Abstract
Engineered zinc-finger nucleases (ZFNs) enable targeted genome modification. Here we describe Context-Dependent Assembly (CoDA), a platform for engineering ZFNs using only standard cloning techniques or custom DNA synthesis. Using CoDA ZFNs, we rapidly altered 20 genes in zebrafish, Arabidopsis, and soybean. The simplicity and efficacy of CoDA will enable broad adoption of ZFN technology and make possible large-scale projects focused on multi-gene pathways or genome-wide alterations.
Engineered zinc-finger nucleases (ZFNs) can be used to introduce targeted alterations into genomes of model organisms, plants, and human cells.1, 2 Repair of ZFN-induced double-strand breaks (DSBs) by error-prone non-homologous end-joining (NHEJ) leads to efficient introduction of insertion or deletion mutations (indels) at the site of the DSB. Alternatively, repair of a DSB by homology-directed repair with an exogenously introduced donor template can promote efficient introduction of alterations or insertions at or near the break site.
Widespread adoption and large-scale use of ZFN technology have been hindered by continued lack of a robust, easy-to-use, and publicly available method for engineering zinc-finger arrays. One approach, known as modular assembly, joins together pre-selected zinc-finger modules into arrays,3 a procedure simple enough to be practiced by any researcher. Some recent reports have demonstrated a high failure rate for this method,4, 5 although the consequent need to construct and test large numbers of ZFNs for any given target gene can be mitigated by using a more limited subset of modules.6 We recently described a robust selection-based method known as Oligomerized Pool ENgineering (OPEN),7 but the labor and expertise required to screen combinatorial libraries have limited its broad adoption.3 Sangamo BioSciences, Inc. has also developed a platform for engineering ZFNs and although some details of this method have been published,8 its practice requires access to a proprietary archive of engineered zinc-finger units.9 Researchers may purchase customized ZFNs made by the Sangamo approach through the Sigma-Aldrich CompoZrR service but the cost of these proteins9 limits the scale and scope of projects that can be performed.
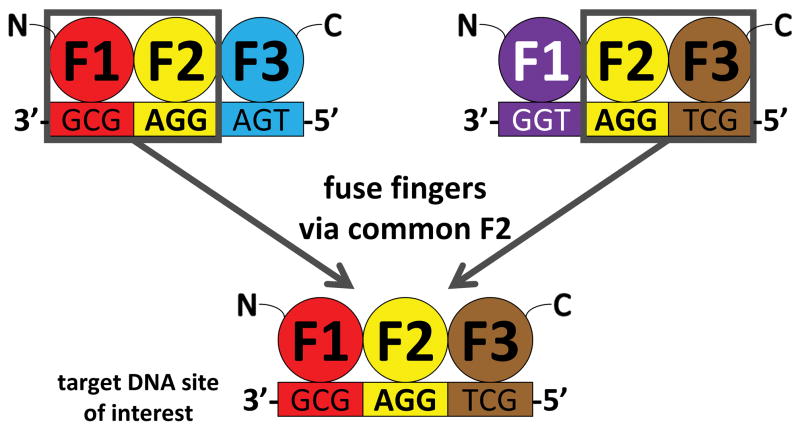
Here we describe Context-Dependent Assembly (CoDA), a publicly available platform of reagents and software that is simple to practice and shows a success rate comparable to selection-based methods such as OPEN. With the CoDA approach, three-finger arrays are assembled using N- and C-terminal fingers that have been previously identified in other arrays containing a common middle finger (Fig. 1). CoDA can be practiced using a large archive consisting of 319 F1 and 344 F3 units (Supplementary Tables 1 and 2) engineered to function well when positioned adjacent to one of 18 fixed F2 units (Methods). Thus, in contrast to modular assembly, CoDA does not treat fingers as independent modules but instead explicitly accounts for context-dependent effects between adjacent fingers,10, 11 thereby increasing the probability that a multi-finger array will function well. CoDA is rapid and requires no specialized expertise; multi-finger arrays can be constructed in one to two weeks or less using standard cloning techniques or commercial DNA synthesis.
