Summary
Human mitochondrial DNA polymerase (Pol γ) is the sole replicase in mitochondria. Pol γ is vulnerable to non-selective anti-retroviral drugs and is increasingly associated with mutations from mitochondriopathic patients. We determined a crystal structure of human Pol γ holoenzyme and, separately, a variant of its processivity factor Pol γB. This is the first atomic structure of any human DNA replicase. The structures reveal that the catalytic subunit Pol γA interacts asymmetrically with the homodimeric Pol γB. A spacer domain of Pol γA, absent in other DNA Pol I family members, is critical for processive DNA synthesis. Pol γ represents a new class of DNA replicase that is structurally and mechanistically different from its eukaryotic and prokaryotic counterparts. The Pol γ structure rationalizes the phenotypes of certain disease-related mutant enzymes that hitherto were inexplicable, and provides a foundation for understanding the molecular basis of toxicity of anti-retroviral drugs targeting HIV reverse transcriptase.
INTRODUCTION
DNA Pol γ, in contrast to the many nuclear DNA polymerases (DNAP) that have specialized functions, is solely responsible for DNA replication and repair in mitochondria. Human mitochondrial DNA (mtDNA) codes for a subset of proteins involving the oxidative phosphorylation electron transfer chain, plus 2 ribosomal rRNAs and 22 tRNAs (Anderson et al., 1981). Accordingly, Pol γ is critically important for mtDNA maintenance, cellular energy supply and viability. Reduced activities of Pol γ lead to mtDNA depletion and impairment of cellular metabolism. Mutations affecting the catalytic subunit Pol γA cause a wide range of genetic syndromes with disease manifestations such as progressive external ophthalmoplegia (PEO, a disorder characterized by slow paralysis of external eye muscle and exercise intolerance), myopathy, epilepsy, neonatal hypotonia, encephalopathy and Alpers’ syndrome (a fatal childhood disease leading to brain and liver failure). In animal studies, homozygous mice with a proofreading defective mutant Pol γA exhibit increased accumulation of point and deletion mutations in mtDNA, as well as premature ageing and a reduced lifespan (Trifunovic et al., 2004). To date, Pol γ mutants have been implicated in more than 30 human diseases (Zeviani and Di Donato, 2004; Chinnery and Zeviani, 2008; Wallace, 2005). The clinical manifestations of many mutations are perplexing as they can be both autosomal dominant and recessive. Understanding the pathology of mitochondrial disorders can thus be a challenge.
Human Pol γ is known to be more susceptible than nuclear DNA polymerases to inhibition by certain Nucleoside Reverse Transcriptase Inhibitors (NRTIs) that target HIV; Pol γ is therefore probably responsible for most cellular toxicity of this class of antiviral drugs. The basis for the high susceptibility of Pol γ to inhibition by NRTIs has thus far been limited to modeling Pol γ with bacteriophage T7 DNAP. The active sites of Pol γ and HIV reverse transcriptase (RT) may exhibit features not found in nuclear DNAPs. However, drug efficacy against HIV is not well correlated with cellular toxicity: Some NRTIs (e.g., zalcitabine (ddC), didanosine (ddI) and stavudine (d4T)) are potent inhibitors of both HIV RT and Pol γ, causing both time-dependent and dose-dependent decreases in mtDNA content and secondary cellular toxicity; whereas others (e.g., tenofovir (PMPA) and abacavir (CBV)) are more selective for HIV RT (see review by Kohler and Lewis, 2007). These observations suggest that significant differences exist in the kinetics of NRTI incorporation into DNA by Pol γ and HIV RT and/or their active site architectures. Such differences can be exploited in the rational design of more selective antiviral agents.
Pol γ, like other DNA replicases, has a catalytic subunit, Pol γA, which possesses both polymerase and proofreading exonuclease activities, and an accessory subunit, Pol γB, which increases enzyme processivity. Pol γB has a novel mode of action. Unlike other processivity factors that enhance processivity by increasing enzyme affinity for template DNA, Pol γB enhances processivity by simultaneously accelerating polymerization rate and suppressing exonuclease activity, in addition to increasing affinity for DNA (Johnson and Johnson, 2001). Reduced exonuclease activity was suggested to help maintain the integrity of the replisome at mitochondrial replication forks (Farge et al., 2007). Structurally, Pol γB resembles class II aminoacyl tRNA synthetases and differs significantly from other processivity factors, including sliding clamps and thioredoxin. The structural basis for the enhanced DNA synthesis processivity by Pol γB is unknown.
