Abstract
Purpose
To evaluate the prognostic significance of expression levels of a single microRNA, miR-181a, in the context of established molecular markers in cytogenetically normal acute myeloid leukemia (CN-AML), and to gain insight into the leukemogenic role of miR-181a.
Patients and Methods
miR-181a expression was measured in pretreatment marrow using Ohio State University Comprehensive Cancer Center version 3.0 arrays in 187 younger (< 60 years) adults with CN-AML. Presence of other molecular prognosticators was assessed centrally. A gene-expression profile associated with miR-181a expression was derived using microarrays and evaluated by Gene-Ontology analysis.
Results
Higher miR-181a expression associated with a higher complete remission (CR) rate (P = .04), longer overall survival (OS; P = .01) and a trend for longer disease-free survival (DFS; P = .09). The impact of miR-181a was most striking in poor molecular risk patients with FLT3-internal tandem duplication (FLT3-ITD) and/or NPM1 wild-type, where higher miR-181a expression associated with a higher CR rate (P = .009), and longer DFS (P < .001) and OS (P < .001). In multivariable analyses, higher miR-181a expression was significantly associated with better outcome, both in the whole patient cohort and in patients with FLT3-ITD and/or NPM1 wild-type. These results were also validated in an independent set of older (≥ 60 years) patients with CN-AML. A miR-181a-associated gene-expression profile was characterized by enrichment of genes usually involved in innate immunity.
Conclusion
To our knowledge, we provide the first evidence that the expression of a single microRNA, miR-181a, is associated with clinical outcome of patients with CN-AML and may refine their molecular risk classification. Targeted treatments that increase endogenous levels of miR-181a might represent novel therapeutic strategies.
INTRODUCTION
Several recent studies have revealed that micro-RNAs, short noncoding RNAs that hybridize to their target mRNAs and repress the expression of the encoded proteins,1 are not only involved in such biologic processes as cellular differentiation, proliferation, and survival, but also play an essential role in the development of solid tumors and acute myeloid leukemia (AML).2–6 In AML, genome-wide microRNA-expression profiling has revealed distinctive microRNA-expression signatures capable of differentiating among specific cytogenetic subtypes, such as core-binding factor (CBF) -AML with t(8;21), CBF-AML with inv(16) or t(16;16), and acute promyelocytic leukemia with t(15;17), and setting them apart from other AML subtypes.7–9 Moreover, microRNA expression signatures have been associated with mutations of NPM1,7,10 FLT3,7,10,11 and CEBPA,7,12 which are genetic alterations known to affect clinical outcome of patients belonging to the largest subset of AML—cytogenetically normal AML (CN-AML).13,14
Furthermore, we have recently demonstrated that deregulated microRNA expression may also be associated with outcome in CN-AML.5,11 Using microRNA-expression profiling in patients with CN-AML with unfavorable molecular features—FLT3-ITD and/or NPM1 wild-type (NPM1wt)—we discovered a prognostic microRNA signature consisting of 12 microRNA probes, five of which corresponded to members of the miR-181 family.5 Although these data provided initial support for the usefulness of microRNAs for assessment of molecular risk in AML, microRNAs have been linked to prognosis in AML mainly in the context of genome-wide profiling. This approach, however, is based on population analysis, and therefore, is relatively difficult to implement for prospectively assessing the molecular risk of individual patients. Thus new strategies are needed to increase the clinical applicability of microRNA expression–based prognostication in AML.
To our knowledge, the independent prognostic impact of expression levels of individual microRNAs, which are relatively easy to measure for molecular risk assessment of individual patients at diagnosis, has not been demonstrated in CN-AML outside of microRNA expression profiles. Thus, we sought evidence here that the expression levels of a single microRNA, miR-181a, could provide prognostic information in patients with CN-AML independently from a comprehensive panel of other established clinical and molecular predictors, and therefore, be readily applicable as a risk-stratification tool. We show that expression of miR-181a is strongly associated with outcome, which suggests that miR-181a expression could be used for individual patients' molecular risk assessment and perhaps as a potential therapeutic target.
PATIENTS AND METHODS
Patients, Treatment, and Cytogenetic Analysis
A total of 187 adult patients younger than 60 years (range, 18 to 59 years) with untreated, primary CN-AML and material available for analysis were included. Patients were treated similarly with intensive induction chemotherapy and consolidation with autologous peripheral blood stem-cell transplantation on Cancer and Leukemia Group B (CALGB) protocols 9621 (n = 89) and 19808 (n = 98).15,16 Of those who achieved a complete remission (CR), 82% received an autologous transplant. Cytogenetic analyses of pretreatment bone marrow (BM) samples were performed by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion study, and centrally reviewed.17,18 All patients gave informed consent for the research use of their specimens, in accordance with the Declaration of Helsinki. No patient received allogeneic stem-cell transplantation in first CR.
A cohort of 122 CN-AML patients age 60 years or older, treated on first-line CALGB protocols (Appendix, online only), constituted an independent validation set for outcome analyses.
Molecular Analyses
The presence or absence of additional molecular markers such as FLT3-ITD, FLT3 tyrosine kinase domain mutations (FLT3-TKD), mutations in the NPM1, CEBPA, WT1, IDH1, and IDH2 genes, MLL partial tandem duplication (MLL-PTD), and BAALC and ERG expression levels were assessed centrally, as previously reported.12,19–29
miR-181aExpression Analyses
For microRNA expression, total RNA was extracted from pretreatment BM or blood mononuclear cells, and biotinylated first-strand complementary DNA was synthesized and hybridized to microRNA microarray chips.5 Images of the microRNA microarray chips were acquired, and calculation, normalization, and filtering of signal intensity for each microarray spot and batch-effect adjustment were performed.5 miR-181a expression was measured using Ohio State University Comprehensive Cancer Center version 3.0 arrays. Log intensities for miR-181a probes were averaged and used as a continuous variable for analyses. To validate measurements of miR-181a expression made using the microRNA microarrays, quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR) was performed in a subgroup of younger patients (Appendix).
Gene Expression Profiling
To gain further insight into the biologic processes associated with miR-181a in CN-AML, we performed gene-expression profiling using the AffymetrixU133 plus 2.0 array (Affymetrix, Santa Clara, CA), and Gene Ontology analysis as reported previously,30 and described in the Appendix.
Definition of Clinical End Points and Statistical Analysis
The main objective of our study was to evaluate the impact of miR-181a expression on outcome (for definition of clinical end points, see Appendix).
The associations of miR-181a expression, considered as a continuous variable, with baseline clinical, demographic, and molecular features were analyzed using one-way analysis of variance. Univariable logistic regression models were constructed to evaluate miR-181a expression for achievement of CR, and univariable Cox proportional hazards models were used to evaluate the associations of miR-181a expression with disease-free survival (DFS) and overall survival (OS). Multivariable logistic regression models were constructed to analyze factors related to the probability of achieving CR, and multivariable Cox proportional hazards models were constructed to analyze factors important for DFS and OS (multivariable analyses are detailed in the Appendix).
