Abstract
Transferring endosymbiotic bacteria between different host species can perturb the coordinated regulation of the host and bacterial genomes. Here we use the most common maternally transmitted bacteria, Wolbachia pipientis, to test the consequences of host genetic background on infection densities and the processes underlying those changes in the parasitoid wasp genus Nasonia. Introgressing the genome of Nasonia giraulti into the infected cytoplasm of N. vitripennis causes a two-order-of-magnitude increase in bacterial loads in adults and a proliferation of the infection to somatic tissues. The host effect on W. pipientis distribution and densities is associated with a twofold decrease in densities of the temperate phage WO-B. Returning the bacteria from the new host species back to the resident host species restores the bacteria and phage to their native densities. To our knowledge, this is the first study to report a host–microbe genetic interaction that affects the densities of both W. pipientis and bacteriophage WO-B. The consequences of the increased bacterial density include a reduction in fecundity, an increase in levels of cytoplasmic incompatibility (CI), and unexpectedly, male-to-female transfer of the bacteria to uninfected females and an increased acceptance of densely infected females to interspecific mates. While paternal inheritance of the W. pipientis was not observed, the high incidence of male-to-female transfer in the introgressed background raises the possibility that paternal transmission could be more likely in hybrids where paternal leakage of other cytoplasmic elements is also known to occur. Taken together, these results establish a major change in W. pipientis densities and tissue tropism between closely related species and support a model in which phage WO, Wolbachia, and arthropods form a tripartite symbiotic association in which all three are integral to understanding the biology of this widespread endosymbiosis.
ALL metazoans are populated by symbiotic bacteria, some of which live in intimate association with their hosts as maternally transmitted infections. Wolbachia pipientis is one of the most prevalent species of bacterial endosymbionts in the animal world that exemplifies this lifestyle. It is maternally transmitted from host ovaries to developing eggs in filarial nematodes and arthropods. In filarial nematodes, W. pipientis are obligate mutualists (Taylor et al. 2005) and assist fertility (Hoerauf et al. 1999) and larval development (Smith and Rajan 2000). Proinflammatory responses in filarial-infected vertebrates indicate that the endosymbiont rather than the nematode is the significant contributor to the acute pathologies of human river blindness (Saint Andre et al. 2002) and elephantiasis (Taylor et al. 2000). In arthropods, W. pipientis are reproductive parasites (Stouthamer et al. 1999; Werren et al. 2008) and, less often, mutualists (Pannebakker et al. 2007; Hosokawa et al. 2010), and occur in ∼66% of all insect species worldwide (Hilgenboecker et al. 2008). Thus, millions of animal species are infected by W. pipientis and over 1 billion people in >80 countries are at risk of filarial/W. pipientis diseases (Ottesen et al. 2008). As one of the great pandemics in the animal world, W. pipientis offer a preeminent model to explore the varied outcomes of infection between animal cells and obligate intracellular bacteria.
Transfer of symbionts between closely related species is a useful tool to determine the role of host genes in regulating a symbiosis. Over time, maternally inherited bacteria may coevolve with their hosts to finely tune their densities, tissue tropism, and interactions with host fitness. Many studies have found that relocating W. pipientis to a new host species causes changes in reproductive parasitism, bacterial titers, and tissue distribution not observed in the donor (Boyle et al. 1993; Bordenstein and Werren 1998; McGraw et al. 2002; Riegler et al. 2004; Ruang-Areerate and Kittayapong 2006).
In addition to host genetic background, temperate phage WO of W. pipientis is hypothesized to regulate infection densities in Nasonia vitripennis wasps owing to its lifecycle as a lytic and lysogenic phage (Bordenstein et al. 2006; Kent and Bordenstein 2010). In that investigation, phage WO was observed in ∼12% of the W. pipientis cells in late pupal testes. These bacteria showed cellular defects consistent with phage-induced mortality, including degraded DNA, a collapsed inner membrane typical of holin enzymes, and lysed membranes associated with phage. Introduction of a new host background could alter lytic development and result in changes in W. pipientis infection densities and tissue tropism from the donor species.
The Nasonia genus is composed of four closely related parasitoid wasp species that are naturally infected with different W. pipientis strains (Raychoudhury et al. 2009). N. vitripennis and N. giraulti laboratory strains and field isolates are coinfected by each of the two major insect-W. pipientis subdivisions, A and B (Werren et al. 1995; Raychoudhury et al. 2009). These infections have been segregated in the lab into strains with single A or B infections (Perrot-Minnot et al. 1996; Bordenstein and Werren 2007). Crosses between the different A infections, and between the single A and B infections within species, are bidirectionally incompatible due to cytoplasmic incompatibility (CI) (Bordenstein and Werren 2007). In addition to Drosophila, the Nasonia genus is one of a few tractable genetic systems for understanding host–W. pipientis interactions because it has many features suited for genetic and functional studies. They have a short generation time, simple husbandry, fertile species hybridizations, a wide array of molecular markers (Werren and Loehlin 2009), and three fully sequenced genomes (Werren et al. 2010).
Here, we describe a comprehensive set of experiments to test the consequences of host genetic background on W. pipientis densities, the processes underlying the changes in density, and the effects of increased bacterial densities on cytoplasmic incompatibility, fitness, behavior, and paternal transmission. To accomplish this, we introgressed the N. giraulti genome into a N. vitripennis A-infected cytoplasm. Results showed a stable increase in W. pipientis density, a reduction in phage WO density, and a vast expansion of the infection into somatic tissues. The ∼100-fold increase in W. pipientis in the new genetic background resulted in male-to-female transfer of W. pipientis and a behavioral mating pathology. Return of the bacteria/bacteriophage combination from the introgression line back to the resident N. vitripennis host restores the native microbial densities. The host genetic effect on microbial densities was not observed with the supergroup-B infected N. vitripennis strain that lacks a lytic phage, suggesting the increase in bacterial densities and reduction in phage WO densities could be specific to the supergroup A strain that harbors the virion-producing phage.
MATERIALS AND METHODS
Strains:
N. vitripennis strains 12.1 (A-infected), 4.9 (B-infected), and 13.2 (uninfected) were derived from the double AB-infected R511 line after a prolonged period of diapause (Perrot-Minnot et al. 1996). Introgression strains for this study were derived by repeatedly backcrossing females of 12.1, 4.9, and 13.2 to uninfected N. giraulti RV2R males (uninfected) for nine, six, and six generations, respectively. These strains are essentially composed of N. giraulti nuclear genomes and N. vitripennis cytoplasms; introgressed strains are IntG12.1 with an A-infected cytoplasm, IntG4.9 with a B-infected cytoplasm, and IntG13.2 with an uninfected cytoplasm. Strains 12.1 and IntG12.1 were subsequently treated with 30 mg/ml tetracycline for three generations to generate W. pipientis-free versions of these lines and are denoted 12.1T and IntG12.1T. The IntG12.1 line was backcrossed to N. vitripennis 12.1 males for seven generations to reintrogress the N. vitripennis genome and is denoted ReInt12.1. All strains were incubated at 25° at constant light during the experiments. To set up crosses, Nasonia wasps were collected as black pupae, sorted into separate virgin male and female stocks, and mated upon eclosure as adults. Copulations were observed for all crosses.
Fluorescent imaging:
Females were presented with one Sarcophaga bullata host pupa embedded halfway in foam plugs to localize egg laying in the host for 24 hr. Embryos to be used in immunofluorescent imaging were then collected and rinsed with heptane into a final 1:1 heptane–methanol mixture and then shaken for ∼5 min. Embryos, which became dechorionated and fixed, settled to the bottom of the methanol layer and were removed with a pipette, transferred to a microcentrifuge tube, and washed three times with methanol for 5 min each. Fixed embryos were then hydrated with TBST (50 mm Tris, 150 mm NaCl, 0.1% Tween, 0.05% NaN3, pH 7.5) through a series of 1:3, 1:1, 3:1 TBST:methanol mixture for 5 min each, followed by TBST and blocked in TBST–BSA (with 1% bovine serum albumin) for 10 min.
Seminal vesicles and malpighian tubules were dissected from 2- to 3-day posteclosion males in TBST and fixed in 3.7% formaldehyde in TBST for 30 min, followed by three 5-min washes in TBST, and blocked in TBST–BSA (with 1% bovine serum albumin) for 10 min.
W. pipientis were labeled using a mouse monoclonal anti-human hsp60 antibody (Sigma), which also labels W. pipientis (Hoerauf et al. 2000). Tissues were incubated with 1:250 primary antibody for 1 hr at room temperature in TBST–BSA and 2 mg/ml RNaseA followed by three 5-min washes in TBST. This was followed by 1 hr at room temperature in 1:500 Alexa Fluor 488 anti-mouse antibody (Invitrogen) followed by three 5-min washes in TBST. DNA was then stained with either 5 μg/ml propidium iodide (embryos) for 20 min, or DAPI (malpighian tubules) for 10 min followed by a brief wash in TBST before mounting in ProLong Gold antifade mounting media (Invitrogen). Confocal images of embryos were obtained using a Leica SP confocal microscope. Seminal vesicles and malpighian tubules were imaged with a Zeiss Axio-Imager Z1 microscope.
