Abstract
The control of cellular proliferation is key in the proper development of a complex organism, the maintenance of tissue homeostasis, and the ability to respond to various hormonal and other inducers. Key in the control of proliferation is the retinoblastoma (Rb) protein which regulates the activity of a family of transcription factors known as E2Fs. The E2F proteins are now recognized to regulate the expression of a large number of genes associated with cell proliferation including genes encoding DNA replication as well as mitotic activities. What has also become clear over the past several years is the intimate relationship between the control of cell proliferation and the control of cell fate, particularly the activation of apoptotic pathways. Central in this connection is the Rb/E2F pathway that not only provides the primary signals for proliferation but at the same time, connects with the p53-dependent apoptotic pathway. This review addresses this inter-connection and the molecular mechanisms that control the decision between proliferation and cell death.
Complex roles for the E2F1 transcription factor
The E2F family of transcription factors regulates cell cycle progression in part through the activation of genes important for G1 to S phase cell cycle transition.1-3 Ectopic expression of E2F proteins induce quiescent cells to enter the cell cycle, consistent with their ability to promote expression of genes required for proliferation.4 E2F regulated genes include cyclins, cyclin-dependent kinases, components of the pre-replication complex such as Mcms and Orc6, and DNA synthesis genes like DNA polymerase, topoisomerases, dihydrofolate reductase (DHFR) and thymidylate synthase (TS).
Recent work has identified the E2F1 protein as a key component of this process of gene control, and likely the key E2F activity for triggering the transition from quiescence to a proliferative state. Indeed, inhibition of E2F1 activity blocks cell cycle entry but does not affect continuing cell cycles.5, 6 Moreover, E2F1, together with Myc, has been shown to be necessary for activation of the E2F program following the stimulation of cell growth.7
The activity of E2F1 is regulated at several levels. First, E2F1, which is absent in quiescent cells, is transcriptionally induced in late G1 following mitogenic stimulation.8, 9 Second, the activity of E2F proteins can be negatively regulated in part through the association of “pocket proteins” like pRb, p107 and p130. These interactions down-regulate E2F transcription by directly blocking the transactivation domain and also by recruiting histone deacetylase complexes to modify chromatin and actively repress transcription.10 Cyclin/Cdk phosphorylation of pocket proteins following mitogenic stimulation reduces their E2F binding affinity, releasing “free” E2F to transcribe proliferative target genes.11 CDK4, cyclin D, and p16, a cyclin/CDK inhibitor, are frequently mutated or deregulated in human cancer, and these mutations have the effect of hyperphosphorylating pRb.12 Tumor promoting viruses like SV40, adeno- and papillomavirus, express proteins capable of promoting E2F dependent proliferation through binding and disruption of Rb inhibition of E2F function.13 Furthermore, mutant alleles of Rb isolated from various tumors are unable to bind E2F, suggesting an important role of this interaction in suppressing tumorigenesis.14 E2Fs clearly are key regulators of cell cycle progression during normal growth stimulation and also in human cancer where function of the RB gene, or upstream regulatory components like Cyclin/Cdks and Cdk inhibitors, has been lost through mutation.
