Abstract
The vitelline membrane (VM), the oocyte proximal layer of the Drosophila eggshell, contains four major proteins (VMPs) that possess a highly conserved “VM domain” which includes three precisely spaced, evolutionarily conserved, cysteines (CX7CX8C). Focusing on sV23, this study showed that the three cysteines are not functionally equivalent. While substitution mutations at the first (C123S) or third (C140S) cysteines were tolerated, females with a substitution at the second position (C131S) were sterile. Fractionation studies showed sV23 incorporates into a large disulfide linked network well after its secretion ceases, suggesting post-depositional mechanisms are in place to restrict disulfide bond formation until late oogenesis, when the oocyte no longer experiences large volume increases. Affinity chromatography utilizing histidine tagged sV23 alleles revealed small sV23 disulfide linked complexes during the early stages of eggshell formation that included other VMPs, namely sV17 and Vml. The early presence but late loss of these associations in an sV23 double cysteine mutant suggests reorganization of disulfide bonds may underlie the regulated growth of disulfide-linked networks in the vitelline membrane. Found within the context of a putative thioredoxin active site (CXXS) C131, the critical cysteine in sV23, may play an important enzymatic role in isomerizing intermolecular disulfide bonds during eggshell assembly.
Keywords: Drosophila, eggshell assembly, vitelline membrane, extracellular disulfides, VM domain, sV23 eggshell protein, cysteine mutants
Introduction
The multi-layered Drosophila eggshell provides an excellent experimental system for studying the assembly of an extracellular matrix in vivo. The eggshell not only provides a protective function, but the vitelline membrane, the oocyte proximal layer, also appears to play an important role in localizing embryonic patterning cues. Torso-like, a protein involved in terminal patterning is localized in the vitelline membrane at the two poles of the egg (Stevens et al., 2003), and several vitelline membrane proteins appear to be substrates of the Drosophila dorsoventral determinant Pipe (Zhang et al., 2009). Spatial regulation of the serine protease cascade that controls dorsal-ventral patterning is dependent upon the Pipe sulfotransferase (Sen et al., 1998). The exacerbation of dorsal/ventral patterning defects by the loss of the vitelline membrane protein Vml in a sensitized pipe genetic background suggests vitelline membrane proteins influence embryonic dorsal-ventral polarity (Zhang et al., 2009). How sulfonated vitelline membrane proteins exert their influence is not known. Vitelline membrane structural components as well as gene products needed for its structural integrity (eg. Nudel, etc) have been identified in numerous studies (Cernilogar et al., 2001; LeMosy and Hashimoto, 2000; Waring, 2000 ). The significance of the components and how they are assembled into a functional unit are not known.
Mutants in which VM assembly is disrupted fall amongst the larger class of female sterile (fs) mutants. Only two VM genes have been identified through fs mutant screens, dec-1 (Bauer and Waring, 1987; Gans et al., 1975) and VM26Ab (Savant and Waring, 1989). In both cases females with null alleles lay flaccid, unfertilized eggs. Surprisingly females homozygous for VmlEPgy2, a Vml null allele, produce eggs that can be fertilized and hatch (Zhang et al., 2009). This finding suggests functional redundancy amongst vitelline membrane proteins and may in part explain the failure to detect genes encoding other VM proteins in previous genetic screens.
Sv23, encoded by VM26Ab, is an abundant vitelline membrane protein that is essential for female fertility. Secreted as a proprotein, sV23 undergoes two post-depositional cleavages (Manogaran and Waring, 2004). During late stage 10 a small C-terminal region is removed; during the later stages of oogenesis a hydrophobic N-terminal prodomain is removed yielding a mature protein consisting of repeats of the octapeptide YSAPAAPS and the signature VM domain, a thirty eight amino acid stretch of amino acids found in many vitelline membrane proteins (Figure 1). By introducing mutated versions of sV23 transgenes into sV23 null females, previous studies have shown that whereas the C-terminal prodomain can be removed without functional consequences, both the hydrophobic N-terminal domain and the signature VM domain are essential (Manogaran and Waring, 2004).
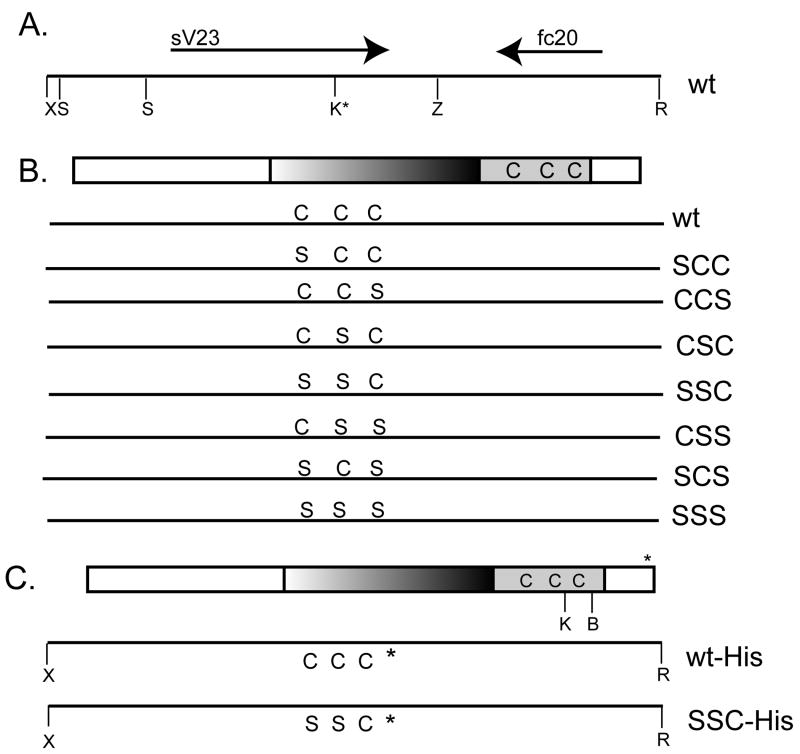
Figure 1.
(A) A wild type (wt) 3.6 kb genomic fragment containing the sV23 open reading frame (ORF-thick rightward arrow) and the fc 20 ORF (VM26Ac) (thick leftward arrow). Key restriction sites used in constructing the sV23 transgene and its mutated derivatives are shown: X (XhoI), S (SalI), Z (XbaI), R (EcoRI), and K* (KpnI engineered into the mutant transgenes). (B) Wild type and mutant sV23 transgenes. The 168 amino acid sV23 open reading frame is shown at the top. The central region consisting of five perfect and three degenerate copies of an octapeptide repeat (PAYSAPAA) and the 38 amino acid VM domain with its three precisely spaced cysteines at positions 123, 131, and 140 are highlighted. The lines below the sV23 ORF show the amino acids at positions 123, 131, and 140 in the context of the 3.6 kb X/R genomic fragment for the wild type allele and seven cysteine substitution mutant transgenes created in this study. The designations on the right are used to denote the single (SCC, CCS, CSC), double (SSC, CSS, SCS), and triple (SSS) sV23 substitution mutations. (C) Histidine tagged wild type and mutant (SSC-His) sV23 transgenes. The top line shows the sV23 ORF with a 6X- histidine tag (*) inserted between R166 and E167. The BamHI restriction site (B) used in creating the SSC-His transgene and the position of the engineered KpnI site (K) relative to the cysteine residues are denoted. The genomic XhoI/EcoRI fragments containing the histidine codons and SSC substitution mutations are shown below.