Fluorescent in situ hybridization (FISH) of 30-μm Nasonia cross-sections was employed to visually show differences in bacterial densities and tissue tropism in the high-density (IntG12.1) and low-density (12.1) strains. Reproductive tissues were dissected within 1 hr after mating directly in 4% PFA on PLUS GOLD adhesion slides. All tissues were fixed for 1 hr and dehydrated using a graded ethanol series. Two W. pipientis-specific 16S oligonucleotide probes, W1 and W2 (Heddi et al. 1999), labeled with Texas Red at the 5′ end were used at 25 ng/ml. Hybridization was performed at 37° in a dark moisture chamber in 0.5 ml hybridization buffer (50% formamide, 5× SSC, 200 g/liter dextran sulfate, 250 mg/liter poly(A), 250 mg/liter salmon sperm DNA, 250 mg/liter tRNA, 0.1 m dithiothreitol (DTT), and 0.5× Denhardt's solution). After an overnight incubation, the slides were washed twice in 1× SSC−10 mm DTT and twice in 0.5× SSC−10 mm DTT at 55°. The slides were rinsed in deionized water, mounted with Vectashield antifade mounting medium containing 1.5 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) and viewed on a Zeiss axiovert compound microscope fitted with epifluorescent optics and photographed with a Cannon G10 fitted with a Zeiss Soligor adapter tube. Confocal images were taken using a Zeiss Axioplan 2 confocal microscope fitted with a HeNe1 laser (543-nm excitation) and DIC.
Quantitative analysis of W. pipientis and phage WO densities:
Genomic DNA was extracted using the Puregene tissue kit (Qiagen, Valencia, CA) from single adult males and females. Real-time quantitative polymerase chain reaction (qPCR) was performed in either an iCycler system (Bio-Rad) or CFX96 Real-Time system (Bio-Rad). Reaction volumes of 25 μl contained 12.5 μl of BioRad SYBR Green Supermix, 10.5 μl sterile water, 0.5 μl of each 10 μm forward and reverse primer, and 2 μl target DNA in single wells of a 96-well plate (Bio-Rad). Selective amplification was performed on a small portion of the Nasonia S6 Kinase (S6K, 133 bp), W. pipientis groEL (groEL, 97 bp), and bacteriophage WO-B (ORF7, 125 bp) genes, as previously described (Bordenstein et al. 2006). A melting curve analysis was performed following each PCR to check for primer dimers and nonspecific amplification. Standard curves for each gene were constructed with a log10 dilution series of PCR products cloned into plasmid vectors using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). qPCR assays were performed in triplicate on all DNA, and data analyses were performed on the average of the three replicates.
Fecundity and survival:
The effect of W. pipientis on fecundity was examined with virgin females that were W. pipientis infected (IntG12.1), antibiotically treated (IntG12.1T), or uninfected (IntG13.2). Virgin females were provided three hosts in a 12 × 75-mm vial for 48 hr, at which time females were removed and transferred to a single host for 8 hr. Limited ovipositioning time prevents developing wasps from becoming resource limited (Bordenstein and Werren 2000). Adult offspring were scored from 80 females during two studies (October 2007 and June 2008) using uninfected, control (IntG13.2) females and antibiotically treated (IntG12.1T) females, respectively. In the haplodiploid Nasonia system, unmated females exclusively produce haploid males. Adult and diapause male F1 offspring were included in offspring production per female.
The effect of W. pipientis on survival under constant environmental conditions without food was also evaluated with infected (IntG12.1) and uninfected, control (IntG13.2) males and females. Upon eclosure, individual insects were transferred to vials and held without food. Deaths were counted at 0, 24, 48, 65, 72, 89, 96, and 120 hr.
Mate discrimination:
The role of W. pipientis in host mate discrimination was assessed by scoring the percentage of successful single-pair copulations observed within a 10-min time span in glass vials. No choice interspecific tests were set up between infected tester N. vitripennis males (12.1) and infected (IntG12.1), antibiotically treated (IntG12.1T) and control, uninfected (IntG13.2) females harboring the N. giraulti genome. Behavioral observations were conducted with 1-day-old individuals. In total, 157 males were mated with uninfected females and 176 were mated with infected females over two replicate experiments in September 2007 and May 2008.
Bidirectional cytoplasmic incompatibility:
Single-pair crosses between introgression and control strains were set up to determine whether gender differences in bacterial loads cause bidirectional CI. Following single-pair copulations, females were hosted on three hosts for 48 hr and transferred to one host for eight hr for egg laying. CI was scored on the basis of the number of adult F1 male and female offspring per female.
Additionally, low-density (12.1) males used in the qPCR experiments were also employed in a study of unidirectional CI to correlate W. pipientis and phage gene copy numbers to levels of CI. Following single-pair copulations with these males, 12.1T tetracycline-cured females were hosted on three hosts for 48 hr and transferred to one host for 8 hr for egg laying. Adult offspring were scored upon death for number of female offspring as a proxy for unidirectional CI.
Male-to-female transfer:
Females were collected immediately after mating to determine the frequency of W. pipientis transmission from an infected male to an uninfected female on the basis of polymerase chain reaction (PCR). DNA from uninfected females was extracted following the methods described below and assessed for transmission of W. pipientis from infected males at time point 0 and/or every 24 hr after mating for 5 days. Primers and PCR conditions are also described below.
Inheritance of paternally transmitted W. pipientis to offspring was tested in two replicate experiments that were carried out in September 2009 (N = 48 families) and October 2009 (N = 40 families). Upon mating with infected males, uninfected mothers in replicate 1 were hosted on two hosts in glass vials for 4 days and then rehosted on two fresh hosts for an additional 5 days in new vials. Females were collected after 9 days and batches of 10 adult offspring from each hosting were collected and frozen at −80° upon emergence. In experimental replicate 2, females were hosted on two hosts for 48 hr and batches of 10 offspring from four life stages (eggs, larvae, yellow-red pupae, and adults) were collected and frozen at −80°. DNA from mothers and offspring was later isolated and PCR tested for W. pipientis transmission. DNA was extracted using a Gentra Puregene tissue kit (Qiagen). PCR was conducted with primers from the W. pipientis-specific 16S rRNA gene (Lo et al. 2002) and bacteriophage WO-specific primers from the gp17 gene from WOCauB1 (Fujii et al. 2004): gp17F (5′CCAGATCAATTAGCATATCTTGCT) and gp17R2 (5′TCTTGGCCGATAAAAACTAAC). PCR conditions for gp17 are as follows: 95° for 2 min, followed by 35 cycles of 94° for 1 min, 53° for 1 min, 72° for 1 min 30 sec, and 72° for 5 min.
Preliminary PCR data indicated that high-density, introgressed males (IntG12.1) transmit W. pipientis to uninfected females immediately after mating but the infection disappears in the mated female after 24 hr. A comparison study with deceased females was then set up to test the female's role in eliminating W. pipientis over time. Female heads were decapitated immediately after mating and the remaining body was held in glass vials. Decapitated females were collected at 0 hr and every 24 hr after mating for 20 days and frozen at −80°. PCR was then used to determine whether the W. pipientis signal disappears in dead females at the same rate as it did in live females.
Statistics:
Pairwise comparisons of adult family size were conducted with the nonparametric Mann–Whitney U-tests (MWU) at α = 0.05 (MiniTab v.12.23, State College, PA), as some distributions were not normal. Fisher's exact test at α = 0.05 was calculated (JMP v. 5.0, SAS Institute) to determine significant deviations from the null hypothesis in discrete datasets. Correlation coefficients were calculated using the nonparametric Spearman's ρ (JMP v. 5.0). A Mantel Cox log-rank test was used to compare survival distributions.
RESULTS
Host variation in infection of the somatic tissues:
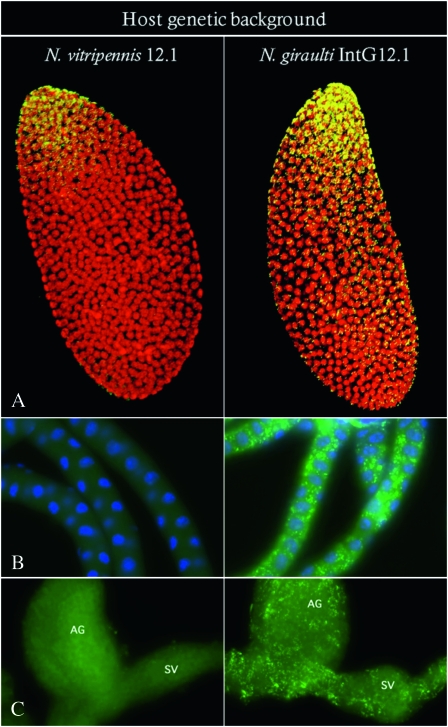
Genome replacement by genetic introgression of the N. giraulti genome into a N. vitripennis A-infected cytoplasm for nine generations in April 2006 led to a stable and heritable increase in W. pipientis densities in the introgression line (IntG12.1) compared to the resident A-infected N. vitripennis line (12.1). Extensive proliferation of the infection in N. giraulti IntG12.1 was evident by the titer increase at the posterior end of the embryo and the spread of the bacteria toward the anterior end (Figure 1A). The proliferation of the infection throughout the embryo in the N. giraulti IntG12.1 line placed W. pipientis infections in cells fated to become somatic cells. Consequently, bacterial proliferation into adult tissues not typically infected by the resident N. vitripennis 12.1 infection occurred in all tissues of the N. giraulti IntG12.1 line examined, including malpighian tubules (Figure 1B), accessory glands, and seminal vesicles (Figure 1C).
