It is becoming increasingly clear that the nuclear architecture plays an essential role in the genome maintenance and functions.[1] However, the study of nuclear structures, dynamics, and functions presents a major challenge to biological scientists. The key challenge is the imaging of nuclear components using conventional probes, such as fluorescent dyes or proteins. Such probes provide weak signal and tend to rapidly photobleach (typically < 10 s), which limits both the temporal and spatial imaging resolution. An alternative is to use the bright probes such as quantum dots (QDs). However, to get such relatively large-sized probes into the nucleus requires overcoming the physical barriers of both the plasma membrane and the nuclear envelope without imposing unintended side effects.[2]
With their bright fluorescence, photostability, and nanoscale size, QDs have emerged as an alternative probe that complements fluorescent dyes and proteins.[3–5] One of the most promising applications of QDs is molecular imaging in living cells.[3, 4, 6–11] However, realization of the full potential of QDs for molecular imaging faces several problems, including the relatively large size of QD–biomolecule conjugates and QD–target molecule complexes,[6, 10, 11] the lack of strategies for targeting intracellular biomolecules,[4] the instability of the antibody-mediated targeting,[6, 11] QD multivalency,[10] and the intracellular delivery of QDs.[2–5, 9, 12, 13] In particular, the intracellular delivery of singly-dispersed QDs into the cytoplasm or organelles, such as the nucleus, remains a major challenge for live cell studies.[2–5, 9, 12, 13] Because of this delivery problem, except for a few studies,[8] the use of QDs for molecular imaging in living cells has been mostly limited to visualizing plasma membrane proteins[6, 7, 9–11] and biological processes related to endocytosis.[13, 14] The targeted delivery of QDs into the nucleus of living cells is even more challenging, and the use of QDs within the nucleus is scarce.[2] Several research groups have demonstrated the targeted nuclear delivery of QDs by using nuclear localization signal (NLS) peptides.[2, 12, 16] However, the delivery efficiency strongly depends on the size of NLS-QD complexes and the cell type and cycle.[2] Furthermore, because of the lack of spatial, temporal, and quantitative control, the NSL-mediated delivery has only been demonstrated to track the nucleus with QDs, but not to observe subnuclear events.[2, 12]
In this regard, direct delivery methods, such as microinjection and nanoneedle-based delivery, have shown better performance: they can deliver homogeneously or sparsely dispersed QDs directly into the cytoplasm and the nucleus with no need for endosomal escape.[5, 12, 17] However, the relatively large size and tapered shape of the injection pipette makes microinjection liable to cell damage, especially for the nuclear delivery into small cells. The recently developed nanoneedle-based intracellular delivery method[17–19] has the potential to achieve the best outcome for the targeted nuclear delivery as it is capable of mechanically passing through both the cellular and nuclear membranes with minimal intrusiveness, and locally delivering and releasing the attached probes with sub-microscale precision. However, the previously reported nanoneedle-based method relies on the endogenous regulatory mechanism of cells to break the disulfide bonds in the linker molecules between the cargo and the nanoneedle.[17, 18] This release process is thus not under any external control and takes over 15 minutes as required.[17, 18]
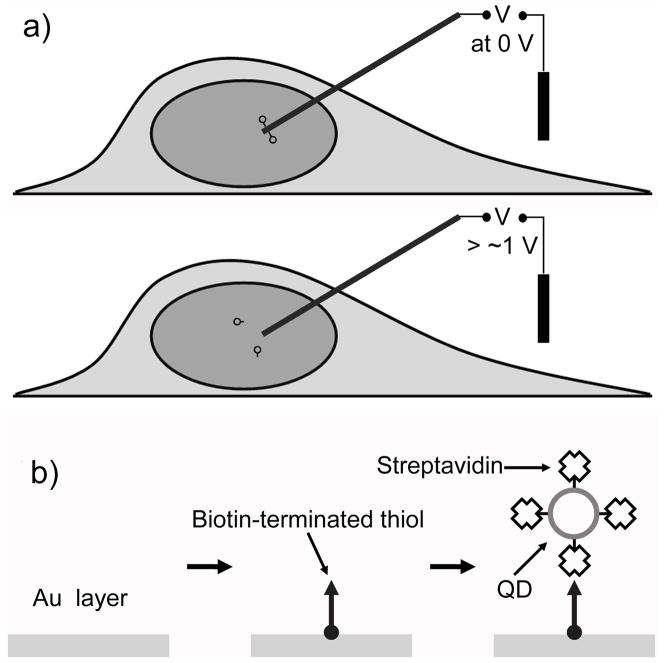
Herein we present a controlled delivery and release method that uses a cargo-carrying nanoneedle (doubly served as a nanoscale electrode) to penetrate into the nucleus of a living cell. To rapidly release the attached cargo inside the nucleus, we exploited an electrochemical means to directly break the incorporated electrochemically-active bonds by applying a small external electrical potential through the nanoneedle (Fig. 1a). The release of the cargo is thus almost instantaneous and is externally controlled by adjusting the magnitude and duration of the applied potential. We further show the use of this active delivery method to deliver singly dispersed QDs into the nucleus.
