Abstract
Background
Pax3 is a key upstream regulator of the onset of myogenesis, controlling progenitor cell survival and behaviour as well as entry into the myogenic programme. It functions in the dermomyotome of the somite from which skeletal muscle derives and in progenitor cell populations that migrate from the somite such as those of the limbs. Few Pax3 target genes have been identified. Identifying genes that lie genetically downstream of Pax3 is therefore an important endeavour in elucidating the myogenic gene regulatory network.
Results
We have undertaken a screen in the mouse embryo which employs a Pax3GFP allele that permits isolation of Pax3 expressing cells by flow cytometry and a Pax3PAX3-FKHR allele that encodes PAX3-FKHR in which the DNA binding domain of Pax3 is fused to the strong transcriptional activation domain of FKHR. This constitutes a gain of function allele that rescues the Pax3 mutant phenotype. Microarray comparisons were carried out between Pax3GFP/+ and Pax3GFP/PAX3-FKHR preparations from the hypaxial dermomyotome of somites at E9.5 and forelimb buds at E10.5. A further transcriptome comparison between Pax3-GFP positive and negative cells identified sequences specific to myogenic progenitors in the forelimb buds. Potential Pax3 targets, based on changes in transcript levels on the gain of function genetic background, were validated by analysis on loss or partial loss of function Pax3 mutant backgrounds. Sequences that are up- or down-regulated in the presence of PAX3-FKHR are classified as somite only, somite and limb or limb only. The latter should not contain sequences from Pax3 positive neural crest cells which do not invade the limbs. Verification by whole mount in situ hybridisation distinguishes myogenic markers. Presentation of potential Pax3 target genes focuses on signalling pathways and on transcriptional regulation.
Conclusions
Pax3 orchestrates many of the signalling pathways implicated in the activation or repression of myogenesis by regulating effectors and also, notably, inhibitors of these pathways. Important transcriptional regulators of myogenesis are candidate Pax3 targets. Myogenic determination genes, such as Myf5 are controlled positively, whereas the effect of Pax3 on genes encoding inhibitors of myogenesis provides a potential brake on differentiation. In the progenitor cell population, Pax7 and also Hdac5 which is a potential repressor of Foxc2, are subject to positive control by Pax3.
Background
During embryonic development, the Pax family of transcription factors play important roles in cell type specification and organogenesis [1]. In vertebrates, Pax3 is a key upstream regulator of skeletal myogenesis. This paired-box homeo-domain transcription factor is present in myogenic progenitor cells of the developing muscle masses and also in the multipotent cells of the somites from which all skeletal muscles in the trunk and limbs derive. Somites form as segments of paraxial mesoderm following a rostral/caudal gradient on either side of the embryonic axis. Initially Pax3 is expressed throughout the epithelial somite and then becomes restricted to the dorsal domain, the dermomyotome, which maintains an epithelial structure. The ventral somite gives rise to bone and cartilage of the vertebral column and ribs, whereas the Pax3 positive cells of the dermomyotome give rise to other mesodermal derivatives, including derm, smooth muscle and endothelial cells, as well as skeletal muscle. Experiments in the chick embryo [2-4] and in the mouse [5] have shown that different cell types derive from a single Pax3 positive cell. Myogenic progenitors delaminate from the edges of the dermomyotome to form the underlying skeletal muscle of the myotome. As development proceeds, the central domain of the dermomyotome where Pax7, the paralogue of Pax3, is also expressed, loses its epithelial structure and these Pax positive cells enter the underlying muscle masses where they constitute a progenitor cell population for all subsequent muscle growth. In the absence of both Pax3 and Pax7, these cells fail to enter the myogenic programme and many of them die [6]. The hypaxial domain of the dermomyotome, where Pax3, but not Pax7, is mainly expressed in the mouse, is an important source of myogenic progenitors. At the level of the limb buds, cells migrate from this domain to form the skeletal muscle masses of the limb. In the absence of Pax3, these cells fail to delaminate and migrate and subsequently undergo cell death [1]. Pax3 therefore controls migration of myogenic progenitor cells from the somite, entry into the myogenic programme and survival.
In order to understand how Pax3 functions in the multipotent cells of the dermomyotome and subsequently in myogenic progenitors, it is necessary to characterize Pax3 targets. During myogenesis in vivo very few targets have been identified. Notably, c-Met has been proposed as a direct Pax3 target [7]. This gene encodes a tyrosine kinase receptor that interacts with HGF, required for the delamination, and probably also the migration, and proliferation of myogenic progenitors [8]. Pax3 activation of the c-Met promoter, although not fully demonstrated in vivo, provides an explanation for the absence of progenitor cell migration and limb myogenesis in Pax3 mutants. This is also consistent with rescue of the ectopic migration seen in Pax3PAX3-FKHR/+ embryos, when c-Met is absent [9]. Entry of Pax3/7 positive progenitor cells into the myogenic programme depends on the myogenic determination factors, Myf5 and MyoD. Analysis of regulatory sequences in the 5' flanking region of Myf5, led to the characterization of an element at -57.5 kb from the gene that is responsible for transcription in the limb buds and older hypaxial somite. Activation of this element depends directly on Pax3 [10]. The MyoD gene is also regulated by a Pax3/7 binding site [11], although this regulation has not been explored in an embryonic context. Pax7 has a more limited expression pattern than Pax3 in the mouse somite, however they probably share many of the same targets, as indicated by the embryonic phenotype of a Pax3Pax7/Pax7 mouse line in which Pax7 replaces Pax3 [12]. Further Pax3/7 targets have been identified using the C2 muscle cell line in which Pax3 or Pax7 was over-expressed [13,14]. In this context the Myf5 regulatory sequence targeted by Pax3 in the embryo was also shown to be a Pax7 target. Id3, which encodes a potential inhibitor of basic-helix-loop-helix transcription factors such as Myf5 or MyoD, was identified as a direct Pax3 target [14]. In the context of human Rhabdomyosarcomas, which result from a chromosomal translocation leading to the expression of a fusion protein, PAX3-FKHR or PAX7-FKHR in which the PAX DNA binding domain is followed by the strong transcriptional activation domain of the FOXO1A (FKHR) factor, a number of microarray screens have been performed on cultured cells (for review see [15]). Examples are provided by cDNA two colour arrays in which the authors identified genes differentially regulated by PAX3 or PAX3-FKHR over-expression in NIH3T3 cells [16], by Affymetrix arrays to find genes induced by PAX3 expression in a human medulloblastoma cell line [17], or by a casting approach of cyclic amplification and selection of cis-regulatory elements bound by human PAX3, PAX3-FKHR or murine Pax3 [18]. Very few target genes were common to these three approaches, probably reflecting the artificial conditions of the screens.
More recently, we have initiated a screen to systematically look for Pax3 targets in the mouse embryo. Since myogenic progenitors tend to die in the absence of Pax3, complicating the interpretation of a screen based on a comparison with material from Pax3 mutants, we used a gain of function approach. This was based on a Pax3PAX3-FKHR-IRESnlacZ/+ (Pax3PAX3-FKHR/+) line that we had made, in which Pax3 targets such as c-Met, are over-activated. Pax3PAX3-FKHR thus constitutes a Pax3 gain of function allele. We had previously shown that in Pax3PAX3-FKHR/Splotch embryos (where Splotch is a spontaneously occurring Pax3 mutant allele) the Pax3 mutant phenotype is not observed, indicating that PAX3-FKHR can replace Pax3, which thus acts as a transcriptional activator in the myogenic context [9]. A Pax3GFP/+ mouse line [6] permitted isolation of Pax3-GFP progenitor cells by flow cytometry, so that the transcriptomes of purified populations of Pax3GFP/+ versus Pax3PAX3-FKHR/GFP cells could be compared by microarray analysis. This screen led to the identification of Sprouty1 and Fgfr4 shown to be a direct Pax3 target, and the demonstration that the self-renewal, versus entry into the myogenic programme, of myogenic progenitors is partly orchestrated by Pax3 modulation of FGF signalling [19]. Dmrt2, was also identified as a direct Pax3 target. This gene encodes a transcription factor, present in the Pax3 positive cells of the dermomytome, which regulates an early epaxial enhancer element of the Myf5 gene, required for the onset of myogenesis in the somite [20]. This screen also revealed that Foxc2 is negatively controlled by Pax3, and that this genetic repression is reciprocal in the epithelial somite and subsequently in the dermomyotome where these genes are co-expressed. Modulation of this equilibrium affects cell fate choices, resulting in Pax3 positive myogenic progenitors or Foxc2 positive vascular progenitors [21].
In this paper, we provide the first documentation of the in vivo gain of function screen for Pax3 targets and present data on other interesting candidates.
Results and Discussion
Experimental strategy and microarray results
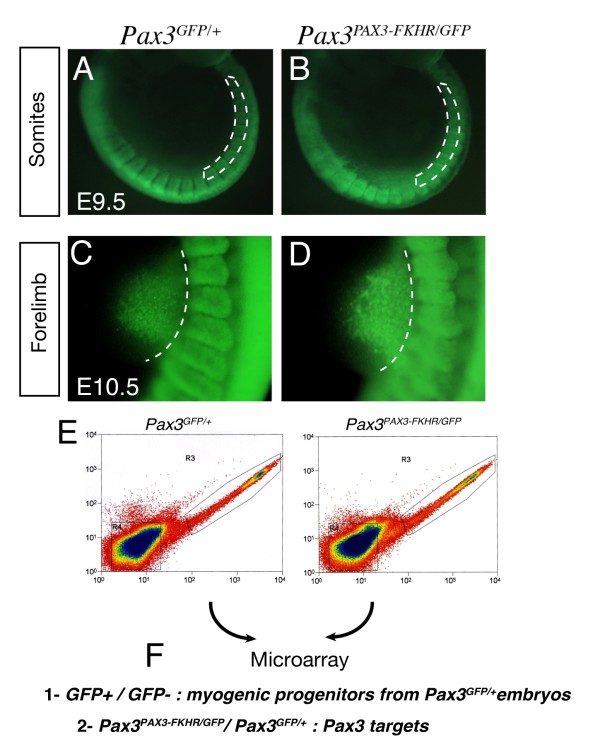
Our strategy depended on the purification of Pax3 positive cells, which was implemented using a mouse line (Pax3GFP/+) in which one allele of Pax3 had been targeted with a GFP coding sequence [6], so that cells expressing this allele could be isolated by flow cytometry. This line was crossed onto the conditional Pax3PAX3-FKHR-IRESnlacZ/+ (Pax3PAX3-FKHR/+) line [9] to produce Pax3PAX3-FKHR/GFP embryos. After a further cross with a PGK-Cre transgenic line [22], this resulted in a Pax3 gain of function genetic background. In the crosses used here, Pax3PAX3-FKHR/GFP embryos had a similar phenotype to Pax3PAX3-FKHR/+ embryos. From our previous analysis of the Pax3PAX3-FKHR/+ line [9], we know that PAX3-FKHR can save the Pax3 mutant phenotype, thus substituting for Pax3. This is accompanied by over-activation of known Pax3 targets such as c-Met, leading to some myogenic abnormalities. We used this allele to generate a gain of function genetic background. Somites in the interlimb region of E9.5 Pax3GFP/+ (Figure 1A) and Pax3PAX3-FKHR/GFP (Figure 1B) embryos were dissected to obtain the dorsal epithelial structure of the dermomyotome, which was cut away from the epaxial domain adjacent to the neural tube. This corresponds to a stage when most myogenic progenitors are still present in the dermomyotome, with cells delaminating from the epaxial dermomyotome to form the early skeletal muscle of the myotome. Care was taken to avoid Pax3 positive cells in the dorsal neural tube, although the presence of migrating neural crest cells from this source, that also express Pax3 [1], could not be excluded. Forelimb buds were dissected from Pax3GFP/+(Figure 1C) and Pax3PAX3-FKHR/GFP (Figure 1D) embryos at E10.5, when Pax3 positive cells had migrated from the hypaxial domain of adjacent somites, but had not yet formed differentiated skeletal muscle in the forelimb buds. In this case, neural crest should be absent, since these cells do not enter the limb buds. After dissection, cells were dissociated from pooled samples of somites or forelimb buds from Pax3GFP/+ and Pax3PAX3-FKHR/GFP embryos and separated by flow cytometry to obtain GFP positive fractions for microarray analysis (Figure 1E). In addition to the comparison of GFP+ cells from Pax3PAX3-FKHR/GFP; Pax3GFP/+embryos, GFP+/GFP- populations were compared from Pax3GFP/+ embryos to identify sequences specific to Pax3 positive myogenic progenitors (Figure 1F). Obtaining enough material is a challenge at these embryonic stages, particularly from the forelimb bud which contains about 1000 Pax3 positive cells at E10.5, so that it was necessary to prepare material from >100 embryos with each genetic background in order to have enough material for cDNA synthesis, sample verification (see Additional file 1 Figure S1) and triplicate Affymetrix chip analyses.