Figure 1. Schematic overview of Context-Dependent Assembly (CoDA).
Zinc fingers are represented as colored spheres (F1 = amino-terminal finger, F2 = middle finger, F3 = carboxy-terminal finger) and 3 bp DNA “subsites” are represented as colored rectangles. Two different three-finger arrays, each engineered to bind different 9 bp target sites and that each share a common middle F2 can be used to create a three-finger array with a new specificity by joining together the amino-terminal finger (F1) from the first array, the middle finger common to both arrays (F2), and the carboxy-terminal finger (F3) from the second array.
To test the CoDA approach, we assembled 181 three-finger arrays and evaluated each for its ability to bind its cognate DNA target site using a well-established bacterial two-hybrid (B2H) reporter assay.4, 7 Previous work has shown that three-finger arrays that fail to activate transcription by more than 1.57-fold in the B2H reporter assay are likely to be inactive as ZFNs in mammalian cells4 and those that activate by three-fold or more have a high probability of functioning efficiently as ZFNs in zebrafish, plant, and human cells.7, 12–15 Of the 181 CoDA arrays we tested using the B2H reporter assay, <8% (14 arrays) activated transcription by <1.57-fold and >76% (139 arrays) activated transcription by >3.00-fold (Supplementary Fig. 1 and Supplementary Table 3). These failure and success rates (as predicted by the B2H reporter assay) are comparable to what we have previously observed with three-finger arrays made by OPEN (Supplementary Note). Because so few (<25%) of the CoDA arrays we tested gave <3.00-fold activation in the B2H reporter assay, our results suggest that one could potentially skip the B2H reporter assay step and move directly to testing in the final desired cell type of interest.
We compared the efficacy of CoDA with that of modular assembly by using both approaches to construct three-finger arrays for 26 different nine bp sites and testing these proteins for DNA-binding activity in the B2H reporter assay (Supplementary Table 4). We observed that, for these sites that can be targeted by both methods, CoDA outperforms modular assembly (Supplementary Fig. 2 and Supplementary Note). The most likely explanation for the relatively higher success rates of CoDA is its explicit consideration of context-dependent activities between fingers.10, 11 We note that these differences in success rates become potentially more pronounced when one considers that two functional arrays must be engineered to create dimers of ZFNs.
We applied CoDA to engineer ZFNs for endogenous gene targets in zebrafish and plants. Using CoDA zinc-finger arrays that activated transcription at least three-fold in the B2H reporter assay, we constructed ZFN pairs for 24 gene targets in zebrafish, 13 gene targets in Arabidopsis thaliana, and one target present in two duplicated genes in soybean (Table 1). CoDA ZFNs induced targeted indel mutations with high efficiencies in 12 out of 24 zebrafish target sites (≤ 1% to 16.7%; Table 1 and Supplementary Fig. 3), in six out of 13 Arabidopsis gene targets (1.1% to 8.4%; Table 1 and Supplementary Fig. 4), and in a target site present in two duplicated soybean genes in transformed root tissue (18.8% and 10.7%; Table 1 and Supplementary Fig. 4).
Table 1.
