of guanine nucleotides inhibits preribosomal rRNA synthesis and causes nucleolar disruption in mammalian cells (Huang et al. 2008). Because low amounts of total ribosomal material were consistently recovered from gua1–G388D cells (Figure 5A), we analyzed the levels of mature ribosomal subunits present in cells grown at 28°. Quantification of total ribosomal subunits in low-Mg2 sucrose gradients, where polysomes and 80S ribosomes dissociate into free subunits, revealed a deficit in 60S relative to 40S ribosomal subunits in gua1–G388D cells. A marked decrease in the total amount of the 60S ribosomal subunits per A260 units of total cell extract (∼30% reduction), and a moderate decrease in the amount of 40S subunits (∼20% reduction) were observed in the mutant under these conditions (Figure 5C, left). Moreover, whereas a 60S/40S ratio of ∼1.25 was consistently observed in the WT at 28°, the 60S/40S ratio was only ∼1.07 in the gua1–G388D mutant.
We also observed a large reduction in the total amount of mature 25S rRNA, and a more moderate decrease in the amount of 18S rRNA, per A260 units of total RNA, in gua1–G388D compared to the WT cells grown at 28° and after 3 hr incubation at 37° (Figure 5C, right). Measurements of the 25S/18S ratios with an Agilent Bionalyzer showed a nearly constant ratio in the WT of 1.0 to 1.1 that was reduced in gua1–G388D cells to 0.7 at both temperatures, similar to the reductions observed in the 60S/40S subunit ratio (Figure 5C, left). These data reveal a significant shortage of both ribosomal subunits and an imbalance of 60S relative to 40S subunits in the gua1–G388D mutant, which most likely also contributes to its decreased rate of general protein synthesis and Slg− phenotype on medium lacking guanine.
Together, our data show that translation of GCN4 mRNA is constitutively derepressed, and that general translation is strongly inhibited in gua1–G388D mutants, at the level of initiation and elongation, as a consequence of reduced de novo synthesis of guanine nucleotides. They also suggest that translation initiation is an important rate-limiting step for cell growth and division under stressful conditions that reduce the availability of guanine nucleotides. Our genetic data further indicate that the de novo pathway is indeed functioning at low levels in gua1–G388D cells.
Evidence that TC and MFC accumulate in gua1–G388D cells:
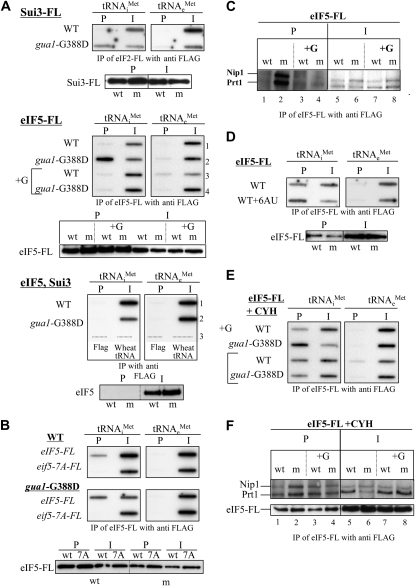
The current model for translation initiation proposes that Met–tRNAiMet is recruited to the 40S ribosomal subunit in the TC it forms with eIF2 · GTP, and that delivery of TC to the 40S ribosome is promoted by its interactions with eIF1, eIF3, and eIF5, constituents of the MFC, either free in the cytoplasm or when these factors are bound to the 40S subunit (Sonenberg and Hinnebusch 2009). To determine whether the reduction in guanine nucleotides reduces the rate of TC or MFC formation, we analyzed the amount of tRNAiMet associated with Sui3, the β-subunit of eIF2 (TC), and eIF5 (MFC) in gua1–G388D and WT cells (materials and methods). We employed pairs of isogenic gua1–G388D and GUA1 strains with a chromosomal deletion of SUI3 and a functional plasmid-borne SUI3–FLAG (SUI3–FL) allele, or with a chromosomal deletion of the gene-encoding eIF5 (TIF5) and a functional TIF5–FLAG allele (TIF5–FL) (Asano et al. 1999).
Unexpectedly, the amounts of tRNAiMet that co-immunoprecipitated with the Sui3–FL and eIF5–FL protein, relative to the tRNAiMet levels in WCE, were ∼10-fold and ∼11-fold higher, respectively, in extracts of the gua1–G388D mutants than in the isogenic GUA1 strains (Figure 6A, top, columns 1 and 2 and middle, columns 1 and 2, rows 1 and 2). Western analysis of the bait proteins confirmed that relatively similar amounts of Sui3–FL and eIF5–FL were immunoprecipitated from the gua1–G388D and WT WCEs (Figure 6A, bottom of the SUI3–FL and eIF5–FL sections), thus indicating that gua1–G388D cells exhibit markedly higher levels of tRNAiMet associated with eIF2 and in complexes containing eIF5.
