Abstract
Peroxisome proliferator-activated receptors (PPARs) are nuclear hormone receptors that have been implicated in a variety of biologic processes. The PPARδ isotype was recently proposed as a downstream target of the adenomatous polyposis coli (APC)/β-catenin pathway in colorectal carcinogenesis. To evaluate its role in tumorigenesis, a PPARδ null cell line was created by targeted homologous recombination. When inoculated as xenografts in nude mice, PPARδ −/− cells exhibited a decreased ability to form tumors compared with PPARδ +/− and wild-type controls. These data suggest that suppression of PPARδ expression contributes to the growth-inhibitory effects of the APC tumor suppressor.
The peroxisome proliferator-activated receptors (PPARs) are nuclear hormone receptors originally identified by their ability to mediate the transcriptional effects of peroxisome proliferators (reviewed in ref. 1). Subsequently, PPARs have been implicated in many normal and disease-related processes, including lipid metabolism, inflammation, embryo implantation, diabetes, and cancer (reviewed in ref. 2). Similar to other nuclear hormone receptors, PPARs heterodimerize with another nuclear hormone receptor, retinoid X receptor, and exert their effects via regulation of gene transcription upon binding of ligand. To date, three isotypes have been identified, α, γ, and δ (the last is also known as PPARβ and NUC-1). Although all three isotypes of PPARs have been shown to modulate lipid metabolism, isotype-specific functions have also been described. For example, on the basis of experiments with specific agonists and knock-out mice, PPARγ has been shown to be involved with insulin resistance (3). PPARγ has also been implicated in a variety of neoplastic processes, including colorectal cancer, although its role as a promoter or suppressor of tumor growth remains controversial because of conflicting results in mouse versus human systems (4–6).
Recently PPARδ was identified as a potential downstream target of the adenomatous polyposis coli (APC)/β-catenin/T cell factor-4 (TCF-4) tumor suppressor pathway (7). In normal colorectal tissue, APC binds to β-catenin and inhibits its ability to form a bipartite transcription complex with TCF-4 (8–10). In the majority of colorectal cancers, APC is inactivated by truncating mutations, thus giving rise to elevated levels of β-catenin/TCF-4-mediated transcriptional activity. In cancers with intact APC genes, mutations of β-catenin that render it resistant to the inhibitory effects of APC often occur, strongly suggesting that inhibition of β-catenin/TCF-4-mediated transcription is critical to APC's tumor suppressive effects (10–12). Several targets of the β-catenin/TCF-4 transcription complex have been identified, including c-MYC (13) and cyclin D1 (14), which have obvious growth-promoting properties. PPARδ was identified as another potential target of this pathway after it was shown to be down-regulated upon restoration of APC expression in a human colorectal cancer cell line (7). This down-regulation appears to be direct, as the PPARδ promoter contains TCF-4 binding sites, and PPARδ promoter reporters are repressed by APC as well as stimulated by mutant β-catenin. Consistent with PPARδ's role as an APC/β-catenin target, PPARδ mRNA has been found to be frequently overexpressed in many colorectal cancers (7, 15). Finally, the ability of nonsteroidal antiinflammatory drugs (NSAIDs) to bind PPARδ (16) and potentially inhibit its function (7) suggested that inhibition of PPARδ might contribute to the chemopreventive effects of NSAIDs such as Sulindac. However, definitive evidence for an involvement of PPARδ in colorectal carcinogenesis or chemoprevention has not yet been presented.
To more rigorously define PPARδ's role in colorectal tumorigenesis, we deleted the PPARδ gene in a human colon cancer cell line. No obvious phenotype was observed upon in vitro culture, and no effects on sensitivity to NSAIDs were observed. However, PPARδ null cell lines had a pronounced effect on tumorigenicity when grown as xenografts in nude mice. The PPARδ null lines were defective in establishing tumors, and the few tumors that did arise grew slowly compared with the parental cells. These data provide unambiguous genetic evidence that PPARδ expression can affect tumorigenesis.