Two mechanisms of mtDNA synthesis have been proposed. A conventional synchronous mode where leading and lagging strand synthesis occur simultaneously (Yang et al., 2002), and a displacement mode where synthesis initiating from the OH origin displaces the parental H strand to form a D-loop. Only when the newly synthesized H strand DNA crosses a second origin (OL) does initiation of L strand synthesis occur. The nascent H and L strands are therefore extended asymmetrically (Clayton, 1982). This model was recently modified to allow initiation of L strand synthesis from a number of origins in addition to OL (Brown et al., 2005).
Here we report a crystal structure of human DNA Pol γ holoenzyme. The holoenzyme is a heterotrimer containing one Pol γA subunit and a dimeric Pol γB subunit. The Pol γA active site domain adopts a canonical polymerase configuration. Between the pol and exo domains lies a spacer domain containing a unique fold that interacts primarily with only one Pol γB monomer. This study provides a structural basis for the processivity enhancement in the holoenzyme by the accessory subunit Pol γB, and lays a foundation for understanding the mechanisms of antiviral drug toxicity and mutant Pol γ-related human diseases.
RESULTS AND DISCUSSION
Structure of the catalytic subunit
Successful crystallization of the Pol γ holoenzyme required altered forms of both subunits. An exonuclease-deficient mutant of Pol γA was crystallized with the deletion mutant Pol γB—ΔI4, which lacks a four-helix bundle at the dimer interface (Carrodeguas et al., 2001; Yakubovskaya et al., 2006). DNA polymerase activity (Supplementary material and Fig. S1) of the enzymes used for crystallization is comparable to the activity of the exo- holoenzyme containing wild-type Pol γB (Johnson et al., 2000; Yakubovskaya et al., 2006). The structure of Pol γ was determined to 3.2 Å resolution. Phases were calculated by combining those from single wavelength anomalous diffraction using selenomethionine-substituted Pol γA and osmium-derivatives of holoenzyme, and molecular replacement using human Pol γB (Fan et al., 2006) as a search model. Density modification applied to the initial combined phases significantly improved the quality of electron density maps; the structure was refined to an Rfactor of 28.4% and Rfree 30.3%. Statistics for data collection and refinement are shown in Table 1.
Table 1.
Statistics of data analysis and structural refinement
| Pol γ holoenzyme | Pol γB-ΔI4 | |||
|---|---|---|---|---|
| Native | Se-Met derivative | K2OsO4 soaked | Native | |
| Data collection | ||||
| Resolution (Å) | 50.0 - 3.2 | 50.0 - 4.0 | 50.0 - 4.0 | 50.0 - 3.3 |
| Wavelength (Å) | 0.979 | 0.979 | 1.140 | 1.140 |
| Space group cell dimensions |
P32 | P32 | P32 | P4122 |
| a, b, c (Å) | 138.4, 138.4, 226.4 | 138.94, 138.94, 227.35 | 139.25, 139.25, 227.70 | 64.42,64.42, 260.64 |
| α, β, γ (°C) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Unique reflections |
76667 | 41728 | 41530 | 11917 |
| Completeness b (%) |
100 (100) | 96.3 (82.4) | 94.8 (87.9) | 99.2 (94.7) |
| Redundancy | 5.7 (5.7) | 3.8 (3.7) | 3.9 (3.7) | 8.2 (3.8) |
| RLineara | 0.09 (1.00) | 0.072 (0.658) | 0.070 (0.622) | 0.167 (0.511) |
| I/σI | 21.5 (2.3) | 19.5 (3.0) | 23.3 (1.8) | 5.8 (2.0) |
| SAD FOM | ||||
| Density modification on Combined phases |
0.72 | 0.35 | 0.40 | |
|
| ||||
| Refinement | ||||
| Rworkc (%) | 28.4 | 25.7 | ||
| Rfreed (%) | 30.3 | 29.4 | ||
| No. amino acids | ||||
|
RMS deviations from ideal values |
1850 | 358 | ||
| Bond (Å) Angle (°) |
0.0108 1.57 |
0.0090 1.71 |
||
Rlinear=∑|Ii−<I>|/∑Ii where Ii is the ith measurement and <I> is the weighted mean of all measurements of I.
Values in parentheses are for the highest resolution shell.
Rwork=∑hkl|Fobs(hkl)−Fcalc(hkl)|∑hkl|Fobs(hkl)| for reflections in the working data set.
Rfree is the same as Rwork for 5% of data randomly omitted from refinement.