Figure 1.
Strategy of the screen for Pax3 targets. (A-D) Embryos viewed under a fluorescence microscope from Pax3GFP/+ (A, C) and Pax3PAX3-FKHR/GFP (B, D) mouse lines at E9.5, focussing on the interlimb somites (A, C) and at E10.5 focussing on the forelimb bud (B, D). Dotted lines indicate the region dissected. (E) Isolation of GFP positive cells by flow cytometry from the two genotypes indicated, shown for material from E10.5 forelimb buds. The region R3, outlined in E, was used for transcriptome analysis of GFP positive cells. GFP negative cells were obtained from the R4 window shown in (E). (F) Microarrays were performed on RNA isolated from cells of interlimb somites and forelimb buds (A-D) with the genotypes indicated. The comparisons gave information about sequences that mark Pax3 positive myogenic progenitors (1) and that are candidate Pax3 targets (2).
Raw data were pre-processed to obtain expression values using the RMA (Robust Multichip Analysis) algorithm. Unreliable probe-sets called "absent" by Affymetrix Gene Chip Operating Software (GCOS) software (http://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf website) for at least 2 GeneChips out of 3 were discarded. LPE (Local Pooled Error) tests [23] were performed to identify significant differences in gene expression among Pax3PAX3-FKHR/GFP; Pax3GFP/+ and GFP+; GFP- samples. Benjamini-Hochberg (BH) [24] multiple-test correction was applied to control for the number of false positives with an adjusted 5% statistical significance threshold.
These data are available on the http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22041 website.
Comparisons for E9.5 dermomyotome and E10.5 forelimb bud preparations are presented in Additional file 2 Tables S1-S3. Genes that are up-regulated (A) or down-regulated (B) in both somites and limb buds are shown in Additional file 2 Table S1. Additional file 2 Table S2 shows genes up-regulated (A) or down-regulated (B) for forelimb buds only, whereas Additional file 2 Table S3A and B shows such genes in somites only. Transcripts absent from the Pax3-GFP positive population, but observed in the presence of PAX3-FKHR are not included, since they may be due to non-PAX3 dependent FKHR activity. Transcripts that are present in Pax3-GFP positive cells and not detectable in the presence of PAX3-FKHR were retained. Pax3 transcripts are in this category and indeed provide a control, since the mouse gene is not transcribed in Pax3PAX3-FKHR/GFP embryos. The additional microarray screen, in which the transcriptome of the GFP negative cell population was compared to that of the Pax3-GFP positive population, gives an overview of transcripts that characterise myogenic progenitor cells of the forelimb bud, as shown in Additional file 3 Table S4.
Neural crest markers, such as AP2 gamma (Tcfap2c) or Ascl1 (also named Mash1) are present in the E9.5 dermomyotome lists and are also seen to a minor extent in the E10.5 limb samples, probably indicating the presence of some somitic material. This is also suggested by the presence of markers of differentiating muscle, such as skeletal muscle myosin or troponin, expressed at this stage in the myotome of the somite. The presence of markers of the sclerotome, such as Pax1 [25], probably reflects the inclusion of cells from the ventral somite compartment, perhaps due to some perduration of GFP, since the Pax3GFP allele is expressed throughout the epithelial somite [21]. A gene encoding another typical marker of the sclerotome compartment, Uncx4.1 [26], was present in the list of Pax3 targets (Additional file 2 Table S1A), and also, unexpectedly, in the list of GFP+ specific genes for the limb bud (Additional file 3 Table S4). This may suggest that it is also expressed in myogenic progenitors, and indeed, the expression of Uncx4.1 is compromised in the absence of Pax3 (data not shown).
Experimental validation of sequences of potential Pax3 targets modulated by PAX3-FKHR
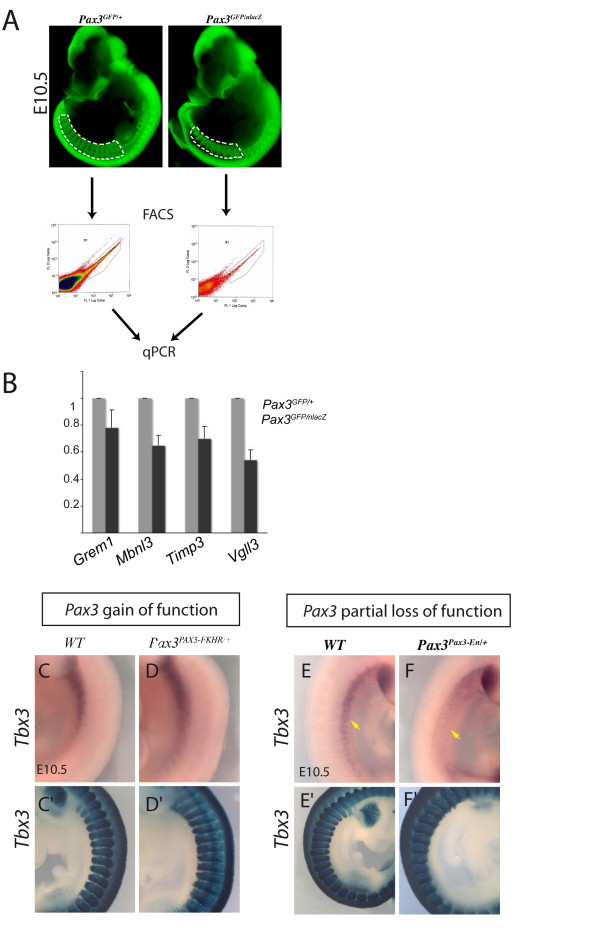
Genes that showed differences in expression on the PAX3-FKHR gain of function genetic background were validated by qRT-PCR on a Pax3 loss of function background. In order to avoid the problem of loss of cells due to cell death in the mutant, the same number of Pax3-GFP positive cells from Pax3GFP/+ and Pax3GFP/nlacZ embryos were analysed, after purification by flow cytometry (Figure 2A). Examples are shown in Figure 2B, for somites at E10.5. This is an important control to confirm that the increase in transcripts seen in the presence of PAX3-FKHR is not due to an effect of the fusion protein, other than that of strong transcriptional activation via Pax3 binding sites. Another control is provided by whole mount in situ hybridisation on different genetic backgrounds, which also demonstrates the localisation of cells that express the gene. In this case, the partial loss of function Pax3Pax3-En-IRESnlacZ/+ (Pax3Pax3-En/+) line [10] was used, which expresses a fusion protein, with the Pax3 DNA binding domains fused to the repression domain of Engrailed. This results in down-regulation of Pax3 targets with the advantage that there is less cell death, as shown by X-gal staining, although migration of progenitor cells to the forelimb bud is compromised. This is illustrated for Tbx3 transcripts on Pax3 gain (Figure 2C, D) and partial loss (Figure 2E, F) of function backgrounds. Tbx3 is also expressed in cardiac neural crest [27], however its expression profile in the somites points to activation in a subdomain of this paraxial mesoderm. Further examples of PCR based analysis of the distribution of Pax3 targets, is shown in Additional file 1 Figure S1. Comparison of Sox2 and Sox10 transcripts in Pax3 positive cells of different somite preparations indicates that these are high in samples that include the neural tube, consistent with expression in neural crest (Additional file 1 Figure S1B). Comparison of expression in the whole somite (Additional file 1 Figure S1A), with the hypaxial domain is also informative, indicating, for example, that Zic1 transcripts are enriched in the whole somite, consistent with an expression mainly in the epaxial domain [28], as seen by immunofluorescence on sections (Additional file 1 Figure S1C), where Zic1 protein is co-expressed with Pax3 in the epaxial dermomyotome as well as in the dorsal neural tube and in Pax3 negative mesenchyme. Pax3 positive neural crest does not appear to express Zic1, in accordance with a recent report on its absence in migratory neural crest cells in the chick embryo [28]. In situ hybridization on sections confirms expression of Zic1 in the epaxial domain of the epithelial dermomyotome (Additional file 1 Figure S1E). Up-regulation of Zic1 transcripts in somites of Pax3GFP/GFP embryos in the epaxial/central domain, which is less affected by cell death, is consistent with negative regulation by Pax3 (Additional file 2 Table S2B).
Figure 2.
Examples of validation on Pax3 loss of function genetic backgrounds. Genes that emerged from the microarray analyses as potential Pax3 targets from the gain of function screen were checked on Pax3 loss of function genetic backgrounds. (A-B) Quantitative PCR analysis of transcripts in Pax3-GFP cells separated by flow cytometry (FACS) from interlimb somites of Pax3GFP/+ and Pax3GFP/nlacZ embryos at E10.5 (A). The same number of cells were analysed for each genotype and the results for Gremlin1, Mbnl3, Timp3 and Vg113 transcripts are presented as histograms relative to the Pax3GFP/+ sample taken as 1 (B). In accordance with the microarray data, these genes are positively regulated by Pax3. (C-F) Whole mount in situ hybridization with a Tbx3 probe on control (C, E), Pax3PAX3-FKHR/+ gain of function (D) and Pax3Pax3-En/+ partial loss of function (F) embryos at E10.5, showing somites in the interlimb region. Tbx3 transcripts are high in the hypaxial somite domain, notably in more anterior somites, and their level depends positively on Pax3, as indicated by the microarray data. In the forelimb buds, there is extensive expression of Tbx3 in posterior mesenchyme which masks transcripts in Pax3 positive myogenic progenitors. In the lower panels, X-gal staining of β-galactosidase from nlacZ (C', E') (Pax3nlacZ/+) or IRES-nlacZ (Pax3PAX3-FKHR-IresnlacZ/+ in D', Pax3Pax3-En-IresnlacZ/+ in F') reporters shows the extent of the somites, notably the hypaxial domain which undergoes cell death in Pax3 mutants (F').