Figure 6.—
Accumulation of initiation factor-tRNAiMet complexes in gua1–G388D cells and in WT cells treated with 6-azauracil (6AU). (A) Methionyl-initiator tRNA (tRNAiMet) associated with flagged-eIF2 (Sui3–FL) and with eIF5–FL accumulates in gua1–G388D cells. WCE were prepared from the following three pairs of isogenic strains: (i) Hm522 (WT) and Hm523 (gua1–G388D), bearing a SUI3–FL-tagged allele, (top); (ii) Hm520 (WT) and Hm521 (gua1–G388D), bearing a TIF5–FL allele (middle); and (iii) H117 (WT) and Hm458 (gua1–G388D), lacking FLAG-tagged genes (bottom). All strains were grown to midlogarithmic phase in liquid SD medium at 28°. Strains Hm520 and Hm521 were also grown in SD containing 130 μm guanine (+G) (middle, lanes 3 and 4). Aliquots of the corresponding WCE were incubated with an anti-FLAG affinity resin for 4 hr at 4°. After intensive washing, RNA from the precipitates (P) and from ∼1% of the immunoprecipitated WCE (I), were extracted and precipitated with ethanol using 50 ng of wheat tRNA as carrier and blotted onto a nylon membrane. Radiolabeled oligonucleotides were used as probes for methionyl-initiator tRNA (tRNAiMet, left) and methionyl-elongator tRNA species (tRNAeMet, right) (materials and methods). The affinity resin and wheat tRNA were independently subjected to the purification process and analyzed with the same probes (bottom, lane 3). Western blot detection of the Flag-tagged proteins showed similar precipitation efficiencies (bottom in SUI3–FL and eIF5–FL sections). (B) An eIF5–7A–FL mutant protein does not co-immunoprecipitate with tRNAiMet. WCE were prepared from transformants of Hm512 (WT) and Hm500 (gua1–G388D) with a low-copy number plasmid containing TIF5–FL (p3147) or the mutant tif5–7A–FL allele (p3148) and co-immunoprecipitation assays conducted as in A. Western blot detection of eIF5–FL and eif5–7A–FL proteins with anti eIF5 antibodies showed similar precipitation efficiencies (bottom). (C) Nip1 and Prt1 subunits of eIF3 co-immunoprecipitate with eIF5 · FL/tRNAiMet complexes in WCE from gua1–G388D cells. WCEs from Hm520 (GUA1, WT) and Hm521 (gua1–G388D, m) were immunoprecipitated as in A. After extensive washing, precipitates (P, lanes 1–4) and ∼5% of the crude extract (I, lanes 5–8) were analyzed by Western using anti-Nip1 and anti-Prt1 antibodies (materials and methods). Growth in minimally supplemented SD containing 130 μm guanine (lanes 3, 4, 7, and 8) is indicated as +G. (D) Accumulation of eIF5 · FL/tRNAiMet complexes in cells treated with 6AU. WCE were obtained from cells of the WT strain Hm520 grown in SD with or without 100 μg/ml of 6-azauracil, and complexes containing eIF5 · FL/tRNAiMet were immunoprecipitated and analyzed by slot blot as above. Western blot detection of eIF5–FL showed similar precipitation efficiencies with and without 6AU (bottom). (E and F) Accumulation of eIF5 · FL/tRNAiMet complexes that contain subunits of eIF3 in WT cells treated with cycloheximide. (E) Detection of tRNAiMet in immunocomplexes with eIF5–FL from WCE of the same strains as in A, middle panels, without and with added guanine (+G), except that 100 μg/ml of cycloheximide (CYH) was added to the cultures prior to the obtention of WCE. (F) The same immunoprecipitates as in E (P, lanes 1–4) and ∼5% of the crude extract (I, lanes 5–8) were analyzed by Western using anti-Nip1, anti-Prt1, and anti-Flag antibodies as in C. Growth in minimally supplemented SD containing 130 μm guanine (lanes 3, 4, 7, and 8) is indicated as +G (top).
The increased association with Sui3–FL and eIF5–FL in gua1–G388D cells was specific for tRNAiMet, as both WT and gua1–G388D cells showed similar low levels of elongator tRNAeMet in the immunoprecipitates (Figure 6A, top and middle, columns 3 and 4). The specificity of the Sui3–FL · tRNAiMet and eIF5–FL · tRNAiMet interactions was further verified in strains bearing untagged SUI3 and TIF5 alleles (Figure 6A, bottom, columns 1 and 2, rows 1 and 2), or when anti-Flag resin alone and ∼50 ng of wheat–tRNA (used as carrier in the purification of total RNAs from the immunoprecipitates) were hybridized with the probe for tRNAiMet (Figure 6A, bottom, row 3). As expected, eIF5 did not immunoprecipitate with anti-Flag from WCEs of untagged gua1–G388D TIF5 and GUA1TIF5 isogenic strains (Figure 6A, bottom of the eIF5, Sui3 section).