Figure 1.
Electrochemically controlled delivery of QDs into a living cell. a) Schematic describing the delivery and release principle of cargo through a membrane penetrating nanoneedle: applying a potential larger than the critical potential (~1.0 V) to the nanoneedle induces desorption of a SAM and thus release of QDs. b) Procedure for surface functionalization of a gold-coated nanoneedle and attachment of streptavidin-coated QDs.
We used an individual boron nitride nanotube (~50 nm in diameter) coated with a thin layer of gold (10–20 nm in thickness) as a nanoneedle (and an electrode), which was then attached onto a conductive sharpened wire for easy handling (See Supporting Figure S1).[17, 20] The nanotube is chemically synthesized and has a uniform diameter, high mechanical strength and resilience, ideal as a membrane-penetrating nanoneedle as reported previously.[17, 20] We attached a limited amount of cargo (streptavidin-conjugated QDs) by conjugating it on a self-assembly monolayer (SAM) formed on the ~ 2 μm long end segment of this gold-coated nanoneedle (Figure 1b). The delivery of such tiny quantities of cargo limited by the nanoscale size of the nanoneedle minimizes their interference with cell dynamics experiments or with cell physiology. For example, excess QDs delivered into cells can present background interferences from out-of-focus QDs, decreasing the signal-to-noise ratio and thus making it difficult to track single QDs, besides interfering with cellular functions.[14] More problematically, QDs are difficult to wash away once they are introduced into cells.[3]
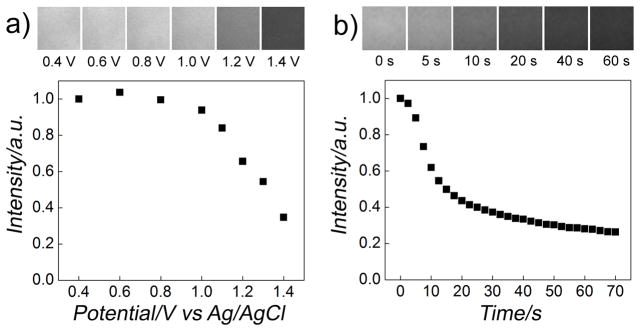
For the controlled release of cargo, we exploited the electrochemical desorption of the SAM from the gold surface (Figure 2).[21–23] A two-electrode configuration was used with the nanoneedle as a working electrode and a Ag/AgCl or Pt wire immersed in the cell medium as a counter/reference electrode.[20] To determine the critical potential (versus the reference electrode in the medium) and the kinetics of the QD release from the gold surface, we measured the fluorescent intensity of QD functionalized gold surfaces in the medium as a function of the increasing electrical potential applied on the gold surface, as shown in Figure 2a; and as a function of time when applying a constant electrical potential sufficient for the release of the QDs (1.4 V versus Ag/AgCl wire in the medium), as shown in Figure 2b. These control experiments showed that the onset of the QD release occurred at an electrical potential of ~1.0 V and the total release could be completed within ~60 s. Because of the different ionic environment in the cytoplasm and the nucleus (i.e., membrane potentials), the actual critical potential and the kinetics of the QD release inside the nucleus would be somewhat different from those obtained from these control experiments. However, because of the small membrane potential (~0.1 V) compared to the applied potential of ~1.0 V, its effect should not be significant for the release of QDs within the nucleus, as shown in our experiment later.