RESULTS
Associations of miR-181a Expression With Clinical and Molecular Characteristics in Patients With CN-AML
At diagnosis, higher expression of miR-181a, analyzed here as a continuous variable, was significantly associated with higher hemoglobin (P = .05) and percentage of circulating blasts (P < .001), French-American-British M1 and M2 subtypes (P < .001) and the absence of extramedullary disease, especially skin and gum involvement (P = .04; Table 1). Higher miR-181a expression was also significantly associated with higher frequency of wild-type NPM1 (P = .003), CEBPA mutations (P < .001), IDH1 mutations (P = .007), and lower ERG (P = .02) and higher BAALC (P = .05) expresser status (Table 1).
Table 1.
Relationship of Clinical and Molecular Characteristics WithmiR-181a Expression in the Whole Group of 187 Younger Patients With Cytogenetically Normal Acute Myeloid Leukemia at Diagnosis
| Characteristic | No. | % | P* |
|---|---|---|---|
| Median age, years | 45 | .08↓ | |
| Range | 18-59 | ||
| Sex | .39 | ||
| Female | 98 | 52 | |
| Male | 89 | 48 | |
| Race | .91 | ||
| White | 163 | 88 | |
| Nonwhite | 23 | 12 | |
| Median hemoglobin, g/L | 9.3 | .05↑ | |
| Range | 4.6-13.6 | ||
| Median platelet count, ×109/L | 58 | .29 | |
| Range | 7-466 | ||
| Median WBC, ×109/L | 27.9 | .13↓ | |
| Range | 0.9-295.0 | ||
| Median blood blasts, % | 62 | < .001↑ | |
| Range | 0-97 | ||
| Median bone marrow blasts, % | 67 | .58 | |
| Range | 21-95 | ||
| FAB | < .001 | ||
| M1/M2 | 92 | 59 | |
| M4/M5 | 56 | 36 | |
| Extramedullary involvement† | .04 | ||
| No | 129 | 70 | |
| Yes | 56 | 30 | |
| FLT3-ITD | .94 | ||
| Negative | 117 | 63 | |
| Positive | 70 | 37 | |
| FLT3-TKD | .06 | ||
| Negative | 167 | 90 | |
| Positive | 18 | 10 | |
| NPM1 | .003 | ||
| Wild type | 67 | 36 | |
| Mutated | 120 | 64 | |
| CEBPA | < .001 | ||
| Wild type | 152 | 83 | |
| Mutated | 32 | 17 | |
| WT1 | .16 | ||
| Wild type | 161 | 88 | |
| Mutated | 22 | 12 | |
| MLL-PTD | .59 | ||
| Negative | 175 | 94 | |
| Positive | 12 | 6 | |
| IDH1 | .007 | ||
| Wild type | 124 | 87 | |
| Mutated | 19 | 13 | |
| IDH2 | .88 | ||
| Wild type | 126 | 88 | |
| Mutated | 17 | 12 | |
| ERG expression | .02 | ||
| Low | 83 | 62 | |
| High | 50 | 38 | |
| BAALC expression | .05 | ||
| Low | 70 | 50 | |
| High | 70 | 50 | |
Abbreviations: FAB, French-American-British classification; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation of the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene.
P values are from the one-way analysis of variance overall F-test, evaluating the presence of any linear relationship between miR-181a expression and the variable tested. For tests with a P value < .20, ↑ indicates that higher values of the continuous variable associate with higher miR-181a expression and ↓ indicates that lower values of the continuous variable associate with higher miR-181a expression; for the categorical variables, those associated with higher miR-181a expression are indicated using bold type.
Primarily extramedullary skin and gum involvement.
Prognostic Value of miR-181a Expression in CN-AML
Patients with higher miR-181a expression had a higher CR rate (odds ratio [OR], 1.38; P = .04). With a median follow-up time for patients alive at the last follow-up visit of 6.5 years (range, 3.1 to 11.0 years), higher miR-181a expressers had a trend for longer DFS (P = .09) and had longer OS (hazard ratio [HR], 0.82; P = .01; Table 2). The prognostic impact of miR-181a expression levels measured using microRNA microarrays was technically validated by outcome analyses in a subgroup of 30 patients for whom miR-181a expression was also determined using real-time RT-PCR (Appendix).
Table 2.
Relationship Between miR-181a Expression and Outcome of Younger Patients With Cytogenetically Normal Acute Myeloid Leukemia
| End Point | OR/HR | 95% CI | P |
|---|---|---|---|
| Analyses in all CN-AML patients | |||
| Complete remission | 1.38 | 1.01 to 1.88 | .04 |
| Disease-free survival | — | — | .09 |
| Overall survival | 0.82 | 0.71 to 0.96 | .01 |
| Analyses in FLT3-ITD and/or NPM1wt patients | |||
| Complete remission | 1.64 | 1.12 to 2.42 | .009 |
| Disease-free survival | 0.66 | 0.53 to 0.84 | < .001 |
| Overall survival | 0.71 | 0.60 to 0.84 | < .001 |
NOTE: An OR greater than 1.0 means a higher complete remission rate for higher values of miR-181a expression. An HR lower than 1.0 means longer survival for higher values of miR-181a expression. The sample size for the entire set was n = 187 for complete remission and overall survival and n = 154 for disease-free survival. The sample size for FLT3-ITD and/or NPM1wt patients was n = 122 for complete remission and overall survival and n = 96 for disease-free survival.
Abbreviations: HR, hazard ratio; OR, odds ratio.
In multivariable analyses (Table 3), higher miR-181a expression levels were associated with an increased rate of CR (OR, 2.36; P = .02), after adjusting for ERG (P = .008) and BAALC expression status (P = .01) and age (P = .01). Higher miR-181a expression was also associated with longer DFS (HR, 0.8; P = .02), after adjusting for CEBPA (P = .005), NPM1 (P < .001), WT1 (P = .003), FLT3-ITD (P < .001) and FLT3-TKD (P = .02) mutational status, and with longer OS (HR, 0.81; P = .01), after adjusting for CEBPA (P < .001), NPM1 (P < .001), WT1 (P < .001), and FLT3-ITD (P = .003) mutational status, and WBC (P = .005).
Table 3.