Endogenous zebrafish and plant genes targeted by CoDA ZFNs
| Gene name | Organism | Target site | Mutant alleles/ Total alleles | Mutation frequency |
|---|---|---|---|---|
| Dcl4a | Soybean | TGCTTCATCacaatGGAGATGAT | 6/32 | 18.8% |
| Dcl4b | Soybean | TGCTTCATCacaatGGAGATGAT | 3/28 | 10.7% |
| MPK8 | Arabidopsis | CTCCACAACatcagGATGACGAA | 7/83 | 8.4% |
| MPK11 | Arabidopsis | CTCTTCGTCctatcgGCAGAGGCG | 3/90 | 3.3% |
| MKK9 | Arabidopsis | GCCAGCGACggtggtGGTGGTGGC | 3/95 | 3.2% |
| MPK15 | Arabidopsis | TTCTTCATCcagatGTTGTTGAG | 2/73 | 2.7% |
| MAPKKK18 | Arabidopsis | CCCTTCCACaacaacGGAGAAGCT | 2/75 | 2.7% |
| GA3OX2 | Arabidopsis | AGCTACGCCgtagccGGAGACGCC | 1/94 | <=1.0% |
| MAPKKK1 | Arabidopsis | GGCACCTCCgatttcGTGGAGGAA | 0/190 | 0.0% |
| MAPKKK12 | Arabidopsis | TCCTCCACCgaatcGACGGCGCT | 0/187 | 0.0% |
| MAPKKK12 | Arabidopsis | TTCCTCCACcgaatcGACGGCGCT | 0/186 | 0.0% |
| MAPKKK4 | Arabidopsis | GTCTCCGCCtaggaGATGCAGAC | 0/190 | 0.0% |
| MPK15 | Arabidopsis | TGCTTCTTCatccaGATGTTGTT | 0/94 | 0.0% |
| MPK4 | Arabidopsis | CTCTTCGTCctatcgGTAGAGGCG | 0/190 | 0.0% |
| TZP | Arabidopsis | TTCGTCTTCgagtcGTCGTTGTT | 0/141 | 0.0% |
| actn1 | Zebrafish | GCCTTCTCCggggcGCAGAAGGT | 10/60 | 16.7% |
| rag2 | Zebrafish | ATCTTCTGCtccaggGGTGAAGGT | 4/52 | 7.7% |
| gad2 | Zebrafish | AGCCGCAGCtctcgGCTGTAGAC | 3/43 | 7.0% |
| lmna | Zebrafish | CTCTTCTCCcccagaGCTGTGGAG | 2/41 | 4.9% |
| apoeb | Zebrafish | CCCCTCAGCccagaTGGGAGGAG | 3/64 | 4.7% |
| trpm7 | Zebrafish | CACACCTGCacacaGATGCTGCT | 2/55 | 3.6% |
| grip1 | Zebrafish | GGCCACCTCcaccaGCAGCGGGC | 3/90 | 3.3% |
| pclo | Zebrafish | CCCCTCTCCtcaaaGCAGATGCA | 3/96 | 3.1% |
| jak3 | Zebrafish | GGCCCCACCaagcctGCTGGAGGA | 1/71 | <=1.0% |
| ago1 | Zebrafish | CTCTGCCGCcacctaGAGGATGGT | 1/96 | <=1.0% |
| slitrk1 | Zebrafish | GCCCACAGCaatggcGGAGCCGCC | 1/96 | <=1.0% |
| bmpr2a | Zebrafish | GACTTCCTCtctgtGCAGTCGGC | 1/117 | <=1.0% |
| bmpr2a | Zebrafish | ACCTCCTGCagtgtGAGGTTGTC | 0/156 | 0.0% |
| cnot1 | Zebrafish | GGCGTCCACgtacgaGCGGAGGAG | 0/93 | 0.0% |
| ctcf | Zebrafish | TTCCTCCTCctgatGCGGAGGCT | 0/96 | 0.0% |
| dicer1 | Zebrafish | TTCTGCAGCtcaatGGAGATGGT | 0/96 | 0.0% |
| dicer1 | Zebrafish | AGCTTCCTCcgccgGAAGTTGAG | 0/96 | 0.0% |
| drosha | Zebrafish | GTCCTCCTCatggcgGTCGATGGT | 0/96 | 0.0% |
| g6pcb | Zebrafish | TCCCACTGCtgattGTAGGTGGA | 0/134 | 0.0% |
| nedd4l | Zebrafish | AACCGCACCacacaGTGGAAGAG | 0/86 | 0.0% |
| nod2 | Zebrafish | AACTACAACattaggGCTGGAGGA | 0/103 | 0.0% |
| rag1 | Zebrafish | GTCCTCCCCttcaaGTCGAATAG | 0/91 | 0.0% |
| th2 | Zebrafish | CTCCTCCTCaaacacGAAGCTGTC | 0/142 | 0.0% |
| tp53 | Zebrafish | AGCAGCTGCatgggGGGGATGAA | 0/107 | 0.0% |
Target sites within each gene are written 5’ to 3’ with the two half-sites targeted by the zinc finger arrays shown in upper case letters and the intervening spacer sequence shown in lower case. Sequences of CoDA ZFN-induced zebrafish and plant gene mutations are shown in Supplementary Figs. 3 and 4, respectively.