Figure 2.
Electrochemically controlled release of QDs from gold surfaces in Dulbecco’s modified Eagle’s medium (DMEM). a) Fluorescence intensity versus applied potential on a gold surface functionalized with QDs via biotin-terminated thiols (biotin-terminated tri(ethylene glycol)hexadecanethiol) (top). The fluorescence of the gold surface was measured after applying a potential for 30 s sequentially. Applying a potential larger than the critical potential (~1.0 V) induced the desorption of the SAM and thus the attached QDs and their diffusion into the bulk solution, decreasing the fluorescence signal. Corresponding fluorescence images at the labeled electrical potentials of the gold surface in gray scale are shown at the top of the panel, showing gradual loss of the fluorescence. b) Fluorescence intensity versus time curve of a gold surface functionalized with QDs at a constant applied potential of 1.4V versus Ag/AgCl wire in DMEM. The fluorescence intensity decreased as the SAM and the attached QDs were desorbed and diffused into the bulk solution. Corresponding fluorescence images at the indicated time are shown at the top of the panel.
We demonstrate this method by delivering QDs into the nucleus of living HeLa cells. We used streptavidin-conjugated QDs with overall diameter of ~15–20 nm (Qdot 655 streptavidin conjugates, Invitrogen). Notably, a previous study indicated that such commercial QDs conjugated with NLS peptides (> ~25 nm in overall diameter including the NLS peptides) did not enter the nucleus by the NLS-mediated delivery.[2] We did the delivery experiment using a micromanipulator (InjectMan NI 2, Eppendorf) integrated in an inverted fluorescence microscope (Leica) as described previously (see Supporting Information).[17] Once the nanoneedle pierced through the plasma membrane and the nuclear envelope and reached the target area, a small electrical potential was applied to the nanoneedle (typically, <1.4 V versus Ag/AgCl wire for 30–90 s) to release the SAM and thus the QDs; either a positive (oxidative) or negative (reductive) potential can be applied to release the QDs.[21–23] Once the desired release was completed, the nanoneedle was retracted from the cell. As the release is electrochemically controlled, we may program the release by adjusting the magnitude and duration of the applied electrical potential. An added benefit of this strategy is that the full electrochemical desorption of the SAM regenerates the gold surface for the reuse of the nanoneedle.
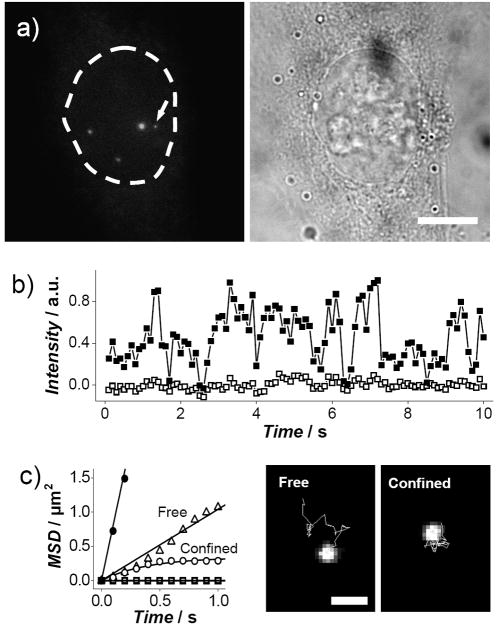
After the delivery, we imaged the target cell using fluorescence microscopy. We detected both slow-moving and fast-moving QDs within the boundary of the nucleus, showing their confinement in the nucleus (Figure 3a and Supporting Information Movie S1). The size limit of the passive diffusion of materials through the nuclear pore complex also excludes the diffusive introduction of the QDs from the cytoplasm into the nucleus.[12] Additionally, when we delivered QDs specifically into the cytoplasm, the QDs were confined in the cytoplasm (see Supporting Figure S3).[17] Because of the substantial reduction of the background signal from the out-of-focus QDs, as a result of delivering a tiny amount of QDs, we could detect single QDs even with a simple epifluorescence microscope. The release of QDs in cell occurred only when an electrical potential above a certain magnitude was applied, confirming both the release of QDs through the electrochemical desorption of the SAM and the initial stable attachment of QDs on the nanoneedle.[17] We did not observe any distinct difference between the tested cells and their neighboring cells during the time span of monitoring the cells (~30 minutes); the tested cells remained viable after the penetration of the nanoneedle and the application of the electrical potential as assessed by the cell shape and the trypan blue assay (see Supporting Information). The charge introduced into the cell via the nanoneedle in the electrochemical release process did not perturb the physiological milieu of the cell (see Supporting Information).