Multivariable Analyses Evaluating miR-181a Expression for Clinical Outcome in Younger Patients With CN-AML
| Variables in Final Models | OR/HR | 95% CI | P |
|---|---|---|---|
| Multivariable analyses in all patients with CN-AML | |||
| CRa | |||
| miR-181a expression | 2.36 | 1.17 to 4.78 | .02 |
| ERG expression; low v high | 5.86 | 1.60 to 21.52 | .008 |
| BAALC expression; low v high | 6.69 | 1.56 to 28.74 | .01 |
| Age | 0.36 | 0.17 to 0.78 | .01 |
| DFSb | |||
| miR-181a expression | 0.80 | 0.66 to 0.97 | .02 |
| CEBPA; mutated v wild type | 0.38 | 0.19 to 0.75 | .005 |
| NPM1; mutated v wild type | 0.42 | 0.24 to 0.75 | < .001c |
| WT1; mutated v wild type | 2.54 | 1.39 to 4.65 | .003 |
| FLT3-ITD; positive v negative | 2.68 | 1.65 to 4.36 | < .001c |
| FLT3-TKD; positive v negative | 2.19 | 1.14 to 4.19 | .02 |
| OSd | |||
| miR-181a expression | 0.81 | 0.69 to 0.95 | .01 |
| CEBPA; mutated v wild type | 0.32 | 0.16 to 0.62 | < .001 |
| NPM1; mutated v wild type | 0.47 | 0.28 to 0.79 | < .001c |
| WT1; mutated v wild type | 2.65 | 1.54 to 4.57 | < .001 |
| FLT3-ITD; positive v negative | 2.39 | 1.46 to 3.93 | .003c |
| WBC | 1.37 | 1.13 to 1.67 | .005c |
| Multivariable analyses in patients with FLT3-ITD and/or NPM1wt | |||
| CRe | |||
| miR-181a expression | 1.61 | 1.07 to 2.42 | .02 |
| Age | 0.53 | 0.33 to 0.85 | .009 |
| DFSf | |||
| miR-181a expression | 0.74 | 0.57 to 0.96 | .02 |
| CEBPA; mutated v wild type | 0.27 | 0.13 to 0.58 | < .001 |
| NPM1; mutated v wild type | 0.33 | 0.14 to 0.79 | .007g |
| FLT3-ITD; positive v negative | 3.05 | 1.30 to 7.14 | .02g |
| Hemoglobin | 0.75 | 0.57 to 0.99 | .04 |
| OSh | |||
| miR-181a expression | 0.74 | 0.61 to 0.90 | .002 |
| CEBPA; mutated v wild type | 0.29 | 0.14 to 0.59 | < .001 |
| NPM1; mutated v wild type | 0.41 | 0.22 to 0.78 | .007g |
| WT1; mutated v wild type | 2.23 | 1.18 to 4.23 | .01 |
| WBC | 1.40 | 1.15 to 1.71 | < .001 |
| Extramedullary involvement; absent v present | 2.45 | 1.27 to 4.71 | .01g |
NOTE. Further details of the multivariable analyses are found in the Appendix (online only). ORs greater than 1.0 mean higher and those less than 1.0 mean lower CR rate for the higher values of the continuous variables and the first category listed for the categorical variables. HRs greater than 1.0 indicate higher and those less than 1.0 indicate lower risk for relapse or death (DFS) or death (OS) for the higher values of the continuous variables and the first category listed for the categorical variables.
Abbreviations: CN-AML, cytogenetically normal acute myeloid leukemia; CR, complete remission; DFS, disease-free survival; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain of the FLT3 gene; HR, hazard ratio; OS, overall survival; OR, odds ratio.
Variables considered in the model based on univariable analyses were miR-181a expression, ERG expression (low vhigh), FLT3-ITD (positive v negative), BAALC expression (low vhigh), age (in 10-year increments), hemoglobin (in 2-unit increments), and WBC (in 50-unit increments).
Variables considered in the model based on univariable analyses were miR-181aexpression, CEBPA (mutated v wild type), ERG expression (low vhigh), WT1 (mutated v wild type), BAALC expression (low vhigh), FLT3-ITD (positive v negative), FLT3-TKD (positive v negative), MLL-PTD (mutated v wild type), NPM1 (mutated v wild type), WBC (in 50-unit increments), extramedullary involvement, and race.
Does not meet the proportional hazards assumption. For DFS, the HR for FLT3-ITD and NPM1 are reported at 9 months; for OS, the HR for NPM1, FLT3-ITD, and WBC are reported at 9 months.
Variables considered in the model based on univariable analyses were miR-181a expression, CEBPA (mutated v wild type), ERG expression (low vhigh), FLT3-ITD (positive v negative), WT1 (mutated v wild type), BAALC expression (low vhigh), NPM1 (mutated v wild type), WBC (in 50-unit increments), age (in 10-year increments), hemoglobin (in 2-unit increments), platelet count, percentage of blood blasts, and extramedullary involvement.
Variables considered in the model based on univariable analyses were miR-181a expression, age (in 10-year increments), hemoglobin (in 2-unit increments), and WBC (in 50-unit increments).
Variables considered in the model based on univariable analyses were miR-181a expression, CEBPA (mutated v wild type), ERG expression (low vhigh), WT1 (mutated v wild type), FLT3-ITD (positive v negative), FLT3-TKD (positive v negative), NPM1 (mutated v wild type), hemoglobin (in 2-unit increments), WBC (in 50-unit increments), and race.
Does not meet the proportional hazards assumption. For DFS, the HR for FLT3-ITD is reported at 1 year, NPM1 is reported at 9 months; for OS, the HR for NPM1 is reported at 1.5 years, extramedullary involvement is reported at 1 year.
Variables considered in the model based on univariable analyses were miR-181a expression, CEBPA (mutated v wild type), ERG expression (low vhigh), WT1 (mutated v wild type), FLT3-ITD (positive v negative), NPM1 (mutated v wild type), hemoglobin (in 2-unit increments), WBC (in 50-unit increments), and extramedullary involvement.
Association of miR-181a Expression Levels With Outcome in Distinct CN-AML Molecular Groups
The presence or absence of FLT3-ITD and NPM1 mutations has been reported to stratify patients with CN-AML into prognostically distinct categories. Patients with NPM1 mutations, but no FLT3-ITD had a more favorable outcome, whereas those with FLT3-ITD and/or NPM1wt had worse prognosis.23 Thus, to better understand the prognostic significance of higher miR-181a expression levels in CN-AML, we analyzed their impact on the aforementioned prognostic subsets. While there was no prognostic impact of miR-181a expression on patients with NPM1 mutations and no FLT3-ITD (n = 65; CR rate, P = .58; DFS, P = .76; and OS, P = .66), we observed that higher miR-181a expression levels were associated with a significantly higher CR rate (OR, 1.64; P = .009), and longer DFS (HR, 0.66; P < .001) and OS (HR, 0.71; P < .001) in patients with FLT3-ITD and/or NPM1wt (n = 122; Table 2).
In multivariable analysis restricted to patients with FLT3-ITD and/or NPM1wt (Table 3), higher miR-181a expression levels were associated with higher odds of achieving a CR (OR, 1.61; P = .02), after adjusting for age (P = .009), with longer DFS (HR = 0.74; P = .02), after adjusting for CEBPA (P < .001), NPM1 (P = .007), and FLT3-ITD (P = .02) mutational status, and hemoglobin levels (P = .04), and with longer OS (HR, 0.74; P = .002), after adjusting for CEBPA (P < .001), NPM1 (P = .007), and WT1 (P = .01) mutational status, WBC (P < .001), and extramedullary involvement (P = .01).