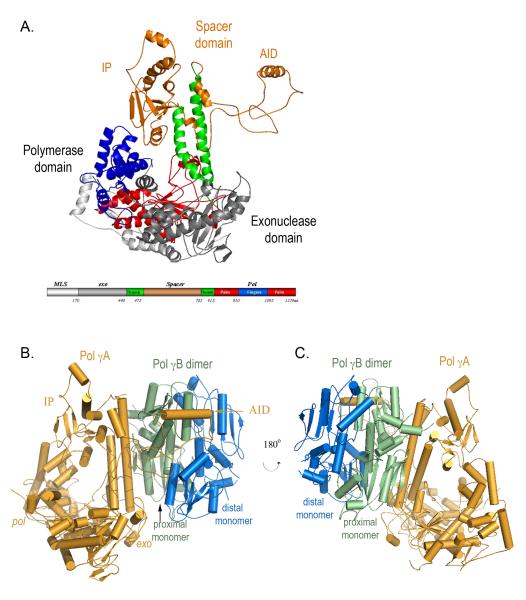
The catalytic subunit Pol γA contains domains for exonuclease (exo) and polymerase (pol) activities separated by a linker or spacer. Pol γA adopts the canonical polymerase ‘right-hand’ configuration with subdomains of ‘fingers’, ‘palm’ and ‘thumb’ that bind template DNA and substrate nucleotide triphosphate, as well as catalyze phosphodiester bond formation (Fig. 1A). The conserved aspartic acids, D890 and D1135 are located in the palm at positions consistent with their known roles in catalysis.
Figure 1.
A) Structure of Pol γA The pol domain shows a canonical ‘right-hand’ configuration with thumb (green), palm (red) and fingers (blue) subdomains, and the exo domain (grey). The spacer domain (orange) presents a unique structure and is divided into two subdomains. Domains are shown in a linear form where the N-terminal domain contains residues 1-170; exo: 171-440; spacer: 476-785; pol: 441-475 and 786-1239. All figures are made with Pymol (DeLano, 2002). B) Structure of the heterotrimeric Pol γ holoenzyme containing one catalytic subunit Pol γA (orange) and the proximal (green) and distal (blue) monomers of Pol γB.
Although the overall fold of Pol γA confirms its classification as a member of the Pol I family, many features of Pol γA are clearly absent in the other enzymes. Most obviously, Pol γA possesses a large spacer domain (~ 400 residues) between the exo and pol domains. In light of the atomic structure, it was necessary to modify the earlier sequence homology-based domain definition of Pol γA because a portion of the originally assigned spacer is actually the thumb subdomain (Fig. 1).
The spacer domain is spatially far from the exo and pol domains and connects to them only through the long helices of the thumb subdomain. The spacer has two obvious subdomains, a globular IP subdomain (Intrinsic Processivity, residues 475-510 and 571-785) and an extended AID subdomain (Accessory Interacting Determinant, residues 511-570) that reaches more than 50 Å away from the main body of Pol γA (Fig. 1). We will show that the IP subdomain explains the intrinsic processivity of Pol γA, and the AID subdomain forms an important interface with Pol γB that is essential for increased processivity of the holoenzyme. A homology search against structures in the protein data bank yields a Z-score of ~0.2, suggesting that the spacer domain has a novel fold (Holm and Sander, 1996).
Holoenzyme formation and subunit interface
In agreement with solution studies (Fig. S2), the crystal form of Pol γ is a heterotrimer containing one catalytic Pol γA subunit (135 kDa) and a dimeric Pol γB—ΔI4 (2 × 50 kDa) with a subunit contact area ~3500 Å2. The deleted helical bundle in Pol γB is distance from the subunit interface and not involved in the subunit interaction with Pol γA (Fig. S3).
The trimeric holoenzyme shows unequal subunit interactions: Pol γA primarily interacts with only one monomer of the Pol γB dimer (Fig. 1B, C). An electron cryo-microscopic analysis of Pol γ at ~17Å resolution came to a similar conclusion (Yakubovskaya et al., 2007), although the subunit interface, modeled onto the crystal structure of Pol γB, appears different from that found in the crystal structure of the holoenzyme. The asymmetrical interactions of Pol γA with the proximal monomer of the Pol γB dimer suggests that a monomeric accessory subunit could be fully functional, which is indeed the case for Drosophila Pol γ (Wernette and Kaguni, 1986).