Materials and Methods
Tissue Culture and Transfections.
The colorectal cancer cell line HCT116 (American Type Culture Collection) and its derivatives were grown in 10% FBS and 1% penicillin/streptomycin in McCoy's 5A modified media and maintained at 37°C in 5% CO2. Cells were transfected with Lipofectamine (GIBCO/BRL), following the manufacturer's protocol and selected by using hygromycin or geneticin at concentrations of 0.1 mg/ml and 0.5 mg/ml, respectively.
Generation of PPARδ Null Cell Lines.
The general strategy for creating the PPARδ null line has been described previously (17) and is outlined in the Results. In brief, the 5′ and 3′ homology arms used for constructing the targeting vectors were PCR amplified from HCT116 genomic DNA, using primers chosen from publicly available genomic sequence databases (GenBank accession no. NT007193). The arms were cloned into vectors containing a hygromycin/thymidine kinase fusion gene that was flanked by loxP sequences. The primers used for deriving the targeting vectors and details of the construction are available from the authors upon request. Screening for homologous recombination events was performed by PCR as previously described (17), with the primer sequences 5′-GCTAGAGGTTTACGTGACCT-3′ for the forward primer and 5′-TATGATACGGCTCAATGATG-3′ for the reverse primer. After Cre-mediated loxP excision, allele-specific primers were used for further genetic verification. The above forward primer was used with the following reverse primers: 5′-CAGTCATAGC TCTGGCATCG-3′ for the wild-type allele and 5′-GTGGGGTATCGACAGAGTGC-3′ for the deleted allele. Lox recombination was mediated by infection with an adenovirus expressing Cre protein (Ad-Cre) prepared as previously described (18).
The PPARδ cDNA construct was created by inserting a hemagglutinin (HA)-tagged PPARδ cDNA from pCMV-HAHA-PPARδ (7) into a modified version of the pMK1059 (pMK-IRES-NEO) vector (19). This modified vector was created by excising pMK1059 with XhoI, blunting the ends, and then inserting the resulting promoter-IRES-NEO fragment into the EcoRV site of the loxP plasmid pBS246 (20). All targeted clones identified by PCR were verified by Southern blotting with 10 μg of genomic DNA digested with the restriction enzyme ApaI and probed with a 400-bp genomic fragment lying just outside the 5′ homology arm.
In Vitro Assays.
In vitro assays for Sulindac sulfide-induced apoptosis were performed as previously described (21). All assays were performed in triplicate, and the triplicates were repeated at least twice. For colony formation assays, cells were plated at varying concentrations into T-25 tissue culture flasks, grown for 1 week, and then stained with crystal violet. For in vitro growth assays, cells were seeded in triplicate in 12-well plates at a concentration of 3 × 104 cells per well. Cells were harvested at days 2, 4, and 6, and viable cells as assessed by trypan blue exclusion were counted in a hemocytometer. Results were expressed as the mean of three wells ± SD of the mean.
In Vivo Assays.
Cells were harvested before inoculation and resuspended in serum-free medium at a concentration of 5 × 107 cells per milliliter. Cells (5 × 106 cells, 0.1 ml) were then inoculated s.c. at the proximal dorsal midline into 3- to 4-week-old female athymic nu/nu mice (Harlan), except for experiment 2, where inoculations were made in the right and left flanks. Tumor sizes in two dimensions were measured twice weekly, and volumes were calculated as previously described, with the formula (L × W2) × 0.5, where L is length and W is width. Mice were housed in barrier environments, with food and water provided ad libitum.
Results
Generation of PPARδ Null Cells.