Cell survival and malignancy
In the absence of Pax3, myogenic progenitor cells undergo apoptosis. This phenotype, in addition to data in adult satellite cells, indicates that Pax3/7 are implicated in cell survival [29]. Our list of Pax3 target genes is not obviously enriched in such genes; however secondary modifications of cell survival proteins are not detected in this approach. Very few genes associated with carcinogenesis emerge as PAX3-FKHR targets. This is in contrast to screens performed in Rhabdomyosarcoma cell lines (for review, see [15]). This may be explained by the physiological level of expression of PAX3-FKHR, similar to that of Pax3 in our screen as well as the in vivo context; Pax3PAX3-FKHR/+ mice do not develop tumours unless a second mutation affecting a tumour suppressor occurs [30]. In Rhabdomyosarcoma, a chromosomal translocation has taken place, potentially affecting genome regulation, and the cells examined are derived from an adult tumour, so that the context is different from that of embryonic cells expressing a Pax3PAX3-FKHR allele.
In this report we concentrate on signalling pathways and transcription factors implicated in myogenesis.
Pax3 modulation of signalling pathways
In Table 1 examples of genes encoding components of signalling pathways that are up- (red) or down- (blue) regulated in the presence of PAX3-FKHR in limbs and/or somites are shown. A number of major signalling pathways lie genetically downstream of Pax3. In some cases, such as FGF or Eph pathways, transcripts for both ligands and receptors are present in Pax3-GFP positive cells, indicative of autocrine signalling, which is subject to Pax3 regulation.
Table 1.
Changes in transcripts for genes implicated in major signalling pathways between Pax3-GFP/+ samples from somites and/or limbs of gain of function Pax3PAX3-FKHR/GFP and control Pax3GFP/+ embryos: UP (bold), DOWN (italics)
| Pax3PAX3-FKHR/GFP vs Pax3GFP/+ | |||
|---|---|---|---|
| RTK | FGF | Ligands/Extracellular matrix | FGF5, FGF12, Sulfatase1 |
| Receptors | Fgfr4, Fgfr3 | ||
| Intracellular components | Dusp4, Fap | ||
| Inhibitors | Spry1, Spry4, Ing5, Spred1 | ||
| Transcriptional components | Etv1, Etv2 | ||
| IGF | Ligand | Igf1 | |
| PDGF | Ligand | Pdgfc | |
| Effector (target) | Csrp1 (Axud1) | ||
| Met | Receptor | c-Met | |
| Eph | Ligands | EphrinA5, EphrinB1 | |
| Receptors | EphA7, EphA3 | ||
| Shh | Ligands/Extracellular matrix | Sulfatase1 | |
| Inhibitors | Hhip | ||
| Transcriptional components | Zic1 | ||
| Wnt | Ligands/Extracellular matrix | Wnt16, Sulfatase1 | |
| Intracellular components | Siah2, Diversin (Ankrd6) | ||
| Inhibitors | Sfrp3 (Frzb1), Dkk1, Dkk2, Diversin, Wif1, Csrp1 (Axud1) | ||
| Transcriptional components | TCF15, TCF7/2 , Nkd2 | ||
| Integrins | Ligands | Lama2 | |
| Intracellular components | Itgβ1bp2 | ||
| Receptors | Itgβ6, Itgβ8 | ||
| Cytokines | Ligands | CXC15, CXC12, Cxcl14, Cxcl5 | |
| Intracellular components | CXCR7, Crlf1 | ||
| Inhibitors | Soc3 | ||
| Notch | Ligands | Dll1, Dll3 | |
| Inhibitors | Fhl1 (Kyot2) | ||
| Transcriptional components | Hes1 | ||
| TGFβ/BMP | Ligands | BMP5, TGFβ2, TGFβinduced, Thbs1 | |
| Inhibitors | Chordin-like1, Follistatin, Gremlin | ||
Transcripts of the enzyme, Sulfatase1, that sulfates extracellular matrix Proteoglycans positively affecting the binding/stability and availability of ligands such as FGF, Wnts and Shh, are up-regulated in the presence of PAX3-FKHR. This is a potentially important level at which signalling, and consequent cell behaviour, is modulated. Indeed in a comparison of quiescent versus in vivo activated Pax3 positive muscle satellite cells, expression of genes such as Sulfatase1, affecting extracellular matrix interaction with growth factors, was strikingly modified [31]. In addition to effectors of signalling pathways, inhibitors depend on Pax3 activity. In some cases, due to feedback regulation, activation of a gene for an inhibitor, may reflect activation of a pathway, however it may also demonstrate an important potential for Pax3 modulation of the outcome of signalling, depending on the myogenic context.
Pathways subject to Pax3 regulation include receptor tyrosine kinase (RTK) pathways, such as FGF, as well as Shh and Wnt signalling which promote myogenesis. Most effectors of RTK signalling pathways are positively regulated by Pax3. Unless specified, regulation is not necessarily direct.
FGF signalling is strikingly affected by Pax3 and Fgfr4 has now been shown to be a direct target [19]. Pax3 regulation of Sprouty1, encoding an intracellular inhibitor of RTK signalling, has consequences for myogenic progenitor self-renewal, versus entry into the myogenic programme, promoted by this pathway in the embryo [19]. More recently, Sprouty1 has been implicated in controlling quiescence of adult satellite cells [32], although it is not notably modulated in vivo in comparisons with activated satellite cells [31]. We investigated Sprouty1 mutant embryos [33], in which a LacZ reporter permitted clearer identification of Sprouty1 expressing cells. In the mutants, expression of the myogenic regulatory genes, Myf5 or MyoD, viewed by whole mount in situ hybridization appears normal (Additional file 1 Figure S2 A-D) and Desmin, which marks muscle cells, is expressed in the myotome as expected (data not shown). However Sprouty2 and Sprouty4 are also expressed in somites [34] (Additional file 1 Figure S2E-F) and may therefore compensate for the absence of Sprouty1. Pax3 regulates multiple components of the FGF signalling cascade, from the ligand/receptor to transcriptional effectors, such as the Ets transcription factors (Etv1, Etv2).
Other RTK pathways, such as IGF and PDGF, also promote myogenesis (for ex [35]); Igf1 and Pdgfc are up-regulated in the presence of PAX3-FKHR. The c-Met gene was one of the first Pax3 targets to be proposed in a myogenic context [7]. In our screen, transcripts for this gene are up-regulated in somites (Additional file 2 Table S2, [9]), but not limb buds (Additional file 2 Table S3), suggesting that Pax3 activation of transcription is confined to the somite (see also Additional file 1 Figure S1), where c-Met is required for delamination [36], although the transcripts (Additional file 3 Table S4) continue to be present in cells that migrate to the limb buds.
A number of genes for Ephrin ligands (EphrinA5, EphrinB1) and receptors (EphA7, EphA3) are regulated by Pax3 in both somites and limbs, suggesting expression in myogenic cells, as well as neural crest [37]. Recently, up-regulation of Eph receptors and ligands has been reported in several Rhabdomyosarcoma cell lines (EphB: [38] EphA [39]). In a myogenic context in vivo, Eph receptors have been implicated in muscle patterning and inervation [40,41]. The EphA signalling pathway may also interfere with FGF/MAPK signalling [42,43].
The notochord and ventral neural tube are sources of Shh signalling. In a myogenic context this impacts the adjacent epaxial dermomyotome where Zic1, for example, is highly expressed (Additional file 1 Figure S1), and where Shh is implicated in the activation of Myf5 [44] as well as in promoting cell survival and proliferation in the somite [45]. Canonical Wnt signalling, from the dorsal neural tube, similarly affects the epaxial somite and Myf5 activation [46], with a potential relay through non-canonical Wnts, such as Wnt11 [47]. Components of these pathways are modulated positively or negatively in the presence of PAX3-FKHR suggesting that Pax3 fine-tunes Wnt and Shh signalling, probably also limiting the spatial extent of their action in the somite (see Additional file 1 Figure S1 for Zic1).
Other signalling pathways, such as Notch, that, like FGF, affect self-renewal/differentiation [48,49] show some modulation by Pax3. This is also the case for signalling through Integrins, where the laminin gene encoding the ligand, Lama2, is up-regulated (Table 1), also seen for Lama1 via Dmrt2, which is a Pax3 target [20], whereas transcripts for the Integrin receptors, Itgβ6 and Itgβ8, are down-regulated (Table 1). Integrins, some of which lie genetically downstream of Myf5, are important for the structure and myogenic regulation of the dermomyotome and for the formation of the basal lamina that contains the myotome [50].
Transcripts for a number of cytokines and their receptors are present in Pax3 positive cells. Some show modulation by Pax3, although this did not include CXCR4, regulated by Lbx1, and important for the migration of a subpopulation of myogenic progenitors into the limb bud [51].
BMP/TGFβ signalling, from the dorsal neural tube and lateral mesoderm, has been shown to antagonise the onset of myogenesis [52]. Again, Pax3 affects this signalling pathway, notably by positively regulating genes encoding both ligands and inhibitors such as Chordin-like1, Follistatin and Gremlin1. In the chick embryo, manipulation of Noggin, produced in the somite, had shown the importance of this inhibitor in permitting MyoD activation and the onset of myogenesis [53]. Gremlin1 is expressed at the extremities of the dermomyotome at E9.5 (Additional file 2 Table S3A) and subsequently declines in the epaxial domain of more mature somites (Additional file 1 Figure S1B, C) to be no longer detectable by E11.5. It is up-regulated in the presence of PAX3-FKHR (Additional file 3 Figure S3A) (Additional file 1 Figure S3 D, E). In Gremlin1 mutant embryos [54], Pax3 expression is normal (results not shown). Somitic expression of Myf5 (Additional file 1 Figure S3F, G) and MyoD (Figure S3 H, I) is also similar to wild type or heterozygote embryos. However, there is some delay in the onset of expression in the hypaxial domain. There is also a delay in MyoD expression in the forelimb bud at E11.5 (Additional file 1 Figure S3), probably reflecting perturbations in signalling pathways within the limb [54]. The lack of a more striking phenotype may reflect compensation by other BMP inhibitors, such as Noggin.
A striking finding of this screen is the variety of genes for inhibitors of signalling pathways that are controlled by Pax3. These include Sprouty1, Sfrp3, Gremlin1 and Hhip, which encode inhibitors of FGF, Wnt, BMP and Shh signalling, respectively (Table 1). This indicates that Pax3 negatively modulates the activity of signalling pathways as well as promoting their activation. The role of the FGF inhibitor, Sprouty, in maintaining the myogenic stem cell population in the face of FGF signalling that promotes entry into the myogenic differentiation programme has been demonstrated [19]. In addition, pathways that negatively impact entry into the myogenic programme, such as BMP/TGFβ, are also abrogated by inhibitors, as illustrated for Gremlin, precisely expressed at the extremities of the dermomyotome where activation of myogenic determination factors is first initiated.