A hallmark of the VM domain is the presence of three precisely spaced cysteine residues (CX7CX8C). Vitelline membrane proteins with VM domains become incorporated into a large disulfide linked network during late oogenesis. Disulfide bonds are routinely used in extracellular matrices to stabilize networks formed via non-covalent interactions. During eggshell assembly both non-reducible and reducible crosslinks are used to stabilize proteins in the eggshell layers. Both types of cross-links stabilize the innermost VM layer while the outer endochorion layer relies strictly on non-reducible crosslinks (Margaritis, 1985). Peroxidase mediated tyrosine crosslinking occurs in the endochorion layers during stage 14, the terminal stage of oogenesis (Mindrinos et al., 1980). Peroxidase mediated crosslinking occurs post-ovulation in the vitelline membrane, hence disulfide crosslinking during oogenesis likely plays an important role in allowing the egg to resist the mechanical pressures incurred during its passage through the oviduct.
Vitelline membrane proteins are secreted during stages 9-10 in membrane bound vesicles that accumulate in the extracellular space as vitelline bodies. At the end of stage 10 microvilli that project from both the follicle cells and oocyte surfaces recede and vitelline bodies fuse into a continuous layer. As the oocyte continues to grow via nurse cell dumping and hydration, the VM layer thins from 1.7 to 0.4 u (Margaritis, 1985). To retain its elasticity, constraints on the number of intermolecular disulfide bonds that form before the oocyte approaches its final size are likely necessary. Balancing the need for early elasticity and late stabilization suggests regulation of disulfide bond formation in the extracellular environment may be critical for proper vitelline membrane morphogenesis. An emerging concept is that extracellular disulfide bonds need not be inert but rather can act as a dynamic scaffold to present mature proteins in different conformational states that can have significant impact on their function (Hogg, 2003). Within this context we investigated the consequences of mutating, singly and in combination, the three evolutionarily conserved cysteines within the VM domain of sV23 on vitelline membrane assembly and function.
Materials and Methods
Culture conditions and stocks
All stocks were maintained on standard yeast, cornmeal, molasses, and agar medium. The w*/w* and VM26AbQJ42 stocks have been described previously (Manogaran and Waring, 2004); the Df (2L)Exel7024 and VmlEPgy2 stocks are described in Flybase. Transformant lines carrying mutated sV23 transgenes created in this study are described below.
Construction of mutant sV23 transgenes
sV23 transgenes with base pair substitutions were created using a combination of PCR mutagenesis and exchanging selected DNA fragments. All of the sV23 transgenes in this study (Figure 1) consisted of the sV23 open reading frame, ~1.25 kb of 5’ flanking and 1.8 kb of 3’ flanking DNA subcloned into a pCaSpeR4 transformation vector (Manogaran and Waring, 2004). A 1 kb SalI-XbaI wild type genomic fragment containing the sV23 ORF (see Figure 1A) subcloned into a pSP73 plasmid vector was used as a template for inverse PCR reactions. Primers with nucleotide changes at designated positions were used to introduce a strategic KpnI site (silent change) in addition to codon changes that created either the CSS or SSS cysteine mutant transgenes. For sV23-SSC, a ~0.4 kb KpnI-XbaI fragment with C rather than S at the third position was amplified from the 3.6 kb XhoI-EcoRI sV23-CSS fragment, subcloned, and exchanged with its counterpart in an XhoI-XbaI sV23-SSS subclone. The resultant XhoI-XbaI SSC fragment was exchanged with its counterpart in the sV23-CSS pCaSpeR4 transformation vector.
Using the sV23-CSS transgene in pCaSpeR4 as template, a CC containing XhoI-KpnI fragment was amplified, subcloned, and exchanged with its counterpart in an XhoI-XbaI CSS subclone. The resultant XhoI-XbaI CCS fragment was then exchanged with its counterpart in the sV23-CSS pCaSpeR4 transgene, yielding the sV23-CCS transgene.
To create the sV23-SCC transgene, an XhoI-KpnI SC fragment was amplified from an XhoI-XbaI SSC template. The XhoI-KpnI SC fragment was exchanged with its counterpart in an XhoI/XbaI- SSC subclone to yield XhoI-XbaI-SCC. The XhoI/XbaI SCC fragment was exchanged with its counterpart in a delN24-42pCaSpeR4 construct (Manogaran and Waring, 2004) yielding the sV23-SCC transgene.
The sV23-CSC and SCS transgenes were created through fragment exchanges. An XhoI/XbaI CSC fragment was created by exchanging the KpnI-XbaI fragment from an SSC subclone with the KpnI-XbaI fragment from a CSS subclone. Exchanging the KpnI/XbaI fragment from a CSS subclone with the KpnI/XbaI fragment in an XhoI/XbaI SCC subclone created an XhoI/XbaI SCS fragment. Each XhoI/XbaI fragment was then exchanged with its counterpart in delN24-42pCaSpeR4 to yield the sV23-CSC and sV23-SCS transgenes, respectively.
Histidine tagged versions of both wild type sV23 and sV23-SSC were created by inserting 6 histidine codons between the codons specifying amino acids 166 (R) and 167(E) (Figure 1C). Using a wild type KpnI/XbaI subclone as template, a 200 bp 5’ fragment with 6 His codons at its 3’ end and a 400 bp 3’ fragment headed by 6 His codons were generated in two separate PCR reactions. The GAA glu codon following the His-tag was changed to a GAG glu codon in order to create a diagnostic SacII restriction site. After mixing the PCR products a 600 bp fragment was amplified and subcloned. The 0.4 kb His-tagged KpnI/XbaI fragment was excised and exchanged with its counterpart in an sV23-CCS XhoI/XbaI fragment, yielding a wild type XhoI/XbaI fragment bearing the engineered KpnI site and His-codons near the end of the sV23 ORF (see Figure 1C). For sV23-SSC-His, an XhoI/BamHI fragment from sV23-SSC was exchanged with its tagged wild type counterpart, yielding an sV23-SSC-His XhoI/XbaI fragment. Both tagged XhoI/XbaI fragments were introduced into the sV23-pCaSpeR4 transgene as previously described. The sV23 coding sequence of all of the constructs was verified by DNA sequencing. Additional details of the constructions are available upon request.
Recombinant pCaSpeR 4 plasmid DNAs were purified and injected along with a S129A helper plasmid (Beall et al., 2002) into w*/w* preblastoderm embryos. After establishing chromosomal linkage of the transgenes in the transformants, two copies of the each transgene were introduced into sV23 null females via a series of genetic crosses.
Egg laying and fertility tests
Well-fed 2-5 day females were placed in egg collection chambers as described previously (Spangenberg and Waring, 2007). Eggs from single females collected over a 24-hour period were counted and the number of larva that hatched from these eggs over the ensuing 48 hours was recorded.
Protein Analyses
Proteins soluble in Laemmli sample containing 5% beta-mercaptoethanol were separated by SDS-PAGE and transferred to nitrocellulose using a Bio-Rad Mini-Protean system. ECL Western blot signals were developed as previously described (Manogaron and Waring 2004). The sV23, sV17 (VM26Aa), and Cfc106 DEC-1 antisera used in this study have been described previously (Noguerón, 1996; Pascucci et al., 1996); rabbit anti-6-His-Antibody (1/5000 dilution) was obtained from Bethyl Laboratories. For silver staining, gels were processed as described (Zhou et al., 2003).
For fractionation studies, egg chambers were disrupted with a Kontes dounce homogenizer (B-type pestle) in Tris buffer based solutions (50 mM Tris, pH7.4, 150 mM NaCl) as indicated. Following centrifugation at 15,000 × g for 15 minutes pellet (P) and supernatant (S) fractions were recovered. Pellet fractions were resuspended in Laemmli sample buffer and 5% beta mercaptoethanol; supernatant fractions were adjusted to similar concentrations by the addition of an appropriate volume of a concentrated Laemmli solution. Prior to SDS-PAGE, samples were boiled for 3 minutes.