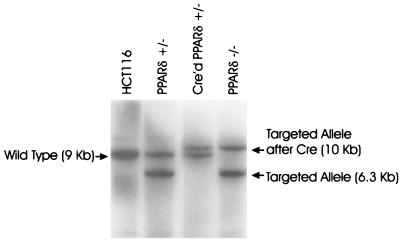
To explore the function of PPARδ in human cancer cells in a rigorous fashion, we chose to disrupt the endogenous gene by targeted homologous recombination in a human colorectal cancer cell line. The HCT116 cell line chosen for these experiments contains a mutated β-catenin gene resulting in increased β-catenin/TCF-4 transcriptional activity and accordingly expresses significant amounts of PPARδ. The first allele of PPARδ was disrupted by homologous recombination with the targeting vector PKO1, depicted in Fig. 1A. Recombination with PKO1 was predicted to result in deletion of 11 kb of the PPARδ gene, including the entire DNA-binding domain. Transfection of HCT116 with this vector and selection with hygromycin resulted in one successful recombination event among 3,000 individual clones screened. This successful recombination event was confirmed by Southern blot analysis (Fig. 2). This PPARδ +/− clone was then used to generate PPARδ −/− cells with a second round of PKO1-mediated recombination. To use the same PKO1 vector in the PPARδ +/− cells, it was necessary to restore hygromycin sensitivity. This was achieved by infection with an adenovirus (Ad-Cre) expressing the Cre protein that mediated excision of the hygromycin resistance gene by virtue of the flanking loxP sequences. Multiple clones were recovered, and all demonstrated loss of hygromycin resistance and excision of the hygromycin gene by Southern blot analysis. One such clone was selected for a second round of PKO1-mediated recombination, as described below.
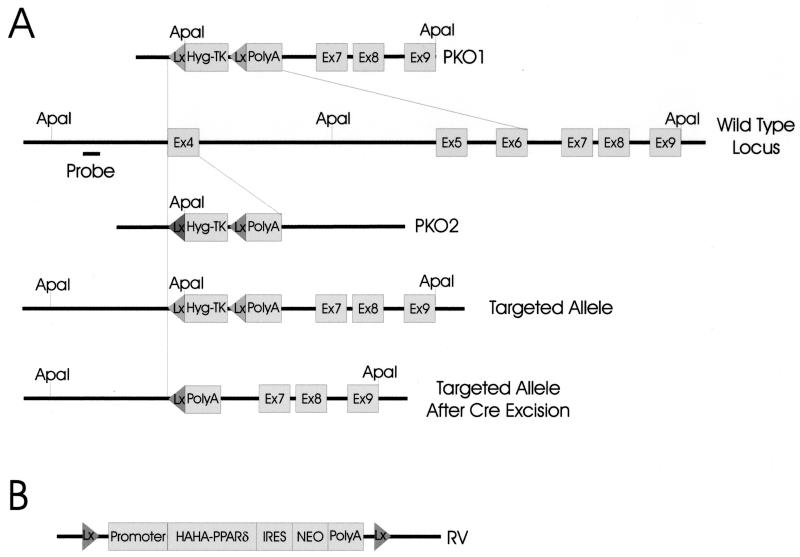
Figure 1.
Strategy for generating PPARδ null somatic cell lines. (A) Constructs used to mediate recombination at the PPARδ locus. PKO1 contained a 1-kb 5′ homology arm and a 4-kb 3′ homology arm and was used to generate an 11-kb deletion encompassing the entire DNA binding domain (exons 4, 5, and 6). PKO2 contained a 1.8-kb 5′ homology arm and a 4-kb 3′ homology arm and was used to delete exon 4. The endogenous PPARδ locus, the locus after targeting with PKO1, and the targeted locus after Cre-mediated excision are illustrated. Lx and Hyg-TK designate LoxP sequences and the hygromycin/thymidine kinase gene, respectively. The thymidine kinase gene was not used for negative selection in the protocols actually applied in this study. ApaI sites and the probe sequence used for Southern blotting are indicated. (B) Schematic of the PPARδ rescue vector. A HAHA-tagged cDNA of PPARδ was cloned into a construct containing an IRES-geneticin resistance gene cassette flanked by LoxP sequences as described in Materials and Methods.
Figure 2.