Our overall per target success rate for obtaining mutations with CoDA ZFNs is 50% (19 out of 38 target sites) in zebrafish and plants, a frequency comparable to our success rate of ~67% (16 out of 24 target sites) with OPEN ZFNs in zebrafish, plants, and human cells (refs. 7, 12–15 and unpublished data). We note that, for CoDA, success rate as calculated per ZFN and per target site is the same, since a single ZFN is synthesized per site. Although we do not know why some CoDA and OPEN ZFNs fail to induce mutations, we hypothesize that chromatin state or DNA methylation of the site or stability or folding of the protein might be responsible. Regardless of the precise mechanism, we recommend that users of CoDA plan to make ZFNs for at least two target sites per gene of interest to increase the likelihood that at least one pair will successfully introduce mutations.
CoDA still possesses some limitations compared to existing methods. Although modular assembly was less efficient than CoDA in our direct comparisons, modular assembly can potentially be used to target sites that CoDA currently cannot5, 6 and one recent report demonstrated a comparable success rate of 23% for modular assembly using a more limited subset of modules.6 In addition, although CoDA accounts for context-dependence between adjacent fingers, it also has some limitations relative to selection-based methods such as OPEN. For example, CoDA constrains the identity of the middle finger (F2) and does not “balance” the effects of all three fingers on affinity and specificity of the final array. In addition, CoDA in its current form guides assembly of arrays to 9 bp target sites, ignoring the identities of the adjacent upstream and downstream bases. Thus, for highly demanding therapeutic applications (e.g.—introduction of alterations into human pluripotent stem cells13), ZFNs made by OPEN may still be preferable to those made by CoDA and it may be necessary to engineer zinc-finger arrays with greater specificities. Nonetheless, our overall results demonstrate that CoDA provides a method for assembling zinc-finger arrays that accounts for context-dependent effects, is easier to perform than OPEN selections, and yields ZFNs that function efficiently for gene modification.
With the current archive of CoDA units, a potential ZFN target site can be found approximately once in every 500 bp of random sequence (Supplementary Note). However, actual targeting range can be higher, depending upon genomic sequences . For example, ~81% of 27,305 unique protein coding transcripts in the zebrafish genome (Ensembl Zv8.57 database) contain one or more potentially targetable ZFN sites (mean of 4.37), a frequency equivalent to one potential site every ~400 bp of transcript coding sequence. By contrast, ~63% of 33,200 unique protein coding transcripts in the Arabidopsis genome (TAIR9 release) contain one or more potential ZFN target sites (mean of 2.45), a frequency equal to one potential site every ~790 bp of transcript coding sequence. To enable users to identify potential CoDA target sites in any given gene sequence, we have updated our publicly available web-based Zinc Finger Targeter (ZiFiT) program (http://bindr.gdcb.iastate.edu/ZiFiT/ or http://www.zincfingers.org/software-tools.htm;Supplementary Note).
In summary, CoDA provides an effective alternative method for using publicly available reagents to engineer ZFNs. With CoDA, dozens of zinc-finger arrays can be rapidly assembled or commercially synthesized in 1 to 2 weeks without the need for labor-intensive selection and moved directly into cells for testing as ZFNs. We note that the rapidity and high success rate of CoDA enabled us to mutate 20 endogenous genes in three different organisms. CoDA will foster broader adoption of ZFN technology and also enable large-scale ZFN projects focused on multi-gene pathways or genome-wide alterations that are difficult to implement using existing methodologies.