Figure 1.—
Immunofluorescence staining of A-group W. pipientis in N. vitripennis and N. giraulti. Images show W. pipientis stained in yellow or green and host DNA stained in red or blue. Density differences and tissue tropism are shown in (A) embryos; posterior end is at the top, (B) malpighian tubules, and (C) ag, accessory glands and sv, seminal vesicles.
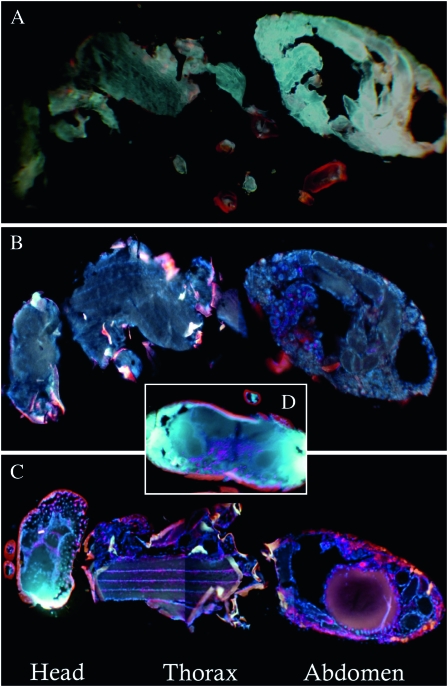
Whole female Nasonia tissue sections hybridized with W. pipientis-specific 16S rRNA fluorescent probes W1 and W2 (Heddi et al. 1999) showed somatic tissue infection throughout the entire N. giraulti IntG12.1 wasp (Figure 2C). Strong W. pipientis signals were also observed in the abdomen, flight muscle tissues in the thorax, and throughout the head (Figure 2, C and D). In contrast, the resident host N. vitripennis 12.1 showed localization only to the abdomen (Figure 2B), and the control unstained N. vitripennis 12.1 yielded little autofluorescence of the probes (Figure 2A).
Figure 2.—
Fluorescent in situ hybridizations of W. pipientis in adults of N. vitripennis and N. giraulti. Comparisons of 30-μm cross-sections from adult females of (A) unstained N. vitripennis 12.1 (control for autofluorescence), (B) stained N. vitripennis 12.1, (C) stained N. giraulti IntG12.1, and (D) a stained N. giraulti IntG12.1 head. Strong W. pipientis signals (red) are observed in IntG12.1 in the head, the flight mussel tissues in the thorax, and the abdomen. Images B–D are counterstained with DAPI (blue) and are presented as composite images from the same individual to enhance resolution of the W. pipientis.
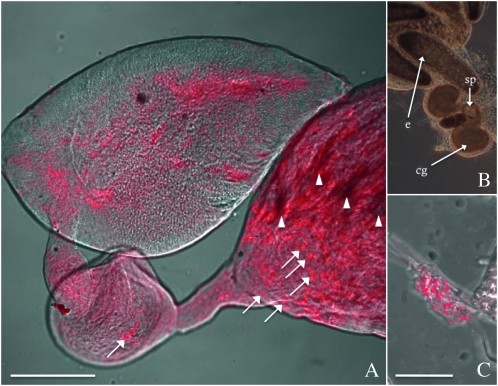
In the male gonads of the new N. giraulti IntG12.1 genetic background, fluorescent imaging of the W1 and W2 probes showed that the bacteria proliferate in the testes, seminal vesicle, and accessory gland (Figure 3A). One notable observation is an unusual signal aggregation or “clumping” of W. pipientis at the base of the testes (Figure 3A, white arrows), suggesting W. pipientis is shed from the sperm by the individualization complexes during spermatogenesis, possibly into waste bags, at high densities (Clark and Karr 2002; Riparbelli et al. 2007). There is a dense fluorescence in the testes that is not present in the accessory gland and seminal vesicles. It could be due to an artifact of autofluorescence or an extremely dense infection in the testes. Control experiments with the testes from cured and low-density N. vitripennis 12.1 males did not show the dense signal in the testes (supporting information, Figure S1). The presence of W. pipientis in the seminal vesicle and accessory gland suggests it could be transmitted paternally through the seminal fluid (see section on male-to-female transfer).
Figure 3.—
Fluorescent in situ hybridizations of W. pipientis in reproductive tissues of N. giraulti. W. pipientis (red) is shown in (A) the testes of an infected IntG12.1 male and (C) the spermatheca of an uninfected female mated to a IntG12.1 male. The ovaries of an uninfected female are also shown in B. e, mature egg at the base of a ovariole; cg, colleterial glands; sp, spermatheca. Arrows indicate W. pipientis aggregation within the testes (A) and may indicate the cellular unit transmitted to the spermatheca of uninfected females (C). Bars, 50 μm.
Host variation in microbial densities and expressivity of CI:
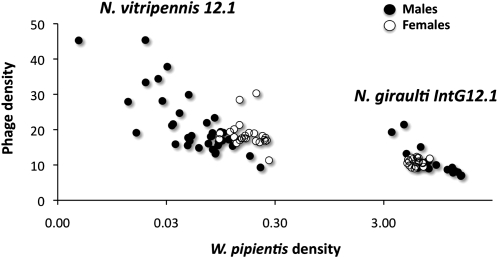
To examine W. pipientis and phage WO-B titer differences between the two host species, we used qPCR of single-copy genes of the N. vitripennis and N. giraulti genomes (S6K), W. pipientis genomes (groEL), and phage WO-B (ORF7). Consistent with the fluorescent imaging, W. pipientis densities were significantly higher in N. giraulti IntG12.1 adults than in N. vitripennis 12.1 adults, for both males (MWU, P < 0.0001) and females (MWU, P < 0.0001) (Figure 4). The mean ± standard error for W. pipientis density in N. giraulti IntG12.1 (males, 8.98 ± 0.82; females, 6.08 ± 0.16) was 128-fold and 36-fold higher than the average W. pipientis density in the resident N. vitripennis 12.1 (males, 0.07 ± 0.01; females, 0.17 ± 0.01). These increases were associated with significant decreases in phage WO-B densities (phage-to-bacteria ratios) for males and females, respectively (Figure 4). There were 2.0-fold and 1.7-fold reductions in phage WO-B densities in the densely infected N. giraulti IntG12.1 relative to N. vitripennis 12.1 for males (MWU, P < 0.0001) and females (MWU, P < 0.0001), respectively. Furthermore, there were significant inverse associations between phage and W. pipientis densities within species in N. vitripennis 12.1 males (ρ = −0.6932, P < 0.0001, Figure 4) and N. giraulti IntG12.1 males (ρ = −0.8455, P < 0.0001, Figure 4), but not females. For instance, the N. vitripennis male with the highest WO-B density (45.36 ORF7 gene copies per W. pipientis groEL gene copies) had the lowest W. pipientis density, and the N. vitripennis male with the lowest WO-B density (9.30) had the highest W. pipientis density. Similarly for N. giraulti, the male with the highest WO-B density (21.43 ORF7 gene copies per W. pipientis groEL gene copies) had the second lowest W. pipientis density, and the male with the lowest WO-B density (6.86) had the second highest W. pipientis density.
Figure 4.—
Inverse relationships between W. pipientis and phage WO-B densities. Bacteriophage WO-B vs. W. pipientis gene copy number in the resident N. vitripennis and novel N. giraulti host genetic backgrounds. Points on the chart denote bacterial and bacteriophage densities in individual males and females.
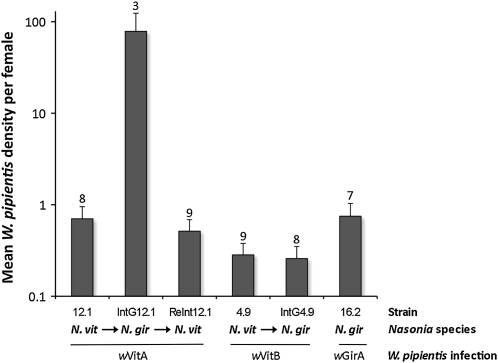
A replicate experiment of the host variation in W. pipientis densities was performed with additional Nasonia strains infected with different bacterial strains (Figure 5). First, the study recapitulated the major density differences described above. Female 12.1 and IntG12.1 infected with the supergroup A strain showed a 112-fold difference (MWU, P = 0.02). Second, the increased bacterial densities appear specific to the A infection as the introgression of the N. giraulti genome into the N. vitripennis cytoplasm carrying the supergroup B infection did not yield different bacterial loads (4.9 vs. IntG4.9, MWU, P = 0.73). The strain-specific increase was further evident by quantifications of the divergent A infection in its resident N. giraulti genome (strain 16.2), in which it exhibited low densities (16.2 vs. 12.1, MWU, P = 0.86). Finally, replacement of the IntG12.1 N. giraulti genome back to a N. vitripennis genome resulted in the return of low W. pipientis densities (ReInt12.1 vs. 12.1, 0.51 vs. 0.70; MWU, P = 0.14) and high phage densities (ReInt12.1 vs. 12.1, 6.1 vs. 5.6; MWU, P = 1.0), indicating the native bacterial and phage densities were restored in the resident N. vitripennis background (Figure 5).