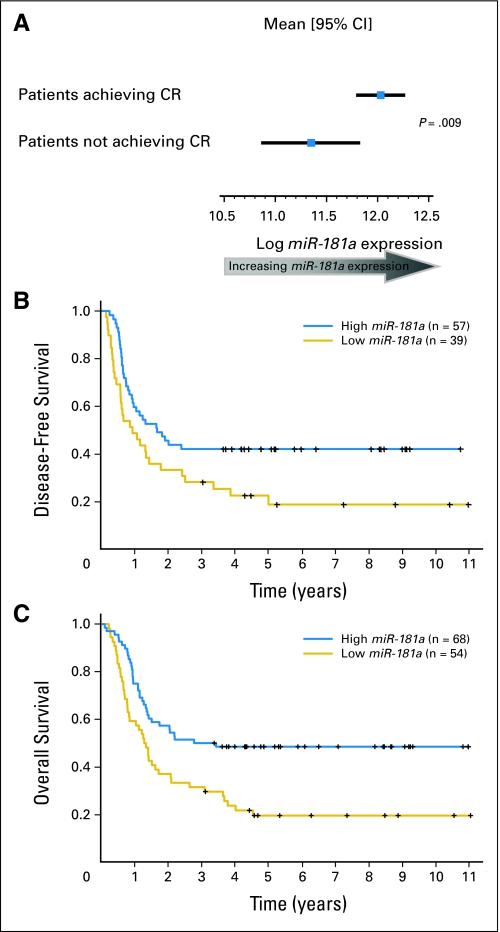
In the aforementioned analyses, we used miR-181a expression values as a continuous variable. To graphically display the relationship between miR-181a expression and achievement of CR, we compared miR-181a expression in patients achieving CR with that of patients experiencing failure with induction therapy within the subgroup of patients with FLT3-ITD and/or NPM1wt (Fig 1A). Furthermore, to graphically display the relationship between miR-181a expression and DFS and OS, we dichotomized miR-181a expression values at the median, and present survival curves for the high and low miR-181a expressers within the subgroup of patients with FLT3-ITD and/or NPM1wt (Fig 1B and 1C).
Fig 1.
Favorable outcome of patients with FLT3-ITD and/or NPM1wt and higher miR-181a expression levels. (A) miR-181a expression in patients who achieved a complete response (CR) versus patients who did not achieve a CR; (B) disease-free and (C) overall survival according to miR-181a expression levels in patients with CN-AML dichotomized into high (above the median miR-181a expression value) or low (at or below the median miR-181a expression value) expression groups.
Importantly, an independent set of older patients with CN-AML with FLT3-ITD and/or NPM1wt (n = 122) was analyzed by microRNA microarray assays to validate the prognostic impact of miR-181a found in younger patients (Appendix). In this validation set, higher expression of miR-181a, used as a continuous variable, did not impact on the CR rate (P = .52), but was associated with longer DFS (P = .04) and with a trend for longer OS (P = .08). In multivariable models for this validation set, miR-181a was independently associated with longer DFS (P = .04) and OS (P = .05), even after adjusting for other clinical and molecular variables (Appendix Table A1, online only).
Biologic Insights
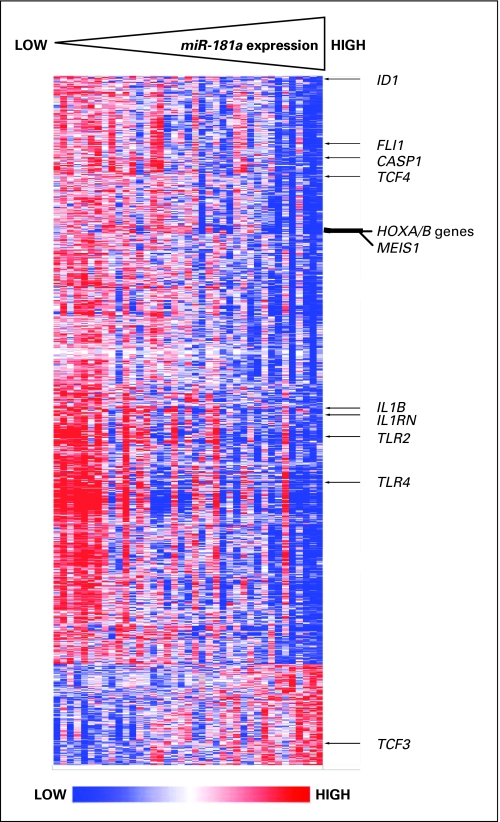
In order to gain insights into the functional contribution of miR-181a expression levels to the poor molecular risk CN-AML subset, we first derived a gene-expression signature associated with miR-181a expression in patients with FLT3-ITD and/or NPM1wt. We observed that the expression of 1,174 probe sets significantly correlated (P < .001) with that of miR-181a; 1,002 probe sets correlated negatively and 172 probe sets positively (Fig 2). Among other genes, we observed a negative correlation of miR-181a expression with the expression of the HOXA and HOXB clusters, as well as the HOX cofactor MEIS1. These genes are important for developmental processes and have also been linked to leukemogenesis and the self-renewal of leukemic stem cells.31,32 We also observed a negative correlation of miR-181a expression with the expression of the transcription coregulator ID1, which is able to prevent hematopoietic differentiation and has recently been associated with adverse outcome in AML33; the FLI1 gene, a known suppressor of erythroid differentiation34; and the transcription factor TCF4, which contributes to neoplastic transformation as a downstream target of the WNT-pathway.35 In contrast, we observed a positive correlation of miR-181a expression with the expression of TCF3, a gene encoding a transcription factor that has been shown to regulate the homeostasis of the hematopoietic stem cell pool and promote differentiation of hematopoietic progenitors.36,37
Fig 2.
Heat map of the derived gene-expression signature correlated with miR-181a expression. Rows represent probe sets and columns represent patients. Probe sets are ordered by hierarchical cluster analysis. Patients are ordered from left to right by increasing miR-181a expression. Expression values of the probe sets are represented by color, with blue indicating expression less than and red indicating expression greater than the median value for the given probe set. Arrows indicate genes that are discussed in the text.
To further understand the potential functional role of miR-181a expression in CN-AML, we performed a Gene Ontology analysis. Biologic processes that relate to cytokine and native immunity-mediated processes, including those involving toll-like receptors (eg, TLR4 and TLR2) and the interleukin pathways (eg, IL1B, IL1RN, and CASP1), were over-represented in the miR-181a-associated gene-expression signature (Table 4).
Table 4.