Paradoxically, E2F1 also has the capacity to induce apoptosis, in part through induction of and cooperation with the pro-apoptotic p53 tumor suppressor.15 Although this activity has been associated with other activator E2Fs (E2F1, E2F2, E2F3), it would appear that E2F1 is the primary component involved in this process and in some circumstances, may mediate the apoptotic effect of the other E2Fs.16-18
Function of E2F1
DNA microarray analysis has identified a diverse group of target genes, including genes encoding proliferative activities as well as putative apoptosis genes.19-22 E2F1 induces apoptosis in part through the transcription of proapoptotic genes such as ARF, p73, APAF1, Casp3, Casp7, Bnip3, Bok, Bim, PUMA, Noxa, and Hrk/DP5 and by down-regulating expression of the anti-apoptotic gene Mcl1.23-33 Interestingly, E2F regulates the expression of Gab2, a positive regulator of the Akt signaling pathway, indicative of a negative feedback loop that must be overcome to induce apoptosis.34 E2F also induces important DNA damage genes like Atm, Nbs1 and Chk2, and these genes appear to be an essential link between E2F deregulation and induced expression and phosphorylation of the p53 tumor suppressor protein.35, 36
Controlling the balance between proliferation and apoptosis
Apoptosis caused by ectopic expression of E2F1 in cultured cells can be inhibited by growth factors.37 This suggested that growth factors stimulate intracellular signaling pathways that counteract the pro-apoptotic function of E2F proteins, in effect raising the threshold required to trigger apoptosis. E2F also induces apoptosis in Drosophila and can be suppressed in vivo through activation of the EGFR/Ras/Raf pathway.38 Using cultured fibroblasts, we have shown that activation of the PI3K but not the MEK pathway is required for growth factor inhibition of E2F1-dependent apoptosis. Likewise, expression of a constitutively active form of Akt inhibited E2F1-directed p53 accumulation and apoptosis induction.17 The PI3K and Akt pathway also inhibits Myc induced apoptosis, although it is unclear whether this might directly affect Myc activity.39 These observations suggest that cells possess an internal “apoptotic threshold” based on the opposing effects of E2F1 or other oncogenes and serum activated survival pathways involving PI3K. In this model, imbalances in E2F level or function can be counteracted by activity of PI3K or Akt. Akt can inhibit apoptosis by directly phosphorylating and inactivating positive regulators of apoptosis like Bad and Caspase-9.40-42 Furthermore, Akt promotes p53 degradation by phosphorylating and activating Mdm2, providing a possible explanation as to how Akt can block E2F1 induction of p53 protein levels.43-45 Considering the large number of apoptotic targets post-translationally regulated by PI3K and Akt, we asked whether PI3K also directly influenced E2F1 transcriptional output to block expression of apoptosis genes during normal growth stimulation.
To determine if PI3K directly influences E2F1 transcriptional output, we utilized DNA microarrays to analyze expression of E2F targets in quiescent rat fibroblasts (REF52) infected with control or E2F1 expressing adenovirus in the presence or absence of 10% serum or LY294002, a PI3K inhibitor. Examination of the genes remaining highly expressed following serum addition revealed significant overrepresentation of cell cycle, DNA synthesis, replication and mitotic genes. Genes repressed by the PI3K pathway, however, appear to be heterogeneous in function, without convincing overrepresentation of any particular functional gene annotation (GO) category. What is true is that the promoters of this PI3K regulated group of genes are very significantly enriched for E2F binding elements.46
We predicted that these serum repressed, E2F1-induced gene products may play a role in apoptosis, which would be consistent with their inhibited expression during normal growth. To test this, we targeted seven of these genes for shRNA degradation and examined their requirement for mediating E2F1 induced apoptosis. Depletion of five of these genes rendered cells resistant to apoptosis induction by E2F1. AMP-activated protein kinase, AMPKα2, a key regulator of cellular energy homeostasis in response to nutrient and ATP depletion, is induced by E2F1 only during serum deprivation and is required for full apoptosis induction. Interestingly, AMPKα2 can directly phosphorylate p53 to regulate growth arrest and may serve as a link between E2F deregulation and p53 induction under conditions of bioenergetic stress.47 Cyp26b1 is a cytochrome p450 enzyme involved in all-trans-retinoic acid metabolism and can regulate germ cell fate outcomes.48 Nr4a3 is an orphan steroid nuclear receptor required for apoptosis induction during T-cell negative selection and is also a direct target of Akt phosphorylation.49 Co-deletion of Nr4a3 and the highly homologous family member Nr4a1 promotes the development of acute myeloid leukemia in knockout mice consistent with a tumor suppressive function.50 Mllt3 is a transcriptional regulator involved in development and cell fate outcomes.51 RNA-binding protein with multiple splicing sites (RBPMS), also called HERMES, is a poorly characterized gene encoding an RNA-binding protein that can also influence germ cell fate determination.52 It is clear why E2Fs must activate such a large number of proliferative target genes, because these products carry out diverse functions in discrete locations like origins of replication, on chromosomes during mitosis and in nucleotide biosynthesis. It is less clear why E2F1 transactivates such a large and diverse number of apoptosis related genes when seemingly many fewer could suffice. It seems likely that the plethora of apoptotic genes may mediate fine-tuned regulation of E2F apoptosis in distinct cell types and developmental stages.