Figure 5.—
Increase in W. pipientis densities is specific to the bacterial genotype. Columns denote the mean W. pipientis densities ± standard error in the native N. vitripennis host (A-infected 12.1 and B-infected 4.9), the native N. giraulti host (A-infected 16.2), and the introgressed N. giraulti (A-infected IntG12.1 and B-infected IntG4.9) and reintrogressed N. vitripennis (A-infected ReInt12.1) genetic backgrounds. Numbers above columns denote female sample size.
Fecundity:
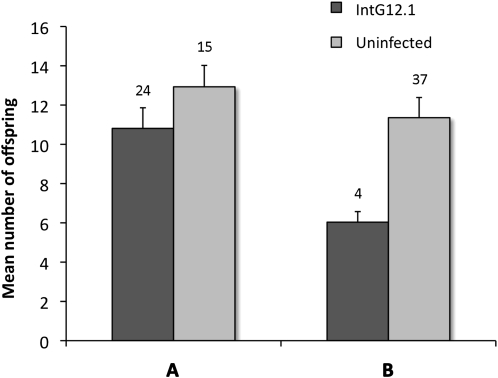
To determine whether the extensive proliferation of W. pipientis densities and tissue tropism in the new N. giraulti host influences fitness, we compared the fecundity of IntG12.1 infected, virgin females (haploid male offspring only), IntG12.1T antibiotically cured virgin females, and IntG13.2 uninfected, control virgin females that went through the same introgression scheme as the infected strain. A Mann–Whitney U-test revealed a marginally nonsignificant decrease (P = 0.07) in comparison to the IntG13.2 uninfected, control females and a significant decrease (P = 0.0007) in mean fecundity per infected female in comparison with IntG12.1T tetracycline-treated females (Figure 6). The difference in the significance of the two datasets was apparently due to the fecundity of infected females (10.83 vs. 6.03, MWU, P = 0.0002), as uninfected females yielded similar offspring numbers (12.93 vs. 11.36, MWU, P = 0.17).
Figure 6.—
Fecundity cost in infected N. giraulti IntG12.1 females. High densities of W. pipientis in N. giraulti IntG12.1 results in reduced fecundity relative to IntG13.2 uninfected, control females (A) and IntG12.1T tetracycline-cured females (B). Bars represent means ± standard errors. Numbers above columns denote female sample size.
Starvation resistance:
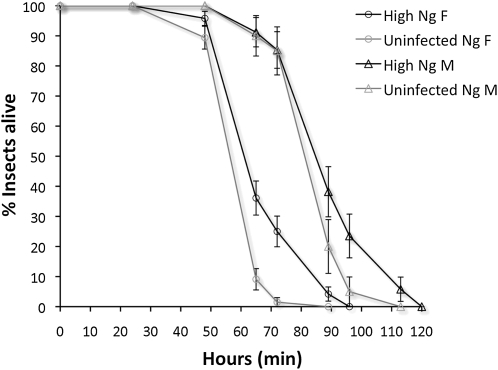
Increases in W. pipientis densities and infection of somatic tissues in the N. giraulti genetic background may also affect the host's duration of life, similar to the rapidly proliferating wMel Popcorn strain in Drosophila melanogaster that causes early mortality (Min and Benzer 1997). We measured survival of individual IntG12.1 infected and IntG13.2 uninfected males and females under constant environmental conditions without food (Figure 7). Survival of starved wasps was slightly extended in infected males and significantly extended in infected females (Mantel Cox log-rank test, χ2 = 19.3, d.f. = 1, P = 0.00001). The time points that were significantly different occurred at 65 and 72 hr (Fisher's exact test, P = 0.0002 and P = 0.0001, respectively). Further, males survived longer than females, for both infected (Mantel Cox log-rank test, χ2 = 1282, d.f. = 1, P < 0.00001) and uninfected wasps (Mantel Cox log-rank test, χ2 = 55.1, d.f. = 1, P < 0.00001).
Figure 7.—
Survival curves of infected and uninfected N. giraulti introgression males and females. Survival under starvation conditions was higher in infected N. giraulti females relative to IntG13.2 uninfected, control females. The fraction of live individuals was counted at 0, 24, 48, 65, 72, 89, 96, and 120 hr. Each point denotes mean ± standard error. Labeling denotes male (M) and female (F).
Host mating behavior:
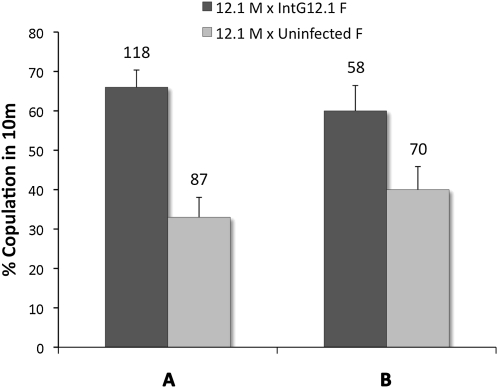
We hypothesized that increases in W. pipientis densities in the N. giraulti background, especially into the brain or sensory organs of the head region (Figure 2, C and D), could affect mate discrimination. We therefore measured the frequency of interspecific matings between N. vitripennis 12.1 males and IntG12.1 infected, IntG12.1T tetracycline-treated, or IntG13.2 uninfected, control females in the introgressed N. giraulti genetic background. Figure 8A shows a highly significant increase (χ2, P < 0.0001) in interspecific mating percentage between IntG12.1 infected females (66% acceptance, N = 118, IntG12.1) vs. the IntG13.2 uninfected, control females (33% acceptance, N = 87, IntG13.2).
Figure 8.—
Interspecific female mate discrimination is reduced in IntG12.1 infected females. Mate discrimination is reduced in infected N. giraulti females in comparison to IntG13.2 uninfected, control females (A) and IntG12.1T antibiotically treated females (B). Labeling denotes male (M) and female (F). Bars represent means ± standard errors, and sample sizes are shown above the bars.
The same increase in interspecific mating was also evident between IntG12.1 infected females (60% acceptance, N = 58, IntG12.1) and IntG12.1T antibiotically treated females (40% acceptance, N = 70, IntG12.1T) (χ2, P = 0.02). Overall, the presence of high W. pipientis densities in females resulted in less discrimination and therefore more interspecific copulations. Similar effects were not observed in low-density 12.1 N. vitripennis females who showed strong interspecific mate discrimination irrespective of whether they were infected (4.9% acceptance, N = 82) or antibiotically treated (7.9% acceptance, N = 38) (χ2, P = 0.51) when paired with N. giraulti males.
CI:
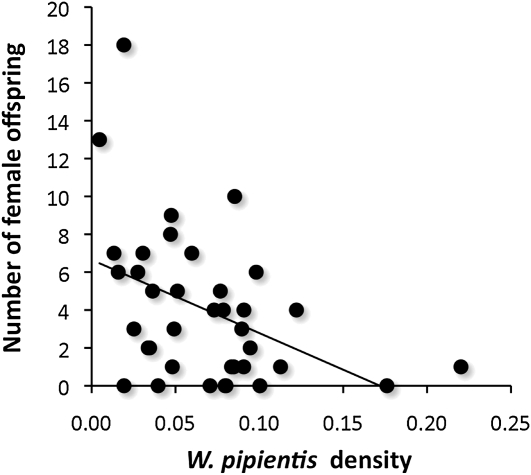
CI is a postmating incompatibility between infected males and uninfected females or females harboring a different infection from the male. The bacterial density model of CI posits that variation in incompatibility levels positively correlates with bacterial densities in the host (Breeuwer and Werren 1993). While investigations of the bacterial density model of CI traditionally test for a positive correlation between W. pipientis titers and the expressivity of CI, as is observed in crosses between N. vitripennis 12.1 infected males and uninfected females in this study (ρ = −0.38, P = 0.021, Figure 9), an extension of the model described in the original article was that differences in bacterial densities between males and females could lead to different incompatibility types that cause bidirectional incompatibility. For instance, males with high-density infections in testes could be incompatible with females that have low densities in their eggs, or conceivably eggs with significantly higher bacterial densities than the testes may be predisposed to a cell cycle asynchrony in the relative timing of the male and female pronuclei. Here we take advantage of the high- and low-density Nasonia lines in this study to explicitly test this prediction.
Figure 9.—
W. pipientis densities correlate with CI levels. Points denote the W. pipientis density and total number of female offspring from single-pair matings between N. vitripennis 12.1 males and uninfected females. Line represents the least squares regression.