Online Methods
Identification of CoDA finger units
To identify “fixed” F2 fingers for various three bp target subsites, we analyzed the amino acid sequences of F2s from a collection of three finger arrays previously identified from OPEN selections performed for over 130 different nine bp sites (references 7, 12–15 and M. Maeder et al., unpublished data). From this analysis, we identified F2 units for 18 different 3 bp subsites that occurred in at least two or more different contexts. The F1 and F3 units found adjacent to these F2 units were also chosen as CoDA units because they had been selected to work well together. To obtain additional F1 and F3 CoDA units for other 3 bp subsites, we performed a series of OPEN selections in which we interrogated combinatorial three-finger array libraries composed of a fixed F2 unit and randomized F1 and F3 fingers for binding to specific 9 bp target sequences. From these selections we analyzed the amino acid sequences of three-finger arrays that activated transcription three-fold or more in the B2H reporter assay to identify additional F1 and F3 finger units that worked well when positioned adjacent to a specific fixed F2 CoDA unit. For selections that yielded multiple three-finger array clones, we chose F1 and F3 finger units that occurred the most frequently in multiple distinct arrays and/or that were found in three-finger arrays that gave the highest fold-activation in the B2H reporter assay. OPEN selections were performed essentially as described7, 16 but with the modification that a beta-lactamase antibiotic resistance gene was used for selection instead of the HIS3 gene. This modified version of OPEN enabled selections to be performed with higher throughput and will be described in a subsequent publication (Goodwin et al., unpublished data). Each of the three-finger arrays from which the F1, F2, and F3 units were derived was determined to be active in a well-established bacterial two-hybrid (B2H) reporter assay.
Construction of zinc finger arrays by modular assembly
Construction of plasmids encoding the modularly assembled zinc finger arrays used in this study has been previously described.4
Construction of zinc finger arrays by CoDA
To assemble CoDA zinc finger arrays, DNA fragments encoding a F1–F2 cassette or a F3 cassette were amplified by PCR from plasmids using primer pair OK1424/OK1427 or OK1428/OK1429, respectively. The resulting PCR products were digested with DpnI to degrade template plasmid DNA and purified using a QIAGEN PCR purification kit. The cassettes were then fused together and amplified in a single PCR step using primer pair OK1430/OK1432. PCR product encoding a three-finger array was then purified using a QIAGEN PCR purification kit, treated with Pfu polymerase in the presence of dTTP nucleotide to create overhangs, phosphorylated with T4 polynucleotide kinase, and ligated to a B2H expression plasmid (pMG414) in which the zinc finger array is expressed as a fusion to a fragment of the yeast Gal11P protein.16 All plasmids were sequence-verified using primer OK61. (Sequences of all primers are provided in Supplementary Table 5).
B2H reporter assay
Zinc finger arrays made by modular assembly or CoDA were each tested for binding to its cognate target site by measuring its ability to activate transcription in the B2H reporter assay as previously described.16, 17 All assays were performed in triplicate.
Zebrafish gene mutation analysis
Injection of zebrafish embryos, isolation of genomic DNA, limited-cycle PCR amplification of the locus of interest, TOPO cloning of PCR fragments, and transformation of E. coli were performed as previously described.12, 18 Resulting colonies were assessed for gene mutations by one of two methods: (1) direct sequencing of individual clones or (2) screening of three pooled clones for alterations in PCR fragment size using fluorescent-based analysis as previously described18 followed by identification of specific mutations by direct sequencing.
Arabidopsis gene mutation analysis
ZFN transgene expression constructs, Arabidopsis transformation methods, induction of ZFN expression in Arabidopsis seedlings by β-estradiol, and isolation of Arabidopsis genomic DNA were as previously described.14 ZFN recognition sites in the Arabidopsis genomic DNA were amplified by PCR, the resulting fragments TOPO-cloned and DNA from individual colonies subjected to DNA sequence analysis to identify mutations at the ZFN recognition site.