Southern blot analysis of PPARδ engineered cell lines. HCT116, parental cells; PPARδ +/−, HCT116 cells after successful recombination with PKO1; Cre'd PPARδ +/−, PPAR +/− cells after Cre-mediated LoxP excision of the Hyg-TK gene; PPARδ −/−, PPARδ +/− cells after successful recombination of the remaining PPARδ locus with PKO1.
After screening approximately 5,000–6,000 clones and failing to detect a successful recombination, we considered the possibility that complete loss of PPARδ might be lethal or severely growth inhibitory. Indeed, complete disruption of the PPARδ gene in mice can result in decreased embryo viability (22). We therefore devised a rescue strategy. In brief, the vector RV, which constitutively expressed a HA-tagged PPARδ that could be subsequently removed by Cre-mediated recombination (Fig. 1B), was transfected into the HCT116 PPARδ +/− cells. PPARδ +/− clones with the RV (PPARδ +/−/RV) were then selected for their resistance to geneticin, which ensured expression of the PPARδ cDNA because of the modified internal ribosomal entry site (IRES) sequence (19) separating the PPARδ cDNA and the geneticin resistance gene (Fig. 1B). Expression of the HA-PPARδ protein in PPARδ +/−/RV clones was demonstrated by Western blotting with an anti-HA antibody (data not shown). Transfection of a representative PPARδ +/−/RV clone with the PKO1 vector resulted in one successful recombination event after approximately 1,000 clones were screened. Southern blotting confirmed that homologous recombination had deleted the remaining endogenous PPARδ allele, resulting in the line PPARδ −/−/RV, as shown in Fig. 2.
PPARδ Null Cell Lines Are Viable in Vitro.
The PPARδ −/−/RV cell line was then used to test whether deletion of PPARδ was lethal or severely growth inhibitory. The PPARδ −/−/RV cells could easily be rendered PPAR null by Cre-mediated excision after infection with Ad-Cre. As a control for nonspecific effects, a sister PPARδ +/−/RV clone was similarly infected with Ad-Cre. Surprisingly, clones that had excised the PPARδ RV were recovered in equal numbers from both the PPARδ −/−/RV and PPARδ +/−/RV clones, indicating that PPARδ is not required for in vitro growth. Excision of the PPARδ cDNA was confirmed by both PCR and Western blot analysis (data not shown). There were no obvious differences in the growth properties between the two groups of recovered clones in vitro, although both PPARδ +/− and PPARδ −/− clones grew slightly more slowly than wild-type HCT116 cells (see below).
To rule out the possibility of clonal variability, additional PPARδ null cell lines were sought. Unfortunately, the low recombination frequency of the targeting vector significantly hampered our ability to generate more clones. To circumvent this problem, a second-generation targeting vector (PKO2) was constructed that had a longer 5′ homology arm (1.8 kb vs. 1.0 kb) and deleted only the first coding exon of the PPARδ gene encompassing 200 bp, as shown in Fig. 1A. Preliminary experiments in our laboratory suggested that decreasing the size of the deleted region might increase the recombination frequency of targeted homologous recombinations. We therefore hoped that recombination rates with PKO2 might be superior to those observed with PKO1. Indeed, transfection of wild-type HCT116 resulted in 11 successful recombinations among 200 clones tested, representing an increase in targeting efficiency of ≈150-fold. Likewise, transfection of PPARδ +/−/RV resulted in two successful recombinations among 200 clones tested. The two PPARδ −/−/RV clones were confirmed by Southern blotting and subjected to infection with Ad-Cre to generate PPARδ null cells (data not shown). As with the original null clone, viable clones could be recovered at similar frequencies, demonstrating that deletion of the PPARδ gene was not lethal.
PPARδ Is Not Required for Sulindac-Mediated Apoptosis.
