Abstract

The mycobacterial cell wall component lipoarabinomannan (LAM) has been described as one of the key virulence factors of Mycobacterium tuberculosis. Modification of the terminal arabinan residues of this lipoglycan with mannose caps in M. tuberculosis or with phosphoinositol caps in Mycobacterium smegmatis results in distinct host immune responses. Given that M. tuberculosis typically persists in the phagosomal vacuole after being phagocytosed by macrophages, we performed a proteomic analysis of that organelle after treatment of macrophages with LAMs purified from the two mycobacterial species. The quantitative changes in phagosomal proteins suggested a distinct role for mannose-capped LAM in modulating protein trafficking pathways that contribute to the arrest of phagosome maturation. Enlightened by our proteomic data, we performed further experiments to show that only the LAM from M. tuberculosis inhibits accumulation of autophagic vacuoles in the macrophage, suggesting a new function for this virulence-associated lipid.
Keywords: Phagosome, autophagosome, membrane proteome, mycobacterial lipoglycans, mannose-capped LAM
Short abstract
LAM has been described as one of the key virulence factors of Mycobacterium tuberculosis. Modification of the terminal arabinan residues of this lipoglycan with mannose caps in M. tuberculosis or with phosphoinositol caps in Mycobacterium smegmatis results in distinct host immune responses. Given that M. tuberculosis typically persists in the phagosomal vacuole after being phagocytosed by macrophages, we performed a proteomic analysis of that organelle after treatment of macrophages with LAMs purified from the two mycobacterial species.
Introduction
Mycobacterium tuberculosis is an extraordinarily successful human pathogen with the ability to replicate within the normally hostile environment of host macrophages. After being phagocytosed by the macrophage, M. tuberculosis resides in a membrane-bound vacuole, the phagosome, which normally undergoes maturation into the phagolysosome that is essential for eliminating invading microbes and for antigen presentation.1 However, M. tuberculosis is able to arrest phagosomal maturation by interfering with Ca2+ signaling and trafficking of the Rab family of small GTPases, two important processes for organelle membrane fusion.2,3 When residing within a phagosome, the live bacilli secrets specific proteins such as tyrosine phosphatases to reduce the phagosomal level of phosphatidylinositol 3-phosphate or inhibit host proteins regulating vacuolar sorting, which all lead to impaired phagolysosomal fusion.4−6 Arrest of phagosomal maturation (or block of phago-lysosome fusion) is critical for M. tuberculosis persistence in host macrophages.1
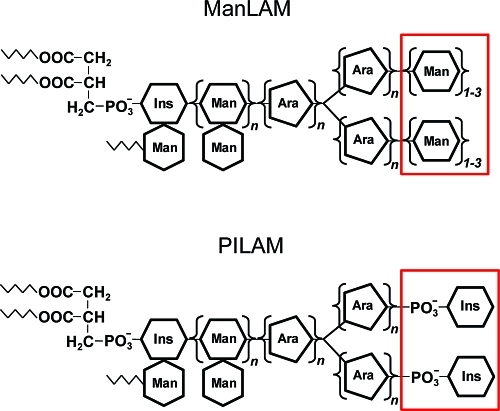
Apart from secreted proteins, the exotic cell wall components of pathogenic mycobacteria are thought to be key modulators of host immune processes, but in most cases, their molecular effects on host cells are not well understood.7 The best characterized are unique mycobacterial lipoglycans termed lipoarabinomannans (LAM), which are noncovalently associated with the bacterial plasma membrane and extend to the exterior of the cell wall.8,9 LAMs are large, extensively glycosylated, phosphatidylinositol derivatives with heterogeneous modifications. Notably, pathogenic M. tuberculosis produces mannose-capped lipoarabinomannan (ManLAM) structures,10 whereas the fast-growing nonpathogenic species Mycobacterium smegmatis synthesizes LAM molecules capped with phosphatidyl-myo-inositol (termed PILAM)11 (Figure 1).
Figure 1.
Mycobacterial lipoglycans ManLAM and PILAM. The structures differ only in their terminal motifs capping the arabinan moiety. Abbreviations: Ins, inositol; Man, mannose; Ara, arabinose.
The different terminal structures of the two LAM variants profoundly affect their biological activities. For instance, macrophages show significantly reduced phagosome−lysosome fusion after engulfing ManLAM-coated microbeads.12 ManLAM interacts with the mannose receptor and blocks the rise in cytosolic Ca2+ that would otherwise accompany mycobacterial entry into macrophages.13,14 Free ManLAM can insert into host cell membranes, leading to membrane reorganization and disruption of signaling pathways.15 By contrast, PILAM has none of these effects. Instead, it is an agonist of Toll-like receptor 2, which results in secretion of various cytokines and activation of apoptotic pathways,16−18 though the notion of its proinflammatory activity has been challenged recently.19
Motivated by reports suggesting different effects of ManLAM and PILAM on macrophages, we sought to probe how the two mycobacterial lipoarabinans affect the molecular properties of host phagosomes. Previously, we developed a method to purify the membrane fraction from latex bead-containing (LBC) phagosomes in order to profile its proteome using mass spectrometry.20 Here we modified our method by combining Tube-Gel digestion21 and iTRAQ labeling22 to study quantitative changes in the macrophage phagosomal proteome upon exposure of cells to ManLAM or PILAM. Escherichia coli-derived lipopolysaccharide (LPS) was used as a third stimulus in order to identify LAM-specific effects. From a total list of 823 proteins found in the phagosomal membranes, we identified 47 proteins that were significantly up- or down-regulated (>1.25-fold, p < 0.05) by exposure of macrophages to ManLAM but not the other two lipoglycans. The list of down-regulated proteins included the autophagy marker LC3. Motivated by this observation, we investigated the effects of mycobacterial LAMs on LC3 recruitment to the phagosome as well as autophagy activation in cultured macrophages. ManLAM inhibited chemical-induced accumulation of autophagosomes, suggesting a previously unrecognized function of this virulence factor in undermining host defense responses.
Experimental Procedures
Reagents
The rat anti-LAMP1 mAb and mouse anti-syntaxin 6 mAb were obtained from BD Biosciences. The goat anti-EEA1 pAb and goat anti-cathepsin D pAb were obtained from Santa Cruz Biotechnology. The rabbit anti-LC3B pAb and mouse anti-tubulin mAb were obtained from Sigma. The mouse anti-transferrin receptor mAb was obtained from Zymed Laboratories. All secondary Abs for Western blots were obtained from SouthernBiotech. FITC-conjugated lectin Concanavalin-A was obtained from EY Laboratories. Acrylamide/bisacrylamide solution (40%, 29:1), ammonium persulfate (APS), and tetramethylenediamine (TEMED) were obtained from Bio-Rad.
Preparation of Lipoglycan-Coated Latex Beads
Latex beads (1.0 μm microspheres, Polysciences) were washed twice in 0.05 M carbonate−bicarbonate buffer (pH 9.6) by centrifugation at 15 000g for 5 min in 1.5-mL presiliconized low-retention microtubes (Fisher). The beads in each tube were then resuspended in 900 μL of carbonate−bicarbonate buffer before a certain amount of a specific lipoglycan was added. Usually, 180 μg of ManLAM or PILAM, or 1.8 μg of LPS was used to coat a total of 4.55 × 109 latex beads. The beads and lipoglycans were incubated for 1 h at 37 °C on an Eppendorf Thermomixer (shaking at 1400 rpm). Control beads were prepared by incubation with buffer alone. The beads were centrifuged and the supernatant was depleted. The beads were washed once and then incubated in 1 mL of PBS buffer containing 5% BSA (Sigma, endotoxin tested) for 0.5 h at 37 °C to block nonspecific binding sites. The latex beads were washed with 0.5% BSA in PBS, resuspended in 1 mL of RPMI-1640 cell culture media, and stored at 4 °C until use (stable for 1 week). The presence of mannose residues on latex beads after ManLAM coating was confirmed by immunofluorescence microscopy using FITC-conjugated Con A. The other lipids were coated onto beads using the same protocol, which was presumed to be effective for all lipoglycans.