Implications for specificity of E2F1 transcriptional control
E2F1 specificity appears to arise from a centrally located domain called the “marked box”, which when used to replace the homologous region of E2F3 allows the resulting E2F chimera to induce the same target genes and cause apoptosis like E2F1.17 The proapoptotic function of the E2F1 marked box is facilitated in part through the binding of Jab1/Csn5, which promotes E2F1 dependent apoptosis but not proliferation.53
The concept that cell proliferation signaling events are coupled with apoptotic processes was first postulated for the transcription factor c-Myc. Like E2F1, Myc can promote cell proliferation but can also trigger apoptotic pathways.54 Thus, normal proliferation must involve a balance in these events, promoting proliferation but at the same time suppressing the apoptotic events. Various lines of study point to the PI3K pathway as critical for this suppression providing a so-called “survival function” .17, 39 The work we detail here now provides evidence for a dual role of E2F1 through the activation of distinct target genes. While we do not yet understand the mechanistic basis for their divergent transcription control, we believe other past work on mechanism of E2F specificity is relevant. In particular, this work has described a role for combinatorial transcription control involving interaction of E2Fs with other transcription factors as critical for promoter specificity. This involves a capacity of the E2F protein to physically interact with the partner, dependent on the marked box domain of the E2F protein. Importantly, the E2F1 marked box domain is sufficient to confer apoptotic activity to E2F3 protein which otherwise would not induce apoptosis.17, 55 As such it appears that the capacity to induce the apoptotic program is dependent on the combinatorial transcription factor specific mechanism.
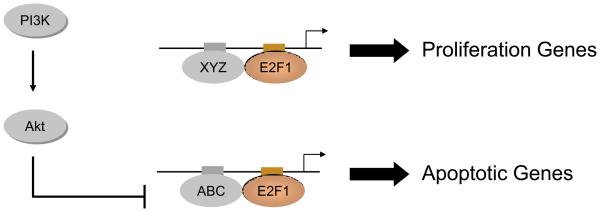
Given this, we suggest a model to account for the E2F1 specificity observed in this work. As depicted in Figure 2, E2F1 target promoters would be regulated by one of two E2F1-containing combinatorial complexes, consistent with previous work that defines a critical role for cooperative protein interactions in mediating E2F function.56, 57 In this model, specificity of E2F1 transcription control is dictated by the nature of the promoter complex reflecting the presence of unique binding sites for the cooperative proteins. Thus, in this example, genes with promoters containing a binding site for factor “XYZ” together with an E2F site might encode the proliferation genes while the apoptotic genes would have promoters with an “ABC” site. If the “ABC” factor were a substrate for the Akt kinase, and if phosphorylation of “ABC” inhibited the capacity of the protein to bind to DNA or to cooperate with E2F1, then it would be possible to selectively inhibit the E2F1 transcriptional program, blocking the expression of the apoptotic genes but not the proliferation genes. While this is speculation at this stage, it does represent a testable model for further investigation.
Figure 2.
A molecular mechanism to explain the transcriptional specificity underlying the dual role of E2F1 in proliferation and apoptosis. A potential mechanism whereby PI3K activity blocks the E2F1 apoptotic program but not the proliferative program could reflect a mechanism of E2F1 transcriptional specificity that involves a role for cooperative transcription factors in the formation of a stable and functional promoter complex. In this model, the proliferation genes are regulated by one E2F1 complex whereas the apoptotic genes are regulated by a distinct E2F1 complex. Specificity could be achieved if phosphorylation of the ‘ABC’ partner affected its capacity to interact with E2F1 or to form a functional complex.