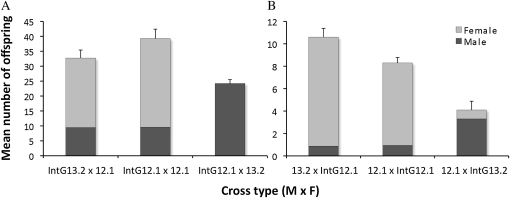
In the haplodiploid Nasonia, W. pipientis-induced CI results in a reduction in the number of diploid females produced since haploid males are produced without fertilization. Therefore, the predicted trend for the bacterial density model is that incompatible crosses yield low numbers of female offspring, i.e., high CI inducers. Results indicate that differences in bacterial density do not cause major effects on bidirectional CI. Crosses between low-density 12.1 N. vitripennis females and either IntG12.1 high-density or IntG13.2 control, uninfected N. giraulti males (Figure 10A) showed no statistical difference in mean number of male offspring (9.56 vs. 9.45, MWU, P = 0.86), female offspring (29.69 vs. 23.27, MWU, P = 0.33), and total family size (39.25 vs. 32.72, MWU, P = 0.23). However as expected for unidirectional CI, the IntG12.1 high-density males were completely incompatible with uninfected females with a complete reduction in female offspring (0 vs. 23.27, P = 0.0001) and correlated increase in the number of males (24.22 vs. 9.45, MWU, P = 0.002) in comparison to the control cross with uninfected males.
Figure 10.—
The bacterial density model of CI does not explain bidirectional CI. Experimental and control crosses are shown for testing incompatibility between (A) high-density males and low-density females and (B) low-density males and high-density females. Data are represented as the average number of male and female offspring produced per cross ± the standard error for total offspring.
Crosses in the reciprocal direction between high-density IntG12.1 N. giraulti females and either low-density or control, uninfected N. vitripennis males (Figure 10B) showed no statistical difference in the number of males (0.96 vs. 0.87, MWU, P = 0.71) and a slight, significant difference in the number of female offspring (7.50 vs. 9.73, MWU, P = 0.02), which caused a difference in total family size (8.46 vs. 10.60, MWU, P = 0.01). These results specify a marginal increase in CI between a high-density IntG12.1 N. giraulti female and a low-density 12.1 N. vitripennis male. As expected in the control incompatible cross, unidirectional CI between the low-density 12.1 males and uninfected IntG13.2 females caused a strong but incomplete reduction in female offspring (0.80 vs. 9.73, MWU, P < 0.0001) and increase in males (3.30 vs. 0.87, MWU, P = 0.0002), which is consistent with previous experiments on these lines (Bordenstein et al. 2003).
Male-to-female transfer of W. pipientis:
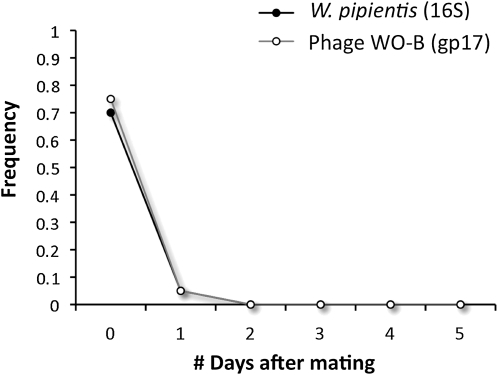
Finally, to test for transfer of the infection from an infected male with unusually high bacterial densities to an uninfected female, two sets of intraspecific crosses were set up using the IntG12.1 and 12.1 infected males with their antibiotically treated female counterparts. In the resident, low-density 12.1 N. vitripennis males (N = 40), no transfer of W. pipientis DNA was observed on the basis of the absence of PCR-amplifiable DNA in uninfected females immediately after mating. However, the high-density IntG12.1 N. giraulti males showed clear evidence of transfer of W. pipientis DNA to uninfected females. Immediately after mating, 14 out of 20 uninfected females yielded a PCR product for W. pipientis (16S rRNA gene) and 15 out of 20 yielded a product for bacteriophage WO (gp17). Subsequent PCRs from 2 through 5 days after mating indicated no detectable W. pipientis DNA, suggesting that most transferred W. pipientis or W. pipientis DNA persists for <24 hr after mating (Figure 11). Fluorescent in situ hybridizations with the W. pipientis 16S rRNA-targeting probes in freshly mated females confirmed transfer of W. pipientis cells in the female's spermatheca (4 out of 9 samples, Figure 3C). No W. pipientis fluorescence was observed in six, negative uninfected control spermathecae. Further, no W. pipientis or bacteriophage WO PCR product was detected in DNA from 10 pooled offspring of uninfected females mated to high-density IntG12.1 N. giraulti males (N = 55 families, 0/550 offspring, 95% confidence interval from 0 to 0.67%), indicating that on a laboratory scale, paternal transmission to offspring is rare or does not occur.
Figure 11.—
Male-to-female transfer of W. pipientis upon transfer to new host species. W. pipientis transferred from infected, IntG12.1 males to uninfected females is transiently detectable in young females. Points denote mean frequency of PCR product detected in uninfected females over 5 days in 20 female replicates per day.
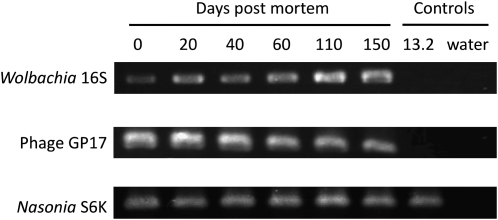
Detection of the transferred W. pipientis cells in the spermathecae of IntG12.1T uninfected females and in the testes of IntG12.1 infected males showed an aggregation of bacterial cells clumped together (Figure 3, A and C). Control crosses between tetracycline-cured IntG12.1T N. giraulti males and females (N = 6) did not show this signal, ruling out the alternative that the aggregate is due to nonspecific binding of the probe. To preliminarily discern the role of a live female in eliminating paternally transmitted W. pipientis, crosses were set up similar to above, but the uninfected females were decapitated immediately after mating. Comparisons of these dead females at 24 hr time points (N = 5 females for each time point) showed that dead females recapitulated the high frequency of paternally transmitted W. pipientis in the first 24 hr, but W. pipientis and phage DNA were still detectable in decapitated, IntG12.1T uninfected females even after 20 days (W. pipientis, 2/5 individuals; bacteriophage, 5/5 individuals). To test whether W. pipientis DNA normally remains in infected wasps postmortem, we examined dead N. vitripennis females from infected 12.1 wasps between 0 and 150 days after death. All template DNA across the time points yielded PCR amplicons from W. pipientis, phage WO-B, and Nasonia (Figure 12).
Figure 12.—
Amplifiable DNA from W. pipientis, phage WO-B, and Nasonia in dead, infected females. PCR products from infected 12.1 N. vitripennis females spanning 0–150 days postmortem. Negative controls are derived from the uninfected 13.2 N. vitripennis and double distilled water.
DISCUSSION
Transfer of symbionts to new host species can decouple host–symbiont coadaptations within the resident species background. Within N. vitripennis, the supergroup A strain of W. pipientis has a relatively low level of infection. Upon introgression of the N. giraulti host genome into the A-infected N. vitripennis cytoplasm, the bacterial concentrations approach seemingly virulent numbers, inundating tissues throughout the entire body cavity. The new strain is therefore an excellent candidate to study W. pipientis–host interactions with a strong host genetic basis. The influence of W. pipientis infections in different host species varies widely in both nature and intensity (Breeuwer and Werren 1993; Bordenstein and Werren 1998; Grenier et al. 1998; Poinsot et al. 1998; Fujii et al. 2001). For some strains, the type of reproductive parasitism expressed can alternate depending on the host species (Sasaki and Ishikawa 2000; Jaenike 2007). For others, host genetic background has been shown to influence W. pipientis density and the degree of CI (Boyle et al. 1993; Bordenstein and Werren 1998; Sinkins et al. 2005; Zabalou et al. 2008), as well as to redefine the types of tissues able to support W. pipientis replication (Min and Benzer 1997; Dobson et al. 1999; Cheng et al. 2000; Frydman et al. 2006).
Compared to the low-density 12.1 N. vitripennis, the IntG12.1 N. giraulti host bears a relative bacterial load that is two orders of magnitude higher and a relative phage/bacteria ratio that is twofold lower (Figure 4). The perturbations in microbial densities associated with the N. giraulti host were not observed with the N. vitripennis B infection, which does not harbor a phage with lytic activity (Bordenstein et al. 2006). Thus, the differences in density and somatic tissue infection are due to a specific interaction between the A infection/phage combination of N. vitripennis and the Nasonia nuclear genomes. It is also possible that mitochondria (mt) are involved in the interaction since the N. vitripennis cytoplasm with both W. pipientis and mt is placed into a N. giraulti genetic background. The Nasonia mtDNA have one of the highest rates of substitution among all animal mitochondria (Oliveira et al. 2008). Breakdown between cytonuclear interactions could conceivably alter the homeostasis of the endosymbiosis. For example, the mitochondrial heat shock protein 60 is upregulated in Onchocerca volvulus filarial nematodes following W. pipientis removal (Pfarr et al. 2008). Reversal of the introgression from N. giraulti back into N. vitripennis recapitulates the low W. pipientis densities and high phage densities native to this species. The genetic pathways involved in this tripartite interaction between invertebrates, bacteria, and phage remain an important, future direction of study.