An asymmetric heterotrimer provokes the question whether the human holoenzyme could also be an A2B2 tetramer, which could position two polymerases at a replication fork - a necessary requirement for coupling leading and lagging strand DNA synthesis. We thus modeled a tetrameric enzyme, adding a second Pol γA to the heterotrimer by following the symmetry operator constraining the Pol γB dimer. The modeled tetramer reveals steric clashes between the AID subdomains of the two Pol γAs that preclude formation of an actual A2B2 tetramer. Although this result could be used as support for the displacement model of mtDNA replication (Clayton, 1982), it should be noted that a trimeric holoenzyme does not rule out other mechanisms for positioning two polymerases at a replication fork (Yang et al., 2002),
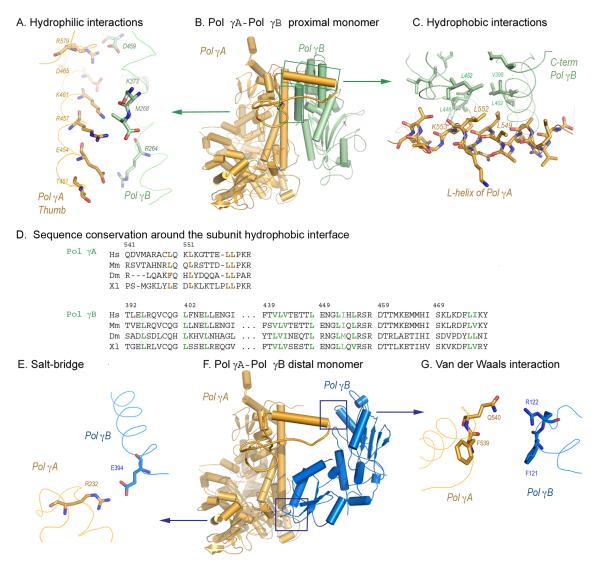
The only contact regions between Pol γA and the distal Pol γB monomer are a salt bridge (2.8 Å) between R232 of Pol γA and E394 of Pol γB (Fig. 2E), and a weak van der Waals contact (5.3 Å) between Pol γA Q540 and Pol γB R122 (Fig. 2G). A R232G substitution, together with T251I and P587L, has been reported in a child with neonatal generalized hypotonia (Ferrari et al., 2005). Healthy siblings of this patient carried T251I and P587L, suggesting that the R232G substitution is associated with disease. This clinical case suggests that either R232 is critical for Pol γA activity or that the contact between Pol γA and the distal Pol γB monomer is important for human holoenzyme function.
Figure 2.
The major Pol γ subunit interfaces. Panels A-C: Pol γA- Pol γB proximal monomer interactions, the distal monomer is omitted for clarity. A) Charge-charge interactions between the thumb domain of Pol γA and the C-terminal domain of Pol γB. B) L-shaped support between Pol γA and the proximal monomer of Pol γB. C) Hydrophobic interactions between the L-helix of Pol γA and a hydrophobic core of Pol γB. Mutated residues L549, L552 and K553 are shown. D) Sequence alignments of residues involved in hydrophobic interactions between Pol γA and Pol γB. Panels E-G: Pol γA- Pol γB distal monomer interactions, the proximal monomer is omitted for clarity. E) The salt-bridge (2.8 Å) between Pol γA R232 and the distal Pol γB E394. F) Pol γA-Pol γB distal monomer. G) The weak van der Waals interaction (5.3 Å) between Pol γA and the distal Pol γB monomer.
In contrast to its limited interaction with the distal monomer, Pol γA makes extensive interactions with the proximal Pol γB monomer (Fig. 2B). Examination of the subunit interface shows two major areas of hydrophilic interactions: between Pol γB R264, K373 and D459 and the Pol γA thumb domain area (E454-D469, and R579) (Fig. 2A), and between Pol γB (D253 and D277) and Pol γA (K1198, R1208 and R1209). In addition, hydrophobic interactions occur between a Pol γB hydrophobic core (V398-L406, V441-L455) in the C-terminal region and Pol γA AID subdomain L-helix (V543-L558) (Fig. 2C). Pol γA AID causes the steric clash in the modeled A2B2 tetramer; in the absence of stabilizing hydrophobic forces for a second Pol γA monomer the holoenzyme is therefore heterotrimeric. In turn, the modeling suggests that the hydrophobic interface is dominant in subunit interaction.