Cell Treatment with Lipoglycan-Coated Beads, Phagosome Isolation, and Membrane Fractionation
The murine macrophage cell line RAW 264.7 was cultured as a monolayer in RPMI-1640 (GIBCO, formulated with HEPES and glutamine) supplemented with 10% fetal bovine serum and 100 units/mL penicillin/streptomycin. Cells were first chilled with 4 °C PBS for 5 min in order to synchronize phagocytosis. Four populations of cells were incubated (2 h, 37 °C) with latex beads coated with a specific bacterial lipoglycan or control beads without lipid coating at a multiplicity of infection (MOI) of 50:1 to generate phagosomes. Cells were then washed with ice-cold PBS three times. Each cell had internalized 5−10 microbeads on average as determined by microscopy. After gentle cell lysis using a Dounce homogenizer to reach 90−95% breakage, the bead-containing phagosomes were isolated by ultracentrifugation on a sucrose gradient as described by Desjardins and co-workers.23
The latex bead-containing (LBC) phagosome fraction was collected from the top of the sucrose gradient (10−20% interface) and resuspended in PBS containing protease inhibitor cocktail (Calbiochem). The LBC-phagosome fraction was washed by ultracentrifugation at 40 000g for 20 min. Each LBC-phagosome pellet from a given treatment was resuspended in 1.5 mL of 0.2 M Na2CO3 (pH 11.0) containing proteases inhibitors. The LBC-phagosomes were disrupted by passaging the suspension 5−7 times through the needle of a 1-mL syringe (25G). The resulting sample was kept on ice for 30 min before the membrane fraction was pelleted by centrifugation for 45 min at 200 600g at 4 °C. The pellet was washed with a low-salt buffer (10 mM triethylammonium bicarbonate (TEAB), Sigma). A third centrifugation was performed at 100 000g for 15 min to acquire the final membrane pellet. The phagosomal pellet samples were stored at −80 °C prior to protein extraction.
Protein Extraction from Phagsomal Membranes
A solution of 4% SDS in 400 mM TEAB buffer (50 μL) was added to the membrane pellet. The resuspended solution was agitated for 30 min at 10 °C on a Thermomixer to extract proteins from the membrane fraction. After centrifugation at 14 000g for 10 min, the supernatant was collected and diluted with additional ddH2O so that the final protein extract was in 1% SDS and 100 mM TEAB buffer (4-fold dilution). For measuring protein concentration, we diluted the extracts by another 5-fold with 50 mM Tris-HCl buffer before carrying out the BCA assay (Pierce).
Trypsin Digestion of Phagosomal Membrane Extracts and iTRAQ Labeling
For in-solution digestion, 4 μg of protein derived from the phagosome membrane extract was diluted either 5-fold or 20-fold in 100 mM TEAB, resulting in a final SDS concentration of 0.2% or 0.01%, respectively. Each protein sample was treated with TCEP (5 mM, 37 °C for 30 min) and iodoacetamide (15 mM, for 30 min in the dark) to reduce and alkylate cysteine residues, then digested with trypsin overnight (E/S = 1:20 w/w). In addition to this single enzyme digestion, we tested the efficiency of dual enzyme digestion with both Lys-C and trypsin. A 5-fold diluted sample was first digested with Lys-C (Promega) for 4 h at 37 °C (E/S = 1:80 w/w) before the trypsinization under the same condition as above. All digests were acidified with 2% formic acid and desalted with a C18 Ziptip (Millipore).
To test the efficiency of in-tube digestion, 5 μg of protein derived from the phagosome membrane extract was diluted to 14 μL with 1% SDS in 100 mM TEAB buffer. Cysteine residues were reduced and alkylated as described earlier. The protein sample was then mixed with 5 μL of acrylamide/bisacrylamide (40%, 29:1), 0.7 μL of 1% APS, and 0.3 μL of TEMED. Gel solidification was complete after 30−40 min. The gel impregnated with membrane proteins was cut into small pieces and washed three times with 50% acetonitrile (ACN) in 100 mM TEAB buffer. The gel pieces were then dehydrated with pure ACN and dried using a SpeedVac. A total of 0.5 μg of trypsin (in 25 mM TEAB buffer, E/S = 1:10 w/w) was absorbed by the dried gel, which was then incubated overnight at 37 °C. Peptides were extracted from the gel with 50 μL of 25 mM TEAB buffer, 100 μL of 0.1% TFA, and 150 μL of ACN in 0.1% TFA, sequentially. The solutions were combined and concentrated using a SpeedVac.
For Tube-Gel digestion of phagosome membrane proteins before iTRAQ labeling, 20 μg of protein from each extract from cells treated with different lipid-coated beads was incorporated into gels using the same protocol, except that 10 mM methyl methanethiosulfonate (MMTS) replaced iodoacetamide and the reagent amounts were increased 4-fold. BSA (1 μg) was added to each sample before gel solidification. The residual protein solution on top of the gel was removed and subjected to a second solidification process. Two gels from the same protein sample were combined before further treatment described above. Each peptide extract was concentrated to roughly 20 μL, and 1 M TEAB buffer was added to adjust the final buffer concentration to 400 mM. Four-plex iTRAQ (Applied Biosystems) reagents were reacted with individual protein digests according to the manufacturer’s protocol. The phagosome membrane protein preparation and digestion procedures were performed twice to acquire two biological replicates.
Protein Identification by 2D LC−MS/MS and Data Analysis
For comparison of the in-solution and Tube-Gel digestion efficiencies, 0.5 μg of each protein digest was analyzed by LC−MS using ESI Linear Ion Trap Mass Analyzer (LTQ, Thermo, Inc.) in a data-dependent acquisition mode. LC separation was performed on a house-made microcapillary column (0.100 × 100 mm, 3 μm Magic C18 packing material, Michrom Bioresources) at a flow rate of 300 nL/min using buffers 0.1% formic acid (FA) in water (A) and 0.1% FA in ACN (B). The gradient was 2−10% B for 5 min, 10−40% B for 60 min, 40−90% B for 10 min, and 90% B for 10 min. The major parameters for LTQ data acquisition were: scan range, 400−2000 m/z; precursor ion selection, six most abundant peaks per scan for MS/MS; minimal ion signal, 500; and normalized collision energy, 35.0%.
The iTRAQ-labeled peptide mixtures from Tube-Gel digestion of two experimental replicates were separated by two-dimensional liquid chromatography and analyzed by ESI-Q-TOF mass spectrometry. Briefly, the peptide mixture was separated by off-line strong cation exchange (SCX) chromatography using an Ultimate HPLC with a UV detector (Dionex-LC Packings, Sunnyvale, CA). Labeled samples were resuspended in SCX running buffer (5 mM KH2PO4, 25% ACN, 0.1% FA) and loaded onto a PolyLC Polysulfethyl A column (2.1 mm × 200 mm, The Nest Group, Southborough, MA). Peptides were eluted with increasing concentrations of 800 mM KCl, 5 mM KH2PO4, 25% ACN, and 0.1% FA using a three step gradient: 0−10% for 10 min, 10−25% for 30 min, and 25−100% for 10 min. Fifteen fractions were collected at a flow rate of 300 μL/min according peaks observed by UV absorption at 214 nm.