GO Terms of Biological Processes Significantly Overrepresented in the miR-181a-Expression Profile
| GO ID | GO Terms | Percentage of Members of the GO Term Present in the miR-181a Profile | P |
|---|---|---|---|
| 50715 | Positive regulation of cytokine secretion | 83.33 | < .001 |
| 50706 | Regulation of interleukin-1 beta secretion | 80 | < .001 |
| 50716 | Positive regulation of interleukin-1 secretion | 80 | < .001 |
| 50704 | Regulation of interleukin-1 secretion | 80 | < .001 |
| 50718 | Positive regulation of interleukin-1 beta secretion | 80 | < .001 |
| 50707 | Regulation of cytokine secretion | 77.78 | < .001 |
| 45123 | Cellular extravasation | 66.67 | < .001 |
| 50701 | Interleukin-1 secretion | 66.67 | .001 |
| 50702 | Interleukin-1 beta secretion | 66.67 | .001 |
| 7159 | Leukocyte adhesion | 66.67 | .002 |
| 50663 | Cytokine secretion | 66.67 | < .001 |
| 9595 | Detection of biotic stimulus | 62.5 | < .001 |
| 50709 | Negative regulation of protein secretion | 60 | .003 |
| 30593 | Neutrophil chemotaxis | 60 | < .001 |
| 45408 | Regulation of interleukin-6 biosynthetic process | 57.14 | .002 |
| 45576 | Mast cell activation | 57.14 | .004 |
| 30149 | Sphingolipid catabolic process | 55.56 | < .001 |
| 42226 | Interleukin-6 biosynthetic process | 50 | .003 |
| 32635 | Interleukin-6 production | 50 | .003 |
| 50714 | Positive regulation of protein secretion | 50 | < .001 |
| 46466 | Membrane lipid catabolic process | 50 | < .001 |
NOTE. Shown are significantly overrepresented GO terms with ≥ 50% of their assigned members represented in the gene expression signature associated with higher miR-181a expression. Gray shading identifies terms associated with genes encoding proteins in the interleukin-1β and toll-like receptor pathways (eg, IL1B, IL1BRN, CASP1, TLR2, TLR4, etc).
Abbreviation: GO, Gene Ontology.
DISCUSSION
We report here that expression levels of miR-181a constitute a strong prognostic factor in younger patients with CN-AML enrolled on similar CALGB first-line treatment protocols. We show that higher levels of miR-181a expression directly correlate with higher odds of achieving a CR and lower risk of experiencing relapse and/or death in patients with CN-AML. This study is the first to demonstrate that a single noncoding RNA associates with clinical outcome in CN-AML, even in the context of other well-established molecular markers including CEBPA and NPM1 mutations, that were recently recognized by the WHO classification as defining markers for novel provisional AML entities,38 and FLT3-ITD. Furthermore, we technically validated these results by using quantitative RT-PCR.
The prognostic impact was most striking in patients with FLT3-ITD and/or NPM1wt, which are associated with adverse outcome. These patients constitute approximately 65% of all CN-AML and one third of all AML patients younger than 60 years.13 Notably, in this group, when other molecular prognostic markers were considered in multivariable models, higher expression of miR-181a was the only molecular marker that independently associated with higher odds of achieving CR, thereby suggesting a potential impact of this microRNA on mechanisms of resistance to chemotherapy-induced apoptosis. Higher expression of miR-181a was also associated with longer DFS after adjusting for the impact of NPM1, CEBPA, and FLT3-ITD mutational status and hemoglobin levels, and OS after adjusting for the impact of NPM1, CEBPA, and WT1 mutational status, extramedullary involvement, and WBC. These results were validated by demonstrating the positive prognostic impact of higher miR-181a expression in an independent validation set of older patients with CN-AML.
Recently, a modified prognostic classification of CN-AML has been recommended by an international expert panel on behalf of the European LeukemiaNet, in which the intermediate I prognostic category also includes patients with FLT3-ITD and/or NPM1wt, but only those who lack CEBPA mutations; patients with FLT3-ITD and/or NPM1wt and CEBPA mutations are classified in the favorable category.39 When we analyzed the prognostic significance of miR-181a expression in this European LeukemiaNet intermediate I prognostic category (n = 92), higher miR-181a expression levels were still associated with a significantly higher CR rate (OR, 1.56; P = .04), and longer DFS (HR, 0.72; P = .03) and OS (HR, 0.77; P = .01). Altogether, these data support a pivotal role of miR-181a expression levels for the response to treatment of patients with CN-AML, and suggest that since miR-181a expression provides additional prognostic information it can be used to further refine this newly devised molecular-risk classification of CN-AML.39 Moreover, the identification of low levels of miR-181a as an adverse prognostic factor provides opportunity for potential therapeutic intervention with agents capable of increasinglow endogenous levels of miR-181a and/or with synthetic miR-181a compounds.
But how do changes of miR-181a expression levels in myeloid blasts affect the aggressiveness of the disease in patients with CN-AML? The biologic role of microRNAs may vary according to their expression in distinct cell populations of normal or neoplastic tissues. miR-181a has been described as a tumor suppressor in gliomas,40 but also has been found elevated in hepatocellular carcinoma cells with features of hepatic cancer stem cells.41 Currently, relatively little is known about the function of miR-181a in normal or malignant hematopoiesis. Previous studies reported that miR-181 regulated B-cell development and influenced T-cell sensitivity to antigens by modulating T-cell receptor signaling strength.42,43 Furthermore, miR-181a may also play a regulatory role in earlier steps of hematopoiesis.44 Recently, it was shown that higher levels of miR-181 are expressed during early erythroid differentiation.45 In line with these findings, in this study, we observed a positive correlation between miR-181a expression and hemoglobin levels, and a negative correlation between miR-181a expression and expression of FLI1, a known suppressor of erythroid differentiation.35 Furthermore, we found a negative correlation of miR-181a expression with the expression of ID1, an inhibitor of hematopoietic differentiation, and TCF4, a transcription factor promoting neoplastic transformation.35 We also observed a negative correlation of miR-181a expression with the expression of the HOXA and HOXB clusters, as previously reported.45 In contrast, we observed a positive correlation between miR-181a expression and TCF3, a transcription factor that seemingly promotes development of hematopoietic progenitors and contributes to regulating hematopoietic cell differentiation.37
In an effort to further understand how changes in miR-181a expression affect the aggressiveness of the disease, response to treatment, and outcome of patients with CN-AML, we used a Gene Ontology analysis. We show an over-representation of cytokine and native immunity-mediated processes in the miR-181a-associated gene-expression signature. The expression of the TLR4, TLR2, IL1B, IL1RN, and CASP1 genes was negatively correlated with miR-181a expression, and we find some of these genes, namely TLR4 and IL1B and CASP1 to be predicted to be direct targets of miR-181a. Of these genes, TLR4 and IL1B have previously been implicated in human cancer.47–50 TLR4 has been shown to promote tumor growth and interfere with response to chemotherapy in ovarian cancer,46 and to contribute to the development of cytopenias in myelodysplastic syndromes.47 In addition, TLR4 signaling has also been linked to blocking myeloid differentiation of hematopoietic stem and progenitor cells in severe sepsis.48 IL-1β has been previously shown to be produced in an autocrine fashion and to stimulate the proliferation of AML blasts.49,50 It is, therefore, tempting to speculate that high expression of miR-181a associates with a less aggressive disease by downregulating genes like TLR4 and IL1B, that modulate the innate immune response to microbial pathogens in the normal host, but also when upregulated may support survival and proliferation of malignant blasts in AML patients.47–50 However, the mechanisms through which the changes in levels of miR-181a expression contribute to different degrees of disease aggressiveness in patients with CN-AML and why miR-181a expression differs among individual patients remain to be elucidated.