Affinity Chromatography
Stage 10 egg chambers were disrupted in lysis buffer (400 mM NaCl, 50 mM Tris, pH 7.4, 2% Triton X-100) with a dounce homogenizer. Following low speed centrifugation (1000 × g, 10 min.) the pellet was resuspended in lysis buffer with a dounce homogenizer, and centrifuged as above. After three such cycles, the pellet was resuspended in DNase I buffer (50 mM Tris, pH 7.5, 10 mM MgCl2) containing DNase I (20 ng/ul) and RNase A (20 ng/ul) and incubated at 37° for 30 min. An enriched eggshell pellet was obtained by centrifugation at 15,000 × g for 15 min. After rinsing in DNaseI buffer, the enriched eggshell pellet was resuspended in 90 ul of denaturing buffer (8M urea, 100 mM NaH2PO4, 10 mM Tris, adjusted to pH 8.0 by the addition of NaOH) containing 2% Triton X-100 and incubated for one hour at room temperature. Following high speed clarification (210,000 × g, 30 min) the supernatant was added to a 100 ul Protino® Ni-IDA 150 packed column (Machery-Nagel) pre-equilibrated with denaturing buffer containing 2% Triton X-100 at room temperature. After a 2 hr incubation period, unbound proteins were removed by gravity flow with 6 column volumes of denaturing buffer. Bound histidine-tagged proteins were eluted with denaturing buffer containing 250 mM imidazole. After applying 6 column volumes of elution buffer, 4 column volumes of denaturing buffer containing 2% SDS were added to remove residual proteins from the column.
Mass spectrometry
Elution fractions were analyzed at the Protein & Nucleic Acid Shared Facility HRC at the Medical College of Wisconsin. Briefly, samples were incorporated into a polyacrylamide gel matrix and digested with trypsin. The tryptic peptides recovered were analyzed by LTQ LC/MS mass spectrometry. Visualize software, version 1.13, designed by Brian D. Halligan (Medical College of Wisconsin) was used for data analyses.
RNA analyses
RNA was extracted from stage 10 egg chambers of Vm26AbQJ42 origin that included the transgenes indicated. Briefly, total RNA from ~100 egg chambers was extracted with 100 ul of TRIzol reagent and purified using the Trizol® Plus RNA purification system (Invitrogen). Approximately 1 ug of RNA was reverse transcribed with the Promega Reverse Transcription System and sV23 specific primers (sense 5’-ATGGCATTCAACTTTGGTCACCTC-3’ and antisense 5’-TCAGATCTCAAGTCGGATCCGTTTCGATCC-3’) were used to amplify a 537 bp sV23 PCR product. The conditions used for the PCR reactions were: 2’ at 95°, 30 cycles of 95° for 1’, 50° for 1’, 72° for 2’, followed by a final extension at 72° for 15’. PCR products were purified using Promega’s Wizard®SV Gel and PCR Clean Up System and digested with KpnI. The digestion products were separated on 2% agarose gels.
Results
Cys131 is critical for sV23 function
The evolutionary conservation of the number and spacing of cysteine residues within the VM domain suggests that these residues are critical for the integration/stabilization of vitelline membrane proteins within the eggshell. To address functional issues, we focused on cysteine residues within the VM domain of sV23 since previous studies established that sV23 is necessary for the production of turgid, fertile eggs. To determine the number and position of cysteines within the sV23 VM domain that are critical for its function, a series of cysteine to serine substitution mutant sV23 transgenes were created. Single serine substitutions at each cysteine residue (Vm26AbC123S (SCC), Vm26AbC131S (CSC), and Vm26AbC140S (CCS), all three combinations of double cysteine substitutions (SSC; SCS; and CSS), as well as the triple substitution mutant (SSS) were created (Figure 1B). To ensure that the mutant transgenes were the only source of sV23 protein, each mutant transgene was crossed into sV23 protein null females (either homozygous Vm26AbQJ42 or Vm26AbQJ42 / Df (2L)Exel7024). If the engineered mutation was tolerated, the sV23 transgene was expected to provide a functional source of sV23 and thus rescue the sterility of sV23 null mutant females. As shown in Table 1 only transgenes bearing single cysteine substitutions at either the first or third position were tolerated. The hatching rate of eggs derived from CCS females (50.5 and 52%) was comparable to the wild type control, CyO/ Vm26AbQJ42 (54%). The lower hatching rate of the SCC eggs (30.6%) may indicate a small functional distinction between the first and third cysteines or differences in their expression levels (see below). In marked contrast to the turgid eggs laid by the CCS and SCC females, most of the eggs laid by CSC females were either collapsed or only slightly turgid, and all failed to hatch. This indicates that the second cysteine within the sV23 VM domain plays a critical role in eggshell assembly. Consistent with this finding, all eggs laid by double cysteine mutants that included a substitution at the second position collapsed and failed to hatch. The severely compromised hatchability of SCS eggs (3.6%) suggests that although critical, a single cysteine at the second position in sV23 is not sufficient to support the assembly of a functional eggshell. The appearance of the SCS eggs was variable. Like the other cysteine double mutants, about half of the eggs collapsed; the remainder were slightly turgid, but clearly distinguishable from wild type eggs. As expected, eggs laid by females with an sV23 transgene bearing substitutions at all three cysteines (SSS) collapsed and failed to hatch.
Table 1.
Rescue of Vm26AbQJ42 female sterility by Vm26Ab transgenes
| Genotype | # of flies | Eggs laid | Larvae | Hatch rate | Egg morphology |
|---|---|---|---|---|---|
| CyO/fs | 4 | 223 | 129 | 54 | Turgid |
| Df/fs | 4 | 336 | 0 | 0 | Collapsed |
| Df/fs;ccs/ccs | 5 | 380 | 192 | 50.5 | Turgid |
| fs/fs;ccs/ccs | 4 | 193 | 101 | 52 | Turgid |
| Df/fs;scc/scc | 4 | 333 | 102 | 30.6 | Turgid |
| fs/fs;csc/csc | 3 | 139 | 0 | 0 | Collapsed |
| Df/fs;csc/csc | 4 | 205 | 0 | 0 | Collapsed |
| Df/fs;css/css | 4 | 143 | 0 | 0 | Collapsed |
| Df/fs;ssc/ssc | 4 | 155 | 0 | 0 | Collapsed |
| fs/fs;scs/scs | 7 | 111 | 4 | 3.6 | Intermediate |
| fs/fs;sss/sss | 12 | 183 | 0 | 0 | Collapsed |
Two copies of each mutant transgene were expressed in either homozygous Vm26AbQJ42 (fs/fs) or heteroallelic Vm26AbQJ42/ Df(2L)7024 (Df/fs) sV23 protein null mutant females. The number of females of each genotype analyzed is shown. Eggs laid within a 24 hr. period were counted and the number of larvae that hatched over the ensuing 48 hours was recorded. Egg morphology was assessed by light microscopy. The intermediate phenotype denotes a mixture of eggs within the population, with most being either collapsed or with a marked reduction in turgidity relative to wild type eggs.