Pax3 modulation of genes implicated in transcription
Examples of genes involved in the control of transcription that show up- or down-regulation in Pax3 positive (Pax3-GFP) cells in the presence of PAX3-FKHR, compared to controls, are presented in Table 2, 3, 4. This is divided into three sections for differentially regulated genes in both somites (E9.5) and forelimbs (E10.5) (Table 2) or only in forelimbs (Table 3) or only in somites (Table 4). Transcriptional effectors of signalling pathway (see Table 1) have been removed from Table 2, 3, 4.
Table 2.
Changes in transcripts for transcription factors common to somites and forelimbs between Pax3-GFP/+ samples from gain of function Pax3PAX3-FKHR/GFP and control Pax3GFP/+ embryos: UP (bold), DOWN (italics), FC (fold change)
| Gene Title | Gene Symbol | FC limb | FC somite |
|---|---|---|---|
| vestigial like 3 (Vito2) | Vgll3 | 8.41 | 2.22 |
| developing brain homeobox 1 | Dbx1 | 6.38 | 5.19 |
| transcription factor AP-2, gamma | Tcfap2c | 4.45 | 1.53 |
| transcription elongation regulator 1-like | Tcerg1l | 3.61 | 2.21 |
| Unc4.1 homeobox | Uncx4.1 | 3.36 | 1.66 |
| PR domain containing 8 | Prdm8 | 3.05 | 1.93 |
| T-box 3 | Tbx3 | 2.30 | 2.16 |
| paired box gene 7 | Pax7 | 2.12 | 1.54 |
| histone deacetylase 5 | Hdac5 | 1.97 | 1.58 |
| myogenic factor 5 | Myf5 | 1.90 | 1.85 |
| nescient helix loop helix 2 | Nhlh2 | -2.06 | -0.59 |
| brachyury | T | -2.17 | -0.55 |
| homeo box A4 | Hoxa4 | -2.64 | -1.95 |
| forkhead box C2 | Foxc2 | -3.17 | -1.47 |
| homeo box B1 | Hoxb1 | -4.49 | -2.33 |
| paired box gene 3 | Pax3 | -4.91 | -1.73 |
Table 3.
Changes in transcripts for transcription factors from forelimbs only between Pax3-GFP/+ samples from gain of function Pax3PAX3-FKHR/GFP and control Pax3GFP/+ embryos: UP (bold), DOWN (italics), FC (fold change)
| Gene Title | Gene Symbol | FC limb |
|---|---|---|
| ladybird homeobox 1 homolog corepressor 1 | Lbxcor1 | 3.58 |
| nuclear receptor subfamily 3, group C, member 1 | Nr3c1 | 2.91 |
| distal-less homeobox 5 | Dlx5 | 2.81 |
| myocardin | Myocd | 2.64 |
| jumonji domain containing 1C | Jmjd1c | 2.58 |
| runt related transcription factor 1 | Runx1 | 2.27 |
| tet oncogene 1 | Tet1 | 2.20 |
| H6 homeo box 3 (Nkx5-1) | Hmx3 | 2.00 |
| homeo box A9 | Hoxa9 | -1.82 |
| homeo box A10 | Hoxa10 | -2.01 |
| nuclear receptor subfamily 0, group B, member 1 | Nr0b1 | -2.50 |
| single-minded homolog 2 | Sim2 | -3.16 |
| SRY-box containing gene 2 | Sox2 | -4.04 |
| zinc finger protein of the cerebellum 1 | Zic1 | -4.45 |
| forkhead box G1 | Foxg1 | -11.40 |
Table 4.
Changes in transcripts for transcription factors from somites only between Pax3-GFP/+ samples from gain of function Pax3PAX3-FKHR/GFP and control Pax3GFP/+ embryos: UP (bold), DOWN (italics), FC (fold change)
| Gene Title | Gene Symbol | FC somites |
|---|---|---|
| paired-like homeodomain transcription factor 2 | Pitx2 | 3.00 |
| ladybird homeobox homolog 1 | Lbx1 | 2.67 |
| trans-acting transcription factor 5 | Sp5 | 2.55 |
| zinc finger protein 568 | Zfp568 | 2.46 |
| inhibitor of DNA binding 4 | Id4 | 2.39 |
| fos-like antigen 2 | Fosl2 | 2.34 |
| Kruppel-like factor 4 | Klf4 | 2.28 |
| nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | 2.26 |
| doublesex and mab-3 related transcription factor like family A2 | Dmrta2 | 1.86 |
| inhibitor of DNA binding 2 | Id2 | 1.82 |
| ISL1 transcription factor, LIM/homeodomain | Isl1 | 1.75 |
| Kruppel-like factor 11 | Klf11 | 1.60 |
| inhibitor of DNA binding 1 | Id1 | 1.57 |
| zinc finger, CCHC domain containing 12 | Zcchc12 | 1.55 |
| zinc finger, MYND domain containing 11 | Zmynd11 | 1.54 |
| LIM homeobox protein 2 | Lhx2 | 1.54 |
| transcription factor AP-2 beta | Tcfap2b | 1.50 |
| basonuclin 2 | Bnc2 | 1.45 |
| doublesex and mab-3 related transcription factor 2 | Dmrt2 | 1.45 |
| mohawk homeobox | Mkx | 1.43 |
| histone cluster 2, H3c1 | Hist2h3c1 | -1.42 |
| nuclear receptor co-repressor 2 | Ncor2 | -1.43 |
| forkhead box C1 | Foxc1 | -1.45 |
| runt-related transcription factor 1; translocated to 1 | Runx1t1 | -1.46 |
| homeo box C8 | Hoxc8 | -1.47 |
| zinc finger and BTB domain containing 16 | Zbtb16 | -1.47 |
| similar to COUP-TFI/nuclear receptor subfamily 2, group F, member 1 | Nr2f1 | -1.47 |
| T-box 22 | Tbx22 | -1.47 |
| homeo box C5 | Hoxc5 | -1.51 |
| homeo box C6 | Hoxc6 | -1.51 |
| myocyte enhancer factor 2C | Mef2c | -1.52 |
| basic helix-loop-helix family, member e22 | Bhlhe22 | -1.55 |
| achaete-scute complex homolog 1 (Drosophila) | Ascl1 | -1.56 |
| single-minded homolog 1 (Drosophila) | Sim1 | -1.58 |
| Meis homeobox 1 | Meis1 | -1.58 |
| dachshund 1 (Drosophila) | Dach1 | -1.72 |
| chromodomain helicase DNA binding protein 8 | Chd8 | -2.04 |
| myogenic factor 6 | Myf6 | -2.26 |
| myogenin | Myog | -2.35 |
| paired-like homeobox 2b | Phox2b | -2.39 |
Pitx genes, such as Pitx2, which is positively regulated by Pax3, have been implicated in myogenesis [55]. Very few Pitx target genes have been identified to date. Recently, in zebrafish, a member of the Shroom family, encoding an actin binding protein implicated in epithelial organization [56,57], has been reported to be a direct target of Pitx factors [58]. Interestingly, Shroom2, like Pitx2, is up-regulatd by Pax3 in the somite (Additional file 2 Table S2). The Pax3-Pitx2-Shroom2 cascade may be implicated in the maintenance of the epithelial organization of the hypaxial dermomyotome in the mouse embryo.
Lbx1 is another gene that is implicated in myogenesis (Table 4). Previous observations on decreased Lbx1 expression in Pax3 mutants were difficult to interpret because of cell death in the hypaxial somite, where the gene is expressed at limb levels [1], however the gain of function result shown here indicates that Pax3 lies genetically upstream of Lbx1. This result was confirmed by whole mount in situ hybridization on Pax3Pax3-En/+ partial loss of function embryos at E9.5 (Figure 3B), when apoptosis due to perturbation of Pax3 function is minimal [10] as shown by X-gal staining of Pax3 expressing cells (Figure 3D). Lbx1 is also expressed in cells migrating to the limbs, but was not modified in the limb bud microarrays suggesting that, like c-Met (which appears as a potential Pax3 target in the dermomyotome but not in the limb), this aspect of its expression is not Pax3 dependent. This result suggests that Pax3 regulates the expression of Lbx1 and c-Met to control cell delamination from the hypaxial dermomyotome of the somite, but not later during progenitor cell migration to the limb buds, thus dissociating its function in delamination and migration. In Pax3 positive cells in the forelimb bud, Lbxcor1, which encodes a co-repressor of Lbx [59] is up-regulated (Table 3), suggesting that by this stage repression of Lbx activity is important. In the limbs, the gene for the cytokine receptor, CXCR4, that is required for the development of a subset of myogenic cells, depends on Lbx1 [51]in vivo. Results in vitro show that expression of CXCR4 is regulated by PAX3-FKHR in Rhabdomyosarcoma cell lines [60] and by Pax7 in C2C12 cell lines [13]. However the level of CXCR4 transcripts is not altered in the presence of PAX3-FKHR in our screen in the embryo, although they are present in Pax3-GFP positive cells, as expected (Additional file 3 Table S4).
Figure 3.
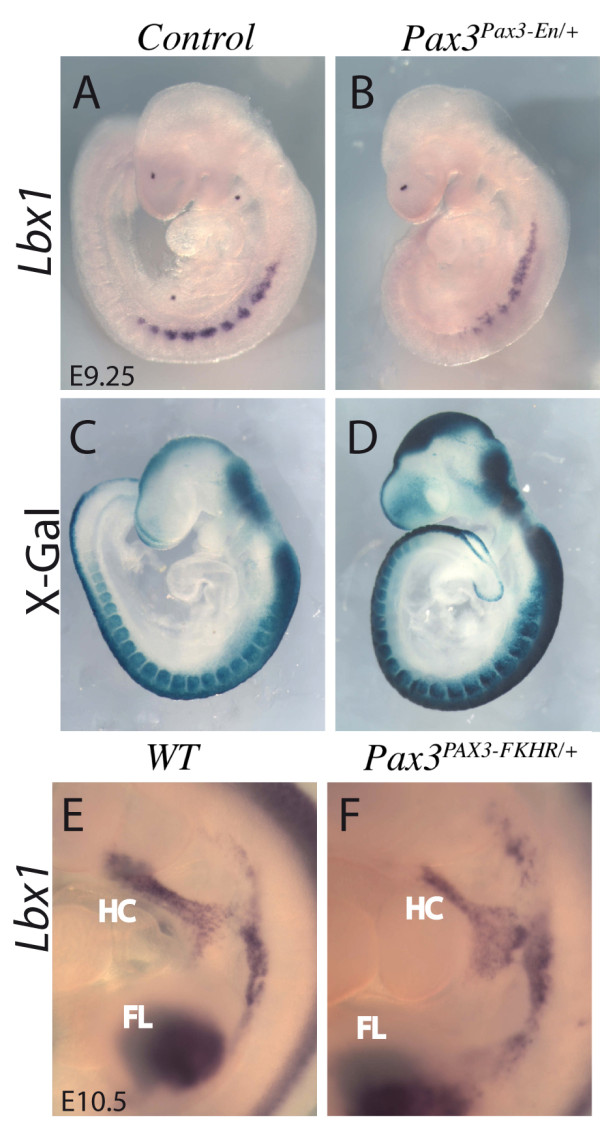
Lbx1 expression depends on Pax3. (A, B, E, F) Whole mount in situ hybridisation with an Lbx1 probe of control (A, E), Pax3Pax3-En/+ partial loss of function (B) and Pax3PAX3-FKHR/+ gain of function embryos (F) at E9.25 (A, B) and E10.5 (E, F), showing down-regulation of Lbx1 transcripts when Pax3 activity is reduced, and some up-regulation of its expression in the presence of PAX3-FKHR in somites. In the lower panels (C, D) X-gal staining of β-galactosidase from the IresnlacZ reporter in Pax3IresnlacZ/+ (C) or Pax3Pax3-En-IresnlacZ/+ (D) shows the extent of the somites, notably the hypaxial domain.