In summary, we report here for the first time that the expression of a single microRNA, miR-181a, associates with clinical outcome in CN-AML. Moreover, it does so independently from other validated clinical and genetic variables, thus adding information useful for a better risk-stratification of patients with CN-AML. High miR-181a expression levels identify those patients with CN-AML who despite having molecular features associated with adverse outcome, such as NPM1wt and/or FLT3-ITD, might not need intensive treatment, such as allogeneic stem-cell transplantation. Moreover, for those patients with low miR-181a expression levels, it is hoped that the development of reliable methods of delivery of this microRNA directly to the leukemia cells and/or identification of agents capable of increasing endogenous levels of miR-181a may provide new therapeutic options. Further prospective studies should be done to confirm our findings. Establishment of standardized methods of microRNA quantification will allow prospective classification of patients according to their miR-181a levels. Finally, the combination of miR-181a-associated gene-expression profiling and Gene Ontology analyses provide insights into the leukemogenic role of genes that are either direct or indirect targets of miR-181a, and therefore should also be investigated as potential therapeutic targets in patients with CN-AML with low miR-181a expression.
Acknowledgment
We thank Donna Bucci of the CALGB Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services, Yue-Zhong Wu for sample processing and laboratory work, and Lisa J. Sterling and Colin G. Edwards for data management.
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study:
The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, and Nyla A. Heerema (Grant no. CA77658); Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Wendy L. Flejter, and Mark J. Pettenati (Grant no. CA03927); North Shore–Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R. K. Koduru (Grant no. CA35279); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (Grant no. CA02599); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett, Michael S. Watson, and Jaime Garcia-Heras (Grant no. CA77440); University of Massachusetts Medical Center, Worcester, MA: William W. Walsh, Vikram Jaswaney, Kathleen E. Richkind, Michael J. Mitchell, and Patricia Miron (Grant no. CA37135); Vermont Cancer Center, Burlington, VT: Hyman B. Muss, Elizabeth F. Allen, and Mary Tang (Grant no. CA77406); Dana-Farber Cancer Institute, Boston, MA: Eric P. Winer, Ramana Tantravahi, Paola Dal Cin, and Cynthia C. Morton (Grant no. CA32291); Eastern Maine Medical Center, Bangor, ME: Harvey M. Segal and Laurent J. Beauregard (Grant no. CA35406); University of North Carolina, Chapel Hill, NC: Thomas C. Shea and Kathleen W. Rao (Grant no. CA47559); Christiana Care Health Services Inc, Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, and Jeanne M. Meck (Grant no. CA45418); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff and Thuluvancheri K. Mohandas (Grant no. CA04326); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Mazin B. Qumsiyeh (Grant no. CA47577); University of Puerto Rico School of Medicine, San Juan, PR: Eileen I. Pacheco, Leonard L. Atkins, Cynthia C. Morton, and Paola Dal Cin; Weill Medical College of Cornell University, New York, NY: John Leonard, Prasad R. K. Koduru, and Andrew J. Carroll (Grant no. CA07968); University of Chicago Medical Center, Chicago, IL: Gini Fleming, Diane Roulston, Katrin M. Carlson, Yanming Zhang, and Michelle M. Le Beau (Grant no. CA41287); University of Iowa Hospitals, Iowa City, IA: Gerald H. Clamon and Shivanand R. Patil (Grant no. CA47642); University of California at San Diego: Barbara A. Parker, Renée Bernstein, and Marie L. Dell'Aquila (Grant no. CA11789); Ft. Wayne Medical Oncology/Hematology, Ft Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Paola Dal Cin, and Cynthia C. Morton (Grant no. CA 12,449); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (Grant no. CA04457); Western Pennsylvania Hospital, Pittsburgh, PA: John Lister and Gerard R. Diggans; Georgetown University Medical Center, Washington, DC: Minnetta C. Liu and Jeanne M. Meck (Grant no. CA77597); Rhode Island Hospital, Providence, RI: William Sikov, Shelly L. Kerman, and Aurelia Meloni-Ehrig (Grant no. CA08025); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano and Constance K. Stein (Grant no. CA21060); Virginia Commonwealth University MB CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (Grant no. CA52784); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim H. Huang (Grant no. CA12046); Long Island Jewish Medical Center CCOP, Lake Success, NY: Kanti R. Rai and Prasad R. K. Koduru (Grant no. CA11028); University of Minnesota, Minneapolis, MN: Bruce A. Peterson and Betsy A. Hirsch (Grant no. CA16450); Medical University of South Carolina, Charleston, SC: Mark R. Green and G. Shashidhar Pai (Grant no. CA03927); Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John A. Ellerton and Marie L. Dell'Aquila (Grant no. CA35421); Minneapolis VA Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (Grant no. CA47555); University of Illinois at Chicago: David J. Peace and Maureen M. McCorquodale (Grant no. CA74811); University of Nebraska Medical Center, Omaha, NE: Anne Kessinger and Warren G. Sanger (Grant no. CA77298); University of California at San Francisco: Charles J. Ryan and Kathleen E. Richkind (Grant no. CA60138); Walter Reed Army Medical Center, Washington, DC: Brendan M. Weiss and Digamber S. Borgaonkar (Grant no. CA26806).
Definition of Clinical End Points
Complete remission (CR) was defined as bone marrow (BM) cellularity higher than 20% with maturation of all cell lines, lower than 5% leukemic blasts, undetectable Auer rods, and recovery of leukocyte (≥ 1,500/μL) and platelet (> 100,000/μL) counts with no leukemic blasts in the blood, all of which had to persist for at least 1 month. Relapse was defined as ≥ 5% BM blasts, reappearance of circulating blasts or development of extramedullary leukemia (Cheson BD, Cassileth PA, Head DR, et al: J Clin Oncol 8:813-819, 1990). Overall survival (OS) was measured from the date the patient was enrolled onto the study until the date of death, and patients alive at last follow-up were censored. Disease-free survival (DFS) was measured from the date of CR until the date of relapse or death; patients alive and relapse-free at last follow-up were censored. Pretreatment CNS, spleen, liver, skin, nodes, gum, or mediastinal mass involvement constituted extramedullary disease.
WBC, BM, and peripheral blood blasts.
Forty-four percent of the 423 cytogenetically normal acute myeloid leukemia (CN-AML) patients enrolled on the two protocols (Cancer and Leukemia Group B [CALGB] 9621 and 19808) were part of our study. WBC, BM, and peripheral blood (PB) blasts were significantly higher in the patients we studied (P < .001 for all three variables) versus the patients we did not. This is a result of selection criteria used to identify patients with adequate blast counts for reliable molecular studies. To explore this potential bias, we thoroughly evaluated WBC and blast percentage (BM and PB) in all our outcome multivariable models to ensure that the outcome association of our main variable, miR-181a expression, did not change. miR-181a stayed significant in all of the models that contained these variables (CR and DFS for WBC; CR, DFS, OS for BM and PB blasts) and neither WBC nor percent blasts (BM or PB) became significant for outcome.
miR-181a expression analyses.
miR-181a expression was measured using the Ohio State University Comprehensive Cancer Center version 3.0 microRNA microarray chip. For quantification, the two miR-181a probes on the chip were averaged. These two probes correlated very strongly (r = .98, P < .001), and no outliers were observed.
BM samples were used to study miR-181a levels in the majority of our patients (72%). When we compared miR-181a levels across BM and blood samples, we did not see any differences across the tissue types.