The effects of mutations can be manifested at many different levels, hence establishing a definitive relationship between a mutated residue and the function of a protein can be problematic. Many mutations induce protein misfolding with consequent alterations in trafficking through the secretory pathway. By disrupting cysteine residues and their potential to form intra- or intermolecular disulfide bonds, the secretion of sV23 from the follicle cells may be compromised. To verify that the amount of sV23 secreted was sufficient to support the production of fertile eggs, sV23 accumulation was analyzed by Western blot analysis in each of the mutant transgenic lines. Figure 2 shows a dilution series in which the signal intensity of sV23 from egg chambers carrying different mutant transgenes was compared with wild type sV23. Synthesis and accumulation of sV23 occurs during stages 9 and 10 of oogenesis, a period encompassing approximately 20 hours. At the end of stage 10 sV23 begins to undergo the first of two post-depositional cleavages. To minimize differences in signal intensity due to loss of epitopes, yet monitor sV23 accumulation at near peak levels, late stage 10 egg chambers were chosen for the comparison. Egg chamber proteins were extracted with Laemmli sample buffer in the presence of a reducing agent and separated by SDS-PAGE. The Western blot signals from the dilution series shown in Figure 2 were quantified using Image J software. Although the fertility of VM26AbQJ42/ VM26AbQJ42; CCS/CCS females was comparable to wild type, sV23-CCS protein accumulated at ~33% the level of wild type sV23. Accumulation in the other fertile single substitution mutant, SCC, was approximately 25% of wild type. Importantly, in the sterile single substitution mutant sV23-CSC levels were at least 50% of wild type. Although accumulation of the sV23-CSC protein was elevated relative to the other substitution derivatives, the higher accumulation level of this mutant derivative per se does not appear to have an adverse effect since fertility was not compromised when two copies of this transgene were present in wild type flies (+/+ or VM26AbQJ42/CyO). Thus the sterility of the sV23-CSC mutant, despite sV23 accumulation levels commensurate with fertility, argues that the second cysteine plays a distinct and critical role in vitelline membrane assembly.
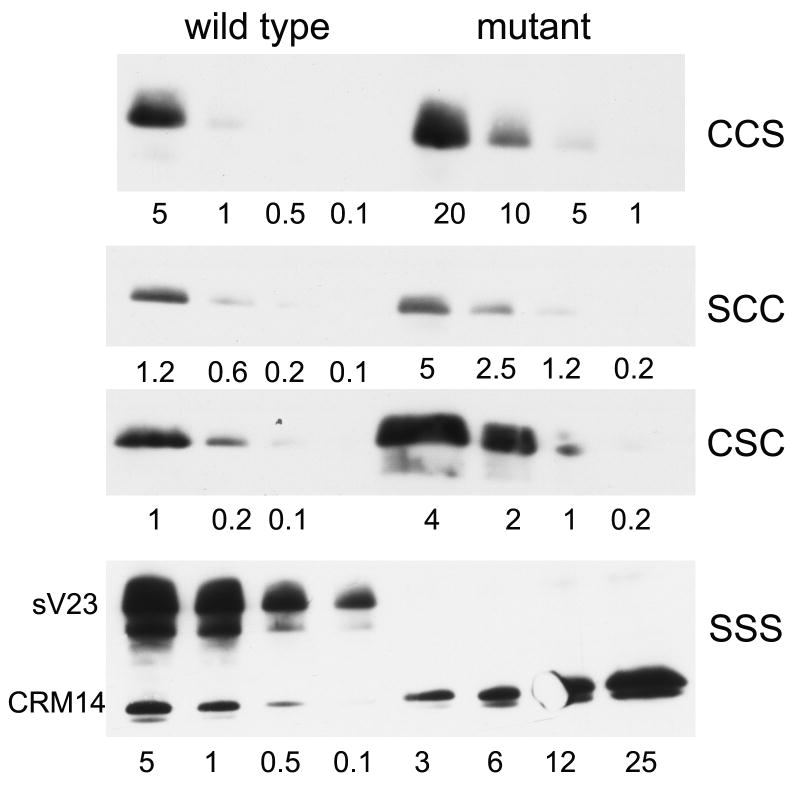
Figure 2.
Accumulation of sV23 in mutant and wild type stage 10 egg chambers. Late stage 10 egg chambers (5-25) were resuspended in Laemmli sample containing 5% beta mercaptoethanol and boiled to generate soluble protein extracts. Protein representing the number of egg chambers indicated below each lane was obtained by diluting the original extract appropriately. Each mutant series, designated on the right, was run in parallel with wild type egg chambers (left). Western blots were developed with sV23 antiserum. The sV23 signals are shown in the three upper panels. The SSS panel also includes CRM-14, a small vitelline membrane protein that is recognized by our sV23 antiserum. The exposure times varied from blot to blot.
At least one cysteine is required for accumulation of sV23 protein
Accumulation levels of sV23 in the double cysteine mutants fell in ranges compatible with fertility (20-30% - data not shown). sV23 accumulation in the triple substitution mutant, sV23-SSS, was severely compromised however. sV23-SSS was not detected in extracts from a range of 3 to 25 mutant egg chambers. In contrast, a vitelline membrane protein that cross reacts with our sV23 antiserum, CRM-14, was detected with signal intensities commensurate with the number of egg chambers used for the protein determination. This suggests that at least one cysteine is needed for the production of stable sV23 protein. To ensure that the lack of sV23-SSS protein did not reflect a deficiency at the RNA level, RT-PCR was used to compare the accumulation of sV23 RNA from the transgene with that from the endogenous VM26AbQJ42 locus (Figure 3). Total RNA from stage 10 egg chambers was reverse transcribed and a 537 bp sV23-specific fragment was amplified using the primers indicated. To distinguish the transgene product from the endogenous product we made use of the ectopic KpnI restriction site that was engineered into the mutant transgenes. After KpnI digestion, the 415 bp fragment derived from the transgene was easily resolved from the endogenous 537 bp product. Image J software was used to determine the relative ratios of the transgene and endogenous products. As shown in Figure 3 the intensity of the sV23-SSS fragment was about 1.6 times that of the endogenous product (accumulation of sV23 RNA from the VM26AbQJ42 mutant allele is much less than wild type, presumably due to the mutation in the AUG initiation codon (Fokta, 2000) that renders it untranslatable). In comparison, the intensity of the sV23-CCS fragment was about 0.8 that of the endogenous VM26AbQJ42 product. Thus sV23-SSS RNA accumulates at approximately twice the level of sV23-CCS RNA. This shows that the lack of sV23-SSS protein is not due to a deficiency at the RNA level and suggests that at least one cysteine in the sV23 VM domain is necessary for its accumulation.
Figure 3.
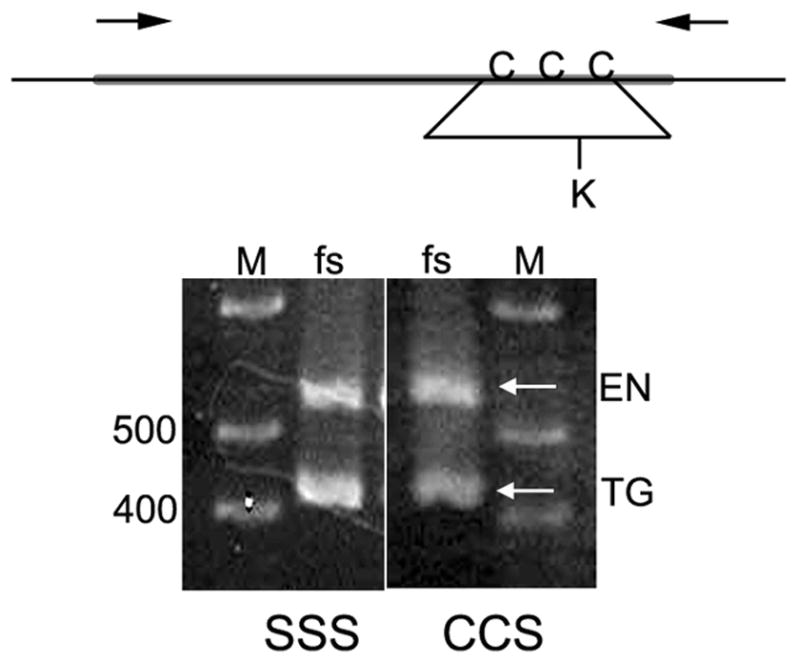
Relative accumulation of sV23 RNA from mutant transgenes and the endogenous Vm26AbQJ42 locus. The schematic shows the sV23 ORF (thick line) along with the 5’ and 3’ untranslated regions. The approximate positions of the cysteine residues within the VM domain and the primers used for amplification are indicated. The K below the line depicts an ectopic Kpn I restriction site that was engineered into the VM domains of all of the cysteine mutant transgenes. RNA from homozygous Vm26AbQJ42 mutant egg chambers carrying two copies of either the sV23-SSS or sV23-CCS transgene, was reversed transcribed, amplified, and digested with KpnI. The arrows indicate the positions of the endogenous (EN) sV23 gene products and the transgene products (TG) from either sV23-SSS or sV23-CCS. The marker lanes (M) show the positions of 400 and 500 bp fragments. The expected length of the full length PCR product was 537 bp; KpnI digestion of the transgene products yielded fragments 415 and 122 bp in length. Only the 415 bp product is shown in this figure.