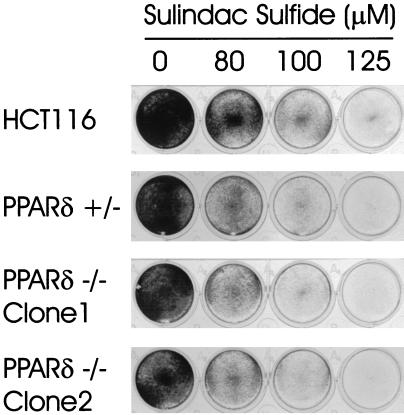
We had previously suggested that inhibition of PPARδ by NSAIDs might contribute to their chemopreventive effects. One potential in vitro surrogate of NSAID chemoprevention is their ability to induce apoptosis. Consistent with a possible role of PPARδ in apoptosis, we have previously demonstrated that overexpression of PPARδ could partially rescue cells from Sulindac-induced apoptosis (7). To determine whether PPARδ is required for Sulindac-mediated apoptosis, wild-type HCT116, two independently derived PPARδ −/− clones, and a sister PPARδ +/− heterozygote clone were analyzed for their response to Sulindac. Cells were treated with Sulindac at various concentrations and then stained with crystal violet to assess the number of surviving cells after 72 h. There was no appreciable difference in cell viability between these four cell lines (Fig. 3), indicating that PPARδ is not required for NSAID-mediated apoptosis.
Figure 3.
PPARδ is not required for Sulindac-mediated apoptosis. HCT116, PPARδ +/−, and PPARδ −/− clones were treated for 72 h with 0, 80, 100, and 125 μM Sulindac sulfide, and surviving colonies were assessed by crystal violet staining. Treatments were performed in triplicate; representative wells are shown.
PPARδ Is Required for Efficient Tumorigenicity.
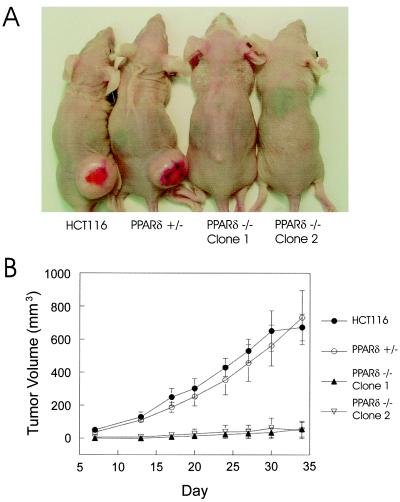
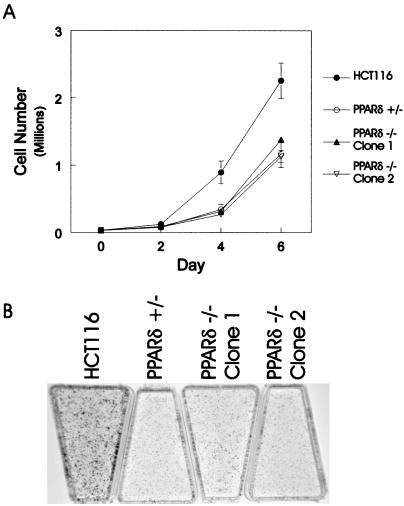
Although the above data conclusively demonstrated that PPARδ is not required for in vitro cell growth, we questioned whether the absence of this receptor had any effect on tumorigenicity. To address this hypothesis, wild-type HCT116, PPARδ +/−, and PPARδ −/− cells were injected s.c. and grown as xenografts in nude mice. There was a dramatic difference in tumor establishment and growth of the PPARδ −/− cells compared with wild-type HCT116 and the PPARδ +/− cells. This in vivo growth deficiency was observed in five independent experiments (Table 1). Although there was no appreciable difference in tumor establishment or growth between wild-type HCT116 and the PPARδ +/− cells, both of the PPARδ −/− cell lines yielded fewer tumors per mouse compared with wild-type and PPARδ +/− cells (Table 1 and Fig. 4). In addition, the few tumors that did arise in the mice were smaller than their wild-type or PPARδ +/− control counterparts (Table 1 and examples in Fig. 4A). It is important to note that the two PPARδ null cell lines used in these experiments were independently derived. The fact that both lines were defective in growth in nude mice indicated that this in vivo phenotype depended on disruption of the PPARδ gene and was not the result of clonal variability. To further assess this issue, a total of four additional independently derived PPARδ null clones were tested for their in vivo tumorigenicity, and all displayed similar defects in tumor growth relative to the wild-type and PPAR (+/−) cells (data not shown). The in vivo growth defects of the PPARδ null cells did not simply reflect general alterations in growth, as all lines grew well in culture. Quantitative analysis demonstrated that the PPARδ +/− and PPARδ −/− cells were indistinguishable in their in vitro growth properties, although parental HCT116 cells generally grew somewhat better than the modified cells (Fig. 5).