Since miR-181b expression was also part of the reported prognostic microRNA signature, in CN-AML with unfavorable molecular features—FLT3-ITD and/or NPM1 wild-type (NPM1wt) — we also analyzed the prognostic impact of miR-181b expression.5 We found that the expression levels of miR-181b did not provide any additional information, with respect to outcome. Therefore, we focused on miR-181a expression levels.
In order to validate the microRNA microarray-based miR-181a quantification, we performed real-time RT-PCR for miR-181a in a subgroup of 30 patients, 15 of whom had high and 15 low miR-181a expression levels according to the microarray data. miR-181a expression was normalized to an internal control, U48 small nucleolar RNA expression. Primers, probes, and amplification conditions will be provided on request. Outcome analysis of these 30 patients analyzed by RT-PCR reproduced the results obtained using the microRNA microarray. Using a median cutoff value for the real-time RT-PCR data to define high and low expressers, patients in the high miR-181a expression group had a significantly better DFS (P < .001) and OS (P < .001) than those in the low miR-181a expression group, thereby confirming the outcome results we observed using the microRNA microarray data for these 30 patients.
Multivariable models.
Multivariable logistic regression models were constructed to analyze factors related to the probability of achieving CR, and multivariable Cox proportional hazards models were constructed to analyze factors important for DFS and OS. Factors examined for inclusion in the CR models were miR-181a, ERG and BAALC expression, FLT3-ITD, MLL partial tandem duplication (MLL-PTD), WT1 and NPM1 mutation status, age, hemoglobin level, platelet count, WBC, percentage of blood and BM blasts, race, sex, induction regimen (ADE versus ADEP), and extramedullary involvement. For OS and DFS, factors examined for model inclusion were miR-181a, ERG and BAALC expression, FLT3-ITD, FLT3 tyrosine kinase domain mutations (FLT3-TKD), MLL-PTD and WT1, NPM1, and CEBPA mutation status, age, hemoglobin level, platelet count, WBC, percentage of blood and BM blasts, race, sex, and extramedullary involvement. Of the above factors, those significant at α = .20 from the univariable models (see footnotes in Table 3) were used in a limited backward selection procedure to build multivariable models, by retaining the main variable miR-181a throughout the model building. Variables remaining in the final models were significant at α = .05.
To further explore the impact of miR-181a expression level in the presence of other prognostic markers, we fitted additional multivariable models for outcome. All molecular markers were retained in these models irrespective of statistical significance. High miR-181a expression levels remained significantly associated with favorable outcome for all three end points. For the achievement of CR, the only molecular marker significant in the model was miR-181a expression (P = .01). For DFS, miR-181a expression (P = .02), CEBPA mutation status (P = .005), FLT3-ITD (P < .001), FLT3-TKD (P = .02), NPM1 mutation status (P < .001), and WT1 mutation status (P = .001) were all significant in the model. For OS, miR-181a expression (P = .03), FLT3-ITD (P < .002), and WT1 mutation status (P < .001) were the only significant predictors.
For all Cox models, the proportional hazards assumption was checked for each variable individually. If the proportional hazards assumption was not met for a particular variable for a given end point, an artificial time-dependent covariate was included in the model for that end point, which requires estimating the hazard ratios at specific time points for these variables as opposed to being able to provide one hazard ratio for the entire time period analyzed. For achievement of CR, estimated odds ratios (OR), and for survival end points, hazard ratios (HR) with their corresponding 95% CI were obtained for each significant prognostic factor Bradburn MJ, Clark TG, Love SB, et al: Br J Cancer 89:605-611, 2003).
Missing data.
Missing data were handled using a complete case analysis approach (Allison PD: Missing Data. Thousand Oaks, CA, SAGE Publications, 2002). This was required only for developing a CR model for the whole patient set.
Validation analysis of prognostic significance of miR-181a expression levels.
Older patients included in the validation set were treated on one of the following CALGB first-line treatment protocols: 8525 (Mayer RJ, Davis RB, Schiffer CA, et al: N Engl J Med 331: 896-903, 1994), 8923 (Stone RM, Berg DT, George SL, et al: Blood 98:548-553, 2001), 9420 (Lee EJ, George SL, Caligiuri M, et al: J Clin Oncol 17:2831-2839, 1999), 9720 (Baer MR, George SL, Dodge RK, et al: Blood 100:1224-1232, 2002), and 10201 (Marcucci G, Moser B, Blum W, et al: J Clin Oncol 25:360s, 2007 (suppl; abstr 7012). Per protocol, patients enrolled on this study did not receive stem-cell transplantation in first CR.
In this set of older patients with CN-AML with FLT3-ITD and/or NPM1wt (n = 122), miR-181a expression levels were measured by microRNA microarray assays. miR-181a expression was used as a continuous variable in univariable and multivariable analyses. Due to the large number of predictors relative to the number of events in the analyzed data set, individual variables were evaluated along with miR-181a expression and based on these bi-variable results the final models were developed (Appendix Table A1).
Gene-expression profiling.
As for the miRNA expression profiling, total RNA was extracted from pretreatment BM or blood mononuclear cells. Using AffymetrixU133 plus 2.0 GeneChips (Affymetrix, Santa Clara, CA), RNA samples from a group of patients with CN-AML treated on CALGB 9621 were analyzed (including data normalization and computation of expression intensities), as previously reported.30 Expression values were logged (base 2) before analysis. A filtering step was performed to remove probe sets that did not display significant variation in expression across arrays. In this procedure, a χ2 test was used to test whether the observed variance in expression of a probe set was significantly larger than the median observed variance in expression for all probe sets using α = .01 as the significance level. A total of 19,871 probe sets met the filtering criterion and were included in subsequent analyses. Thirty-nine patients with FLT3-ITD and/or NPM1 wild-type and enrolled on CALGB 9621 were studied in the gene-expression profiling studies detailed above. These patient samples were used for the identification of the gene-expression profile associated with miR-181a expression. For this purpose, univariable correlation tests (using Pearson's correlation) were performed between the median miR-181a expression level and expression values of each Affymetrix probe set, using a univariable significance level of α = .001.
Analyses were performed using BRB-ArrayTools version 3.8.0 Beta 2 Release (R. Simon and A.P. Lam, National Cancer Institute, Bethesda, MD) and using the R version 2.9.0 (R Foundation for Statistical Computing, Vienna, Austria). For in silico target prediction of miRNAs, the online applications miRBase Targets Version 5 and Targetscan Release 5.0 were used.
Gene ontology analysis.
We used GenMAPP version 2.1 and MAPPFinder version 2.0 to assess which biologic processes (as designated by the Gene Ontology project at www.geneontology.org) were over-represented among the genes that constituted the signature. An over-represented biologic process is one that has more associated genes (also referred to as members) in the gene-expression signature than is expected by chance. In our analysis, we considered only biologic processes that were represented by ≥ five members among the genes that could be analyzed in our microarray expression database. MAPPFinder uses a permutation procedure to determine over-represented biologic processes. An α level of .005 was used for identifying such biologic processes. Furthermore, we only report the over-represented biologic processes for which at least half of their members (ie, genes) analyzed in our microarray expression database were identified as part of the signature associated with miR-181a expression.