As mentioned before, addition of guanine refills the internal pool of GMP restoring the growth rate of the gua1–G388D cells (Figure 4A). Consistently, guanine addition also reduced the amount of tRNAiMet that co-immunoprecipitated with eIF5–FL from gua1–G388D cells nearly to the levels seen in the isogenic WT (Figure 6A middle, rows 1–4). Thus, the increased association of tRNAiMet with eIF2 and eIF5 in the gua1–G388D mutant can be attributed to the defective de novo synthesis of guanine nucleotides.
To confirm that the eIF5–FL · tRNAiMet association specifically reflects the intracellular levels of MFC that are formed in vivo, isogenic gua1–G388D and GUA1 strains carrying either TIF5–FL or the tif5–7A–FL mutant allele, whose product is defective in forming the MFC (Asano et al. 2000), were analyzed as above in strains with wild-type chromosomal TIF5. As expected, no detectable amounts of tRNAiMet were associated with the mutant eIF5–7A–FL protein (Figure 6B). Thus, the elevated association of tRNAiMet with eIF5–FL in gua1–G388D cells likely occurs in the context of the MFC. It was noticeable that the association of wild-type eIF5–FL and tRNAiMet was only ∼6-fold higher in gua1–G388D than in WT strains, rather than the ∼10-fold difference seen in Figure 6A. This difference can be due to the presence of chromosomal untagged eIF5 in these strains, which can form MFC that is not precipitated with anti-Flag antibodies.
In an effort to confirm that the MFC accumulates in gua1–G388D cells, we measured the amounts of eIF3 that are associated with eIF5–FL. To that end, immunoprecipitates from WCEs of gua1–G388D TIF5–FL and GUA1TIF5–FL isogenic strains were analyzed by Western blotting using antisera against the Nip1 and Prt1 subunits of eIF3 (materials and methods). As shown in Figure 6C, a much greater amount of Nip1 and Prt1 were present in the immunoprecipitates of gua1–G388D (lane 2) relative to WT cells (lane 1), but a similar amount was detected when mutant cells were grown in the presence of guanine (lanes 3 and 4). These data indicate that the MFC accumulates when guanine is not provided to gua1–G388D cells.
The accumulation of eIF2 · tRNAiMet and eIF5 · tRNAiMet complexes in gua1–G388D cells suggests that TC and MFC are assembled but not utilized in translation initiation when guanine nucleotide synthesis is substantially impaired. Consistent with this interpretation, treatment of WT cells with 100 μg/ml of 6AU leads to a significant ∼2.5-fold accumulation of tRNAiMet bound to eIF5–FL (Figure 6D). This result indicates that the inhibition of the IMPD dehydrogenase by 6AU (Figure 1) has the same qualitative effect as the gua1–G388D mutation leading to an in vivo accumulation of MFC (Figure 6A, middle). Presumably, the reduction of guanine nucleotide synthesis by gua1–G388D is greater than that in WT cells treated with 6AU, because the accumulation of MFC was less pronounced in the last case.
We wished to determine to what extent accumulation of the MFC merely reflects the reduced rate of protein synthesis that occurs in gua1–G388D cells, leading to a buildup of initiation intermediates. To address this, we analyzed whether completely inhibiting both the last step of initiation and elongation of translation with cycloheximide (Hartwell and McLaughlin 1968) produces an accumulation of MFC similar to that observed in gua1–G388D cells. As shown in Figure 6E, addition of cycloheximide to WT cells at a concentration that impairs polysome runoff during preparation of the WCE (100 μg/ml, Figure 5A) resulted in a small accumulation (approximately twofold) of tRNAiMet associated with eIF5–FL relative to the untreated WT control (Figure 6A, middle). These immunocomplexes contain tRNAiMet, eIF5, and the Nip1 and Prt1 components of eIF3 (Figures 6, C and F) and thus likely correspond to the MFC. Addition of guanine reduces the yield of MFC in the gua1–G388D mutant, but not in the WT cells treated with cycloheximide, confirming that low rates of guanine nucleotide synthesis in gua1–G388D cells leads to a greater accumulation of MFC than occurs with a complete block of translation. Thus, the reduction in the rate of elongation probably accounts for only a small part of the accumulation of the MFC that occurs in gua1–G388D cells.
The gua1–G388D cells are defective in biogenesis and aminoacylation of tRNAiMet:
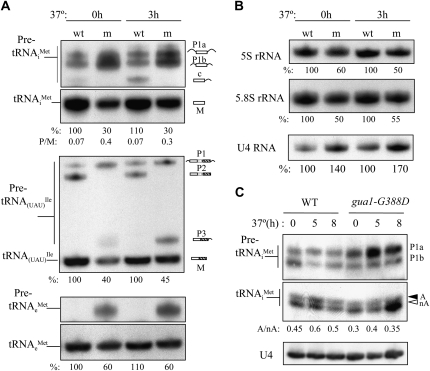
When we quantified the amount of tRNAiMet that immunoprecipitates with the Flag-tagged forms of Sui3 or eIF5 in the aforementioned experiments, we often observed that the amount of tRNAiMet present in WCEs of gua1–G388D cells (input) was lower than that seen in WT cells (e.g., Figure 6A, top, column 2, rows 1 and 2). This observation prompted us to analyze the steady-state levels of tRNAiMet and of other Pol III transcripts in gua1–G388D mutants. Total RNAs were extracted from gua1–G388D and WT cells growing logarithmically at 28° in SD or after transferring to 37° and analyzed by Northern blotting using radioactive oligonucleotides to probe for several Pol III transcripts (tRNAiMet, tRNAeMet, tRNA(UAU)Ile, and 5S rRNA), the Pol I transcript 5.8S rRNA and the Pol II transcript U4 RNA.