Inverse association between phage WO-B and W. pipientis:
We established that W. pipientis densities within infected males of each species correlate inversely with phage WO-B densities. We also showed that the dramatic increase of W. pipientis densities in the new host species was associated with a significant reduction in relative phage densities. These data corroborate our previous findings on how the phage and Wolbachia densities are intertwined in N. vitripennis (Bordenstein et al. 2006). There are at least three possible explanations for why the phage densities change after movement to a new host species.
First, a tritrophic interaction between the phage, W. pipientis, and insect nuclear genome may govern a reduction in phage densities in the novel host species, which ultimately results in higher relative W. pipientis densities. This reticulated control of the symbiosis could, in theory, arise by selection on arthropods to utilize the phage as a natural therapy against W. pipientis infection. This hypothesis is based on two arguments: (i) in the absence of symbiont-induced fitness advantages, the phage and arthropod have a common selective pressure in killing the bacteria—the phage uses it for replication and the host arthropod could suppress fitness costs or expressivity of cytoplasmic incompatibility; (ii) furthermore, ankyrin repeats are found within both the W. pipientis and phage genomes (Masui et al. 2000; Wu et al. 2004; Iturbe-Ormaetxe et al. 2005; Klasson et al. 2008; Klasson et al. 2009), indicating the potential occurrence of protein–protein interactions between the phage and insect genome. While these ankyrin repeats have been suggested to be causative factors in the induction of reproductive parasitism, they may alternatively facilitate a direct interaction between Nasonia and the phage that mediates the regulation of lytic activity and a reduction in W. pipientis densities. Similarly, host level selection could also explain its absence from the genome of mutualistic W. pipientis in filarial nematodes (Foster et al. 2005). Selection at the host level to maintain the beneficial symbiosis could select against a lytic phage of a mutualistic infection, which is required for host oogenesis and larval development (Bordenstein et al. 2006). Strict vertical transmission, genome reduction, and absence of coinfections of W. pipientis in filarial nematodes are also adequate reasons for the absence of bacteriophage in this species.
A second explanation for the inverse association between phage WO-B and W. pipientis is that the W. pipientis cells are stably replicating in the new N. giraulti background, leading to a suppression of the bacterial interactors that elicit lytic phage development. Perhaps the new host is more permissive for W. pipientis growth. The basic rationale here is that if phage WO-B responds to bacterial stress responses by entering lytic development, then by extension, the W. pipientis in N. vitripennis could be more stressed than the same infection in N. giraulti. Many temperate phages “sense” stressed bacteria, thereby inducing prophage excision, replicating as virions, and triggering bacterial lysis. There may be many ways by which a sensing feedback mechanism leads to phage induction, and one of the most common is the SOS response. Curiously, key SOS genes are present in the Wolbachia genome, including recA, uvrA, uvrB, uvrD, and a cI-like repressor; lexA, however, is missing.
Third and finally, stochastic changes in phage densities during the introgression rather than host genotype could also explain the change in phage densities. However, the IntG12.1 strain has now been maintained in the laboratory since April 2006, indicating a stable host genome interaction that is not subject to random fluctuations in bacterial density. In addition, it is the second time we have generated the strain and observed an increase to complete CI, which is indicative of higher densities, although the densities were not measured in that experiment (Bordenstein and Werren 1998).
Fitness:
The dramatic 100-fold increase in infection densities in the naive host species is likely to modify the traditional costs and benefits associated with the symbiosis. Previous studies indicate fecundity effects of W. pipientis vary widely. Some studies suggest there is no effect of W. pipientis on fecundity under laboratory conditions in D. melanogaster (Clark and Karr 2002; Reynolds and Hoffmann 2002) or in N. vitripennis (Bordenstein and Werren 2000), while others have found harmful effects (Hoffmann et al. 1990; Nigro 1991; Stouthamer and Luck 1993; Calviti et al. 2010). In our study, a decrease in fecundity was found in the high-density IntG12.1 N. giraulti females. This negative fecundity effect was not observed within N. vitripennis where W. pipientis densities are lower and restricted to the host reproductive tissues. Thus, the high-density infection in the introgression line may have intensified effects on fecundity in Nasonia that were not seen in comparisons with lower densities (Bordenstein and Werren 2000).
In contrast to fecundity, survival of starved wasps was slightly extended in Nasonia harboring high W. pipientis densities. This finding was unexpected since we anticipated the high densities in somatic tissues would yield a decrease in survival. Previous studies showed that W. pipientis did not influence adult starvation resistance in the quiescent wMel infection of D. melanogaster (Harcombe and Hoffmann 2004). However, the rapidly proliferating wMel Popcorn strain in laboratory-reared D. melanogaster causes early mortality (Min and Benzer 1997). Possible reasons for the increase in survival include (i) an increase in immunity to other bacterial or viral parasites, as evident from recent findings of RNA viral resistance mediated by W. pipientis (Hedges et al. 2008; Teixeira et al. 2008); (ii) a life history tradeoff in which decreased fecundity in infected females may lead to an increased energy investment in adult survival; in this study, fecundity was significantly reduced, but not measured over the lifetime of the female; and (iii) mitochondrial haplotype differences between the experimental lines; IntG12.1 and IntG13.2 are derived from N. vitripennis lines 12.1 and 13.2 in a segregation experiment conducted in 1996 (Perrot-Minnot et al. 1996). Further work is necessary to determine the importance of these mechanisms.
Other recent studies suggest the fitness interactions between host and W. pipientis are more complex than previously recognized. For instance, loss of insulin-like ligands produced in the Drosophila brain greatly extended lifespan in the presence of W. pipientis, demonstrating a specific interaction between the insulin/IGF signaling (IIS) pathway and the bacteria in lifespan regulation (Gronke et al. 2010). Coevolution between host and W. pipientis results in an intricate merger of signaling pathways, which can result in highly specific phenotypic effects unique to each association.
Bidirectional cytoplasmic incompatibility:
The positive association between W. pipientis densities and incompatibility levels is characterized as the “bacterial density model” of CI (Breeuwer and Werren 1993), and numerous studies have confirmed positive correlations between CI levels and bacterial densities within eggs, sperm cysts, whole adults, and reproductive tissues (Poinsot et al. 1998; Perrot-Minnot and Werren 1999; Noda et al. 2001; Clark and Karr 2002; Veneti et al. 2003). Extrinsic factors that affect W. pipientis density can modify the nature and degree of CI including host genetic background (Boyle et al. 1993; Bordenstein and Werren 1998; Sinkins et al. 2005) and environmental stresses such as host age (Hoffmann et al. 1986; Sinkins et al. 1995; Feder et al. 1999; Clark et al. 2002), heat treatment (Feder et al. 1999), mating history (Karr et al. 1998), larval crowding (Sinkins et al. 1995), as well as lytic phage proposed as part of the “phage density model” (Bordenstein et al. 2006).
Here we used high- and low-density Nasonia lines to explicitly test a second prediction of the bacterial density model of CI: bidirectional CI may result from gender differences in bacterial densities. We showed that the large differences in bacterial loads between IntG12.1 and 12.1 do not lead to strong bidirectional CI. Slight incompatibility observed between low-density males and high-density females could possibly be due to the dense infection in the egg, which causes a change to the embyronic factors that rescue the paternal genome modification caused by CI. Previous studies demonstrated that rescue is either achieved through correction of cell cycle defects in the male pronucleus or through a compensatory slowing of the female pronucleus cell cycle to restore the cell cycle synchrony with the altered male pronucleus (Tram and Sullivan 2002; Serbus et al. 2008). Nonetheless, results generally indicate that male and female differences in bacterial densities do not single handedly explain the significant differences in compatibility (Poinsot et al. 1998; Perrot-Minnot and Werren 1999; Noda et al. 2001; Clark and Karr 2002; Veneti et al. 2003; Bordenstein and Werren 2007). Thus, these results show that bidirectional incompatibility is more likely due to rapid genetic changes between strains and/or hosts.
Male-to-female transfer of W. pipientis:
The exceptionally high level of infection in the new genetic background leads to colonization of the male seminal vesicles and accessory glands. This change presumably leads to the high rate of male-to-female transfer observed in crosses between infected males and uninfected females (70%). Both PCR-amplifiable DNA and 16S rDNA probes provided evidence for the sexual transmission. Although the infection did not remain within the living female for >24 hr or transfer to offspring, the presence of W. pipientis within an uninfected female could potentiate a rare opportunity for paternal transmission, which is not likely to be captured in small-scale laboratory experiments. Strikingly, dead females who had been decapitated for up to 20 days yielded transferred DNA, suggesting there may be some direct or indirect physiological process that eliminates the W. pipientis within 24 hr after transfer. Alternatively, the dead female could offer a more permissive environment for W. pipientis to inhabit. Experiments are underway to determine the viability of W. pipientis in dead wasps.