The growth of disulfide-linked networks within the vitelline membrane is developmentally regulated
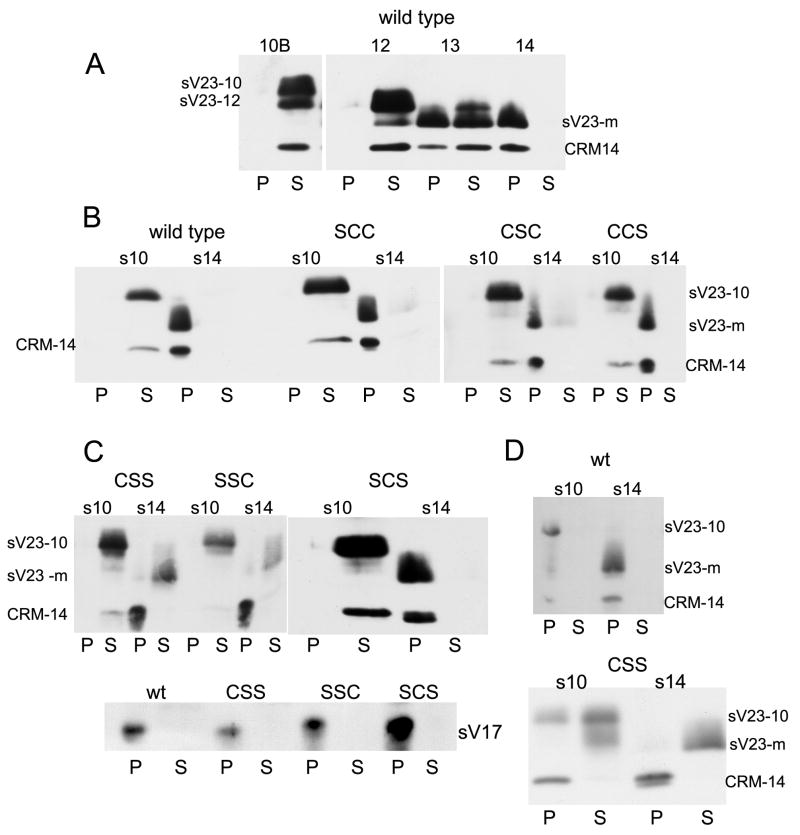
Prior to ovulation, the vitelline membrane is stabilized by the formation of intermolecular disulfide bonds. Although disulfide linked oligomers may form during the initial stages of vitelline membrane formation, based on pelleting behavior, dramatic post-depositional growth of the sV23 disulfide-linked network occurs between stages 10 and 14 (Figure 4A). To track temporal changes in the size of the sV23 disulfide linked network, egg chambers at different developmental stages were homogenized in buffered SDS solutions (1%) in the absence of a reducing agent. Following low speed centrifugation, proteins in the supernatant and pellet fractions were reduced and separated by SDS-PAGE. The Western blot in Figure 4A shows that sV23 and its derivatives, as well as CRM-14, were recovered only in the supernatant fraction during stages 10 and 12. In contrast, the mature proteolytic sV23 derivative (sV23-m) and CRM-14 were only in the pellet fraction in stage 14 egg chambers. Stage 13 appears to be a transitory period when vitelline membrane proteins begin to become part of a large disulfide-linked network(s).
Figure 4.
Fractionation of eggshell proteins. A. Wild type egg chambers at the stages of oogenesis indicated were homogenized in Tris buffered saline (TBS) containing 1% SDS, and separated into pellet (P) and post-15,000 × g supernatant (S) fractions. Western blots of P and S proteins incubated with sV23 antiserum. sV23-10 – sV23 proprotein; sV23-12 – derivative after cleavage of C-terminal amino acids; sV23-m –mature derivative after cleavage of N- and C-terminal amino acids; CRM-14 – VM protein migrating in the 14 kDa range that cross reacts with the sV23 antiserum. (B.) stage 10 or 14 egg chambers from wild type or VM26AbQJ42 females with two copies of sV23 transgenes harboring single cysteine substitution mutations (SCC, CSC, and CCS) fractionated as in A. (C) Fractionation of egg chambers as in (B) except mutant transgenes have double cysteine substitution mutations (CSS, SSC, SCS). The lower panel is an independent Western blot of fractionated stage 14 samples incubated with an sV17 (VM26Aa) antiserum. (D) Stage 10 or 14 egg chambers from wild type (wt) or sV23-CSS females homogenized in TBS and processed as in (A).
Cysteine residues in the sV23 proprotein are only found in the VM domain. To determine whether the incorporation of sV23 into a large disulfide linked network was impaired in any of the cysteine mutants, VM26AbQJ42/ VM26AbQJ42 egg chambers expressing each cysteine mutant transgene were fractionated as above. As shown in Figure 4B, the fractionation behavior of sV23 in all of the single cysteine substitution mutants from stage 10 and 14 egg chambers was similar to wild type. While incorporated into a large network, the sterility associated with the sV23-CSC transgene suggests that the organization of the network in this mutant is aberrant. Surprisingly, as shown in Figure 4C, despite having only a single cysteine residue, sV23-SCS integrates into a large disulfide linked network in stage 14 egg chambers. This was in marked contrast to sV23-CSS and sV23-SSC, which were recovered almost exclusively in the supernatant at stage 14. The behavioral differences between the latter transgene products and sV23-SCS confirm that disulfide-bonding status underlies the fractionation behavior of vitelline membrane proteins under denaturing conditions. Although sV23-CSS and sV23-SSC failed to pellet, other vitelline membrane proteins, CRM-14 and sV17 (Figure 4C), were recovered exclusively in the pellet fraction at stage 14. Taken together these results suggest that 1.) The second cysteine (C131) is sufficient for disulfide bond formation between sV23 and other vitelline membrane proteins and (2.) that other vitelline membrane proteins can incorporate into large disulfide based networks without sV23.
Although large disulfide linked networks within the vitelline membrane are not in place during early eggshell morphogenesis (stage 10), vitelline membrane proteins are integrated into molecular networks that pellet following low speed centrifugation. As shown in Figure 4D when wild type egg chambers were homogenized in buffered saline (TBS) without SDS and fractionated as above, sV23 and CRM-14 were recovered exclusively in the pellet at both stage 10 and 14. When VM26AbQJ42/ VM26AbQJ42 egg chambers with the sV23-CSS transgene were disrupted in TBS, sV23-CSS was recovered in the pellet and supernatant at stage 10 and almost exclusively in the supernatant at stage 14. Although partitioning between the pellet and supernatant fractions varied from experiment to experiment for stage 10 egg chambers, sV23-CSS was only found in the supernatant fraction in stage 14 egg chambers. Thus beyond its incorporation into disulfide-linked networks, integration of sV23-CSS into the eggshell in general is severely compromised, especially in late stage egg chambers.