As shown in Figure 7A (top two panels), levels of the mature initiator tRNAiMet were diminished by ∼70% in gua1–G388D cells relative to WT cells, concomitant with accumulation of immature pre-tRNAiMet precursors containing 5′ and 3′ extensions, in cells grown at 28° or 37°. Because of the defective accumulation of ribosomal subunits and reduced 25S and 18S rRNA levels in gua1–G388D cells (Figure 5C), loading equal A260 units of total RNA in each lane led to a relative increase in the steady-state level of the Pol II transcript U4 in the mutant (Figure 7B, bottom), which was used as an internal standard for normalization of results on tRNA levels. Relative to U4, the 5S and 5.8S rRNAs were also reduced by as much as ∼50% in gua1–G388D cells compared to the WT (Figure 7B). From these data we conclude that gua1–G388D cells exhibit specific defects in processing and accumulation of tRNAiMet.
Figure 7.—
The gua1–G388D mutation impairs biogenesis and methionine aminoacylation of tRNAiMet. (A) Steady-state levels of several tRNA transcripts in gua1–G388D mutants. Total RNA was purified from isogenic strains Hm520 (GUA1, WT) and Hm521 (gua1–G388D, m) that were grown in SD medium to midlogarithmic phase at 28° or after being shifted for 3 hr at 37°, and 15 μg electrophoresed in polyacrilamide/urea gel and analyzed by Northern blot. The blot was sequentially probed using specific radiolabeled oligonucleotides for each tRNA species indicated on the left (materials and methods). Probes for tRNAiMet, tRNA(UAU)Ile, and tRNAeMet detect precursor (P) and mature (M) forms of each tRNA. The precursor tRNA species (pre-tRNA) are indicated on the right as P1a: pre-tRNAiMet containing -5′ and -3′ extensions encoded by IMT2 and IMT3; P1b: pre-tRNAiMet plus -5′ and -3′ extensions encoded by IMT1 and IMT4; P1c: pre-tRNAiMet containing -3′ extensions. P1: pre-tRNA(UAU)Ile containing two exons, an intron and -5′ and -3′ extensions; P2: pre-tRNA(UAU)Ile containing the two exons and the intron; P3: pre-tRNA(UAU)Ile containing a -3′ extension. (B) Steady-state levels of several small RNAs in gua1–G388D mutants. The Pol III 5S rRNA, Pol II U4 RNA and Pol I 5.8S rRNA transcripts were analyzed in the same blot as in (A) with specific oligonucleotides (materials and methods). To control for loading, the blot was probed for U4 rRNA. (C) Aminoacylation of initiator Met–tRNAiMet is reduced in gua1–G388D cells. Northern blot analyses of total RNA isolated under acidic conditions (pH 4.5) from WT (Hm520) and gua1–G338D (Hm521) cells. After polyacrylamide gel electrophoresis and transfer under appropriate conditions, the blot was probed tRNAiMet (materials and methods). To control for loading, the blot was probed for U4 RNA. A black arrow indicates the position of aminoacetylated Met–tRNAiMet and a white arrow that of deacylated tRNAiMet species.
An obvious defect in processing of the primary pre-tRNA (UAU)Ile precursor “P1” was also observed in gua1–G388D cells, concomitant with reductions of ∼60% in the levels of the tRNA (UAU)Ile mature transcript (Figure 7A, middle). The production of an intermediate “P2” pre-tRNA (UAU)Ile precursor requires the splicing of an intron present in the P1 precursor form of this tRNA species, and “P3” contains the second exon plus the 3′ extension present in the primary precursor (O'Connor and Peebles 1991). Whereas P1 accumulates, P2 is undetectable in gua1–G388D cells, indicating that trimming of the P1 extensions is required prior to elimination of the intron from this pre-tRNA species. Accumulation of P3 in the mutant also indicates that processing of the 3′ extension is still impaired after the 5′ end has been removed (5′ processing would mainly occur prior to 3′ elimination by endo- or exonucleolytic processing). A less severe defect in processing of 5′ and 3′ extensions and accumulation of the elongator tRNAeMet (∼40%) was detected in gua1–G388D mutants (Figure 7A, bottom), indicating that the mutation differentially impacts the final levels of distinct tRNA species, with stronger effects on mature initiator tRNA, which presumably contributes to the severe defect in translation initiation in gua1–G388D cells.