Six homeo-domain transcription factors, with their Eya co-activators and Dach co-repressors, are also important upstream regulators of myogenesis [1]. Transcripts for these factors are present in the Pax3-GFP positive cells (Additional file 3 Table S4), but only Dach1 expression is affected by PAX3-FKHR (Table 4); it is down-regulated, in keeping with Pax3 promotion of Six myogenic activity. Manipulation of Dach, which is high in quiescent satellite cells, demonstrates its negative role in activated Pax3-GFP positive cells, retarding their entry into myogenesis [31].
Sim1 and Sim2 transcripts, that mark hypaxial somite domains and migrating myogenic progenitors [61,62], are both negatively regulated by PAX3-FKHR. Sim2 has been shown to prevent epithelial/mesenchymal transitions (EMT) through repression of Slug [63]. When PAX3-FKHR is transfected into NIH3T3 cells, Slug transcripts are up-regulated [16]. Pax3 repression of Sim2 may be necessary to promote delamination of migratory myogenic cells and indeed in Pax3PAX3-FKHR/+ embryos there is premature EMT, accompanied by up-regulation of c-Met [9]. Experiments in the chick embryo have shown that FGF signalling from the myotome triggers the expression of Snail, a known regulator of EMT [64]. In our transcriptome data, neither Snail nor Slug expression was affected and therefore EMT in this context may involve other transcriptional regulators.
Many Hox genes (Hoxa4, a9, a10, Hoxb1 and Hoxc5, c6, c8), present in somites and/or limbs, are down-regulated in Pax3-GFP positive cells in the presence of PAX3-FKHR. This is an intriguing finding. Hox gene regulation at the level of the somites, with consequences for myogenesis, has already been documented [65,66]. Our findings now suggest a reciprocal relationship.
Foxc2, is negatively regulated by PAX3-FKHR [21] (Table 2) and this is also the case to a lesser extent for Foxc1 in somites (Table 4). Reciprocal negative regulation between Pax3 and Foxc2 has been implicated in cell fate choices of multipotent cells in the dermomyotome, such that high Pax3 promotes myogenesis at the expense of vasculogenesis and vice versa [21]. Runx1, and the related gene Runx1t1, are up-regulated in the presence of PAX3-FKHR in forelimb buds and somites respectively (Table 2, 3, 4). Runx1 is a factor that marks endothelial cells, some of which, in the limb, derive from the dermomyotome [2]. This would suggest that Pax3 may contribute to the priming of cells to become endothelial, although it is Foxc2 that promotes the vascular fate. Myocardin is also up-regulated, indicating positive control by Pax3 (directly or indirectly). Myocardin controls smooth muscle differentiation [67] and this may also be indicative of such "priming". However some smooth muscle markers are also expressed in differentiating skeletal muscle cells in the embryo. Unexpected expression of Myocardin in the dorsal somite had already been reported [67].
The gene that encodes the myogenic determination factor, Myf5, is up-regulated by PAX3-FKHR, both in the somite at E9.5 and in E10.5 forelimb buds (Table 2). This is consistent with direct activation by Pax3 of the Myf5 limb regulatory element [10] and of regulation of early Myf5 expression through Dmrt2, which is itself a Pax3 target in the dermomyotome in the epaxial and potentially also the hypaxial domain [20] (Table 4). Interestingly the gene for the transcriptional co-factor Vgll3 (also called Vito-2) is up-regulated (Table 2 see also Figure 2B). Vgll3, expressed at the onset of myogenesis [68], enhances the transcriptional activity of TEF transcription factors that bind to MCAT motifs, present in many skeletal muscle specific genes [69]. In this context, Six homeodomain proteins, in addition to their upstream role in concertation with Pax3 at the onset of myogenesis, also, unlike Pax3, directly activate differentiation genes. Down-regulation of the gene for the Six co-repressor, Dach1 will also promote differentiation. However genes encoding transcription factors associated with myogenic differentiation, such as Myogenin, Mrf4, Mef2c and downstream muscle genes, such as Myosins or Troponins are down-regulated in the presence of PAX3-FKHR (Additional file 2 Table S3B). Activation of myogenic differentiation may be prevented by negative regulation in Pax3 expressing cells of Meis1 (Table 4), which encodes a protein, required for chromatin accessibility in a myogenic context, as shown for MyoD [70]. Mbnl3 (Muscleblind-like 3), up-regulated by PAX3-FKHR (Additional file 2 Table S3A, Figure 2B), encodes a protein that inhibits MyoD dependent gene expression, thus antagonising differentiation [71]. In this context, Myocardin in concertation with Hdac5, also modulated by Pax3 (see below), represses MyoD/Myf5 activation of the Myogenin promoter [72], thus preventing skeletal muscle differentiation. Myocardin expression, also detected in the dermomyotome [72], may prevent premature differentiation of Myf5 expressing cells in the hypaxial domain. Furthermore differentiation will be reduced by the positive effect of Pax3 on the expression of Id1, Id2 and Id4 (Table 4), encoding helix-loop-helix proteins which complex with basic helix-loop-helix factors such as MyoD, interfering with DNA binding [73]. Id3 did not emerge from our screen, but this gene had been identified as a Pax3 target in cultured muscle cells [14]. Overexpression of Pax7 in cultured muscle cells identified Id2, as well as Id3, as a target [13]. In the embryo, targeting of Pax3 alleles with a Pax7 coding sequence [12] showed that Pax7 can replace Pax3 and therefore that these genes share common targets. These observations indicate that while Pax3 is required for entry into the myogenic programme, it also acts as a brake on muscle differentiation and indeed continued high level of expression of Pax3 retards the onset of differentiation in muscle satellite cells [74].
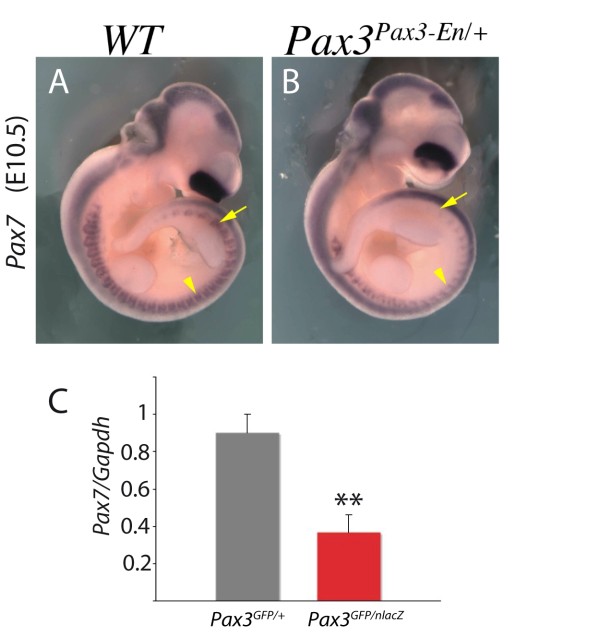
Transcripts of Pax7, the paralogue of Pax3, are higher in the presence of PAX3-FKHR in both limbs and somites (Table 2). Down-regulation of Pax7 in somites, when Pax3 activity is reduced, is seen by whole mount in situ hybridization with a Pax7 probe on Pax3Pax3-En/+ embryos (Figure 4B). When quantitative PCR is performed on RNA isolated from the same numbers of Pax3-GFP cells purified by flow cytometry from Pax3GFP/+ and Pax3GFP/nlacZ embryos, the reduction of Pax7 transcripts in the absence of Pax3 is striking (Figure 4C, D). This observation on Pax3 dependence of Pax7 expression is important in understanding Pax3/7 regulation of muscle stem cell fate. This in vivo result is in contrast to in vitro observations where PAX3-FKHR overexpression in human Rhabdomyosarcoma cell lines leads to a down-regulation of PAX7 expression [60]. It has also been reported that Pax7 is up-regulated in the somites of Pax3 mutant (Splotch) embryos [75], however this was not a quantitative analysis, and somite disorganisation in the absence of Pax3 complicates the interpretation of in situ hybridisation.
Figure 4.
Pax7 expression depends on Pax3. (A, B) Whole mount in situ hybridisation with a Pax7 probe of wild-type (WT) (A) and Pax3Pax3-En/+ partial loss of function (B) embryos at E10.5, showing down-regulation of Pax7 transcripts when Pax3 activity is reduced. (C) GFP positive cells were isolated by flow cytometry (see Figure 2) from the interlimb somites of Pax3GFP/+ and Pax3GFP/nlacZ embryos at E10.5. The same number of cells in each case was used for RNA preparation and quantitative PCR analysis, with Pax7 and Gapdh primers; mRNA levels are shown relative to Gapdh transcripts with the heterozygote control level taken as 1. Histograms represent mean fold change for four different biological samples analyzed in duplicate at each stage; error bars, standard error to the mean (SEM); **p < 0.005.
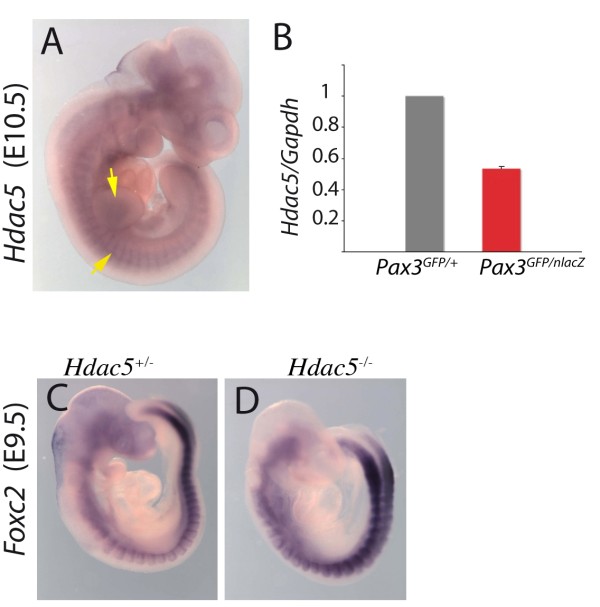
Finally, the gene encoding the histone deacetylase, Hdac5, a class II histone deacetylase, that acts as a negative regulator of transcription, is up-regulated in Pax3-GFP positive cells in the presence of PAX3-FKHR in somites and limb buds (Table 2). Expression of Hdac5 at these sites of myogenesis is shown by whole mount in situ hybridization in Figure 5A and confirmation that it is positively regulated by Pax3 is shown by qRT-PCR on RNA from the same number of Pax3-GFP positive cells (Figure 5B). Pax3 acts mainly as a transcriptional activator in the myogenic context [9] and therefore genes that are down-regulated in a Pax3 gain of function context are probably indirectly regulated by Pax3. Foxc2 is an example of such a gene [21]. Hdac5 is a candidate negative intermediary. In Hdac5 mutants, Foxc2 is up-regulated (Figure 5D), indicating that Pax3 repression of Foxc2 may be mediated by Hdac5, which is itself positively regulated by Pax3.