Table 1.
In vivo growth of PPARδ null cells
| Experiment no. | Frequency | All tumors | Established tumors |
|---|---|---|---|
| 1 (28 days) | |||
| PPARδ +/+ | 2/2 | 1218 ± 1095 | 1218 ± 1095 |
| PPARδ +/− | 2/2 | 987 ± 49 | 987 ± 49 |
| PPARδ −/− clone 1 | 1/2 | 2 ± 2 | 4 ± NA |
| 2 (30 days) | |||
| PPARδ +/+ | 4/4 | 1306 ± 680 | 1306 ± 680 |
| PPARδ +/− | 4/4 | 2333 ± 803 | 2333 ± 803 |
| PPARδ −/− clone 1 | 4/4 | 374 ± 46 | 374 ± 46 |
| 3 (33 days) | |||
| PPARδ +/+ | 10/10 | 1617 ± 248 | 1617 ± 248 |
| PPARδ −/− clone 1 | 4/10 | 271 ± 107 | 609 ± 90 |
| 4 (34 days) | |||
| PPARδ +/+ | 5/5 | 1922 ± 823 | 1922 ± 823 |
| PPARδ −/− clone 2 | 2/5 | 86 ± 64 | 214 ± 115 |
| 5 (34 days) | |||
| PPARδ +/+ | 6/6 | 673 ± 80 | 673 ± 80 |
| PPARδ +/− | 5/5 | 732 ± 164 | 732 ± 164 |
| PPARδ −/− clone 1 | 2/5 | 56 ± 48 | 141 ± 66 |
| PPARδ −/− clone 2 | 1/5 | 49 ± 49 | 243 ± NA |
In vivo growth of PPARδ null cells. Five experiments are presented measuring the growth of various PPARδ null lines as described in Materials and Methods. The Frequency column indicates the fraction of inoculation sites in which a tumor grew. The All tumors column indicates the average volume (mm3) ±SE of tumors for all inoculation sites, whereas the Established tumors column indicates the average volume (mm3) ±SE for only those tumors that were clearly detectable on the date of the final measurement, as indicated in parentheses next to the experiment number. NA, not applicable.
Figure 4.
PPARδ is required for efficient tumorigenicity. HCT116, PPARδ +/−, and PPARδ −/− cells were injected as xenografts in nude mice and sacrificed after 34 days as described in Materials and Methods. (A) Representative mice at day 34. (B) Growth curves of HCT116, PPARδ +/−, and PPARδ −/− xenografts. Each data point for HCT116 represents the average tumor size of six mice, and each data point for the PPARδ +/− and PPARδ −/− cell lines represents the average tumor size of five mice. Error bars represent the SEM. Tumors that did not grow were assigned a volume of zero.
Figure 5.
In vitro growth characteristics of PPARδ-modified cells. (A) In vitro growth assay comparing growth kinetics of parental HCT116, PPARδ +/−, and PPARδ −/− clones. Each data point represents the average of three wells, and the bars represent the SD from the mean. (B) Colony formation assay demonstrating no difference in colony number between parental HCT116, PPARδ +/−, and PPARδ −/− clones. Note that colony numbers were roughly equivalent among all four cell lines, although the parental HCT116 colony sizes were slightly larger, consistent with the growth curves in A.