We next reasoned that reduced rates of guanine nucleotide synthesis in gua1–G388D cells could be coupled with a decrease of S-adenosyl-methionine (SAM) and aminoacylated methionyl-tRNA species, because GTP is a precursor of tetrahydrofolate, which is required for the methylation of homocysteine to form methionine (Freese et al. 1984). To determine the levels of charged Met–tRNAiMet relative to uncharged tRNAiMet, total RNAs were extracted at pH 4.5 from gua1–G388D and WT cells growing logarithmically at 28° and after being transferred to 37° for 5 hr and 8 hr, and analyzed by Northern blot (materials and methods). As shown in Figure 7C, the ratios of aminoacylated to nonaminoacylated tRNAiMet species dropped ∼30% in gua1–G388D relative to WT cells, at both 28° and 37°, which would be consistent with a reduction in the synthesis of methionine in the mutant. Since SAM is the principal methylating agent for many macromolecules, reduced SAM synthesis might evoke deficient methylation of tRNA species, which could account for defects in tRNA processing observed in gua1–G388D cells. It is remarkable that the TC and MFC, which presumably contain eIF2 bound to mature Met–tRNAiMet, accumulate by factors of ∼10 (Figure 6A) despite the reductions in mature tRNAiMet abundance (Figure 7A) and aminoacylation in gua1–G388D relative to WT cells.
DISCUSSION
We have identified a substitution mutation (G388D) in the GMP-synthetase Gua1 that impairs guanine nucleotide biosynthesis, leading to slow growth (Slg−) and constitutive derepression of GCN4 translation (Gcd−), which are suppressed by providing the mutant cells with an external supply of guanine. The G388D mutation maps to the GMPSase domain of the Gua1 enzyme and is predicted to reduce but not abolish this activity, because extra copies of the hypomorphic gua1–G388D allele revert all of the mutant phenotypes, and Δhpt1 gua1–G388D cells (lacking the salvage pathway for GMP synthesis) are viable. We obtained evidence that G388D reduces the total guanine nucleotide pool (GMP + GDP + GTP) in vivo by ∼40%, and leads to accumulation of XMP, the Gua1 substrate. Interestingly, the levels of GMP, GDP, and GTP were not reduced equivalently: whereas GMP was moderately reduced, GDP was undetectable, and GTP was actually 1.5- to 1-7-fold higher in gua1–G388D cells than in wild type.
The unexpected high GTP and undetectable GDP levels observed in gua1–G388D cells might be attributable to a massive reduction in the metabolic rate. Consequently, there would be a very low rate of GTP to GDP turnover by the many G proteins that participate in the myriad aspects of cell metabolism, and any GDP that is generated would be rapidly converted back to GTP. In wild-type cells, GTP is continuously converted to GDP by the turnover of activated G proteins to yield a steady-state level of GDP that exceeds that of GTP. In contrast to our findings, it was shown previously that the GTP pool drops dramatically by ∼90% in a leaky gua1 mutant subjected to guanine starvation for 24 hr (Freese et al. 1984). At present, we have no explanation for this discrepancy in results between the two studies.
Similar to all other known mutations conferring Slg− and Gcd− phenotypes, gua1–G388D cells exhibit a general defect in the rate of translation initiation; however, an elongation defect was also detected, manifested by a slower rate of polysome runoff when initiation was blocked by glucose starvation. The reduction in average polysome size and abundance indicates that the initiation defect is relatively more severe during guanine starvation of gua1–G388D cells. In addition, a marked reduction in the amounts of mature 60S and 40S subunits likely impair all steps of translation in the mutant. Together, these defects provoke a ∼50% decrease in the rate of incorporation of 35S-methionine into total protein (Table 4).
Because GTP is required for all phases of protein synthesis, it would be natural to assume that genetically impairing GMP synthesis would affect translation by decreasing the amount of GTP available for this process. However, because GTP levels are elevated, it is possible that the different G proteins that participate in translation occur primarily in the GTP-bound state in gua1–G388D cells, and that the process is impeded by one or more unknown signal transduction pathways that target both the initiation and elongation steps as a response to reduced synthesis of guanine nucleotides. Consistent with this interpretation, tRNAiMet-containing complexes with the protein compositions of TC and MFC accumulate in excess of ∼10-fold in gua1–G388D relative to their levels in isogenic WT cells (Figure 6). The apparent accumulation of TC and MFC is even more striking considering that pre-tRNAiMet processing, and the accumulation and aminoacylation of mature tRNAiMet were all reduced in the mutant, which should compromise assembly of TC. These findings suggest that translation initiation is blocked downstream of the assembly of TC and MFC, and that the underutilization of these complexes leads to their abnormal accumulation in gua1–G388D cells.