Early sV23 disulfide-linked complexes include other vitelline membrane proteins
The integration of sV23-SCS into a large disulfide linked network indicates sV23 can form disulfide bridges with other vitelline membrane proteins. To begin to investigate the complexity of the sV23 disulfide-linked network, we created a histidine (His-) tagged version of sV23 in order to isolate sV23-containing complexes by nickel affinity chromatography. While the need for strong denaturants to extract vitelline membrane proteins precludes investigating non-covalent associations, proteins linked by disulfide bonds can be recovered under denaturing conditions.
Previous mutagenesis studies showed that small deletions were tolerated in the sV23 C-terminus, but not in the N-terminal prodomain. Reasoning that insertion of a 6X His-tag would be least detrimental at the C-terminus, we inserted histidine codons into a wild type sV23 gene between the codons specifying R166 and E167 (Figure 1C). To verify its function, the transgene was tested in VM26AbQJ42/ VM26AbQJ42 females. Accumulation data and fertility tests showed that VM26AbQJ42/ VM26AbQJ42 females carrying two copies of a sV23-His transgene produced sV23 at near wild type levels (~80%) and laid turgid eggs with fertility rates comparable to wild type (data not shown). By inserting the His-tag within the C-terminal prodomain, sV23 disulfide linked complex isolation was restricted to stages 8 through 10 since the C-terminal prodomain is removed via cleavage during late stage 10 and 11.
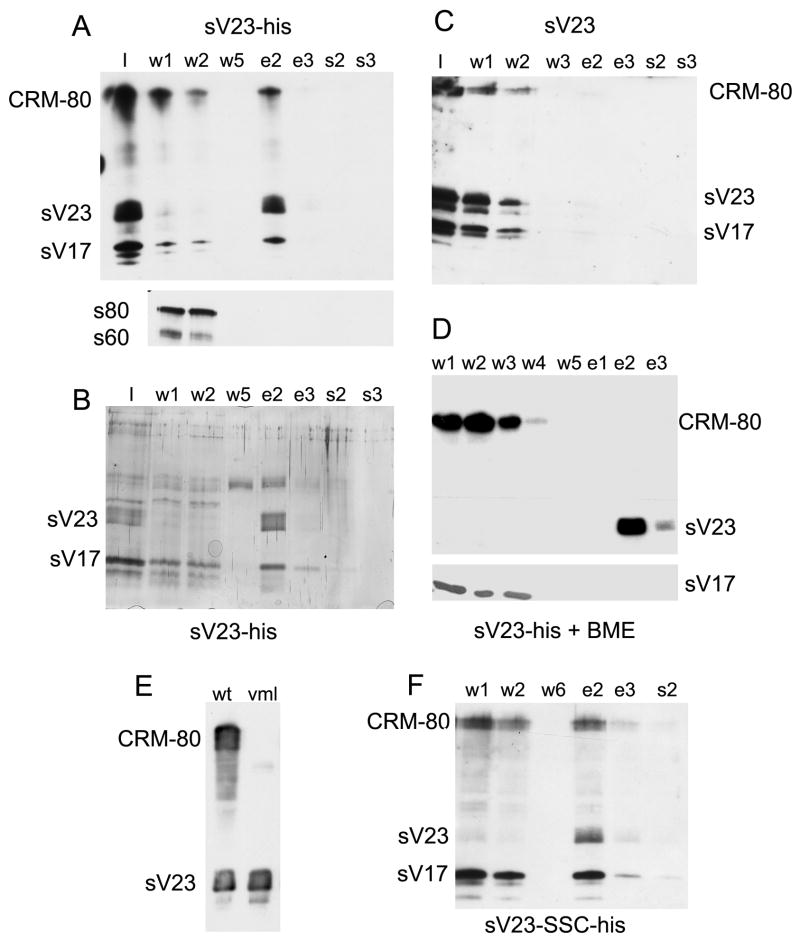
Disulfide-linked complexes containing sV23 were isolated from stage 10 egg chambers by Ni-IDA affinity chromatography. Proteins in enriched eggshell fractions were solubilized with 8M urea containing 2% Triton X-100. Following clarification by high- speed centrifugation, soluble complexes were applied to the affinity matrix. Unbound proteins (w), proteins eluted with imidizole (e), and residual proteins eluted with 2% SDS (s) were analyzed by Western blot analysis and, in some cases, by silver staining. Western blot analysis of fractions derived from egg chambers in which the sV23-His transgene was the only source of sV23 (Figure 5A) showed negligible amounts of sV23 in the unbound fractions (w) and efficient recovery of sV23 in the fractions eluted with imidizole (e). A significant portion of sV17 and a cross reacting species, denoted CRM-80, co-eluted with sV23 suggesting that these two proteins are part of early sV23 disulfide-linked complexes. Immunolocalization studies have shown that dec-1 gene products, which do not possess a VM domain, are localized exclusively within the vitelline membrane in stage 10 egg chambers (Nogueron et al., 2000). Unlike sV17 and CRM-80, the s80 and s60 DEC-1 derivatives were found exclusively in the unbound fractions. To detect other proteins that might co-fractionate with sV23, larger portions of the fractions shown in Figure 5A were run in a parallel gel and the proteins were visualized by silver staining (Figure 5B). Beyond sV23 and a species that co-migrated with sV17, reproducible bands were not apparent. The recovery of CRM-80, sV23, and sV17 exclusively in the unbound fractions when urea soluble complexes of stage 10 eggshells produced by wild type females lacking the His-tagged transgene were incubated with the matrix (Figure 5C) confirms that (1) sV23 binding is dependent upon the presence of the His-tag, and (2) binding of CRM-80 and sV17 is dependent upon sV23 binding. To show that the co-fractionation of sV17 and CRM-80 with sV23 depends on disulfide bonds, enriched eggshells were resuspended in urea in the presence of a reducing agent (5mM beta-mercaptoethanol). As expected, monomeric sV23-His bound efficiently while CRM-80 and sV17 were recovered exclusively in the unbound fractions (Figure 5D).
Figure 5.
Isolation of sV23 disulfide-linked complexes by Ni-affinity chromatography.
(A) Enriched eggshells from 900 stage 10 egg chambers from VM26AbQJ42/VM26AbQJ42 females carrying two copies of the sV23-His transgene were resuspended in denaturing buffer (I-input) and applied to a packed 100 ul Ni-IDA affinity column. Six 80 ul wash fractions (w) were used to collect unbound proteins; following the addition of 250 mM imidizole, six 80 ul elution fractions (e) were collected; 4 column volumes of denaturing buffer containing 2% SDS pre-heated to 95° [(s) – 80ul/fraction] were used to collect residual proteins. After the addition of Laemmli sample buffer and β-ME, proteins from a small portion (0.6%) of selected fractions were separated by SDS-PAGE. The upper panel shows a blot incubated simultaneously with sV23 and sV17 antisera. The lower panel shows a blot of the same column fractions incubated with a DEC-1 antiserum (Cfc106). The positions of sV23, sV17, and CRM-80 (cross reacts with the sV23 antiserum) are shown to the left. In the lower panel the positions of the DEC-1 fc106 derivatives, s80 and s60 are indicated. (B) Silver stained gel of 12.5% of each fraction shown in (A) [Note: the band observed in the w5 and e2 fractions was not reproducible]. (C) As in (A) except enriched eggshells were prepared from 200 wild type stage 10 egg chambers and 3% of each fraction was analyzed. (D) As in (A) except 200 stage 10 egg chambers were used to prepare the enriched eggshells and the enriched eggshells were resuspended in denaturing buffer containing 5mM βME. The upper panel shows a blot incubated with the sV23 antiserum; the lower panel shows the reactive region of a parallel blot incubated with the sV17 antiserum. (E) Western blot of SDS soluble proteins from wild type (wt) and VmlEPgy2 (vml) stage 10 egg chambers incubated with the sV23 antiserum. (F) Western blot of sample processed as in (A) above except the enriched eggshells were from 250 stage 10 egg chambers derived from homozygous Vm26AbQJ42 females carrying two copies of a sV23-SSC-His transgene.