Recently there have been a number of animal and plant systems in which paternal leakage of mitochondrial genomes has been demonstrated in hybrids (White et al. 2008). Presumably, there is a hybrid breakdown of mechanisms that recognize and remove paternal cytoplasmic elements such as mitochondria. Therefore, while paternal transfer of W. pipientis is absent or rare in D. melanogaster (Hoffmann et al. 1998), D. simulans (Hoffmann and Turelli 1988), and Aedes albopictus (Dobson et al. 2002) within species, our study suggests more attention to paternal transfer in hybrid zones or hybrids in the lab because the IntG12.1 is a hybrid composed of a N. vitripennis cytoplasm and N. giraulti genome.
Host behavioral pathology:
Finally, previous studies showed that W. pipientis can alter host behaviors. One study found W. pipientis-induced effects on precopulatory behavior where inbred, uninfected female spider mites preferentially mated with uninfected males to avoid W. pipientis-induced CI (Vala et al. 2004). A change in ovipositioning behavior was also reported in that study. Mating behavior influences have also been described in isopods (Moreau et al. 2001) where W. pipientis infections distort sex ratios toward females through the feminization of males. Here, males showed preference for genetic females as opposed to copulations with feminized males.
Interspecific mate discrimination between N. vitripennis and N. giraulti is well characterized and likely plays a role in reinforcing reproductive isolation in sympatric populations because of F1 CI and severe F2 hybrid breakdown between the species (Breeuwer and Werren 1990; Bordenstein and Werren 1998; Drapeau and Werren 1999). Our studies in the laboratory showed that IntG12.1 infected females have reduced interspecific mate discrimination (i.e., more copulations). Similar mating behavior effects in low-density 12.1 N. vitripennis females were not observed. Our results are the opposite of those of a previous study in which the removal of W. pipientis from choosy D. melanogaster strains resulted in a reduction of discrimination (Koukou et al. 2006), indicating that W. pipientis-induced effects (i.e., on pheromones and/or behavior) enhance a preexisting genetic bias in mate discrimination in both males and females.
A simple explanation for the increase in interspecific mating frequency in our study is that the infection in the head regions of N. giraulti females causes a behavioral pathology in which females suffer from a reduced competence to discriminate against interspecific males. This conclusion is based on two observations. First, neurological infections of W. pipientis in Nasonia are abnormal, potentiating a pathology related to behavior. Second, wasps that are infection-free show high levels of discrimination typical of N. giraulti females infected with their native W. pipientis strain (Bordenstein and Werren 1998).
Concluding remarks:
The arthropod–W. pipientis association is an ancient symbiosis, yet new symbiotic combinations between different W. pipientis strains and host species constantly evolve as the infection promiscuously transfers to new arthropod species. A conflict or compromise between the endosymbiont and host must ensue, which determines the outcome of the symbiosis. Here we have shown that the two-order magnitude increase in relative W. pipientis densities in a new host species leads to reduced fecundity, higher levels of CI, and unexpectedly, male-mediated transfer of the bacteria to females and a behavioral pathology. Results from this study also raise the idea that the host may have direct or indirect effects on biological entities one trophic level down from W. pipientis, i.e., the bacteriophage WO, which could alter densities of the endosymbiont. The host genetic mechanisms within Nasonia that permit/suppress a hyperinfection to take a permanent hold, and the question of whether the molecular interactions with the host involve both the bacteria and phage, remain an important topic of investigation.
Acknowledgments
We thank Bethany Kent for designing the phage gp17 primers, Lisa Funkhouser for reviewing the manuscript, and two anonymous reviewers for helpful feedback. This work was supported by award National Science Foundation (NSF) DEB-0821936 to John Werren and by awards NSF IOS-0852344 and National Institutes of Health R01 GM085163-01 to S.R.B.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.120675/DC1.
References
- Bordenstein, S. R., and J. H. Werren, 1998. Effects of A and B Wolbachia and host genotype on interspecies cytoplasmic incompatibility in Nasonia. Genetics 148 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein, S. R., and J. H. Werren, 2000. Do Wolbachia influence fecundity in Nasonia vitripennis? Heredity 84(Pt 1): 54–62. [DOI] [PubMed] [Google Scholar]
- Bordenstein, S. R., and J. H. Werren, 2007. Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. Heredity 99 278–287. [DOI] [PubMed] [Google Scholar]
- Bordenstein, S. R., J. J. Uy and J. H. Werren, 2003. Host genotype determines cytoplasmic incompatibility type in the haplodiploid genus Nasonia. Genetics 164 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein, S. R., M. L. Marshall, A. J. Fry, U. Kim and J. J. Wernegreen, 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, L., S. L. Oneill, H. M. Robertson and T. L. Karr, 1993. Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260 1796–1799. [DOI] [PubMed] [Google Scholar]
- Breeuwer, J. A. J., and J. H. Werren, 1990. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346 558–560. [DOI] [PubMed] [Google Scholar]
- Breeuwer, J. A. J., and J. H. Werren, 1993. Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics 135 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviti, M., R. Moretti, E. Lampazzi, R. Bellini and S. Dobson, 2010. Characterization of a new Aedes albopictus (Diptera: Culicidae)–Wolbachia pipientis (Rickettsiales: Rickettsiaceae) symbiotic association generated by artificial transfer of the wPip strain from Culex pipiens (Diptera: Culicidae). J. Med. Entomol. 47 179–187. [DOI] [PubMed] [Google Scholar]
- Cheng, Q., T. D. Ruel, W. Zhou, S. K. Moloo, P. Majiwa et al., 2000. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14 44–50. [DOI] [PubMed] [Google Scholar]
- Clark, M. E., and T. L. Karr, 2002. Distribution of Wolbachia within Drosophila reproductive tissue: implications for the expression of cytoplasmic incompatibitity. Integr. Comp. Biol. 42 332–339. [DOI] [PubMed] [Google Scholar]
- Clark, M. E., Z. Veneti, K. Bourtzis and T. L. Karr, 2002. The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila. Mech. Dev. 111 3–15. [DOI] [PubMed] [Google Scholar]
- Dobson, S. L., K. Bourtzis, H. R. Braig, B. F. Jones, W. G. Zhou et al., 1999. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem. Mol. Biol. 29 153–160. [DOI] [PubMed] [Google Scholar]
- Dobson, S. L., E. J. Marsland and W. Rattanadechakul, 2002. Mutualistic Wolbachia infection in Aedes albopictus: accelerating cytoplasmic drive. Genetics 160 1087–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau, M. D., and J. H. Werren, 1999. Differences in mating behaviour and sex ratio between three sibling species of Nasonia. Evol. Ecol. Res. 1 223–234. [Google Scholar]
- Feder, M. E., T. L. Karr, W. Yang, J. M. Hoekstra and A. C. James, 1999. Interaction of Drosophila and its endosymbiont Wolbachia: natural heat shock and the overcoming of sexual incompatibility. Am. Zool. 39 363–373. [Google Scholar]
- Foster, J., M. Ganatra, I. Kamal, J. Ware, K. Makarova et al., 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, H. M., J. M. Li, D. N. Robson and E. Wieschaus, 2006. Somatic stem cell niche tropism in Wolbachia. Nature 441 509–512. [DOI] [PubMed] [Google Scholar]
- Fujii, Y., D. Kageyama, S. Hoshizaki, H. Ishikawa and T. Sasaki, 2001. Transfection of Wolbachia in Lepidoptera: the feminizer of the adzuki bean borer Ostrinia scapulalis causes male killing in the Mediterranean flour moth Ephestia kuehniella. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268 855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, Y., T. Kubo, H. Ishikawa and T. Sasaki, 2004. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 317 1183–1188. [DOI] [PubMed] [Google Scholar]
- Grenier, S., B. Pintureau, A. Heddi, F. Lassabliere, C. Jager et al., 1998. Successful horizontal transfer of Wolbachia symbionts between Trichogramma wasps. Proc. R. Soc. Lond. Ser. B Biol. Sci. 265 1441–1445. [Google Scholar]
- Gronke, S., D. F. Clarke, S. Broughton, T. D. Andrews and L. Partridge, 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6 e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe, W., and A. A. Hoffmann, 2004. Wolbachia effects in Drosophila melanogaster: in search of fitness benefits. J. Invertebr. Pathol. 87 45–50. [DOI] [PubMed] [Google Scholar]
- Heddi, A., A.-M. Grenier, C. Khatchadourian, H. Charles and P. Nardon, 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 96 6814–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, L. M., J. C. Brownlie, S. L. O'Neill and K. N. Johnson, 2008. Wolbachia and virus protection in insects. Science 322 702. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow and J. H. Werren, 2008. How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiol. Lett. 281 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerauf, A., K. Nissen-Pahle, C. Schmetz, K. Henkle-Duhrsen, M. L. Blaxter et al., 1999. Tetracycline therapy targets intracellular bacteria in the filarial nematode Litomosoides sigmodontis and results in filarial infertility. J. Clin. Invest. 103 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerauf, A., L. Volkmann, K. Nissen-Paehle, C. Schmetz, I. Autenrieth et al., 2000. Targeting of Wolbachia endobacteria in Litomosoides sigmodontis: comparison of tetracyclines with chloramphenicol, macrolides and ciprofloxacin. Trop. Med. Int. Health 5 275–279. [PubMed] [Google Scholar]