The fact that the gua1–G388D mutation constitutively derepresses GCN4 translation is consistent with the possibility that guanine nucleotide depletion provokes a reduction in the rate of TC recruitment by the reinitiating 40S ribosomes that remain associated with the GCN4 mRNA leader after translating uORF1. As a result, a fraction of these scanning 40S subunits would not reacquire the TC in time to reinitiate translation at uORFs 2–4 (either 60S subunits) but would do so by the time they reach the GCN4 AUG and reinitiate translation there instead. Because TC levels apparently are not reduced by gua1–G388D, the Gcd− phenotype would arise from a reduced rate of TC binding to the reinitiating 40S subunits, or the failure to recognize the AUG codons at uORFs 2–4 after rebinding of TC, rather than a deficit in TC assembly. The reduced concentration of 60S subunits in the mutant could also contribute to the bypass of uORFs 2–4 via impaired subunit joining, as mutations in 60S subunit proteins that reduce 60S subunit levels confer Gcd− phenotypes and constitutively derepress GCN4 translation (Foiani et al. 1991; Martin-Marcos et al. 2007). One interesting, albeit speculative, possibility to explain the Gcd− phenotype would be that low-level GMP synthesis triggers a signal transduction pathway that reduces TC loading on 40S subunits, by decreasing the affinity of eIF2 or tRNAiMet for the 40S, either through modification of one of these components of the 43S preinitiation complex (PIC) or by impairing the function of one or more other factors eIFs -1, -1A, -3, or -5, that enhance TC recruitment. This would be akin to the effects of known mutations in tRNAiMet, eIF2 subunits, or in other eIFs that confer Gcd− phenotypes by this mechanism (Hinnebusch 2005). The decrease in SAM expected in gua1 mutants (Varma et al. 1985), owing to the involvement of GTP in SAM biosynthesis, could reduce methylation of tRNAiMet bases and thereby also impair tRNAiMet function in 40S binding. All of these reactions represent plausible targets for the hypothetical signal-transduction pathway(s) that would modify TC components, other eIFs, or the ribosomal subunits in response to low-level GMP synthesis in gua1–G388D cells. In view of our finding that ATP levels are also markedly reduced in the gua1–G388D mutant, the hypothetical signal-transduction response could be triggered by reduced ATP levels, as a manifestion of energy depletion, rather than a response to diminished guanine nucleotides.
Our finding that tRNAiMet-containing complexes accumulate by ∼10-fold but the GTP level is only ∼1.5-fold higher in gua1–G388D vs. GUA1 cells further suggests that at least a proportion of these complexes are not genuine TC or MFC, and lack GTP bound to eIF2. The Kd for Met–tRNAiMet binding to apo–eIF2 is only an order of magnitude higher than for eIF2 · GTP (Kapp and Lorsch 2004). Hence, eIF2 · tRNAiMet binary complexes might accumulate if eIF2 cannot compete effectively with other G proteins for the available GTP under conditions of guanine nucleotide limitation. Similarly, the Kd for binding nonaminoacetylated tRNAiMet to eIF2 · GTP is only an order of magnitude higher than for methionyl–tRNAiMet (Kapp and Lorsch 2004). Thus, the defect in aminoacetylating tRNAiMet could provoke the formation of defective complexes between eIF2 · GTP and uncharged tRNAiMet in gua1–G388D cells. If these defective complexes compete with authentic TC for binding to 40S subunits, they would be expected to reduce the rate of assembling functional 43S or 48S PICs, contributing to the decrease in general translation initiation and the bypass of uORFs 2–4 during reinitiation on GCN4 mRNA.
Nearly one-half of the eIF2 in yeast cells, presumably in its GDP-bound state, forms a complex with eIF5, which appears to antagonize GTP exchange on eIF2–GDP to render eIF2 recycling rate limiting for translation initiation (Singh et al. 2006); this complex insures an effective inhibition of eIF2 recycling by eIF2B when eIF2α is phosphorylated by Gcn2 (Jennings and Pavitt 2010). However, the eIF2 · GDP/eIF5 complexes identified by Singh et al. (2006) likely differ from those described here, which contain tRNAiMet and accumulate under conditions of guanine nucleotide depletion where GDP is undetectable.
Recently it was reported that depletion of the guanine nucleotide pool by three different means led to increased expression of a large number of genes, most of which participate in amino acid metabolism and are under the transcriptional control of Gcn4 (Saint-Marc et al. 2009); however, neither the Gcn4 dependence nor molecular mechanism of the response was investigated. In light of our data, we suggest that GCN4 mRNA translation was derepressed by the GDP/GTP shortages evoked by the different conditions examined by Saint-Marc et al. (2009) and it would be interesting to determine whether tRNAiMet containing complexes accumulate under those conditions.
We found that reduced synthesis of G nucleotides in gua1–G388D cells impairs the processing of pre-tRNAiMet and precursors of certain elongator tRNA species (Figure 7). The predicted reduction in SAM biosynthesis and hypomethylation of pre-tRNAs might impede their proper folding and processing, as occurs in gcd10 and gcd14 mutants defective in methylation of m1A58 (Anderson et al. 1998; Calvo et al. 1999). It is also interesting to note that Ran-dependent nucleocytoplasmic transport is dependent on both GTP- and GDP-bound forms of the Ran GTPase, and that defects in tRNA maturation have been observed in nucleoporin mutants (Gorlich et al. 1996; Sharma et al. 1996; Izaurralde et al. 1997). Hence, the abnormal GTP/GDP ratio in the gua1–G388D mutant could contribute to the observed defects in tRNA maturation by interfering with Ran function. Defects in Ran-dependent transport of ribosomal processing factors or ribosomal subunit precursors (Thomas and Kutay 2003; Todaka et al. 2005) could likewise be involved in the reduced levels of ribosomal subunits seen in gua1–G388D cells.