Fractions were partially evaporated to remove ACN on a SpeedVac and desalted using C18 MacroSpin columns (The Nest Group, Southborough, MA). Desalted fractions were dried and reconstituted in 0.1% FA. Each fraction was injected onto a PepMap100 trapping column (0.3 mm ×5 mm). Reversed-phase separation was performed on a LC Packings PepMap C18 column (3 μm, 0.075 × 150 mm) at a flow rate of 300 nL/min using buffers 2% ACN, 0.1% FA (A) and 80% ACN, 0.1% FA (B). The gradient was 0−35% B for 100 min, 30−100% B for 10 min, and 100% B for 10 min. The samples from one experiment were injected into a QSTAR Pulsar-I Hybrid Quadrupole TOF (Applied Biosystems, Framingham, MA), while the samples from another replicate were analyzed by a QSTAR Elite Hybrid Quadrupole TOF (Applied Biosystems, Framingham, MA). The major parameters for Q-TOF data acquisition were: MS scan range, 350−1800 m/z; MS/MS scan range, 70−2000 m/z; precursor ion selection, three most abundant peaks per scan for MS/MS; minimal ion counts, 30; and automated collision energy was applied (fragment intensity multiplier = 2.0).
LC−MS/MS data acquired using a LTQ ion trap mass spectrometer (Thermo) were searched against the mouse Swiss-Prot protein database using the SEQUEST algorithm provided with Bioworks 3.2. SEQUEST results were filtered by Xcorr (+1 > 1.8; +2 > 2.5; +3 > 3.5), DeltaCN > 0.8 and the requirement of at least two different peptides from a protein. Parent and product ion mass errors were set at 1.2 and 0.8 Da, respectively. A static modification of carboxyaminomethylation and a variable modification of Met oxidation were specified.
LC−MS/MS data acquired on a Q-TOF for iTRAQ-based quantification were searched against a mouse International Protein Index (IPI) database (mouse, version 3.27, ∼56 000 entries) using the Paragon algorithm within ProteinPilot version 2.0 software (Applied Biosystems). The major parameters in the software were explained by Pierce et al.24 Protein identification was based on a confidence level >95% and at least two different peptides assigned to the protein. A search performed against a concatenated database containing both forward and reversed sequences allowed estimation of the false discovery level (below 1%). The number of transmembrane helices in each protein was predicted using the TMHMM online program (http://www.cbs.dtu.dk/services/TMHMM/).
For relative protein quantification, each protein ratio reported by Protein Pilot was associated with a p-value (evaluating the statistical difference between the observed ratio and unity) and EF (error factor) for each protein ID.25 The EF term indicates that the actual ratio lies between (reported ratio)/(EF) and (reported ratio) × (EF) at a 95% confidence. The following criteria were required to consider a change in protein level significant: the protein ID had a p-value < 0.05 and a meaningful EF (<2), at least two unique peptides were identified, and the fold difference was greater than 1.25 (i.e., the ratio was >1.25 or <0.80) as explained in Results and Discussion.
Immunoblots
Proteins from the LBC-phagosome membrane extracts were separated by SDS-PAGE (4−12% acrylamide for most experiments, 12% acrylamide gel for LC3 detection) and analyzed by Western blot using appropriate antibodies. For each sample, 3 μg of protein was loaded. Mouse tubulin was probed as a loading control.
Live Cell Imaging of Autophagy Marker LC3
RAW264.7 cells stably expressing GFP-fused LC3 (kindly provided by Patrick Fitzgerald, St. Jude’s Children’s Research Hospital, Memphis, TN) were used to investigate LC3 trafficking after internalization of lipoglycan-coated latex beads. The cells were cultured in DMEM (GIBCO, containing high-glucose, glutamine, and sodium pyruvate) supplemented with nonessential amino acids (GIBCO), 10% fetal bovine serum, and 100 units/mL penicillin/streptomycin. Cells were seeded onto 8-well, Nunc Lab-Tek Chambered Coverglass microscopy slides (Fisher) 20 h before images were acquired. After incubation with lipoglycan-coated beads for 2 h, cells were gently washed with PBS twice to remove free beads. The cells were then treated with LysoTracker Red DND-99 (Molecular Probes) (100 nM) for 15 min. The cells were rinsed and incubated in fresh warm media and imaged using a Zeiss 200 M epifluorescence deconvolution microscope. All images were processed using the nearest neighbor deconvolution algorithm in the instrument software Slidebook 4.2 (Intelligent Imaging Innovations). In a separate experiment, the cells were incubated with the autophagy inducer chloroquine (50 μM) and a given bacterial lipoglycan at various concentrations. After 2 h, the cells were washed with PBS and placed in warm media for fluorescence imaging.
Results and Discussion
Quantitative Membrane Proteomics of Phagosomes by Combining Tube-Gel Digestion and iTRAQ Labeling
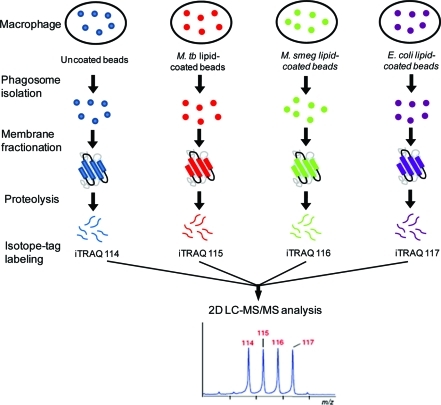
Previously, we reported a workflow for LBC phagosome isolation, membrane fractionation, and profiling of the phagosome membrane proteome by mass spectrometry.20 Here, we modified our platform to include the use of the iTRAQ method for multiplexed protein quantitation.22,26 Figure 2 illustrates the workflow of the entire experiment. First, we coated latex beads with a lipoglycan from M. tuberculosis (ManLAM), M. smegmatis (PILAM), or E. coli (LPS). The efficiency of lipid coating was verified by specific staining of ManLAM-coated beads with FITC-conjugated Concanavalin-A, a lectin that binds terminal mannose residues27 (Supporting Information SI Figure 1).
Figure 2.
Experimental design for quantitative analysis of the phagosomal membrane proteome under different lipoglycan treatments.
LBC-phagosomes were isolated using the method initially reported by Desjardins and colleagues23 and modified in our previous work.20 To enrich membrane-bound components, we fractionated the phagosome based on our previous protocol with one modification: we performed an additional wash of the membrane pellet using a low-salt buffer to remove a greater proportion of highly abundant luminal proteins. This adjustment allowed the identification of a greater number of membrane-associated proteins (vide infra).
A consequence of enriching the samples for membrane-associated proteins is the requirement of high detergent concentrations (>4% SDS) for their solubilization, which poses a problem for in-solution trypsin digestion due to the loss of enzyme activity. Additionally, the in-gel digestion method, though compatible with high detergent concentrations, is not amenable to subsequent chemical labeling reactions as required by the iTRAQ method. We attempted to solve these problems using reduced amounts of SDS (0.2%) in solution as well as a more detergent-resistant protease, Lys-C, prior to trypsin digestion, but neither of these modifications gave satisfactory results.
We turned our attention to the recent method of Tube-Gel digestion, developed by Lu and Zhu, which has proven effective for proteomic analysis of membrane proteins.21,28 An advantage of this method is that detergent can be washed from the protein-impregnated gel prior to trypsinization. Furthermore, the resulting peptide products can be released from the gel into aqueous solution prior to chemical labeling. We modified the Tube-Gel digestion method in three ways to better suit the analysis of phagosomal membrane extracts: (1) NH4HCO3 was replaced with TEAB, a primary amine-free buffer compatible with iTRAQ reagents; (2) the residual protein solution that was excluded from the gel matrix was subjected to a second gel solidification process, so as to minimize sample loss; and (3) we performed the cysteine reduction and alkylation steps prior to gel formation.