- Hoffmann, A. A., and M. Turelli, 1988. Unidirectional incompatibility in Drosophila-simulans - inheritance, geographic-variation and fitness effects. Genetics 119 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A., M. Turelli and G. M. Simmons, 1986. Unidirectional incompatibility between populations of Drosophila-simulans. Evolution 40 692–701. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A., M. Turelli and L. G. Harshman, 1990. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila-simulans. Genetics 126 933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A., M. Hercus and H. Dagher, 1998. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, T., R. Koga, Y. Kikuchi, X. Y. Meng and T. Fukatsu, 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 107 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe, I., G. R. Burke, M. Riegler and S. L. O'Neill, 2005. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 187 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J., 2007. Spontaneous emergence of a new Wolbachia phenotype. Evolution 61 2244–2252. [DOI] [PubMed] [Google Scholar]
- Karr, T. L., W. Yang and M. E. Feder, 1998. Overcoming cytoplasmic incompatibility in Drosophila. Proc. R. Soc. Lond. Ser. B Biol. Sci. 265 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, B. N., and S. R. Bordenstein, 2010. Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson, L., T. Walker, M. Sebaihia, M. J. Sanders, M. A. Quail et al., 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson, L., J. Westberg, P. Sapountzis, K. Nasiund, Y. Lutnaes et al., 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. USA 106 5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukou, K., H. Pavlikaki, G. Kilias, J. H. Werren, K. Bourtzis et al., 2006. Influence of antibiotic treatment and Wolbachia curing on sexual isolation among Drosophila melanogaster cage populations. Evolution 60 87–96. [PubMed] [Google Scholar]
- Lo, N., M. Casiraghi, E. Salati, C. Bazzocchi and C. Bandi, 2002. How many Wolbachia supergroups exist? Mol. Biol. Evol. 19 341–346. [DOI] [PubMed] [Google Scholar]
- Masui, S., S. Kamoda, T. Sasaki and H. Ishikawa, 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51 491–497. [DOI] [PubMed] [Google Scholar]
- McGraw, E. A., D. J. Merritt, J. N. Droller and S. L. O'Neill, 2002. Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. USA 99 2918–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K. T., and S. Benzer, 1997. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 94 10792–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, J., A. Bertin, Y. Caubet and T. Rigaud, 2001. Sexual selection in an isopod with Wolbachia-induced sex reversal: males prefer real females. J. Evol. Biol. 14 388–394. [Google Scholar]
- Nigro, L., 1991. The effect of heteroplasmy on cytoplasmic incompatibility in transplasmic lines of Drosophila simulans showing a complete replacement of the mitochondrial DNA. Heredity 66 41–45. [DOI] [PubMed] [Google Scholar]
- Noda, H., Y. Koizumi, Q. Zhang and K. J. Deng, 2001. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem. Mol. Biol. 31 727–737. [DOI] [PubMed] [Google Scholar]
- Oliveira, D. C., R. Raychoudhury, D. V. Lavrov and J. H. Werren, 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (hymenoptera: pteromalidae). Mol. Biol. Evol. 25 2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen, E. A., P. J. Hooper, M. Bradley and G. Biswas, 2008. The global programme to eliminate lymphatic filariasis: health impact after 8 years. PLoS Negl. Trop. Dis. 2 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannebakker, B. A., B. Loppin, C. P. Elemans, L. Humblot and F. Vavre, 2007. Parasitic inhibition of cell death facilitates symbiosis. Proc. Natl. Acad. Sci. USA 104 213–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Minnot, M. J., and J. H. Werren, 1999. Wolbachia infection and incompatibility dynamics in experimental selection lines. J. Evol. Biol. 12 272–282. [Google Scholar]
- Perrot-Minnot, M. J., L. R. Guo and J. H. Werren, 1996. Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripennis: effects on compatibility. Genetics 143 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr, K. M., U. Heider, C. Schmetz, D. W. Buttner and A. Hoerauf, 2008. The mitochondrial heat shock protein 60 (HSP60) is up-regulated in Onchocerca volvulus after the depletion of Wolbachia. Parasitology 135 529–538. [DOI] [PubMed] [Google Scholar]
- Poinsot, D., K. Bourtzis, G. Markakis, C. Savakis and H. Mercot, 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhury, R., L. Baldo, D. C. Oliveira and J. H. Werren, 2009. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63 165–183. [DOI] [PubMed] [Google Scholar]
- Reynolds, K. T., and A. A. Hoffmann, 2002. Male age, host effects and the weak expression or nonexpression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80 79–87. [DOI] [PubMed] [Google Scholar]
- Riegler, M., S. Charlat, C. Stauffer and H. Mercot, 2004. Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: investigating the outcomes of host-symbiont coevolution. Appl. Environ. Microbiol. 70 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli, M. G., R. Giordano and G. Callaini, 2007. Effects of Wolbachia on sperm maturation and architecture in Drosophila simulans riverside. Mech. Dev. 124 699–714. [DOI] [PubMed] [Google Scholar]
- Ruang-areerate, T., and P. Kittayapong, 2006. Wolbachia transinfection in Aedes aegypti: a potential gene driver of dengue vectors. Proc. Natl. Acad. Sci. USA 103 12534–12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Andre, A. V., N. M. Blackwell, L. R. Hall, A. Hoerauf, N. W. Brattig et al., 2002. The role of endosymbiotic Wolbachia bacteria in the pathogenesis of river blindness. Science 295 1892–1895. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., and H. Ishikawa, 2000. Transinfection of Wolbachia in the mediterranean flour moth, Ephestia kuehniella, by embryonic microinjection. Heredity 85(Pt 2): 130–135. [DOI] [PubMed] [Google Scholar]
- Serbus, L. R., C. Casper-Lindley, F. Landmann and W. Sullivan, 2008. The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42 683–707. [DOI] [PubMed] [Google Scholar]
- Sinkins, S. P., H. R. Braig and S. L. Oneill, 1995. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc. R. Soc. Lond. Ser. B Biol. Sci. 261 325–330. [DOI] [PubMed] [Google Scholar]
- Sinkins, S. P., T. Walker, A. R. Lynd, A. R. Steven, B. L. Makepeace et al., 2005. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 436 257–260. [DOI] [PubMed] [Google Scholar]
- Smith, H. L., and T. V. Rajan, 2000. Tetracycline inhibits development of the infective-stage larvae of filarial nematodes in vitro. Exp. Parasitol. 95 265–270. [DOI] [PubMed] [Google Scholar]
- Stouthamer, R., and R. F. Luck, 1993. Influence of microbe-associated parthenogenesis on the fecundity of Trichogramma deion and T. pretiosum. Entomol. Exp. Appl. 67 183–192. [Google Scholar]
- Stouthamer, R., J. A. J. Breeuwer and G. D. D. Hurst, 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53 71–102. [DOI] [PubMed] [Google Scholar]
- Taylor, M. J., H. F. Cross and K. Bilo, 2000. Inflammatory responses induced by the filarial nematode Brugia malayi are mediated by lipopolysaccharide-like activity from endosymbiotic Wolbachia bacteria. J. Exp. Med. 191 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M. J., C. Bandi and A. Hoerauf, 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60 245–284. [DOI] [PubMed] [Google Scholar]
- Teixeira, L., A. Ferreira and M. Ashburner, 2008. The bacterial symbiont Wolbachia induces resistance to RNA Viral Infections in Drosophila melanogaster. PLoS Biol. 6 2753–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram, U., and W. Sullivan, 2002. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296 1124–1126. [DOI] [PubMed] [Google Scholar]
- Vala, F., M. Egas, J. A. J. Breeuwer and M. W. Sabelis, 2004. Wolbachia affects oviposition and mating behaviour of its spider mite host. J. Evol. Biol. 17 692–700. [DOI] [PubMed] [Google Scholar]
- Veneti, Z., M. E. Clark, S. Zabalou, T. L. Karr, C. Savakis et al., 2003. Cytoplasmic incompatibility and sperm cyst infection in different Drosophila Wolbachia associations. Genetics 164 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H., and D. W. Loehlin, 2009. The parasitoid wasp Nasonia: an emerging model system with haploid male genetics. Cold Spring Harb. Protoc. 2009: pdb emo134. [DOI] [PMC free article] [PubMed]
- Werren, J. H., W. Zhang and L. R. Guo, 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261 55–63. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., L. Baldo and M. E. Clark, 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6 741–751. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., S. Richards, C. A. Desjardins, O. Niehuis, J. Gadau et al., 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D. J., J. N. Wolff, M. Pierson and N. J. Gemmell, 2008. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 17 4925–4942. [DOI] [PubMed] [Google Scholar]
- Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy et al., 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalou, S., A. Apostolaki, S. Pattas, Z. Veneti, C. Paraskevopoulos et al., 2008. Multiple rescue factors within a Wolbachia strain. Genetics 178 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]