In summary, our results indicate that genetically reducing the activity of the GMP synthase (Gua1) in budding yeast has myriad effects on cell biology and metabolism. A reduction in the total G nucleotide pool, with a precipitous drop in the GDP/GTP ratio, plus a decrease in ATP levels, are among the most direct effects of the gua1–388D mutation. These alterations in nucleotide levels evoke defects in ribosome and tRNA biogenesis and decreased rates of both the initiation and elongation phases of protein synthesis, leading to a reduced rate of general translation, and a specific derepression of GCN4 mRNA translation. Contrary to our expectations, the defects in general and GCN4-specific translation do not appear to arise from impaired assembly of the eIF2·tRNAiMet.GTP ternary complex. Rather, the apparent accumulation of TC and MFC in the gua1–388D mutant argues for a block further downstream in the initiation pathway that decreases the rate of TC consumption. Future studies are required to identify the exact steps in the initiation pathway that are downregulated on reduction of GMP synthesis and also the hypothetical regulatory mechanisms that mediate this multifaceted response.
Acknowledgments
We thank D. Martín-Zanca for helpful suggestions regarding this work and J. R. Gutiérrez for help with the procedure of yeast metabolite extractions. D.I.-G. was supported by a postgraduate fellowship (BFI02.95), granted by the Department of Education, Universities, and Research of the Basque government (Spain). M.A.S. was supported by Project #SA098A09, granted by the Department of Universities and Research of the Junta de Castilla y León Government (JCyL) (Spain). M. Tamame was supported by Projects PET2008-0283-01, granted by the Spanish Ministerio de Ciencia e Innovación (MICINN), and CSI007A10-2, granted by JCyL. The work was supported partly by the Intramural Research Program of the National Institutes of Health.
References
- Alani, E., L. Cao and N. Kleckner, 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., L. Phan, R. Cuesta, B. A. Carlson, M. Pak et al., 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, K., T. Krishnamoorthy, L. Phan, G. D. Pavitt and A. G. Hinnebusch, 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, K., J. Clayton, A. Shalev and A. G. Hinnebusch, 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 14 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe, M. P., S. K. De Long and A. B. Sachs, 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Bremer, S., N. T. Vethe, H. Rootwelt and S. Bergan, 2009. Expression of IMPDH1 is regulated in response to mycophenolate concentration. Int. Immunopharmacol. 9 173–180. [DOI] [PubMed] [Google Scholar]
- Breton, A., B. Pinson, F. Coulpier, M. F. Giraud, A. Dautant et al., 2008. Lethal accumulation of guanylic nucleotides in Saccharomyces cerevisiae HPT1-deregulated mutants. Genetics 178 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, P. V., and K. E. Kwast, 2000. Oxygen dependence of expression of cytochrome C and cytochrome C oxidase genes in S. cerevisiae. Adv. Exp. Med. Biol. 475 197–208. [DOI] [PubMed] [Google Scholar]
- Calvo, O., R. Cuesta, J. Anderson, N. Gutierrez, M. T. Garcia-Barrio et al., 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta, R., A. G. Hinnebusch and M. Tamame, 1998. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 148 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier, B., and G. R. Fink, 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89 6746–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever, T. E., W. Yang, S. Astrom, A. S. Bystrom and A. G. Hinnebusch, 1995. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol. Cell. Biol. 15 6351–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin, G., M. Kermorgant, P. P. Slonimski and H. Boucherie, 1994. Cloning and sequencing of the GMP synthetase-encoding gene of Saccharomyces cerevisiae. Gene 139 127–132. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques, M., and B. Daignan-Fornier, 2001. Transcriptional regulation of the yeast gmp synthesis pathway by its end products. J. Biol. Chem. 276 1523–1530. [DOI] [PubMed] [Google Scholar]
- Exinger, F., and F. Lacroute, 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22 9–11. [DOI] [PubMed] [Google Scholar]
- Foiani, M., A. M. Cigan, C. J. Paddon, S. Harashima and A. G. Hinnebusch, 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese, E. B., Z. Olempska-Beer, A. Hartig and E. Freese, 1984. Initiation of meiosis and sporulation of Saccharomyces cerevisiae by sulfur or guanine deprivation. Dev. Biol. 102 438–451. [DOI] [PubMed] [Google Scholar]
- Gardner, W. J., and R. A. Woods, 1979. Isolation and characterisation of guanine auxotrophs in Saccharomyces cerevisiae. Can. J. Microbiol. 25 380–389. [DOI] [PubMed] [Google Scholar]
- Gonzalez, B., J. Francois and M. Renaud, 1997. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 13 1347–1355. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., N. Pante, U. Kutay, U. Aebi and F. R. Bischoff, 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Guetsova, M. L., K. Lecoq and B. Daignan-Fornier, 1997. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics 147 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima, S., and A. G. Hinnebusch, 1986. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6 3990–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima, S., E. M. Hannig and A. G. Hinnebusch, 1987. Interactions between positive and negative regulators of GCN4 controlling gene expression and entry into the yeast cell cycle. Genetics 117 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., and C. S. McLaughlin, 1968. Temperature-sensitive mutants of yeast exhibiting a rapid inhibition of protein synthesis. J. Bacteriol. 