Implications for oncogenesis
These results point to a role for PI3K activity in regulating the balance of E2F1 proliferative and apoptotic transcriptional output. In light of the frequent deregulation of both the Rb/E2F and the Pten/PI3K/Akt pathways and the universal suppression of apoptosis in human cancer, we examined the extent to which PI3K repressed E2F1 apoptotic genes displayed aberrant expression in human cancer datasets. Hierarchical clustering of the E2F apoptotic and proliferative target genes from two breast and an ovarian tumor datasets indicated that each of these tumor types can be subdivided into two major groups based on their expression profile. One group displayed high levels of E2F1 proapoptotic target gene expression with modest expression of the E2F1 proliferative targets. In contrast, the second group exhibited an inverse pattern, poorly expressing the apoptotic gene products while robustly expressing proliferative targets. These distinct tumor populations also reflected activation of the PI3K pathway, using an expression signature developed to predict the pathway.58 The tumor samples with high predicted PI3K activation corresponded to the same tumors with poor expression of the E2F1 apoptotic targets. And conversely, tumors with low predicted PI3K activity generally display high expression of the E2F1 pro-apoptotic target genes.
The patients displaying low-level expression of the PI3K repressed genes had a worse survival rate, higher rate of recurrence and had generally progressed to late stage tumors. In contrast, the patients expressing this set of E2F apoptotic genes generally survived longer and had fewer advanced tumors. Mutations in Her2, PTEN and PI3K occur with high frequency in breast cancer, and generally these mutations are mutually exclusive indicative of largely overlapping function. Our prediction of PI3K pathway activation indicates a near uniform activation of PI3K in advanced stage tumors (stage 3 and 4), and this will be interesting to determine empirically. These observations suggest potential therapeutic opportunities. For example, it is possible that pharmacologic inhibition of PI3K in tumors with an activated PI3K and E2F pathways will restore expression of the E2F1 pro-apoptotic genes and potentially suppress tumorigenesis by restoring apoptosis. Some of the PI3K repressed targets may represent additional therapeutic targets. For example, AMPKα2, one of the PI3K repressed target genes and a regulator of cellular energy homeostasis during energy deprivation, can further enhance apoptosis induction when stimulated with the AMP analog AICAR. Metformin, a commonly prescribed therapy for type 2 diabetes, also activates AMPK.59 Interestingly, type 2 diabetes patients treated with Metformin display lower cancer-related mortality than patients treated with sulfonylurea substitutes.60 This indicates a potential for combinatorial treatment with an inhibitor of PI3K to derepress AMPKα2 along with an activator of AMPK, like Metformin, to further enhance apoptosis. To the extent that a residual E2F1 apoptotic program remains intact in a tumor, however, targeting this pathway may elicit a new effectiveness in tumor specific therapy.
Figure 1.
A balance in cell signaling networks that couples proliferation control with the control of apoptosis. While the activation of E2F1 leads to the activation of genes responsible for DNA replication and execution of S phase, it also triggers a p53 response and the potential for apoptosis. In this model, the growth-dependent activation of PI3K activation serves to block the apoptotic component of the E2F1 program.