Figure 5.
The Hdac5 gene is positively regulated by Pax3 and negatively affects Foxc2. (A) Whole mount in situ hybridisation at E10.5 with an Hdac5 probe. Arrows point to expression at sites of myogenesis in the somites and forelimb bud. (B) GFP positive cells were isolated by flow cytometry (see Figure 2) from interlimb somites of Pax3GFP/+ and Pax3GFP/nlacZ embryos and the same numbers of cells were used for RNA preparation and quantitative PCR analysis of Hdac5 and Gapdh transcripts; mRNA levels are shown relative to Gapdh transcripts with the heterozygote control (Pax3GFP/+) level taken as 1. Histograms represent mean fold change for two different biological samples analyzed in duplicate at each stage; error bars, standard error to the mean (SEM). (C-D) Whole mount in situ hybridisation with a Foxc2 probe on Hdac5+/- (C) and Hdac5-/- (D) embryos at E9.5, showing up-regulation of Foxc2 in somites in the absence of Hdac5.
Conclusions
We have identified sequences that are potential Pax3 targets, thus giving insight into Pax3 orchestration of progenitor cell behaviour prior to, and at the onset of, myogenesis. Many components of signalling pathways, including inhibitors as well as activators, emerge from the screen, demonstrating how Pax3 modulates their impact on progenitor cell behaviour and progression towards muscle. This is also evident from transcriptome analysis of chromatin remodelling and transcription factors/co-factors. Pax3 regulated sequences modulate initial cell fate decisions in the multipotent Pax3 positive stem cells of the dermomyotome. In this case Hdac5, positively regulated by Pax3, negatively impacts Foxc2 expression. Foxc1 is also down-regulated indirectly by Pax3. In this stem cell context, Pax3 positively regulates Pax7 also implicated in reciprocal repression with Foxc1/c2 [21]. Entry into the myogenic programme is promoted by down-regulation by Pax3 of the gene for the Six1/4 co-repressor Dach2 and also by the previously demonstrated activation of the myogenic determination gene Myf5, which in this case has been shown to be direct [10]. Genes for myogenic differentiation factors and downstream muscle proteins are mainly down-regulated by Pax3, acting negatively on the gene for the chromatin remodelling factor Meis and positively on the Id gene family of myogenic inhibitors as well as on Myocardin. Pax3, either directly or indirectly, is thus acting as a brake on muscle differentiation, while priming entry into the myogenic programme. Regulation of myogenic progenitor cell behaviour, both at the level of signalling pathways and of transcriptional control, is modulated by balanced up- and down-regulation of genes that lie genetically downstream of Pax3.
Methods
Mouse lines
The following mouse lines were used: Pax3GFP/+, Pax3PAX3-FKHR-IRESnlaZ/+ (referred to as Pax3PAX3-FKHR/+), Pax3Pax3-En-IRESnlacZ/+ (referred to as Pax3Pax3-En/+), Pax3nLacZ/+, Hdac5nlacZ/+, Sprouty1lacZ/+, Gremlin1+/- and the PGK-Cre transgenic line. Embryos were genotyped as described previously: Pax3GFP/+ [6,10], Pax3PAX3-FKHR and Pax3nLacZ [9], Pax3Pax3-En [10], PGK-Cre [22], Hdac5nlacZ/+ [76], Sprouty1lacZ/+ [33], Gremlin1+/- [54]. The targeted Pax3 lines used in this analysis have been bred for many generations on a C57 BL6/DBA2 genetic background.
For the screen, Pax3GFP/+ mice were crossed with PGK-Cre transgenic mice to obtain Pax3GFP/+; PGK-Cre females. These females were crossed with Pax3PAX3-FKHR-IRESnlacZ/+ males to obtain embryos with one Pax3GFP allele and one floxed Pax3PAX3-FKHR-IRESnlacZ allele [9].
Preparation of embryonic material for in situ gene expression and micro-array analysis
Embryos were collected after natural overnight mating and dated, taking Embryonic day (E) 0.5 as the day after the appearance of the vaginal plug. Briefly, embryos were fixed in 4% para-formaldehyde at 4°C, overnight for in situ hybridization, 2 hours for immuno-detection and 15 minutes for X-Gal staining.
Embryos were dissected in DMEM medium. For tissue preparation from E9.5 embryos, somites were dissected from the interlimb region and the more hypaxial domain separated from the neural tube and epaxial extremity of the somites. An effort was made to take the epithelial dermomyotome, viewed by Pax3-GFP fluorescence under the microscope. The forelimb buds at E10.5 were separated from the adjacent somites under a fluorescence microscope. Only 1000 cells were collected per limb bud from a Pax3GFP/+ embryo, so that a total of 490 embryos were dissected, 125 of which were Pax3PAX3-FKHR/GFP and 107 were Pax3GFP/+. The genotype was revealed by GFP fluorescence and characteristic head and neural tube abnormalities in Pax3PAX3-FKHR/GFP embryos [9], as well as by β-galactosidase activity shown by X-gal staining of the rest of the embryo (from the Pax3PAX3-FKHR allele). The limb buds and hypaxial somites were pooled according to their genotype and then dissociated by passage through a 2 ml syringe and filtered before the flow cytometry sorting.
Triplicate samples of each population were prepared, representing a starting material of a minimum of 100,000 cells per sample. GFP+ cells were separated by flow cytometry using a MoFlo cell sorter (Beckman-Coulter USA). The gates for positive and negative GFP cells were determined using an equivalent sample isolated from wild type embryos. Analysis was done with the Summit software version 3.4.
RNA isolation and microarray analysis
Total RNA was extracted and purified after DNase 1 (Amersham) treatment using the RNeasy Mikro kit (Qiagen). RNA and cRNA quality was monitored on Agilent RNA Pico LabChips (Agilent). cRNA obtained from 100 ng of RNA was amplified by using the GeneChip Expression Two-Cycle 3'amplification system (Affymetrix). Fragmented biotin-labeled cRNA samples were hybridized on GeneChip Mouse Genome 430_2 arrays, according to the manufacturer's protocol (http://www.affymetrix.com/support/downloads/manuals/expression analysis technical manual.pdf). The Affymetrix 430.2.0 mouse array that contains 45,000 probe sets was used. Each probe set consists of 22 probes of 25 bp, with 11 perfect matches and 11 mismatches. For each experimental group (Pax3+/+, Pax3GFP/+ and Pax3PAX3-FKHR/GFP), three biological replicates were hybridized. The generation of cell intensity files and the quality control of hybridizations were performed with GeneChip Operating Software (Affymetrix).
Statistical analysis of microarray data
Statistical analyses of data were performed as described previously [31].
Raw data were pre-processed using the Robust Multichip Analysis (RMA) algorithm in order to correct the background, to adjust the intensity distribution over the arrays and to convert probe intensity summarisation into a unique probe set signal. Unreliable probe-sets called "absent" by Affymetrix GCOS software for at least 2 GeneChip out of 3 were discarded. Local Pooled Error (LPE) tests [23] were performed in order to identify significant differences in gene expression between Pax3-GFP positive cells from Pax3PAX3-FKHR/GFP and Pax3GFP/+ embryos on the one hand and between GFP+ and GFP- samples from Pax3GFP/+ embryos on the other hand. The Benjamini-Hochberg (BH) multiple correction test [24] was applied to control for the number of false positive with an adjusted 5% statistical significance threshold. The fold changes of the differentially expressed genes after p-value adjustement were analyzed by filtering the data set with a threshold of 1.5 (Log2 ratio = 0.5). Significantly regulated genes in Pax3PAX3-FKHR/GFP samples, that are common to both limb and somite extracts, are represented in Additional file 2 Table S1. Transcripts that are specifically up- or down-regulated in the limb or in the trunk, respectively, are referred to as "somite only" in Additional file 2 Table S2 and "limb only" in Additional file 2 Table S3.
Genes that are specifically transcribed in the GFP positive fraction (absent in GFP negative fraction) are represented in Additional file 3 Table S4, again as common to somites and limbs (A), somite specific sequences (B) and limb specific sequences (C).
Accession Numbers
The complete microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE22041 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22041).
In situ hybridization
Whole-mount in situ hybridizations with digoxigenin-labeled probes were performed as described in [77]. In situ hybridization for Pax7 transcripts was carried out as described in [12] and for transcripts of Foxc2 as described in [21]. The Tbx3 probe was as described in [27]. The Zic1 probe was synthesized using the image clone Image 4314316 (Open Biosystems) and linearised by EcoR1 and transcribed using T3 polymerase. The mouse Grem1 cDNA (containing the complete coding region and 3-UTR) was isolated by RT-PCR from cDNA of RNA prepared from C57BL/6 mouse embryos at E9.5. The Grem1 cDNA was subcloned into pBS digested with EcoRl and BamHl and transcribed using T3 polymerase for in situ hybridization.
When needed, the whole-mount stained embryos were embedded into gelatin-sucrose, frozen and sectioned, as described in [19].
Quantitative and semi-quantitative real-time PCR
RNA was extracted from embryonic material (interlimb somites) and reverse transcribed using SuperScript II kit (Invitrogen) for qRT-PCR and SuperscriptIII kit for semi-quantitative RT-PCR. All PCR reactions were carried out in duplicate (triplicate for the standard curves) using the Power Sybergreen Mix (Applied Biosystems) and a 7500 thermal cycler (Applied Biosystems). All qPCR results are expressed as relative ratios of the target cDNA to Gapdh transcripts normalized to that ratio in the reference condition, which always corresponds to heterozygote Pax3GFP/+ embryos. Primers used for detecting specific transcripts were designed with Primer3 (see Additional file 4 Table S5).
Immunofluorescence
Fluorescent co-immunohistochemistry on sections was carried out as described previously [19]. The following antibodies were used: anti-Zic1 (Abcam, ab7524-25), 1/500; anti-Pax3 (DSHB), 1/250. Images were acquired with Apotome Zeiss and Axiovision 4.6 software at the Pasteur imaging center (Imagopole, Institut Pasteur).
Mouse work was carried out in accordance with the regulations of the French Ministry of Agriculture, as practised by the Ministry accredited mouse animal house of the Pasteur Institute under the supervision of scientists and technicians with the official authorisation to experiment on mice. The authors have paid attention to the ARRIVE and MIQE guidelines, in reporting their work.