Figure 3.
Delivery and tracking of single QDs inside a nucleus. a) Delivery of QDs into the nucleus of a living HeLa cell: fluorescence image (left, bright spots showing delivered QDs) and the corresponding bright-field images (right) of the cell on a focal plane. The dotted line indicates the boundary of the nucleus. Scale bar, 10 μm. b) Typical time trace of the fluorescence intensity (■) of a slow-moving QD, indicated by the arrow in (a), showing the blinking pattern, plotted with the background signal (□) of neighboring areas. c) MSD versus time data of QDs in the nucleus, showing three types of characteristic motions: free diffusion (△ and ●), confined diffusion (○) and virtually stationary during the observation (□) (left). The solid lines are the fit of free and confined diffusion models.[9, 17] The freely-diffusing QDs in the plot have D values of 0.3 and 2 μm2/s. Tracking of QDs in the nucleus, showing freely diffusing and confined QDs (right). Scale bar, 1 μm.
To determine whether this method can deliver singly-dispersed QDs, we measured the fluorescence intensity of slow-moving QDs. The detection of fast-moving QDs, especially those with low fluorescence, was still difficult due to their random diffusion in the three-dimensional environment of cell (this also made difficult the absolute quantification of the already tiny amount of delivered QDs). The acquired blinking behavior, typically only present for single QDs, indicated that the QDs were mostly single (Fig. 3b and Supporting Information Movie S2).[3, 6, 9, 17] Small QDs clusters were also seen near the release site, mostly less mobile, resulted probably from their trapping to intracellular structures upon release. To demonstrate the capability of tracking single QDs and determine the dynamic behavior of the QDs in the nucleus, we applied the single-molecule tracking technique (Figure 3c). The mean-square displacement (MSD) of moving QDs showed that QDs could freely diffuse in the nucleoplasm at a similar rate (with a diffusion coefficient D of ~0.01–2 μm2/s, mean = 0.5 ± 0.7 μm2/s, n = 8) as in the cytoplasm (D = ~0.1–4 μm2/s);[17] but, these values might be underestimated as overall it was more difficult to track fast-diffusing QDs than slow-diffusing ones. The tracking of the diffusive QDs can also be used to probe the local physical properties within the nucleus by bio-microrheology (the nuclear delivery of probe particles has been a major obstacle in nuclear bio-microrheology measurement).[24] The viscosity of fluid can be related to the diffusion coefficient according to the Stokes–Einstein relation, D = kT/(6πηr), where k is the Boltzmann’s constant, T is the absolute temperature, η is the viscosity, and r is the hydrodynamic radius of the particle (QDs). Taking r to be ~ 12.8 nm for QDs (estimated from the diffusion coefficient of QDs in aqueous solution of ~17 μm2/s,[25]), the “nanoscale” viscosity inside the nucleus was determined, from the locally measured diffusion coefficient, to be ~5–2000 cP. These result suggests that the local viscosity in the region where QDs travel (probably, chromatin-poor domains) is similar to that of the cytoplasm that we measured previously,[17] favorable for molecular transport processes through diffusion within the nucleus.[1, 26–28] It also indicates that the physical environment of the nucleus is heterogeneous, consistent with published results.[1, 29]