Using phagosomal membrane extracts as substrates, we compared the efficiency of this modified Tube-Gel digestion protocol to in-solution methods and analyzed the peptide products by LC−MS. The total number of proteins identified using different digestion protocols in three separate experiments are shown in Supporting Information Table 1. The modified Tube-Gel protocol identified three times as many proteins as the best in-solution procedure (using dual enzymes and 0.2% SDS). A recent publication by Han et al. also highlights the robustness of the Tube-Gel method for processing membrane proteins prior to iTRAQ labeling.29
The digested peptides derived from untreated, ManLAM-, PILAM-, or LPS-treated samples were labeled with iTRAQ114, iTRAQ115, iTRAQ116, or iTRAQ117, respectively. The pooled peptide mixture was subjected to 2D LC−MS/MS analysis both for protein identification and relative quantitation. Biological replicates were analyzed either on a QSTAR Pulsar I or Elite mass spectrometer, and the combined data sets produced a total of 823 nonredundant proteins (Supporting Information Table 2A). Notably, 540 of these proteins were not found in our earlier phagosome membrane proteomics study.20 A summary of overlapping and unique protein IDs from both data sets is present in Supporting Information Table 3. We attribute the superior performance of the new method to (1) the additional low-salt wash, which depleted more soluble proteins and enriched membrane proteins accordingly (Supporting Information Figure 2B), and (2) the reduced sampling handling required of the Tube-Gel method, which minimized contamination with keratins.
Changes of Phagosomal Membrane Proteins in Response to Lipoglycans
The iTRAQ isotopic labels were used to calculate relative protein levels from lipoglycan-treated or untreated samples. Supporting Information Table 2B provides a list of 658 quantitative ratios (lipoglycan treated:untreated) measured in two experimental replicates. The criteria for selecting ratios that indicate significant changes in protein abundances are described in Experimental Procedures. Applying these filters allowed us to identify four protein subsets that were regulated by (i) ManLAM alone, (ii) ManLAM and PILAM but not LPS, (iii) ManLAM and LPS but not PILAM, or (iv) all three lipoglycans. Because ManLAM has been implicated in M. tuberculosis virulence, we focused our studies on the phagosomal proteins that were specifically regulated by this lipoglycan.
Apart from the p-value filter, we applied a threshold of 1.25-fold difference because: (1) the average technical and biological variations for iTRAQ-based quantitation have been reported to be ±11% and ±25%, respectively;30 (2) precedents have established that in some cases changes in protein level as low as 1.2-fold can be biologically relevant;31,32 (3) after applying these criteria, we found 42 proteins specifically regulated by ManLAM (Table 1), with the remaining unchanged proteins covering 94% of all the proteins quantified in this study. This value is higher than the percent of unchanged proteins found in an analysis of biological replicates using iTRAQ.30 Therefore, our criteria defined a suitable variation tolerance for identifying potentially regulated proteins.
Table 1. Quantitative Changes of Phagosomal Proteins Regulated by ManLAM Treatmenta.
| biological process | protein name | ManLAM | PILAM | LPSb |
|---|---|---|---|---|
| Down-Regulated Proteins | ||||
| Membrane receptor/Signaling | Mannose-6-phosphate receptor | 0.75 | 1.04 | 1.30 |
| CD63 | 0.67 | 1.06 | 1.27* | |
| Scavenger receptor class B member 2 | 0.66 | 1.05 | 1.23* | |
| Membrane structure and lipid rafts | Lysosomal membrane glycoprotein 1 | 0.60 | 0.94 | 1.08* |
| Lysosomal membrane glycoprotein 2 | 0.76 | 1.02 | 1.02 | |
| Transmembrane protein 55A | 0.79 | 0.80 | 0.89 | |
| Vesicular and protein trafficking | Early endosome antigen 1 | 0.55 | 0.96 | 1.04 |
| Vesicle transport through interaction with t-SNAREs homologue 1B | 0.75 | 1.03 | 1.28 | |
| Rab-7A | 0.71 | 1.08 | 1.14 | |
| Rab-7B | 0.65 | 0.86 | 1.12* | |
| Molecular transport | Immune regulator 1, ATPase, H+ transporting | 0.72 | 0.99 | 1.31* |
| ATPase, H+ transporting, lysosomal V0 subunit a | 0.75 | 0.95 | 1.04* | |
| Vacuolar ATP synthase subunit B | 0.74 | 1.04 | 1.42 | |
| Vacuolar ATP synthase subunit E1 | 0.65 | 0.92 | 1.35 | |
| Vacuolar ATP synthase subunit S1 | 0.69 | 0.89 | 0.99 | |
| Vacuolar ATP synthase subunit C | 0.79 | 1.09 | 1.40 | |
| Vacuolar ATP synthase subunit F | 0.79 | 1.25 | 1.34 | |
| Vacuolar ATP synthase subunit d | 0.71 | 0.98 | 1.15* | |
| Vacuolar ATP synthase catalytic subunit A | 0.79 | 0.96 | 1.28 | |
| Isoform 1 of Reticulon-4 | 0.78 | 0.88 | 0.94 | |
| Hydrolases | Cathepsin Z | 0.74 | 0.97 | 0.98 |
| Lysosomal acid phosphatase | 0.75 | 1.15 | 1.23 | |
| Metabolism | Collectin subfamily member 12 | 0.79 | 0.92 | 0.88 |
| Uncharacterized | Zinc finger, FYVE domain containing 26 | 0.60 | 0.93 | 1.27 |
| UP-Regualted Proteins | ||||
| Membrane receptor/Signaling | Transferrin receptor protein 1 | 1.34 | 0.96 | 1.05 |
| Vesicular and protein trafficking | Vacuolar protein sorting-associated protein 41 homologue | 1.50 | 0.96 | 1.04* |
| Rab-5A | 1.33 | 1.10 | 0.91 | |
| Rab-5C | 1.29 | 0.96 | 0.98 | |
| Rab-14 | 1.67 | 1.17 | 1.13* | |
| Signaling | Isoform 1 of Sequestosome-1 | 1.39 | 1.06 | 1.12 |
| Isoform 2 of Sequestosome-1 | 1.82 | 1.10 | 1.19 | |
| Iron homeostasis | Ferritin H subunit | 1.50 | 1.28 | 1.06* |
| Protein folding | Calnexin | 1.53 | 1.01 | 1.40 |
| Protein folding/antigen presentation | Heat-shock protein 90B1 | 1.48 | 0.77 | 1.31 |
| Hydrolases | Tripeptidyl-peptidase 1 | 1.32 | 1.21 | 1.08 |
| Metabolism | N-acetylglucosamine-6-sulfatase | 1.26 | 1.09 | 1.20 |
| d-3-phosphoglycerate dehydrogenase | 1.58 | 1.10 | 1.20* | |
| Alpha-N-acetylglucosaminidase | 1.37 | 1.08 | 1.07 | |
| Cell mobility and cytoskeleton | Vimentin | 1.82 | 1.12 | 1.26 |
| Titin | 1.28 | 0.99 | 1.20 | |
| Uncharacterized | 24 kDa protein | 1.30 | 0.99 | 1.02 |
| Ddost | 1.31 | 0.98 | 1.18 | |
The protein ratios represent the relative abundance of a specific protein in the phagosome bearing latex beads coated with ManLAM, PILAM, or LPS vs uncoated beads. Macrophage cells were treated by latex beads coated with three different bacterial lipoglycans or uncoated beads, and proteins were extracted from phagosomal membranes for quantitative comparison using iTRAQ isotopic labels. The protein ratios indicate relative changes of protein levels in the phagosomes under a specific lipoglycan treatment in comparison with no treatment. Forty-two proteins were found to be significantly changed only by ManLAM treatment, which were summarized in this table.