Authors' contributions
ML conducted experiments, discussed results and wrote a first draft. TS conducted and discussed experiments. BR performed the statistical analyses of the data. AC helped with the flow cytometry sorting. JL and AZ provided transgenic embryos. FR made the Pax3 alleles and initiated the study when in MB's laboratory and continued to supervise the work of ML thereafter. MB directed the work, discussed results and wrote the manuscript.
The manuscript has been approved by all the authors.
Supplementary Material
Supplementary Figures S1-S3. Figure S1: Examples of transcript validation and analysis of the expression of Zic1, negatively regulated by Pax3. (A, B) Examples of the analysis of transcripts in different Pax3-GFP cell populations isolated from Pax3GFP/+ embryos at E10.5. (A) RT-PCR analysis of RNA extracted from FACs sorted GFP positive and GFP negative cells from Pax3GFP/+ embryos, as indicated in Figure 1 showing that transcripts for the neural tube markers, Sox2 and Sox10, which are potential Pax3 targets in neural crest, since the transcripts are enriched in whole somite preparations which include neural tissue. (B) RT-PCR analysis of transcripts in GFP positive cells from the hypaxial versus whole somites of Pax3GFP/+ embryos. c-Met, Six1, Sim1 and Zic1 transcripts are shown, with HPRT transcripts as a control. Zic1 expression is higher in the whole somite preparation, consistent with a more epaxial location. (C) Immunohistochemistry with Zic1 and Pax3 antibodies on a transverse section of an interlimb somite of an E10.5 embryo, showing Zic1 protein mainly detected in Pax3 positive epithelial cells of the epaxial dermomyotome (Ep), as well as in the dorsal neural tube (NT) and in mesenchymal cells between the neural tube and somites. Zic1 is mainly absent from Pax3 positive migratory neural crest in this region. (D, E) Whole mount in situ hybridization with a Zic1 probe on posterior somites of control Pax3GFP/+ (D) and mutant Pax3GFP/GFP (E) embryos at E11, showing up-regulation in the absence of Pax3. Cell death is extensive in the hypaxial domain of more anterior somites by this stage, but despite some loss of cells in immature posterior somites, Zic1 hybridisation is still higher in the Pax3 mutant. (F, G) Transverse sections of immature posterior somites from Pax3GFP/+ (F) and Pax3GFP/GFP (G) embryos at E11. In this tail region, immature somites have not yet undergone cell death. In the control (F), transcripts are concentrated in the more central and epaxial domain whereas in the Pax3 mutant (G) they are detected throughout the somite. NT, neural tube. Figure S2: The onset of myogenesis in Sprouty1 mutants and reduction of Sprouty2 expression in the absence of Pax3. (A-D) Whole mount in situ hybridisation on Sprouty1lacZ/+ (A, C) and Sprouty1lacZ/- (B, D) embryos at E10 (somite stages (ss) are indicated), using Myf5 (A, B) and MyoD (C, D) probes. FL, forelimb bud. (E, F) In situ hybridisation with a Sprouty2 probe on E9.5 (E), and with a Sprouty4 probe on E10.5 (F), embryos showing interlimb somites and limb buds. There is extensive Sprouty expression in the distal limb buds, but positive Sprouty4 positive myogenic progenitors in the proximal forelimb bud are detectable at E10.5 (arrow). Figure S3: Expression of Gremlin1 and perturbation of MyoD activation in Gremlin1 mutant embryos. (A, B, D-I) Whole mount in situ hybridisation on wild type (A, B), control (PGK-Cre) (D, D'), Pax3PAX3-FKHR/+ (E, E'), Gremlin1+/- (Grem1+/-) (F, H) and Gremlin1-/- (G, I) embryos at the stages indicated (ss, somite stage), using Gremlin1 (Grem1) (A-E'), Myf5 (F, G) and MyoD (H, I) probes. (C) Transverse section of the embryo shown in (B) at the level indicated by the white bar in (B). Note the expression of Grem1 in the hypaxial domain of the dermomyotome (Hyp). NT, neural tube; DA, dorsal aorta; FL, forelimb bud; HL, hindlimb bud. Arrows and arrowheads point to differences in expression in forelimb buds and more posterior somites, respectively.
Tables S1-S3. In all tables, the probe name corresponds to the probe set designed by Affymetrix. In some cases, multiple probe sets correspond to the same transcript, the name of which is indicated in the gene title and gene symbol columns. When a probe set has not yet been annotated, a - sign has been assigned or the corresponding Riken number. The Fold change (FC) corresponds to the difference in signal intensities between Pax3PAX3-FKHR/GFP and Pax3GFP/+ embryos. This number is not on a logarithmic scale. The Adjusted (Adj) p-Value is also indicated. Table S1: Transcripts up-regulated (Table 1S-A) or down-regulated (Table 1S-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ embryos) in both forelimb and somite extracts. Table S2: Transcripts up-regulated (Table S2-A) or down-regulated (Table S2-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ samples) in forelimb buds. Table S3: Transcripts up-regulated (Table S3-A) or down-regulated (Table S3-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ samples) in somites.
Table S4. Transcripts that are specifically expressed in GFP positive muscle progenitor cells and that are not detected in the GFP negative population of the forelimb bud. This list of transcripts corresponds to myogenic progenitor cell markers. In this table, mean intensity, not fold change ((shown in Tables S1-3), is represented.
Table S5. Sequence of reverse (Rev) and forward (Fwd) primers used for semi-quantitative and quantitative PCR.
Contributor Information
Mounia Lagha, Email: mounia.lagha@gmail.com.
Takahiko Sato, Email: satotakahiko@gmail.com.
Béatrice Regnault, Email: beatrice.regnault@pasteur.fr.
Ana Cumano, Email: cumano@pasteur.fr.
Aimée Zuniga, Email: aimee.zuniga@unibas.ch.
Jonathan Licht, Email: j-licht@northwestern.edu.
Frédéric Relaix, Email: frelaix@gmail.com.
Margaret Buckingham, Email: margaret.buckingham@pasteur.fr.
Acknowledgements
Margaret Buckingham's laboratory is supported by the Institut Pasteur and the CNRS (URA 2578), with grants for work on myogenic stem cells from the AFM and the European Union 7th Framework Programme through EuroSyStem and Optistem. ML was supported by fellowships from the Ministère de l'Education et la Recherche, the AFM and EuroSyStem. TS was an Optistem postdoctoral fellow. FR's laboratory is supported by the INSERM Avenir programme and a project grant from the AFM.
References
- Buckingham M, Relaix F. The Role of Pax Genes in the Development of Tissues and Organs: Pax3 and Pax7 Regulate Muscle Progenitor Cell Functions. Annu Rev Cell Dev Biol. 2007;23:645–73. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Kardon G, Campbell JK, Tabin CJ. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev Cell. 2002;3(4):533–545. doi: 10.1016/S1534-5807(02)00291-5. [DOI] [PubMed] [Google Scholar]
- Ben-Yair R, Kalcheim C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 2005;132(4):689–701. doi: 10.1242/dev.01617. [DOI] [PubMed] [Google Scholar]
- Ben-Yair R, Kalcheim C. Notch and bone morphogenetic protein differentially act on dermomyotome cells to generate endothelium, smooth, and striated muscle. J Cell Biol. 2008;180(3):607–618. doi: 10.1083/jcb.200707206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esner M, Meilhac SM, Relaix F, Nicolas JF, Cossu G, Buckingham ME. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133(4):737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci USA. 1996;93(9):4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Brohmann H. Genes that control the development of migrating muscle precursor cells. Curr Opin Cell Biol. 2000;12(6):725–730. doi: 10.1016/S0955-0674(00)00159-9. [DOI] [PubMed] [Google Scholar]
- Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17(23):2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20(17):2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev Cell. 2008;15(4):534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18(9):1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10(1):77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20(14):3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado GE, Barr FG. Fusions involving PAX and FOX genes in the molecular pathogenesis of alveolar rhabdomyosarcoma: recent advances. Curr Mol Med. 2007;7(1):47–61. doi: 10.2174/156652407779940440. [DOI] [PubMed] [Google Scholar]
- Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA. 1999;96(23):13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanil CS, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem. 2001;276(52):49299–49309. doi: 10.1074/jbc.M107933200. [DOI] [PubMed] [Google Scholar]
- Barber TD, Barber MC, Tomescu O, Barr FG, Ruben S, Friedman TB. Identification of target genes regulated by PAX3 and PAX3-FKHR in embryogenesis and alveolar rhabdomyosarcoma. Genomics. 2002;79(3):278–284. doi: 10.1006/geno.2002.6703. [DOI] [PubMed] [Google Scholar]
- Lagha M, Kormish JD, Rocancourt D, Manceau M, Epstein JA, Zaret KS, Relaix F, Buckingham ME. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 2008;22(13):1828–1837. doi: 10.1101/gad.477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Rocancourt D, Marques L, Thorsteinsdottir S, Buckingham M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010;6(4):e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham ME. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell. 2009;17(6):892–899. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7(2):105–112. doi: 10.1023/A:1008868325009. [DOI] [PubMed] [Google Scholar]
- Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19(15):1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Stat Methodol. 1995;27:289–300. [Google Scholar]
- Peters H, Wilm B, Sakai N, Imai K, Maas R, Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126(23):5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Voss AK, Thomas T, Yokota Y, Gruss P. Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development. 2000;127(11):2251–2258. doi: 10.1242/dev.127.11.2251. [DOI] [PubMed] [Google Scholar]
- Mesbah K, Harrelson Z, Theveniau-Ruissy M, Papaioannou VE, Kelly RG. Tbx3 is required for outflow tract development. Circ Res. 2008;103(7):743–750. doi: 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Rhodes LS, Merzdorf CS. The zic1 gene is expressed in chick somites but not in migratory neural crest. Gene Expr Patterns. 2006;6(5):539–545. doi: 10.1016/j.modgep.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Montarras D. Skeletal muscle stem cells. Curr Opin Genet Dev. 2008;18(4):330–6. doi: 10.1016/j.gde.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, Depinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18(21):2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Francois S, Regnault B, Czarny B, Dive V, Cumano A, Montarras D, Buckingham M. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4(2):77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6(2):117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP. et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8(2):229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126(20):4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Montarras D, Aurade F, Johnson T, J II, Gros F, Pinset C. Autonomous differentiation in the mouse myogenic cell line, C2, involves a mutual positive control between insulin-like growth factor II and MyoD, operating as early as at the myoblast stage. J Cell Sci. 1996;109(Pt 3):551–560. doi: 10.1242/jcs.109.3.551. [DOI] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376(6543):768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Davy A, Soriano P. Ephrin signaling in vivo: look both ways. Dev Dyn. 2005;232(1):1–10. doi: 10.1002/dvdy.20200. [DOI] [PubMed] [Google Scholar]
- Berardi AC, Marsilio S, Rofani C, Salvucci O, Altavista P, Perla FM, Diomedi-Camassei F, Uccini S, Kokai G, Landuzzi L. et al. Up-regulation of EphB and ephrin-B expression in rhabdomyosarcoma. Anticancer Res. 2008;28(2A):763–769. [PubMed] [Google Scholar]