This spatially and now temporally controlled cellular delivery of QDs serves as the first step towards providing alternative new strategies to study biology within the nucleus of living cells, which would otherwise be technically challenging or even impossible. For example, in combination with effective molecular targeting strategies of nanoparticles, which are already under active research,[4, 6, 10, 11] this method can potentially enable the use of nanoparticles, such as quantum dots and magnetic nanoparticles, as molecular probes for simultaneous imaging and manipulation of single biomolecules (a major challenge in single-molecule studies)[30] in the nucleus and for observation of subnuclear structures and events.[29] Technically, it is also possible to apply this method for the direct intracellular delivery of biomolecules, such as proteins and DNA, which has been demonstrated previously outside cells.[21, 23] For this applications, although a recent study showed that proteins were still active after an electrical potential-induced release from a TiO2 electrode (after applying a much higher electrical potential of 5 V than that used in our study),[31] the effect of the applied electrical field on the functionality of the delivered biomolecules needs a more thorough examination, especially when the biomolecules are directly conjugated on the nanoneedle without a layer of insulating linker molecules. Finally, a major concern in the application of this method for cellular studies is its limited throughput. One way to overcome that is to automate the delivery process or adapt it to an array of nanoneedles[32, 33]. For example, by integrating this electrochemical release concept with the delivery method demonstrated in a most recent study that showed the use of a substrate patterned with a large array of vertical and fixed Si nanowires for the efficient delivery of DNAs, RNAs, peptides, proteins and small molecules into cells[33], a spatially and temporally-programmed release and delivery into cells at a large scale may be realizable, if in such applications, the spatial precision (the sub-cellular precision) of the delivery is not of the chief requirement.
Experimental Section
Surface functionalization of nanoneedles
The gold-coated nanoneedle was prepared using a boron nitride nanotube as described previously.[17, 20] A self-assembled monolayer (SAM) of biotin-terminated thiols was formed on the surface of the gold-coated nanoneedle by incubating the nanoneedle in 0.5 mM biotin-terminated tri(ethylene glycol)undecanethiol (Nanoscience instruments) or biotin-terminated tri(ethylene glycol)hexadecanethiol (Asemblon) in ethanol for 12 hours. Streptavidin-coated QDs (Qdot 655 streptavidin conjugates, Invitrogen) were conjugated by incubating the nanoneedle in 40 nM streptavidin-coated QDs in borate buffer (50 mM, pH 9.0) containing 1% bovine serum albumin for 30 minutes. A more detailed procedure is described previously.[17]
Cell culture and imaging
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C under 5% CO2. Images were acquired using a Leica inverted epifluorescence microscope with a 63× 1.32 numerical aperture oil-immersion objective, a charge-coupled device camera (C4742-95-12ERG, Hamamatsu), and a QD filter set for QD 655 (Chroma). The acquisition time for QD imaging was 50–500 ms. For the QD imaging inside the nucleus, to avoid the possibility of detecting QDs at the top or bottom of the nucleus, the nucleus envelop in the bright-field mode was identified and the cell on the same focal plane in the fluorescence mode was imaged.[17]
QD tracking
QDs were tracked in sequential fluorescence images of a cell using ImageJ.[34] The mean-square displacement (MSD) was computed as described previously.[17]
Cell viability tests
Cell viability was examined using the trypan blue assay (0.4%, Invitrogen). After the penetration of the nanoneedle into cells and the application of a potential (1.4 V for 60 s), the cells were incubated at 37 °C under 5% CO2 for about 10 minutes. After the incubation, individual cells were visually examined in serum-free medium with trypan blue (0.08%). The cells excluded the dye, indicating their viability (n = 5). To track individual cells, cells were placed on gridded cell culture dishes for the viability tests.
Supplementary Material
Footnotes
The work is supported by NIH grant GM072744 and NSF grant CBET-0933223 and CBET-0731096.