As an independent means to identify proteins in sV23 disulfide linked complexes, the peak elution fraction shown in Figures 5A and B (equivalent to material derived from 900 stage 10 egg chambers), as well as the peak elution fraction from an independent experiment (~ 200 egg chambers) were analyzed by LC-mass spectrometry. Multiple peptides from only one gene product, Vml, were identified by these analyses. Vml possess a VM-like domain that includes the three evolutionarily conserved cysteines. Its large central region consists of 30 perfect plus additional imperfect copies of an octapeptide repeat (PSYSAPAA) almost identical in sequence to the octapeptide repeat found in sV23 (PAYSAPAA). With a predicted molecular weight of 56.1 kD, it is likely that CRM-80 is Vml. To confirm the identity of CRM-80, a Western blot of extracts from stage 10 egg chambers from wild type females or females with a P-element insertion in the Vml open reading frame (P{EPgy2}CG2879VmlEY21650) was incubated with the sV23 antiserum. As expected and as shown in Figure 5E, the CRM-80 signal was absent in the Vml mutant.
To determine if the complexity of sV23 disulfide linked complexes was altered when the ability of sV23 to form disulfide bonds was compromised, a double cysteine mutant transgene, sV23-SSC, was His-tagged at the same position as the wild type gene. Western blot analysis showed that when two copies of a His-tagged sV23-SSC transgene was the only source of sV23 (VM26AbQJ42/ VM26AbQJ42; sV23-SSC-His), sV23 accumulated at approximately 40% of wild type levels (not shown). Surprisingly nickel affinity chromatography of enriched eggshells from these flies showed that significant amounts of Vml and sV17 continue to co-fractionate with sV23-SSC-His (Figure 5F). Since only one cysteine residue is available for disulfide bond formation, sV23-SSC-His must form a disulfide bond with at least one other species. Direct disulfide linkages between sV23-SSC-His and Vml or sV17 cannot be inferred from this data. While previous data indicated that sV23-SSC was excluded from sV17 containing disulfide networks at stage 14, these data suggest that at earlier stages sV23-SSC and sV17 are included in the same disulfide linked complexes.
Discussion
Vitelline membrane proteins are co-packaged into secretory vesicles that are constitutively released in the form of vitelline bodies into the extracellular space between the oocyte and overlying follicle cells. Vitelline bodies coalesce and eventually form a continuous layer at stage 10B of oogenesis. As residual nurse cell cytoplasm is transferred to the oocyte during stages 10B to 12, the oocyte volume increases and the vitelline membrane thins. Significant volume changes are not incurred during stages 13 and 14. Fractionation of wild type eggshells under denaturing conditions showed a transition in pelleting behavior of vitelline membrane proteins during stage 13. This suggests constraints are imposed on disulfide bonding within the vitelline membrane layer during stages 11 and 12 when elasticity is needed as the vitelline membrane thins.
Aside from signal peptides, all of the cysteine residues in the vitelline membrane proteins followed in this study, sV23, sV17, and Vml, reside in the VM domain. The co-elution of sV17, Vml, and His-tagged sV23 following affinity chromatography of eggshells solubilized in denaturing buffer suggests that small disulfide linked complexes are in place in stage 10 egg chambers. Since secretion of these VMPs is complete by the end of stage 10, the late stage growth of the disulfide-linked network(s) suggests temporally regulated post-depositional changes in the availability of free cysteines, the rate of covalent assembly, or intermolecular disulfide bonding partners. The behavior of the sV23-SSC mutant protein is consistent with post-depositional disulfide exchange. The stage 10 affinity purification data showed a sizeable fraction of sV17 and Vml co-eluted with sV23-SSC-His (Figure 5F) in a cysteine dependent manner. While the formation of direct disulfide bonds between sV23 and sV17 or Vml cannot be inferred, the data showed that in stage 10 egg chambers sV23-SSC containing disulfide linked complexes include sV17 and Vml. When stage 14 egg chambers from either the sV23-SSC or sV23-CSS mutants were disrupted in denaturing solvents, sV23 was recovered almost exclusively in the supernatant fraction after low speed centrifugation. With a single cysteine available for intermolecular disulfide bonds, a compromise in the ability of sV23-SSC to incorporate into a large disulfide linked network in late stage egg chambers was not surprising. The recovery of sV17, along with VM-CRM14, exclusively in the pellet fraction (Figure 4C) was unexpected however, since substantial amounts of sV17 were included in sV23-SSC disulfide linked complexes at stage 10. Taken together these data suggest rearrangements in intermolecular disulfide bonds occur in the vitelline membrane as eggshell morphogenesis progresses.
Extracellular remodeling of protein disulfide bonding patterns has been associated with regulatory roles (Hogg, 2003). A complex intramolecular disulfide exchange controls the activity of thrombospondin-1 (TSP-1), an extracellular glycoprotein that participates in cell-cell and cell-matrix communication. In turn TSP-1 reduces the size and biological activity of the platelet adhesion factor vWF in the vasculature by facilitating reversible cleavage of the disulfide bonds that create large vWF multimers (Xie et al., 2001). Remodeling of disulfide bonds in the extracellular domains of cell surface receptors, including integrin alpha1beta3, CD4, and the tumor necrosis factor receptor CD30, induces conformational changes that strengthen the interactions with specific ligands (Hogg, 2003; Jordan and Gibbins, 2006).
Disulfide exchange in the extracellular environment requires cleavage and reformation of disulfide bonds. Mass spectrometric analysis of enriched eggshell preparations from Drosophila ovaries revealed a putative GMC oxidoreductase (Fakhouri et al., 2006). Its stage specific expression in the follicle cells suggests it may play a role in VM morphogenesis. Typically disulfide bonds are reduced, oxidized, and rearranged by thiol-disulfide oxidoreductases that function in the endoplasmic reticulum. Secreted forms have been reported for a variety of cells however (Hogg, 2003; Jordan and Gibbins, 2006). In extracellular environments thioredoxin and protein disulfide isomerases (PDI) are thought to act as reductants.
The classic redox active site motif of thioredoxin/PDI is CXXC. Non-classical motifs (CXXS and SXXC) have been found however in the thioredoxin domains of a testis specific PDI (van Lith et al., 2005), a subset of the Arabidopsis thaliana thioredoxin h family (Serrato et al., 2008), and in a novel endoplasmic reticulum folding assistant of the thioredoxin family, ERp44 (Anelli et al., 2002). Using CXXS as a query, analyses of the Escherichia coli, Campylobacter jejuni, Methanococcus jannaschii, and Saccharomyces cerevisiae genomes revealed a high proportion of proteins known to use the CXXS motif for redox function. A strong correlation was established between the conservation of the CXXS sequence and proteins with possible redox functions (Fomenko and Gladyshev, 2002).
While the secretion of classical thioredoxins or PDIs into the space between the follicle cells and oocyte where the vitelline membrane forms has not been reported, the VM domains of sV23, Vml, and fc20 (VM26Ac) have an SXXC motif that includes the first cysteine C123. In addition sV23 has a motif, CXXS at C131, the second cysteine (Figure 6). Vitelline envelope proteins in the mosquito, Aedes aegypti, also possess a VM-like domain in which the spacing of the three cysteines is strictly conserved (Figure 6). Notably in two of these proteins the CXXS motif at the second cysteine is conserved along with the proline residue that precedes the serine (CXPS) in sV23. Given the critical nature of the second cysteine of sV23 in building a functional eggshell (Table 1) and the evolutionary conservation of CXXS in the mosquito proteins, we speculate that this thioredoxin like motif may play an essential redox role in isomerizing disulfide bonds during vitelline membrane morphogenesis. Noteworthy in this regard, of the Drosophila vitelline membrane proteins with a VM domain, to date female sterile mutations have only been reported for the sV23 gene. The fertility of VmlEPgy2/ VmlEPgy2 females is particularly surprising given the striking structural similarity of Vml and sV23. Both proteins are headed by a short N-terminal domain that include a potential furin cleavage site, followed by a central region consisting of tandem repeats of an octapeptide (P A/S YSAPAA) and a C-terminal VM domain. Unlike sV23, the thioredoxin-like motif at the second cysteine is missing in Vml as well as other VM domain containing eggshell proteins.