The ratio with an asterisk is the mean of two replicate measurements.
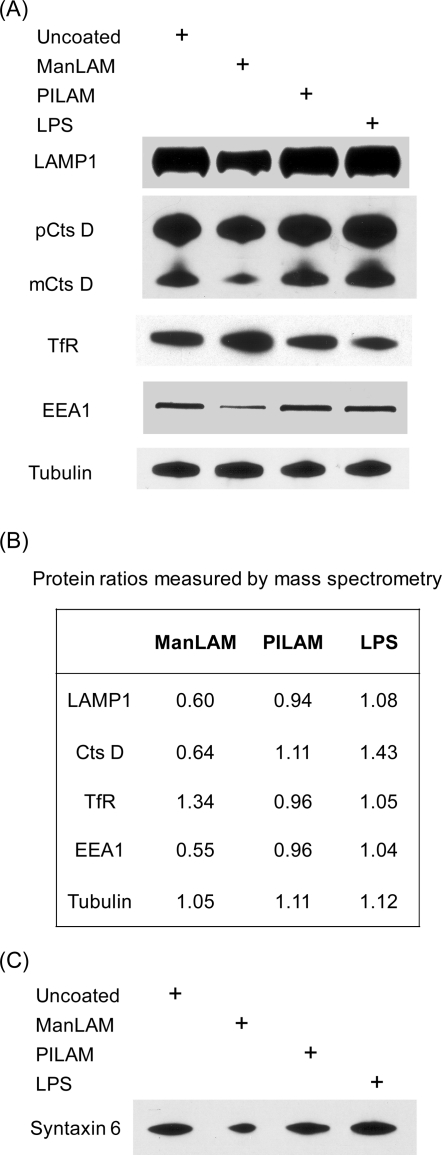
Of the 42 proteins found to be regulated by ManLAM, 24 were down-regulated compared to samples from untreated macrophages. Lysosome-associated membrane protein 1 (LAMP1), the late endosome membrane marker CD63, and the late endosome-specific small GTPase Rab7 were all in this group, a finding that agrees with previous microscopy studies showing diminished recruitment of these proteins to phagosomes containing ManLAM-coated beads13 or live M. tuberculosis.3 In addition, lysosomal enzymes such as the cysteine protease cathepsin D (CatD), and subunits of lysosomal vacuolar ATPase (v-ATPase) were down-regulated by 30−40% (Table 1). Loss of v-ATPase, a proton transporter, could explain the reduced LysoTracker staining of phagosomes containing ManLAM-coated beads reported earlier.15 Interestingly, we noticed that the phagosomes containing ManLAM-coated beads partially share the features of those harboring live M. tuberculosis. Russell group has reported earlier that mycobacterium phagosomes exclude v-ATPase to restrict vacuole acidification33 as well as reduce acquisition of the mature form of CatD.34 In Western blot analysis, we also observed reduced production of the mature CatD in the ManLAM-containg phagosome (Figure 3A), though to a less extent than that observed in the live mycobacterium phagosome.34 In summary, our proteomic data are consistent with classical cell-based or biochemical assays and suggest that ManLAM contributes to phagosome maturation arrest.
Figure 3.
Effect of ManLAM on selected proteins involved in phagosome maturation. (A) Immunoblot of phagosomal membrane extracts from cells treated with different lipoglycans using antibodies specific for endosomal markers and a lysosomal protease. (B) Relative ratios of specific proteins measured in our proteomic study. Each ratio represents the changes of a particular protein in the presence of ManLAM-, PILAM-, or LPS-coated beads relative to the uncoated beads. Ratios for LAMP1 and CatD are shown as the mean values from two experiments. Tubulin was used as loading control. (C) Immunoblot of syntaxin 6 in phagosome membrane extracts under different lipoglycan treatments. Note: this protein was not identified in the proteomic analysis.
Among the down-regulated species, we were particularly interested in a protein of unknown function containing a zinc finger FYVE domain (IPI00554920.5). This domain is known to bind phosphatidylinositol 3-phosphate (PI3P),35 a regulatory phospholipid that mediates membrane trafficking, endosomal protein sorting, and multisubunit enzyme assembly via multiple effectors.36−39 Importantly, PI3P plays an essential role in phagosomal acquisition of lysosomal constituents, and it is excluded from phagosomes containing live mycobacteria.4 Early endosome antigen 1 (EEA1), a protein that is known to be required for phagolysosome biogenesis and that also contains a FYVE domain, was also down-regulated by ManLAM. By analogy, the uncharacterized protein may be a new PI3P-binding effector involved in phagosome maturation and is possibly modulated by the M. tuberculosis lipoglycan.
Eighteen proteins increased in abundance when cells were treated with ManLAM-coated beads (Table 1). The small GTPase Rab14 is critical for stimulating vesicle fusion specifically between phagosomes and early endosomes rather than late endosomes.40 Overexpression of this regulatory protein is known to prevent phagosomes containing dead mycobacteria from maturing into bactericidal phagolysosomes.40 ManLAM-induced upregulation of Rab14 in phagosomes may promote phagosome-early endosome fusion thereby stalling the maturation process. Another protein in this group, vacuolar protein sorting-associated protein 41 (Vam2p homologue), is also known to concentrate in early endosomes.41 The protein interacts with multiple SNARE family members, which are important vesicle fusion regulators.41 The observation that ManLAM up-regulates at least two known early endosomal proteins suggests that its host phagosome resembles early endosomes, a proposal that been previously put forth for M. tuberculosis-containing phagosomes.33,34,40,42,43
Biochemical Validation of ManLAM-Regulated Proteins
We performed immunoblotting to validate quantitative changes of certain phagosomal proteins in response to ManLAM treatment. LAMP1, CatD, and EEA1 were down-regulated in the presence of ManLAM-coated beads compared to samples treated PILAM-coated, LPS-coated or uncoated beads (Figure 3A). The observed down-regulation of EEA1 concurs with Deretic and co-workers’ report that EEA1 is depleted from phagosomes containing live M. tuberculosis.44 By contrast, transferrin receptor (TfR), an early endosome marker, was found to be up-regulated in phagosomes containing ManLAM-coated beads. This coincides with a previous report by Clemens and Horwitz, showing that only the phagosomes harboring live mycobacterium acquire transferrin added exogenously.43 These trends revealed by immunoblotting matched those observed by quantitative mass spectrometry (Figure 3B).
The reduced levels of EEA1 in phagosomes containing ManLAM-coated bead may have broad consequences for vesicle fusion and membrane trafficking. EEA1 is recruited to organelles via its association with PI3P on organelle membranes45 and also interacts with syntaxin 6, a SNARE protein that participates in the vesicular traffic between the TGN and the endocytic system including phagosomes.46,47 Inhibition of EEA1’s function causes reduced accumulation of syntaxin 6 in phagosomes.12 Likewise, we considered the possibility that syntaxin 6 is depleted from phagosomes bearing ManLAM-coated beads. Indeed, Western blot analysis showed that syntaxin 6 was down-regulated in the presence of ManLAM-coated beads (Figure 3C) while four other syntaxins (Stx3, 7, 8, 12) and two syntaxin-binding proteins (Stxbp2, 5) identified by our proteomic analysis did not exhibit reduced levels upon ManLAM treatment (Supporting Information Table 2B). Loss of syntaxin 6 from phagosomal membranes may cause a global reduction in the delivery of lysosomal components, including v-ATPase and CatD, from the TGN to phagosomes.