Table A1.
Multivariable Analyses Evaluating miR-181a Expression for Clinical Outcome in Older Cytogenetically Normal Acute Myeloid Leukemia Patients With FLT3-ITD and/or Wild-Type NPM1
| End Point and Variables in Final Models | HR | 95% CI | P |
|---|---|---|---|
| DFS* | |||
| miR-181a expression | 0.64 | 0.46 to 0.90 | .04† |
| NPM1; mutated v wild-type | 0.28 | 0.11 to 0.71 | .03† |
| Platelets | 1.45 | 1.14 to 1.85 | .002 |
| WBC | 1.85 | 1.32 to 2.61 | < .001 |
| OS* | |||
| miR-181a expression | 0.78 | 0.63 to 0.95 | .05† |
| FLT3-TKD; positive v negative | 5.89 | 1.68 to 20.58 | .006 |
| NPM1; mutated v wild-type | 0.49 | 0.24 to 0.98 | .03† |
| Platelets | 1.20 | 1.01 to 1.35 | .005 |
| WBC | 1.15 | 1.02 to 1.30 | .02 |
NOTE. HRs greater than 1.0 indicate higher and those less than 1.0 indicate lower risk for relapse or death (DFS) or death (OS) for the higher values of the continuous variables and the first category listed for the categorical variables.
Abbreviations: HR, hazard ratio; DFS, disease-free survival; OS, overall survival; FLT3-TKD, tyrosine kinase domain of the FLT3 gene.
Variables considered in the models were miR-181aexpression, CEBPA (mutated v wild-type), FLT3-ITD (positive v negative), FLT3-TKD (positive v negative), NPM1 (mutated v wild-type), WBC (in 50-unit increments), platelet count (in 50-unit increments), and age (in 10-year increments).
Does not meet the proportional hazards assumption. For DFS, the HRs for miR-181aexpression and NPM1 mutation status are estimated at 18 months; for OS, the HRs for miR-181aexpression and NPM1mutation status are estimated at 24 months.
Footnotes
Supported in part by Grants No. CA101140, CA114725, CA31946, CA33601, CA16058, CA77658, CA129657, and CA140158 from the National Cancer Institute, The Coleman Leukemia Research Foundation, and the Deutsche Krebshilfe–Dr. Mildred Scheel Cancer Foundation (H.B.).
Presented in part at the 45th Annual Meeting of the American Society of Clinical Oncology, Orlando, FL, May 29-June 2, 2009.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Sebastian Schwind, Guido Marcucci,Clara D. Bloomfield
Financial support: Guido Marcucci, Clara D. Bloomfield
Administrative support: Michael A. Caligiuri, Guido Marcucci,Clara D. Bloomfield
Provision of study materials or patients: Bayard L. Powell, Jonathan E. Kolitz, Andrew J. Carroll, Michael A. Caligiuri, Richard A. Larson, Guido Marcucci, Clara D. Bloomfield
Collection and assembly of data: Sebastian Schwind, Kati Maharry, Michael D. Radmacher, Dean Margeson, Susan P. Whitman, Christopher Hickey, Heiko Becker, Klaus H. Metzeler, Peter Paschka, Claudia D. Baldus, Shujun Liu, Ramiro Garzon, Andrew J. Carroll, Guido Marcucci, Clara D. Bloomfield
Data analysis and interpretation: Sebastian Schwind, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Kelsi B. Holland, Guido Marcucci, Clara D. Bloomfield
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 5.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 6.Marcucci G, Mrózek K, Radmacher MD, et al. MicroRNA expression profiling in acute myeloid and chronic lymphocytic leukaemias. Best Pract Res Clin Haematol. 2009;22:239–248. doi: 10.1016/j.beha.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Jongen-Lavrencic M, Sun SM, Dijkstra MK, et al. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 8.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLOS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garzon R, Volinia S, Liu C-G, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 15.Kolitz JE, George SL, Dodge RK, et al. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B study 9621. J Clin Oncol. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 16.Kolitz JE, George SL, Marcucci G, et al. A randomized comparison of induction therapy for untreated acute myeloid leukemia (AML) in patients < 60 years using P-glycoprotein (Pgp) modulation with Valspodar (PSC833): Preliminary results of Cancer and Leukemia Group B study 19808. Blood. 2005;106:122a–123a. abstr 407. [Google Scholar]
- 17.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 18.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 19.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 20.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 21.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcucci G, Maharry K, Wu Y-Z, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 24.Paschka P, Marcucci G, Ruppert AS, et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 26.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 28.Marcucci G, Maharry K, Whitman SP, et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 29.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radmacher MD, Marcucci G, Ruppert AS, et al. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: A Cancer and Leukemia Group B study. Blood. 2006;108:1677–1683. doi: 10.1182/blood-2006-02-005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Argiropoulos B, Humphries RK. HOX genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 32.Erklund EA. The role of HOX genes in malignant myeloid disease. Curr Opin Hematol. 2007;14:85–89. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- 33.Tang R, Hirsch P, Fava F, et al. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009;114:2993–3000. doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- 34.Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, et al. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14:439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- 35.Kolligs FT, Nieman MT, Winer I, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with ß-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 36.Graf T. Differentiation plasticity of hematopoietic cells. Blood. 2002;99:3089–3101. doi: 10.1182/blood.v99.9.3089. [DOI] [PubMed] [Google Scholar]
- 37.Semerad CL, Mercer EM, Inlay MA, et al. E2A protein maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 39.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 40.Shi L, Cheng Z, Zhang J, et al. Hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 41.Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 43.Li QJ, Chau J, Ebert PJ, et al. MiR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Georgantas RW, III, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debernardi S, Skoulakis S, Molloy G, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 46.Wang AC, Su QB, Wu FX, et al. Role of TLR4 for paclitaxel chemotherapy in human epithelial ovarian cancer cells. Eur J Clin Invest. 2009;39:157–164. doi: 10.1111/j.1365-2362.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 47.Maratheftis CI, Andreakos E, Moutsopoulos HM, et al. Toll-like receptor 4 is upregulated in hematopoietic progenitor cells and contributes to increased apoptosis in myelodysplastic syndromes. Clin Cancer Res. 2007;13:1154–1160. doi: 10.1158/1078-0432.CCR-06-2108. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez S, Chora A, Goumnerov B, et al. Dysfunctional expansion of hematopoietic stem cells and block of myeloid differentiation in lethal sepsis. Blood. 2009;114:4064–4076. doi: 10.1182/blood-2009-04-214916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cozzolino F, Rubartelli A, Aldinucci D, et al. Interleukin 1 as an autocrine growth factor for acute myeloid leukemia cells. Proc Natl Acad Sci U S A. 1989;86:2369–2373. doi: 10.1073/pnas.86.7.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Cimadevilla JC, Beauchemin V, Villeneuve L, et al. Coordinate secretion of interleukin-lβ and granulocyte-macrophage colony-stimulating factor by the blast cells of acute myeloblastic leukemia: Role of interleukin-1 as an endogenous inducer. Blood. 1990;76:1481–1489. [PubMed] [Google Scholar]