Figure 6.

Alignment of the VM domains from Drosophila VM proteins and related domains found in Aedes aegypti vitelline envelope proteins (Edwards et al., 1998). The three precisely spaced cysteines are highlighted as well as Ser residues found within the context of putative redox motifs. The line highlights the evolutionarily conserved CXXS motif that includes the critical C131 of sV23.
Switches in disulfide bonding partners may underlie the growth of the sV23 disulfide network. Interestingly of the three sV23 double cysteine mutants tested, only the mutant with cysteine at the second position, sV23-SCS, was able to incorporate into a large disulfide network with the other VMPs suggesting the second cysteine has a greater propensity to form disulfide bonds with other VMPs than either the first or third cysteines. The first and third cysteines can form bridges with other VMPs since sV23-CSC was incorporated into a large disulfide linked network (Figure 4). Whether the bridges formed under these circumstances are normal is questionable however since these females lay sterile eggs with structurally compromised eggshells.
Temporally regulated formation of higher order disulfide networks involving other VMPs can occur in the absence of sV23. While the putative redox active second cysteine may be essential for proper incorporation of sV23 into higher order networks, other mechanisms must be in place to restrict temporal growth of the network(s) in general. Along with regulated growth of the network(s), remodeling of disulfide bonds within the vitelline membrane may also play an allosteric role in late eggshell morphogenesis by changing the presentation of proteins and how they interact. While reversible phosphorylation has widespread regulatory significance in the cell, perhaps reversible disulfide bond formation will prove to have widespread regulatory significance in the extracellular environment.
Acknowledgments
We thank Stephan Jacobsen for assistance with the RNA analyses and Bassam Wakim at the Protein and Nucleic Acid Core Facility, Biochemistry Department, Medical College of Wisconsin for the mass spectrometry. Marquette University fellowship support to T.W. and funding from the National Institutes of Health (R15GM062816) to G.L.W. are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–44. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer BJ, Waring GL. 7C female sterile mutants fail to accumulate early eggshell proteins necessary for later chorion morphogenesis in Drosophila. Dev Biol. 1987;121:349–58. doi: 10.1016/0012-1606(87)90171-0. [DOI] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–7. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Fabbri F, Andrenacci D, Taddei C, Gargiulo G. Drosophila vitelline membrane cross-linking requires the fs(1)Nasrat, fs(1)polehole and chorion genes activities. Dev Genes Evol. 2001;211:573–80. doi: 10.1007/s00427-001-0192-1. [DOI] [PubMed] [Google Scholar]
- Edwards MJ, Severson DW, Hagedorn HH. Vitelline envelope genes of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1998;28:915–925. doi: 10.1016/s0965-1748(98)00083-6. [DOI] [PubMed] [Google Scholar]
- Fakhouri M, Elalayli M, Sherling D, Hall JD, Miller E, Sun X, Wells L, LeMosy EK. Minor proteins and enzymes of the Drosophila eggshell matrix. Dev Biol. 2006;293:127–41. doi: 10.1016/j.ydbio.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokta F. Biology. Marquette; Milwaukee: 2000. Regulatory elements of the sV23 gene of Drosophila melanogaster; p. 160. [Google Scholar]
- Fomenko DE, Gladyshev VN. CxxS: fold-independent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci. 2002;11:2285–96. doi: 10.1110/ps.0218302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans M, Audit C, Mason M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–4. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- Jordan PA, Gibbins JM. Extracellular disulfide exchange and the regulation of cellular function. Antioxid Redox Signal. 2006;8:312–24. doi: 10.1089/ars.2006.8.312. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Hashimoto C. The nudel protease of Drosophila is required for eggshell biogenesis in addition to embryonic patterning. Dev Biol. 2000;217:352–361. doi: 10.1006/dbio.1999.9562. [DOI] [PubMed] [Google Scholar]
- Manogaran A, Waring GL. The N-terminal prodomain of sV23 is essential for the assembly of a functional vitelline membrane network in Drosophila. Dev Biol. 2004;270:261–71. doi: 10.1016/j.ydbio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Margaritis LH. Structure and physiology of the eggshell. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Vol. 1. Pergamon; Elmsford, NY: 1985. pp. 153–230. [Google Scholar]
- Mindrinos MN, Petri WH, Galanopolous VK, Lombard MF, Margaritis LH. Crosslinking of the Drosophila chorion involves a peroxidase. Wilhelm Roux’s Arch Dev Biol. 1980;189:187–196. doi: 10.1007/BF00868677. [DOI] [PubMed] [Google Scholar]
- Noguerón MI. Department of Biology. Marquette University; Milwaukee, WI: 1996. Processing and Distribution of dec-1 Eggshell Products in Drosophila melanogaster; p. 99. [Google Scholar]
- Nogueron MI, Mauzy-Melitz D, Waring GL. Drosophila dec-1 eggshell proteins are differentially distributed via a multistep extracellular processing and localization pathway. Dev Biol. 2000;225:459–70. doi: 10.1006/dbio.2000.9805. [DOI] [PubMed] [Google Scholar]
- Pascucci T, Perrino J, Mahowald AP, Waring GL. Eggshell assembly in Drosophila: processing and localization of vitelline membrane and chorion proteins. Dev Biol. 1996;177:590–8. doi: 10.1006/dbio.1996.0188. [DOI] [PubMed] [Google Scholar]
- Savant SS, Waring GL. Molecular analysis and rescue of a vitelline membrane mutant in Drosophila. Dev Biol. 1989;135:43–52. doi: 10.1016/0012-1606(89)90156-5. [DOI] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Stevens L, Stein D. Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity. Cell. 1998;95:471–81. doi: 10.1016/s0092-8674(00)81615-3. [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Guilleminot J, Meyer Y, Vignols F. AtCXXS: atypical members of the Arabidopsis thaliana thioredoxin h family with a remarkably high disulfide isomerase activity. Physiol Plant. 2008;133:611–22. doi: 10.1111/j.1399-3054.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- Spangenberg DK, Waring GL. A mutant dec-1 transgene induces dominant female sterility in Drosophila melanogaster. Genetics. 2007;177:1595–608. doi: 10.1534/genetics.107.080168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. The Drosophila embryonic patterning determinant torsolike is a component of the eggshell. Curr Biol. 2003;13:1058–63. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- van Lith M, Hartigan N, Hatch J, Benham AM. PDILT, a divergent testis-specific protein disulfide isomerase with a non-classical SXXC motif that engages in disulfide-dependent interactions in the endoplasmic reticulum. J Biol Chem. 2005;280:1376–83. doi: 10.1074/jbc.M408651200. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. Int Rev Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Xie L, Chesterman CN, Hogg PJ. Control of von Willebrand factor multimer size by thrombospondin-1. J Exp Med. 2001;193:1341–9. doi: 10.1084/jem.193.12.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Stevens LM, Stein D. Sulfation of eggshell components by Pipe defines dorsal-ventral polarity in the Drosophila embryo. Curr Biol. 2009;19:1200–5. doi: 10.1016/j.cub.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZD, Liu WY, Li MQ. Chromium (III) enhanced diamine silver staining of proteins and DNA in gels. Biotechnol Lett. 2003;25:1801–4. doi: 10.1023/a:1026280519327. [DOI] [PubMed] [Google Scholar]