Taken together, the proteomic and biochemical data are consistent with the proposal that ManLAM undermines trafficking from the TGN to the phagosome, which is reported to depend on the production of PI3P by the kinase Vps34.48 Interestingly, we also observed LAMP1 down-regulated by ManLAM treatment, yet this protein is thought to be delivered to phagosomes in a PI3P-independent manner.12,35 Therefore, ManLAM may affect multiple trafficking pathways that contribute to maturation of the phagosome and its proteome.
ManLAM Diminishes Translocation of the Autophagosome Marker LC3 to Phagosomal Membranes and Inhibits Autophagosome Accumulation
A connection was recently revealed between the pathways of phagosome maturation and autophagy activation.20 Autophagy serves as a means for the removal of intracellular bacteria and viruses, apart from its primary function of maintaining cytoplasmic homeostasis.49−52M. tuberculosis survival in infected macrophages is suppressed by artificial induction of the autophagy response.53 We previously observed that the autophagosomal marker LC3 is enriched on LBC phagosomal membranes upon autophagy activation.20 LC3 is not only a widely used marker but also an essential component of the autophagy machinery.54 Upon the induction of autophagy, the 18-kDa cytosolic precursor LC3-I is cleaved at its C-terminus and conjugated to phosphatidylethanolamine, generating a 16-kDa form termed LC3-II.55 Lipid-modified LC3-II integrates into the membranes of autophagosomes and undergoes either recycling or degradation when autophagosomes fuse with lysosomes.56
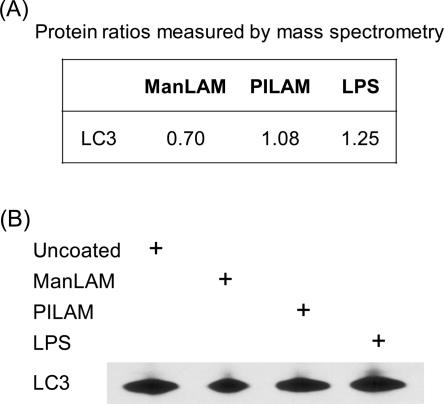
Given the importance of the autophagy pathway in the macrophage response to M. tuberculosis and the role of LC3 in this process, we sought to determine whether ManLAM affects the cellular distribution of LC3. Our proteomic data suggested that LC3 levels were reduced by about 30% in phagosomes containing ManLAM-coated beads (Figure 4A), an observation that was confirmed by Western blot analysis of phagosomal membrane extracts prepared in the same manner (Figure 4B). By contrast, LC3 was up-regulated in phagosomes containing LPS-coated beads and essentially unchanged in phagosomes containing PILAM-coated beads (Figure 4).
Figure 4.
The level of LC3 in phagosomal membranes is reduced by ManLAM treatment. (A) Proteomic quantitation; (B) and immunoblot of phagosomal membrane extracts using an anti-LC3 antibody.
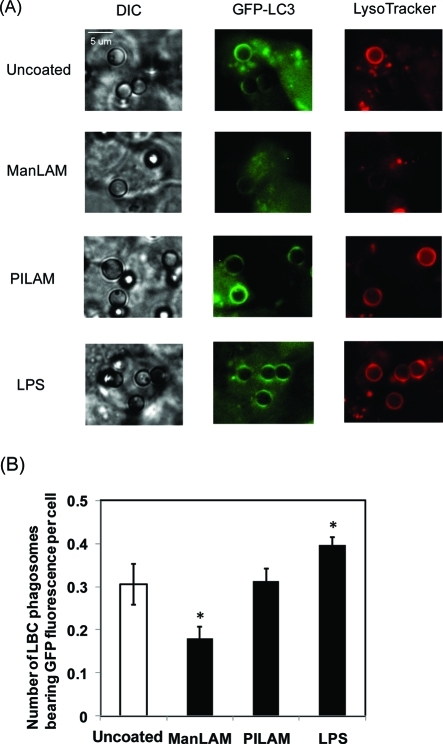
To determine the effects of ManLAM treatment on subcellular localization of LC3, we monitored a GFP-LC3 fusion protein in RAW264.7 murine macrophages upon treatment with various lipid-coated and uncoated beads (Figure 5A). The average number of microbeads bearing bright fluorescence of GFP per cell provided a metric of the relative amount of LC3 in the phagosomes (Figure 5B). Basal levels of GFP-LC3 were observed in phagosomes containing uncoated beads, reflecting the endogenous low autophagy activity in resting macrophages. Additionally, these phagosomes stained with LysoTracker, indicating successful fusion with lysosomes to become acidic vacuoles. Phagosomes bearing PILAM-coated beads showed similar levels of GFP-LC3 fluorescence and LysoTracker staining as in the presence of uncoated beads. In contrast, phagosomes containing ManLAM-coated beads exhibited weaker GFP-LC3 fluorescence than observed with uncoated beads, as well as weaker LysoTracker staining. LPS-coated beads affected LC3 translocation to phagosomes in an opposite manner compared to ManLAM-coated beads. In response to LPS-coated beads, GFP-LC3 levels were increased in phagosomes, consistent with previous observations that LPS activates the autophagy pathway.57
Figure 5.
ManLAM reduces GFP-LC3 fluorescence in LBC phagosomes. (A) Translocation of GFP-LC3 to LBC phagosomes under different lipoglycan treatment. RAW cells stably expressing GFP-LC3 were allowed to internalize latex beads (3 μm) coated with a lipoglycan for 2 h. Control cells were treated with lipid-free beads. LysoTracker was added in the last 15 min to stain acidic vacuoles. (B) Quantitation of GFP-LC3 colocalization with phagosomes containing beads coated with different lipoglycans. A total of 200−250 cells were sampled in each experiment to find out the average number of LBC phagosomes bearing GFP fluorescence in each cell. The data are mean values ± SD from three separate experiments. *P < 0.05.
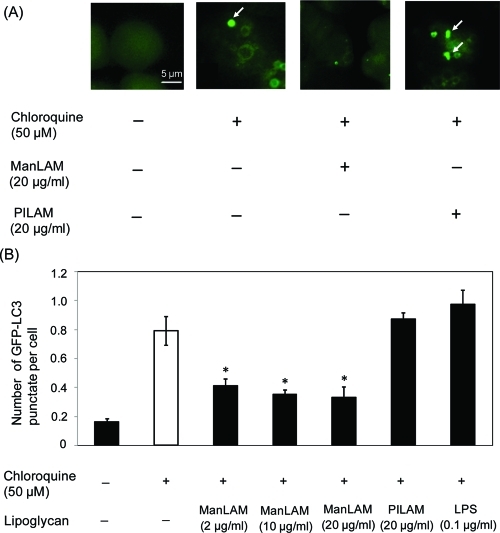
Next, we investigated the effects of the bacterial lipids on LC3 distribution in the presence of chloroquine, an anti-inflammatory drug that causes accumulation of autophagosomes by preventing their fusion with lysosomes.58 The drug has been previously employed to facilitate measurements of autophagic flux in vivo.58 RAW264.7 cells stably expressing GFP-LC3 were treated by chloroquine in the absence or presence of a given lipoglycan suspended in medium. Trypan blue staining was performed to confirm that the chemical and lipid treatments did not affect cell viability (Supporting Information Figure 3). Cells treated with chloroquine alone showed punctate fluorescence derived from GFP-LC3-II (Figure 6A), an indicator of elevated autophagy activity.55,57,59 Such punctate fluorescence was barely observed in untreated cells due to lysosomal degradation of autophagosomal LC3 in the absence of chloroquine.58 Cells treated with PILAM and chloroquine demonstrated a distribution of GFP-LC3-II that was similar to that observed with chloroquine treatment alone. However, GFP-LC3II fluorescence was significantly more diffuse in cells that were treated with ManLAM (Figure 6A). Quantitation of the average number of GFP-LC3-II puncta (>1 μm) per cell in the presence of the various bacterial lipids is shown in Figure 6B. Similar data were obtained from cells treated with three different doses of ManLAM, suggesting that the minimal effective concentration could be even lower than the 2 μg/mL used in our assay. Collectively, these data indicate that ManLAM, but not PILAM or LPS, suppresses the accumulation of autophagosomes induced by chloroquine. Thus, ManLAM appears to interfere with the endogenous formation of autophagic vacuoles, an early stage of autophagy activation.