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
References
- 1.Misteli T, Soutoglou E. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen F, Gerion D. Nano Lett. 2004;4:1827–1932. [Google Scholar]
- 3.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 5.Delehanty J, Mattoussi H, Medintz I. Anal Bioanal Chem. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- 6.Howarth M, Takao K, Hayashi Y, Ting AY. Proc Natl Acad Sci U S A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 8.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 9.Bannai H, Levi S, Schweizer C, Dahan M, Triller A. Nat Protocols. 2007;1:2628–2634. doi: 10.1038/nprot.2006.429. [DOI] [PubMed] [Google Scholar]
- 10.Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY. Nat Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roullier V, Clarke S, You C, Pinaud F, Gouzer G, Schaible D, Marchi-Artzner V, Piehler J, Dahan M. Nano Lett. 2009;9:1228–1234. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 12.Derfus AM, Chan WCW, Bhatia SN. Adv Mater. 2004;16:961–966. [Google Scholar]
- 13.Ruan G, Agrawal A, Marcus AI, Nie S. J Am Chem Soc. 2007;129:14759–14766. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 14.Tekle C, van Deurs B, Sandvig K, Iversen TG. Nano Lett. 2008;8:1858–1865. doi: 10.1021/nl0803848. [DOI] [PubMed] [Google Scholar]
- 15.Stephens DJ, Pepperkok R. Proc Natl Acad Sci U S A. 2001;98:4295–4298. doi: 10.1073/pnas.081065198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozenzhak SM, Kadakia MP, Caserta TM, Westbrook TR, Stone MO, Naik RR. Chem Commun. 2005:2217–2219. doi: 10.1039/b418454h. [DOI] [PubMed] [Google Scholar]
- 17.Yum K, Na S, Xiang Y, Wang N, Yu MF. Nano Lett. 2009;9:2193–2198. doi: 10.1021/nl901047u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Kis A, Zettl A, Bertozzi CR. Proc Natl Acad Sci U S A. 2007;104:8218–8222. doi: 10.1073/pnas.0700567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SW, Nakamura C, Kotobuki N, Obataya I, Ohgushi H, Nagamune T, Miyake J. Nanomedicine. 2008;4:215–225. doi: 10.1016/j.nano.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Yum K, Cho HN, Hu J, Yu MF. ACS Nano. 2007;1:440–448. doi: 10.1021/nn700171x. [DOI] [PubMed] [Google Scholar]
- 21.Mali P, Bhattacharjee N, Searson PC. Nano Lett. 2006;6:1250–1253. doi: 10.1021/nl0609302. [DOI] [PubMed] [Google Scholar]
- 22.Canaria CA, So J, Maloney JR, Yu CJ, Smith JO, Roukes ML, Fraser SE, Lansford R. Lab Chip. 2006;6:289–295. doi: 10.1039/b510661c. [DOI] [PubMed] [Google Scholar]
- 23.Huang S, Schopf E, Chen Y. Nano Lett. 2007;7:3116–3121. doi: 10.1021/nl071643g. [DOI] [PubMed] [Google Scholar]
- 24.Weihs D, Mason TG, Teitell MA. Biophys J. 2006;91:4296–4305. doi: 10.1529/biophysj.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grünwald D, Hoekstra A, Dange T, Buschmann V, Kubitscheck U. ChemPhysChem. 2006;7:812–815. doi: 10.1002/cphc.200500632. [DOI] [PubMed] [Google Scholar]
- 26.Phair RD, Misteli T. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 27.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Proc Natl Acad Sci U S A. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handwerger KE, Gall JG. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Neuman KC, Nagy A. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y-Y, Roy P, Paramasivam I, Schmuki P. Angew Chem. 2010;122:361–364. doi: 10.1002/anie.200905111. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed Engl. 2010;49:351–354. [Google Scholar]
- 32.Park S, Kim YS, Kim WB, Jon S. Nano Lett. 2009;9:1325–1329. doi: 10.1021/nl802962t. [DOI] [PubMed] [Google Scholar]
- 33.Shalek AK, Robinsona JT, Karpa ES, Leea JS, Ahnb DR, Yoona MH, Suttona A, Jorgollic M, Gertnera RS, Gujrala TS, MacBeatha G, Yang EG, Park H. Proc Natl Acad Sci USA. 2010;107:1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sbalzarini IF, Koumoutsakos P. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.