Discussion
Since their identification and cloning a decade ago, PPARs have been implicated in numerous biologic pathways. Although various functions for the PPAR α and γ isotypes have been described, less is known about the biological significance of PPARδ. We and others have demonstrated that PPARδ is overexpressed in both human and rodent colorectal tumors (7, 15). However, the functional consequence of this overexpression for tumor growth is not clear. The generation of a PPARδ−/− colon cancer cell line has enabled us to rigorously evaluate the role of this receptor in colorectal tumorigenesis. By comparing isogenic cell lines that differed only in their presence or absence of the PPARδ gene, we have unambiguously shown that this receptor can directly affect the growth and establishment of colorectal tumors in vivo. This observation, in combination with the demonstration that PPARδ is consistently overexpressed in colorectal cancers and can be controlled by TCF-4/β-catenin transcription complexes, suggests that APC mediates part of its growth-suppressive effects through PPARδ.
The recent creation of the PPARδ null mouse has demonstrated that this receptor is involved with lipid metabolism, similar to the other two PPAR isotypes (22). However, PPARδ −/− mice were only generated with a Mendelian pattern of inheritance after backcrossing, suggesting that the absence of PPARδ may be lethal in certain mouse strains. In addition, PPARδ null mice are smaller than their control littermates, and as previously stated, PPARδ is overexpressed in colorectal cancers. These results suggested that PPARδ null cancer cells may not be viable. We therefore developed a rescue strategy that enabled us to conditionally express a PPARδ cDNA before deletion of the second allele. This system allowed us to generate a PPARδ −/− line without concern that we had selected for a viable phenotype by clonal selection. The subsequent isolation of multiple viable PPARδ null clones conclusively proved that this gene was not required for in vitro growth of colorectal cancer cells.
Previous experiments have demonstrated that PPARδ can bind to lipids resembling the substrates and products of cyclooxygenases (16, 23) and that NSAIDs can inhibit PPARδ activity (7). Moreover, we have previously shown that PPARδ overexpression can partially rescue colon cancer cells from the apoptotic effects of NSAIDs (7). However, our current data suggest that PPARδ is not a major mediator of Sulindac-induced apoptosis, as PPARδ −/− cell lines did not display any appreciable difference in sensitivity to this NSAID. The partial rescue from NSAID-mediated apoptosis may have been due to the broadly overlapping effects of growth-regulatory genes within cell circuits (24, 25). These results also emphasize the value of gene disruption experiments, which are considerably more straightforward to interpret than those employing unphysiologic levels of overexpression. However, it remains possible that inhibition of PPARδ contributes to other important chemopreventive effects of NSAIDs. Consistent with this notion, PPARδ null mice appear to be partially resistant to the antiinflammatory effects of Sulindac (22), and products of COX-2 metabolism have recently been shown to act as ligands for PPARδ (15, 26). These issues will become clearer once the relationship between in vitro apoptosis caused by NSAIDs and in vivo chemoprevention become better defined.
Although the absence of PPARδ clearly affects tumorigenicity in the HCT116 cell line, it is difficult to determine whether this effect is unique to this cell line or is generally applicable. However, because the APC pathway is mutated with high frequency in colon cancer and overexpression of PPARδ has been observed in many colorectal cancers, it seems likely that increased expression of PPARδ could generally contribute to the neoplastic process. Furthermore, the fact that PPARδ is a transcription factor suggests that such growth-promoting effects are directly related to expression and/or repression of target genes. As such, a comprehensive analysis of gene expression with PPARδ −/− cells and appropriate controls may allow identification of important growth-regulatory genes. To our knowledge, there is only one other gene whose disruption in a colorectal cancer cell line has been shown to limit tumorigenicity. Sasazuki and colleagues showed that disruption of the mutant K-RAS gene in HCT116 cells resulted in a phenotype very similar to that observed in the PPARδ null cells: continued growth in vitro but poor growth in vivo (27). Such experiments provide critical information for defining appropriate targets for therapy. PPARδ is an especially good target for pharmaceutical development because it is a receptor, and its ligands could potentially be modified to form antagonists. Moreover, it is a member of a family of genes that are eminently “drug targetable” and thereby form the basis for a large number of current drugs used to treat a variety of diseases. Specific pharmacologic inhibitors may be developed against PPARδ and may be predicted to inhibit the in vivo growth of tumors in ways similar to that observed here after genetic disruption of this gene.