Figure 6.
Direct exposure of macrophages to ManLAM reduces chloroquine-induced accumulation of autophagosomes. (A) Macrophages expressing GFP-LC3 were incubated with chloroquine (50 μM) in the presence or absence of either ManLAM or PILAM for 2 h. Arrows, representative LC3 punctate stains. (B) Quantitation of LC3 punctate structures (>1 μm) in cells incubated with chloroquine in the absence or presence of a specific lipoglycan. Three concentrations of ManLAM were tested. The data are mean values ± SD from three separate experiments. *P < 0.05.
Conclusions
Using a new platform for quantitative comparison of the membrane proteomes of macrophage phagosomes, we identified 823 proteins of which 42 were significantly regulated by ManLAM from M. tuberculosis but not PILAM derived from nonpathogenic mycobacteria or E. coli LPS. Several ManLAM-regulated proteins are known to be involved in vesicle trafficking pathways and phagosome maturation. Others have unknown function (e.g., FYVE domain-containing protein), but their regulation by ManLAM suggests a role in membrane trafficking events important for endosomal fusion and interaction with phagosomes.
We also found that ManLAM suppresses the accumulation of LC3-II in both LBC phagosomes and autophagosomes, whereas PILAM has no such effects. Given the established importance of phagocytosis and autophagy in the macrophage response to M. tuberculosis infection, it is possible that ManLAM’s functions include interference in these critical processes of innate immunity.
The direct molecular targets of ManLAM remain an open question. Fratti et al. proposed Vps34, the kinase responsible for PI3P production, as a possible target,44 and indeed phagosomes containing M. tuberculosis show retarded and reduced acquisition of this critical traffic-regulating lipid.45 Delineating the downstream effectors of ManLAM is likely to reveal new players in phagosome maturation, autophagy activation, and other pathways. As well, understanding the mechanisms by which ManLAM undermines the macrophage response may reveal new therapeutic avenues.
Acknowledgments
We thank Dr. Patrick Fitzgerald (St. Jude’s Children’s Research Hospital, Memphis, TN) for the gift of LC3-GFP-expressing RAW cells and Dr. Hu Cang for assistance with proteomic data analysis. This work was supported by a grant from the National Institutes of Health (AI51622) and a grant from the GTL program of the U.S. Department of Energy (DE-AC02-05CH11231).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Vergne I.; Chua J.; Singh S. B.; Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 2004, 20, 367–94. [DOI] [PubMed] [Google Scholar]
- Koul A.; Herget T.; Klebl B.; Ullrich A. Interplay between mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2004, 2 (3), 189–202. [DOI] [PubMed] [Google Scholar]
- Fratti R. A.; Vergne I.; Chua J.; Skidmore J.; Deretic V. Regulators of membrane trafficking and Mycobacterium tuberculosis phagosome maturation block. Electrophoresis 2000, 21 (16), 3378–85. [DOI] [PubMed] [Google Scholar]
- Vergne I.; Chua J.; Lee H. H.; Lucas M.; Belisle J.; Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2005, 102 (11), 4033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M. T.; Belisle J. T. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J. Bacteriol. 2000, 182 (23), 6850–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach H.; Papavinasasundaram K. G.; Wong D.; Hmama Z.; Av-Gay Y. Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 2008, 3 (5), 316–22. [DOI] [PubMed] [Google Scholar]
- Beatty W. L.; Rhoades E. R.; Ullrich H. J.; Chatterjee D.; Heuser J. E.; Russell D. G. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 2000, 1 (3), 235–47. [DOI] [PubMed] [Google Scholar]
- Besra G. S.; Brennan P. J. The mycobacterial cell wall: biosynthesis of arabinogalactan and lipoarabinomannan. Biochem. Soc. Trans. 1997, 25 (3), 845–50. [DOI] [PubMed] [Google Scholar]
- Nigou J.; Gilleron M.; Puzo G. Lipoarabinomannans: from structure to biosynthesis. Biochimie 2003, 85 (1−2), 153–66. [DOI] [PubMed] [Google Scholar]
- Khoo K. H.; Tang J. B.; Chatterjee D. Variation in mannose-capped terminal arabinan motifs of lipoarabinomannans from clinical isolates of Mycobacterium tuberculosis and Mycobacterium avium complex. J. Biol. Chem. 2001, 276 (6), 3863–71. [DOI] [PubMed] [Google Scholar]
- Khoo K. H.; Dell A.; Morris H. R.; Brennan P. J.; Chatterjee D. Inositol phosphate capping of the nonreducing termini of lipoarabinomannan from rapidly growing strains of Mycobacterium. J. Biol. Chem. 1995, 270 (21), 12380–9. [DOI] [PubMed] [Google Scholar]
- Fratti R. A.; Chua J.; Vergne I.; Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (9), 5437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. B.; Azad A. K.; Torrelles J. B.; Kaufman T. M.; Beharka A.; Tibesar E.; DesJardin L. E.; Schlesinger L. S. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J. Exp. Med. 2005, 202 (7), 987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I.; Chua J.; Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J. Exp. Med. 2003, 198 (4), 653–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welin A.; Winberg M. E.; Abdalla H.; Sarndahl E.; Rasmusson B.; Stendahl O.; Lerm M. Incorporation of Mycobacterium tuberculosis lipoarabinomannan into macrophage membrane rafts is a prerequisite for the phagosomal maturation block. Infect. Immun. 2008, 76 (7), 2882–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Pal S.; Das S.; Dasgupta S. K.; Majumdar S. Lipoarabinomannan induced cytotoxic effects in human mononuclear cells. FEMS Immunol. Med. Microbiol. 1998, 21 (3), 181–8. [DOI] [PubMed] [Google Scholar]
- Quesniaux V. J.; Nicolle D. M.; Torres D.; Kremer L.; Guerardel Y.; Nigou J.; Puzo G.; Erard F.; Ryffel B. Toll-like receptor 2 (TLR2)-dependent-positive and TLR2-independent-negative regulation of proinflammatory cytokines by mycobacterial lipomannans. J. Immunol. 2004, 172 (7), 4425–34. [DOI] [PubMed] [Google Scholar]
- Yoshida A.; Koide Y. Arabinofuranosyl-terminated and mannosylated lipoarabinomannans from Mycobacterium tuberculosis induce different levels of interleukin-12 expression in murine macrophages. Infect. Immun. 1997, 65 (5), 1953–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigou J.; Vasselon T.; Ray A.; Constant P.; Gilleron M.; Besra G. S.; Sutcliffe I.; Tiraby G.; Puzo G. Mannan chain length controls lipoglycans signaling via and binding to TLR2. J Immunol 2008, 180 (10), 6696–702. [DOI] [PubMed] [Google Scholar]
- Shui W.; Sheu L.; Liu J.; Smart B.; Petzold C. J.; Hsieh T. Y.; Pitcher A.; Keasling J. D.; Bertozzi C. R. Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (44), 16952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Zhu H. Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol. Cell. Proteomics 2005, 4 (12), 1948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal K.; Choe L. H.; Lee K. H. Shotgun proteomics using the iTRAQ isobaric tags. Briefings Funct. Genomics Proteomics 2006, 5 (2), 112–20. [DOI] [PubMed] [Google Scholar]
- Desjardins M.; Celis J. E.; van Meer G.; Dieplinger H.; Jahraus A.; Griffiths G.; Huber L. A. Molecular characterization of phagosomes. J. Biol. Chem. 1994, 269 (51), 32194–200. [PubMed] [Google Scholar]