Soybean gene mutation analysis
Cotyledons of the soybean variety Bert were transformed using a previously described hairy root transformation protocol.19 The ZFN transgene was induced by application of 10 μM of β-Estradiol (Sigma) on tissue culture media. Hairy root DNA was isolated using the Qiagen DNeasy kit. Transformed roots were screened for the ZFN transgene using the primers (fwd: 5’-TGGATATGTATATGGTGGTAATGC-3’) and (rev: 5’-TTGAGCTTGTGGCGCAGCTCG-3’). Positive roots for the transgene were further screened for mutations by a cleaved amplified polymorphic sequence (CAPS) analysis (fwd: 5’-GTAAAAGATGTTGAAAGAAAGTTGG-3’ and rev: 5’-GCTTTTGACTTGAGCATGATGG-3’) utilizing restriction enzyme MslI, which digests the nucleotide sequence targeted for mutagenesis. A single root was identified as carrying putative mutations in the Dcl4a and Dcl4b genes. The targeted regions of Dcl4a and Dcl4b were amplified by PCR from this root using the CAPS primers. PCR fragments were cloned in pGem T-easy (Promega Corp., Madison, WI) and colony PCR products for 60 clones were subsequently sequenced. Mutations were identified via sequence alignments using MEGA 4.1.20
Identification of potential CoDA ZFN sites in zebrafish and Arabidopsis
Potential ZFN target sites in zebrafish and Arabidopsis were identified from the Ensembl (Zv8.57) and the Arabidopsis information resource (TAIR9) chromosomal assemblies and gene table files. Potential ZFN target sites were defined as those that could be targeted using the CoDA reagents described in this paper and that possess a spacer sequence of 5, 6, or 7 nucleotides that falls entirely within an exon.
Supplementary Material
Supplementary Note Comparisons of CoDA and modular assembly, comparison of mutation frequencies induced by ZFNs made using CoDA and other engineering platforms, predicted targeting range of CoDA ZFNs, modified ZiFiT software.
Acknowledgments
This work was supported by National Institutes of Health grants R01 GM088040 (J.K.J. & R.T.P.), R01 GM069906 (J.K.J.), R24 GM078369 (J.K.J.), R01 CA140188 (J-R.J.Y.), R01 GM081602 (A.J.G.), RC2 MH089956 (A.J.G.), K01 AG031300 (J-R.J.Y), K01 AR055619 (D.M.L.), T32 CA009216 (J.D.S. & J.S.B.); by National Science Foundation grant DBI 0923827 (D.F.V., D.D., & J.K.J); by the Claflin Distinguished Scholar Award (J-R.J.Y.); by the Minnesota Soybean Research and Promotion Council (S.J.C. & R.M.S.), and by Alex’s Lemonade Stand and the Leukemia Research Foundation (D.M.L.).
Footnotes
Author Contributions
J.D.S. and J.K.J. conceived of the CoDA engineering method, J.D.S., S.J.C., D.M.L., R.M.S., A.J.G., D.F.V., R.T.P, J-R.,J.Y., and J.K.J. designed research, J.D.S., E.J.D., M.J.G., L.C., F.Z., D.C., S.J.C., J.B., S.T.-B., Y.Q., C.J.P., E.H., M.L.M., and C.K. performed experiments, J.D.S., D.R., and D.D. identified potential genomic CoDA target sites, and J.D.S., R.M.S., D.F.V., R.T.P, J-R.,J.Y., and J.K.J. wrote the paper.
References
- 1.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 3.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 7:91. doi: 10.1038/nmeth0210-91b. author reply 91–92 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez CL, et al. Unexpected failure rates for modular assembly of engineered zinc–fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson H. The Fate of Fingers. Nature. 2008;455:160–164. doi: 10.1038/455160a. [DOI] [PubMed] [Google Scholar]
- 10.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci U S A. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 12.Foley JE, et al. Rapid Mutation of Endogenous Zebrafish Genes Using Zinc Finger Nucleases Made by Oligomerized Pool ENgineering. PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsend JA, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 18.Foley JE, et al. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govindarajulu M, Elmore JM, Fester T, Taylor CG. Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol Plant Microbe Interact. 2008;21:1027–1035. doi: 10.1094/MPMI-21-8-1027. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note Comparisons of CoDA and modular assembly, comparison of mutation frequencies induced by ZFNs made using CoDA and other engineering platforms, predicted targeting range of CoDA ZFNs, modified ZiFiT software.