Acknowledgments
We thank T. Chan, I. Rhee, E. Carson-Walter, D. Cahill, L. Zawel, S. Zhou, L. Zhang, J. Yu, T.-C. He, and G. Traverso for helpful discussions and technical comments and R. A. Casero for critical reading of the manuscript. B.H.P. thanks Rachel L. Damico for her constant support and encouragement. B.H.P. is a recipient of a Howard Hughes Postdoctoral Fellowship for Physicians, and B.V. is an investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants CA57345 and CA62924. K.W.K. receives research funding from Genzyme Molecular Oncology. K.W.K. and B.V. are consultants to Genzyme. Under a licensing agreement between Johns Hopkins University and Genzyme, K.W.K. and B.V. are entitled to a share of royalties received by the university from sales of licensed technology. The terms of these arrangements are being managed by the university in accordance with its conflict of interest policies.
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- APC
adenomatous polyposis coli
- NSAID
nonsteroidal antiinflammatory drug
- HA
hemagglutinin
- TCF-4
T cell factor-4
- IRES
internal ribosomal entry site
References
- 1.Escher P, Wahli W. Mutat Res. 2000;448:121–138. doi: 10.1016/s0027-5107(99)00231-6. [DOI] [PubMed] [Google Scholar]
- 2.Kersten S, Desvergne B, Wahli W. Nature (London) 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 3.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 4.Sarraf P, Mueller E, Jones D, King F J, DeAngelo D J, Partridge J B, Holden S A, Chen L B, Singer S, Fletcher C, et al. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre A M, Chen I, Desreumaux P, Najib J, Fruchart J C, Geboes K, Briggs M, Heyman R, Auwerx J. Nat Med. 1998;4:1053–1057. doi: 10.1038/2036. [DOI] [PubMed] [Google Scholar]
- 6.Saez E, Tontonoz P, Nelson M C, Alvarez J G, Ming U T, Baird S M, Thomazy V A, Evans R M. Nat Med. 1998;4:1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 7.He T C, Chan T A, Vogelstein B, Kinzler K W. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 10.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 11.Sparks A B, Morin P J, Vogelstein B, Kinzler K W. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 12.Shitoh K, Furukawa T, Kojima M, Konishi F, Miyaki M, Tsukamoto T, Nagai H. Genes Chromosomes Cancer. 2001;30:32–37. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1065>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 13.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 14.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R A, Tan J, Krause W F, Geraci M W, Willson T M, Dey S K, DuBois R N. Proc Natl Acad Sci USA. 2000;97:13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K, Bayona W, Kallen C B, Harding H P, Ravera C P, McMahon G, Brown M, Lazar M A. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 17.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 18.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi M, Yamauchi Y, Tanaka A, Shimamura S. BioTechniques. 1996;21:398–402. doi: 10.2144/96213bm12. [DOI] [PubMed] [Google Scholar]
- 20.Sauer B. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 21.Chan T A, Morin P J, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1998;95:681–686. doi: 10.1073/pnas.95.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters J M, Lee S S, Li W, Ward J M, Gavrilova O, Everett C, Reitman M L, Hudson L D, Gonzalez F J. Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstein I B, Begemann M, Zhou P, Han E K, Sgambato A, Doki Y, Arber N, Ciaparrone M, Yamamoto H. Clin Cancer Res. 1997;3:2696–2702. [PubMed] [Google Scholar]
- 25.Vogelstein B, Lane D, Levine A J. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 26.Lim H, Gupta R A, Ma W, Paria B C, Moller D E, Morrow J D, DuBois R N, Trzaskos J M, Dey S K. Genes Dev. 1999;13:1561–1574. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]