- Pierce A.; Unwin R. D.; Evans C. A.; Griffiths S.; Carney L.; Zhang L.; Jaworska E.; Lee C. F.; Blinco D.; Okoniewski M. J.; Miller C. J.; Bitton D. A.; Spooncer E.; Whetton A. D. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteomics 2008, 7 (5), 853–63. [DOI] [PubMed] [Google Scholar]
- Shui W.; Gilmore S. A.; Sheu L.; Liu J.; Keasling J. D.; Bertozzi C. R. Quantitative proteomic profiling of host-pathogen interactions: the macrophage response to Mycobacterium tuberculosis lipids. J. Proteome Res. 2009, 8 (1), 282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. L.; Huang Y. N.; Marchese J. N.; Williamson B.; Parker K.; Hattan S.; Khainovski N.; Pillai S.; Dey S.; Daniels S.; Purkayastha S.; Juhasz P.; Martin S.; Bartlet-Jones M.; He F.; Jacobson A.; Pappin D. J. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3 (12), 1154–69. [DOI] [PubMed] [Google Scholar]
- Hoffman K. B.; Kessler M.; Ta J.; Lam L.; Lynch G. Mannose-specific lectins modulate ligand binding to AMPA-type glutamate receptors. Brain Res. 1998, 795 (1−2), 105–11. [DOI] [PubMed] [Google Scholar]
- Yu H.; Wakim B.; Li M.; Halligan B.; Tint G. S.; Patel S. B. Quantifying raft proteins in neonatal mouse brain by ‘tube-gel’ protein digestion label-free shotgun proteomics. Proteome Sci. 2007, 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C. L.; Chien C. W.; Chen W. C.; Chen Y. R.; Wu C. P.; Li H.; Chen Y. J. A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Mol. Cell. Proteomics 2008, 7 (10), 1983–97. [DOI] [PubMed] [Google Scholar]
- Gan C. S.; Chong P. K.; Pham T. K.; Wright P. C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 2007, 6 (2), 821–7. [DOI] [PubMed] [Google Scholar]
- Spellman D. S.; Deinhardt K.; Darie C. C.; Chao M. V.; Neubert T. A. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol. Cell. Proteomics 2008, 7 (6), 1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. M.; Tan Y.; Yuan Y. J. Temporal quantitative proteomics of Saccharomyces cerevisiae in response to a nonlethal concentration of furfural. Proteomics 2009, 9 (24), 5471–83. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S.; Schlesinger P. H.; Chakraborty P.; Haddix P. L.; Collins H. L.; Fok A. K.; Allen R. D.; Gluck S. L.; Heuser J.; Russell D. G. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 1994, 263 (5147), 678–81. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S.; Schaible U. E.; Russell D. G. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996, 15 (24), 6960–8. [PMC free article] [PubMed] [Google Scholar]
- Lindmo K.; Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 2006, 119 (Pt. 4), 605–14. [DOI] [PubMed] [Google Scholar]
- Vieira O. V.; Botelho R. J.; Rameh L.; Brachmann S. M.; Matsuo T.; Davidson H. W.; Schreiber A.; Backer J. M.; Cantley L. C.; Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 2001, 155 (1), 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A.; Lippe R.; Christoforidis S.; Gaullier J. M.; Brech A.; Callaghan J.; Toh B. H.; Murphy C.; Zerial M.; Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 1998, 394 (6692), 494–8. [DOI] [PubMed] [Google Scholar]
- Siddhanta U.; McIlroy J.; Shah A.; Zhang Y.; Backer J. M. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3′-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J. Cell Biol. 1998, 143 (6), 1647–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F.; Liu H.; Field S. J.; Akbary H.; Matsuo T.; Brown G. E.; Cantley L. C.; Yaffe M. B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001, 3 (7), 675–8. [DOI] [PubMed] [Google Scholar]
- Kyei G. B.; Vergne I.; Chua J.; Roberts E.; Harris J.; Junutula J. R.; Deretic V. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. Embo J 2006, 25 (22), 5250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. C.; Winistorfer S. C.; Poupon V.; Luzio J. P.; Piper R. C. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell 2004, 15 (3), 1197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via L. E.; Deretic D.; Ulmer R. J.; Hibler N. S.; Huber L. A.; Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 1997, 272 (20), 13326–31. [DOI] [PubMed] [Google Scholar]
- Clemens D. L.; Horwitz M. A. The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J. Exp. Med. 1996, 184 (4), 1349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R. A.; Backer J. M.; Gruenberg J.; Corvera S.; Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 2001, 154 (3), 631–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy G. E.; Owens R. M.; Bennett L.; Russell D. G.; Butcher B. A. Kinetics of phosphatidylinositol-3-phosphate acquisition differ between IgG bead-containing phagosomes and Mycobacterium tuberculosis-containing phagosomes. Cell Microbiol. 2005, 7 (11), 1627–34. [DOI] [PubMed] [Google Scholar]
- Bock J. B.; Klumperman J.; Davanger S.; Scheller R. H. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol. Biol. Cell 1997, 8 (7), 1261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F.; Tang B. L.; Galli T.; Tenza D.; Saint-Pol A.; Yue X.; Antony C.; Hong W.; Goud B.; Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 2002, 156 (4), 653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvera S. Phosphatidylinositol 3-kinase and the control of endosome dynamics: new players defined by structural motifs. Traffic 2001, 2 (12), 859–66. [DOI] [PubMed] [Google Scholar]
- Levine B.; Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007, 7 (10), 767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I.; Amano A.; Mizushima N.; Yamamoto A.; Yamaguchi H.; Kamimoto T.; Nara A.; Funao J.; Nakata M.; Tsuda K.; Hamada S.; Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science 2004, 306 (5698), 1037–40. [DOI] [PubMed] [Google Scholar]
- Ogawa M.; Yoshimori T.; Suzuki T.; Sagara H.; Mizushima N.; Sasakawa C. Escape of intracellular Shigella from autophagy. Science 2005, 307 (5710), 727–31. [DOI] [PubMed] [Google Scholar]
- Paludan C.; Schmid D.; Landthaler M.; Vockerodt M.; Kube D.; Tuschl T.; Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005, 307 (5709), 593–6. [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G.; Master S. S.; Singh S. B.; Taylor G. A.; Colombo M. I.; Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119 (6), 753–66. [DOI] [PubMed] [Google Scholar]
- Tanida I.; Ueno T.; Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36 (12), 2503–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y.; Mizushima N.; Ueno T.; Yamamoto A.; Kirisako T.; Noda T.; Kominami E.; Ohsumi Y.; Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19 (21), 5720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T.; Fujita N.; Yoshimori T. The late stages of autophagy: how does the end begin. Cell Death Differ. 2009, 16 (7), 984–90. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Jagannath C.; Liu X. D.; Sharafkhaneh A.; Kolodziejska K. E.; Eissa N. T. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007, 27 (1), 135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai-Kanai E.; Yuan H.; Huang C.; Sayen M. R.; Perry-Garza C. N.; Kim L.; Gottlieb R. A. A method to measure cardiac autophagic flux in vivo. Autophagy 2008, 4 (3), 322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.; De Haro S. A.; Master S. S.; Keane J.; Roberts E. A.; Delgado M.; Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 2007, 27 (3), 505–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.