96 1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A. G., 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A. G., 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59 407–450. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A. G., and G. R. Fink, 1983. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., Y. Ji, K. Itahana, Y. Zhang and B. Mitchell, 2008. Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption. Leuk. Res. 32 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyle, J. W., R. J. Shaw and D. Reines, 2003. Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 278 28470–28478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R., C. Saint-Marc, S. Chaignepain, R. Katahira, J. M. Schmitter et al., 2003. The yeast ISN1 (YOR155c) gene encodes a new type of IMP-specific 5′-nucleotidase. BMC Biochem. 4 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., U. Kutay, C. von Kobbe, I. W. Mattaj and D. Gorlich, 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, M. D., and G. D. Pavitt, 2010. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp, L. D., and J. R. Lorsch, 2004. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 335 923–936. [DOI] [PubMed] [Google Scholar]
- Kuehner, J. N., and D. A. Brow, 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell 31 201–211. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., T. V. Pestova, B. S. Shin, C. Cao, S. K. Choi et al., 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci.USA 99 16689–16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini, G., A. G. Hinnebusch, C. Chen and G. R. Fink, 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos, P., A. G. Hinnebusch and M. Tamame, 2007. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 27 5968–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, P. P., and A. G. Hinnebusch, 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45 201–207. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts et al., 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger, P., M. Aebi and R. Hutter, 1986. Identification and characterization of four new GCD genes in Saccharomyces cerevisiae. Curr. Genet. 10 657–664. [DOI] [PubMed] [Google Scholar]
- Nielsen, K. H., B. Szamecz, L. Valasek, A. Jivotovskaya, B. S. Shin et al., 2004. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 23 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, J. P., and C. L. Peebles, 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T. V., J. R. Lorsch and C. U. T. Hellen, 2007. The mechanism of translation initiation in eukaryotes, pp. 87–128 in Translational Control in Biology and Medicine, edited by M. B. Mathews, N. Sonenberg and J. W. Hershey. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pinson, B., S. Vaur, I. Sagot, F. Coulpier, S. Lemoine et al., 2009. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 23 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, M., R. C. Wek, C. R. Vazquez de Aldana, B. M. Jackson, B. Freeman et al., 1992. Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 12 5801–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes, R. J., 2006. Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochem. Soc. Trans. 34 786–790. [DOI] [PubMed] [Google Scholar]
- Rolfes, R. J., and A. G. Hinnebusch, 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., P. Novick, J. H. Thomas, D. Botstein and G. R. Fink, 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60 237–243. [DOI] [PubMed] [Google Scholar]
- Saint-Marc, C., B. Pinson, F. Coulpier, L. Jourdren, O. Lisova et al., 2009. Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics 183: 529–538, 521SI–527SI. [DOI] [PMC free article] [PubMed]
- Sarkar, S., A. K. Azad and A. K. Hopper, 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M. E., T. A. Brown and B. L. Trumpower, 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K., E. Fabre, H. Tekotte, E. C. Hurt and D. Tollervey, 1996. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol. Cell. Biol. 16 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton, D., J. B. Smirnova, J. N. Selley, K. Carroll, S. J. Hubbard et al., 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281 29011–29021. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, C. R., B. Lee, T. Udagawa, S. S. Mohammad-Qureshi, Y. Yamamoto et al., 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 25 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N., and A. G. Hinnebusch, 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F., and U. Kutay, 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116 2409–2419. [DOI] [PubMed] [Google Scholar]
- Todaka, Y., Y. Wang, K. Tashiro, N. Nakashima, T. Nishimoto et al., 2005. Association of the GTP-binding protein Gtr1p with Rpc19p, a shared subunit of RNA polymerase I and III in yeast Saccharomyces cerevisiae. Genetics 170 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufano, S., F. del Rey and C. R. Vazquez de Aldana, 2004. Swm1p, a subunit of the APC/cyclosome, is required to maintain cell wall integrity during growth at high temperature in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 234 371–378. [DOI] [PubMed] [Google Scholar]
- Varma, A., E. B. Freese and E. Freese, 1985. Partial deprivation of GTP initiates meiosis and sporulation in Saccharomyces cerevisiae. Mol. Gen. Genet. 201 1–6. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wek, R. C., M. Ramirez, B. M. Jackson and A. G. Hinnebusch, 1990. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol. 10 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]