- Clifford N, Smith LM, Powell J, Gattenlohner S, Marx A, O'Connor R. The EphA3 receptor is expressed in a subset of rhabdomyosarcoma cell lines and suppresses cell adhesion and migration. J Cell Biochem. 2008;105(5):1250–1259. doi: 10.1002/jcb.21926. [DOI] [PubMed] [Google Scholar]
- Araujo M, Piedra ME, Herrera MT, Ros MA, Nieto MA. The expression and regulation of chick EphA7 suggests roles in limb patterning and innervation. Development. 1998;125(21):4195–4204. doi: 10.1242/dev.125.21.4195. [DOI] [PubMed] [Google Scholar]
- Iwamasa H, Ohta K, Yamada T, Ushijima K, Terasaki H, Tanaka H. Expression of Eph receptor tyrosine kinases and their ligands in chick embryonic motor neurons and hindlimb muscles. Dev Growth Differ. 1999;41(6):685–698. doi: 10.1046/j.1440-169x.1999.00468.x. [DOI] [PubMed] [Google Scholar]
- Picco V, Hudson C, Yasuo H. Ephrin-Eph signalling drives the asymmetric division of notochord/neural precursors in Ciona embryos. Development. 2007;134(8):1491–1497. doi: 10.1242/dev.003939. [DOI] [PubMed] [Google Scholar]
- Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135(5):931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP Jr. Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev. 2002;16(1):114–126. doi: 10.1101/gad.940702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP Jr. Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126(18):4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- Borello U, Berarducci B, Murphy P, Bajard L, Buffa V, Piccolo S, Buckingham M, Cossu G. The Wnt/beta-catenin pathway regulates Gli-mediated Myf5 expression during somitogenesis. Development. 2006;133(18):3723–3732. doi: 10.1242/dev.02517. [DOI] [PubMed] [Google Scholar]
- Gros J, Serralbo O, Marcelle C. WNT11 acts as a directional cue to organize the elongation of early muscle fibres. Nature. 2009;457(7229):589–593. doi: 10.1038/nature07564. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc Natl Acad Sci USA. 2007;104(2):537–542. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E, Lenhard DC, Wende H, Erdmann B, Epstein JA, Birchmeier C. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci USA. 2007;104(11):4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajanca F, Luz M, Raymond K, Martins GG, Sonnenberg A, Tajbakhsh S, Buckingham M, Thorsteinsdottir S. Integrin alpha6beta1-laminin interactions regulate early myotome formation in the mouse embryo. Development. 2006;133(9):1635–1644. doi: 10.1242/dev.02336. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 2005;19(18):2187–2198. doi: 10.1101/gad.346205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O, Fan CM, Coltey M, Hirsinger E, Watanabe Y, Breant C, Francis-West P, Brickell P, Tessier-Lavigne M, Le Douarin NM. Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell. 1996;84(3):461–471. doi: 10.1016/S0092-8674(00)81291-X. [DOI] [PubMed] [Google Scholar]
- Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, Pourquie O. Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development. 1997;124(22):4605–4614. doi: 10.1242/dev.124.22.4605. [DOI] [PubMed] [Google Scholar]
- Michos O, Panman L, Vintersten K, Beier K, Zeller R, Zuniga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131(14):3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- L'Honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, Camper S, Drouin J. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307(2):421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99(5):485–497. doi: 10.1016/S0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci. 2005;118(Pt 22):5191–5203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- Chung MI, Nascone-Yoder NM, Grover SA, Drysdale TA, Wallingford JB. Direct activation of Shroom3 transcription by Pitx proteins drives epithelial morphogenesis in the developing gut. Development. pp. 1339–1349. [DOI] [PMC free article] [PubMed]
- Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. J Biol Chem. 2005;280(5):3645–3655. doi: 10.1074/jbc.M411652200. [DOI] [PubMed] [Google Scholar]
- Tomescu O, Xia SJ, Strezlecki D, Bennicelli JL, Ginsberg J, Pawel B, Barr FG. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest. 2004;84(8):1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- Fan CM, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JL. et al. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol Cell Neurosci. 1996;7(1):1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- Coumailleau P, Duprez D. Sim1 and Sim2 expression during chick and mouse limb development. Int J Dev Biol. 2009;53(1):149–157. doi: 10.1387/ijdb.082659pc. [DOI] [PubMed] [Google Scholar]
- Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, Schedin P, Porter WW. Loss of singleminded-2 s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28(6):1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfini MC, De La Celle M, Gros J, Serralbo O, Marics I, Seux M, Scaal M, Marcelle C. The timing of emergence of muscle progenitors is controlled by an FGF/ERK/SNAIL1 pathway. Dev Biol. 2009;333(2):229–237. doi: 10.1016/j.ydbio.2009.05.544. [DOI] [PubMed] [Google Scholar]
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev Cell. 2003;5(3):379–390. doi: 10.1016/S1534-5807(03)00263-6. [DOI] [PubMed] [Google Scholar]
- Vinagre T, Moncaut N, Carapuco M, Novoa A, Bom J, Mallo M. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev Cell. 2010;18(4):655–661. doi: 10.1016/j.devcel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23(7):2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M, Piotrowska I, Schneider A, Gunther S, Braun T. VITO-2, a new SID domain protein, is expressed in the myogenic lineage during early mouse embryonic development. Gene Expr Patterns. 2009;9(3):129–137. doi: 10.1016/j.gep.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Gunther S, Mielcarek M, Kruger M, Braun T. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 2004;32(2):791–802. doi: 10.1093/nar/gkh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132(12):2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Lee KS, Smith K, Amieux PS, Wang EH. MBNL3/CHCR prevents myogenic differentiation by inhibiting MyoD-dependent gene transcription. Differentiation. 2008;76(3):299–309. doi: 10.1111/j.1432-0436.2007.00209.x. [DOI] [PubMed] [Google Scholar]
- Long X, Creemers E, Wang D, Olson E, Miano J. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proc Natl Acad Sci USA. 2007;104(42):16570–16575. doi: 10.1073/pnas.0708253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;106(32):13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Li J, Jin F, Emerson CP, Epstein JA. Pax3 functions in cell survival and in pax7 regulation. Development. 1999;126(8):1665–1674. doi: 10.1242/dev.126.8.1665. [DOI] [PubMed] [Google Scholar]
- Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89(1):127–138. doi: 10.1016/S0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S3. Figure S1: Examples of transcript validation and analysis of the expression of Zic1, negatively regulated by Pax3. (A, B) Examples of the analysis of transcripts in different Pax3-GFP cell populations isolated from Pax3GFP/+ embryos at E10.5. (A) RT-PCR analysis of RNA extracted from FACs sorted GFP positive and GFP negative cells from Pax3GFP/+ embryos, as indicated in Figure 1 showing that transcripts for the neural tube markers, Sox2 and Sox10, which are potential Pax3 targets in neural crest, since the transcripts are enriched in whole somite preparations which include neural tissue. (B) RT-PCR analysis of transcripts in GFP positive cells from the hypaxial versus whole somites of Pax3GFP/+ embryos. c-Met, Six1, Sim1 and Zic1 transcripts are shown, with HPRT transcripts as a control. Zic1 expression is higher in the whole somite preparation, consistent with a more epaxial location. (C) Immunohistochemistry with Zic1 and Pax3 antibodies on a transverse section of an interlimb somite of an E10.5 embryo, showing Zic1 protein mainly detected in Pax3 positive epithelial cells of the epaxial dermomyotome (Ep), as well as in the dorsal neural tube (NT) and in mesenchymal cells between the neural tube and somites. Zic1 is mainly absent from Pax3 positive migratory neural crest in this region. (D, E) Whole mount in situ hybridization with a Zic1 probe on posterior somites of control Pax3GFP/+ (D) and mutant Pax3GFP/GFP (E) embryos at E11, showing up-regulation in the absence of Pax3. Cell death is extensive in the hypaxial domain of more anterior somites by this stage, but despite some loss of cells in immature posterior somites, Zic1 hybridisation is still higher in the Pax3 mutant. (F, G) Transverse sections of immature posterior somites from Pax3GFP/+ (F) and Pax3GFP/GFP (G) embryos at E11. In this tail region, immature somites have not yet undergone cell death. In the control (F), transcripts are concentrated in the more central and epaxial domain whereas in the Pax3 mutant (G) they are detected throughout the somite. NT, neural tube. Figure S2: The onset of myogenesis in Sprouty1 mutants and reduction of Sprouty2 expression in the absence of Pax3. (A-D) Whole mount in situ hybridisation on Sprouty1lacZ/+ (A, C) and Sprouty1lacZ/- (B, D) embryos at E10 (somite stages (ss) are indicated), using Myf5 (A, B) and MyoD (C, D) probes. FL, forelimb bud. (E, F) In situ hybridisation with a Sprouty2 probe on E9.5 (E), and with a Sprouty4 probe on E10.5 (F), embryos showing interlimb somites and limb buds. There is extensive Sprouty expression in the distal limb buds, but positive Sprouty4 positive myogenic progenitors in the proximal forelimb bud are detectable at E10.5 (arrow). Figure S3: Expression of Gremlin1 and perturbation of MyoD activation in Gremlin1 mutant embryos. (A, B, D-I) Whole mount in situ hybridisation on wild type (A, B), control (PGK-Cre) (D, D'), Pax3PAX3-FKHR/+ (E, E'), Gremlin1+/- (Grem1+/-) (F, H) and Gremlin1-/- (G, I) embryos at the stages indicated (ss, somite stage), using Gremlin1 (Grem1) (A-E'), Myf5 (F, G) and MyoD (H, I) probes. (C) Transverse section of the embryo shown in (B) at the level indicated by the white bar in (B). Note the expression of Grem1 in the hypaxial domain of the dermomyotome (Hyp). NT, neural tube; DA, dorsal aorta; FL, forelimb bud; HL, hindlimb bud. Arrows and arrowheads point to differences in expression in forelimb buds and more posterior somites, respectively.
Tables S1-S3. In all tables, the probe name corresponds to the probe set designed by Affymetrix. In some cases, multiple probe sets correspond to the same transcript, the name of which is indicated in the gene title and gene symbol columns. When a probe set has not yet been annotated, a - sign has been assigned or the corresponding Riken number. The Fold change (FC) corresponds to the difference in signal intensities between Pax3PAX3-FKHR/GFP and Pax3GFP/+ embryos. This number is not on a logarithmic scale. The Adjusted (Adj) p-Value is also indicated. Table S1: Transcripts up-regulated (Table 1S-A) or down-regulated (Table 1S-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ embryos) in both forelimb and somite extracts. Table S2: Transcripts up-regulated (Table S2-A) or down-regulated (Table S2-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ samples) in forelimb buds. Table S3: Transcripts up-regulated (Table S3-A) or down-regulated (Table S3-B) in Pax3PAX3-FKHR/GFP (in comparison with Pax3GFP/+ samples) in somites.
Table S4. Transcripts that are specifically expressed in GFP positive muscle progenitor cells and that are not detected in the GFP negative population of the forelimb bud. This list of transcripts corresponds to myogenic progenitor cell markers. In this table, mean intensity, not fold change ((shown in Tables S1-3), is represented.
Table S5. Sequence of reverse (Rev) and forward (Fwd) primers used for semi-quantitative and quantitative PCR.