References
- 1.van den Heuvel S, Dyson N. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–24. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 2.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–52. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 5.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes & Dev. 1998;12:2120–30. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong L-J, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–7. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- 7.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park C-H, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. MolCell. 2001;8:105–13. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 8.Slansky JE, Li Y, Kaelin WG, Farnham PJ. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. 1993;13:1610–8. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes & Dev. 1994;8:1514–25. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 10.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–73. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 11.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–61. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 13.O'Shea CC. DNA tumor viruses - the spies who lyse us. Curr Opin Genet Dev. 2005;15:18–26. doi: 10.1016/j.gde.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. MolCell. 2003;12:639–49. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Nat'l Acad Sci. 1994;91:3602–6. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–50. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallstrom TC, Nevins JR. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci USA. 2003;100:10848–53. doi: 10.1073/pnas.1831408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denchi EL, Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6:661–8. doi: 10.1038/sj.embor.7400452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 2001;15:267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in the control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–99. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–46. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 22.Ma Y, Croxton R, Moorer RL, Cress WD. Identification of novel E2F1-regulated genes by microarray. ArchBiochemBiophys. 2002;399:212–24. doi: 10.1006/abbi.2002.2761. [DOI] [PubMed] [Google Scholar]
- 23.Zhu JW, DeRyckere D, Li FX, Wan YY, DeGregori J. A role for E2F1 in the induction of ARF, p53, and apoptosis during thymic negative selection. 1999;10:829–38. [PubMed] [Google Scholar]
- 24.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–5. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 25.Irwin M, Martin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–8. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 26.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–4. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 27.Stiewe T, Putzer BM. Role of the p53 homologue p73 in E2F1-induced apoptosis. Nature Genetics. 2000;26:464–9. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 28.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nature Cell Biology. 2002;4:859–64. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 29.Tracy K, Dibling B, Spike B, Knabb J, Schumacker P, Macleod K. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez J, Glozak M, Ma Y, Cress W. Bok, Bcl-2-related Ovarian Killer, Is Cell Cycle-regulated and Sensitizes to Stress-induced Apoptosis. J Biol Chem. 2006;281:22729–35. doi: 10.1074/jbc.M604705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas SC, Liu DX, Greene LA. Bim is a direct target of a neuronal E2F-dependent apoptotic pathway. J Neurosci. 2005;25:8349–58. doi: 10.1523/JNEUROSCI.1570-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–34. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 33.Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–69. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- 34.Chaussepied M, Ginsberg D. Transcriptional regulation of AKT activation by E2F. Mol Cell. 2004;16:831–7. doi: 10.1016/j.molcel.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Rogoff HA, Pickering MT, Debatis ME, Jones S, Kowalik TF. E2F1 induces phosphorylation of p53 that is coincident with p53 accumulation and apoptosis. Mol Cell Biol. 2002;22:5308–18. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogoff HA, Pickering MT, Frame FM, Debatis ME, Sanchez Y, Jones S, et al. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol. 2004;24:2968–77. doi: 10.1128/MCB.24.7.2968-2977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–73. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon N, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26:7601–15. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kauffmann-Zeh A, Rodriquez-Viciana P, Elrich E, Gilbert C, Coffer P, Downward J, et al. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–8. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 40.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–41. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 41.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin 3 induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–9. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 42.Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, et al. Regulation of cell death protease caspase 9 by phosphorylation. Science. 1998;282:1318–21. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 43.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung M-C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. 2001;3:973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 45.Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–50. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 46.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–33. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 47.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–92. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 48.Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 49.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16:1865–75. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullican S, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–5. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 51.Pina C, May G, Soneji S, Hong D, Enver T. MLLT3 regulates early human erythroid and megakaryocytic cell fate. Cell Stem Cell. 2008;2:264–73. doi: 10.1016/j.stem.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Shimamoto A, Kitao S, Ichikawa K, Suzuki N, Yamabe Y, Imamura O, et al. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci U S A. 1996;93:10913–7. doi: 10.1073/pnas.93.20.10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallstrom TC, Nevins JR. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes & Dev. 2006;20:613–23. doi: 10.1101/gad.1345006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evan GL, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–28. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 55.Black EP, Hallstrom T, Dressman HK, West M, Nevins JR. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc Natl Acad Sci U S A. 2005;102:15948–53. doi: 10.1073/pnas.0504300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giangrande PH, Hallstrom TC, Tunyaplin C, Calame K, Nevins JR. Identification of the E box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol Cell Biol. 2003;23:3707–20. doi: 10.1128/MCB.23.11.3707-3720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–86. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 59.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–74. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowker SL, Majumdar SR, Veugelers P, Johnson JA. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin. Diabetes Care. 2006;29:254–8. doi: 10.2337/diacare.29.02.06.dc05-1558. [DOI] [PubMed] [Google Scholar]