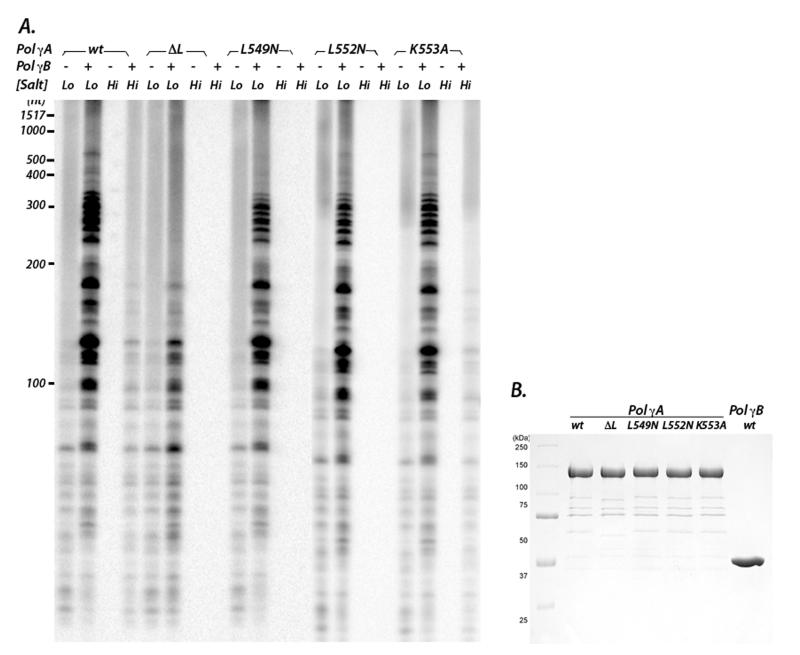
To test this idea, we made four L-helix mutants: L549N and L552N, which reduce hydrophobicity with only minimal structural alteration, a complete deletion (ΔL), and K553A (Fig. 2C-D). The latter change nullifies the electrostatic interaction between Pol γA K553 and E404 of Pol γB. In the absence of Pol γB, all mutants exhibited activities comparable to wild-type Pol γA, demonstrating that the alterations do not disrupt the active site (Fig. 3 and supplementary material). The L-helix is therefore not directly involved in DNA-binding, in agreement with our structural observation that the entire AID subdomain, which includes the L-helix, is connected to the body of Pol γA by flexible linkers and would likely be disordered in the absence of Pol γB.
Figure 3.
A) DNA synthesis activities of Pol γA mutants ΔL, L549N, L552N, K553A were assayed without or with Pol γB at different ionic strengths. Lo and Hi denote 90 mM and 190 mM NaCl, respectively. Denatured products were separated by electrophoresis on an acrylamide gel. B) The purity of Pol γA wt and mutants proteins are shown after SDS-PAGE.
However, the addition of Pol γB reveals a very different outcome. In 90 mM salt, the Pol γA ΔL deletion not only severely lowers the stimulation by Pol γB, but it also reduces the length of product DNA (Fig. 3). In contrast, the L-helix missense mutants had little effect. At higher ionic strength (190 mM salt), wild-type Pol γA is inactive but retains considerable activity when complexed to Pol γB. However, holoenzymes containing Pol γA L549N, L552N, or the ΔL mutant are completely inactive in high salt. This salt-dependent reduction of activity strongly suggests that the mutations disrupt the hydrophobic interactions between Pol γA and Pol γB. Interestingly, holoenzyme containing K553A appeared equally active as wild-type; simply disrupting one electrostatic interaction between Pol γA and Pol γB therefore has only a minor effect when the hydrophobic interactions in this region are preserved. These data provide strong support to our conclusion that hydrophobic interactions between the Pol γA L-helix and the C-terminal domain of Pol γB are the dominant attractive forces that stabilize the AID subdomain so that it can support processive DNA synthesis by the holoenzyme.
Although there is low overall sequence similarity in the Pol γA L-helix, residues involved in the hydrophobic interaction with Pol γB are conserved and are predicted to be α-helical in the mouse, Drosophila and Xenopus proteins (Fig. 2D). The hydrophobic residues on the respective Pol γB proteins are also conserved. Conservation of interacting residues in both subunits suggests that all these mitochondrial holoenzymes likely possess a common subunit interface that involves an AID subdomain.
Interestingly, the spacer containing AID is not only missing in the non-processive DNA polymerases, it is also largely absent in fungal Pol γA (Fig. S4). Perhaps not coincidentally, these enzymes also lack a Pol γB-type processivity factor. It seems likely that the ancestor of human Pol γA first acquired a spacer domain, which then allowed a Pol γB-like protein to interact, and the interacting domains subsequently co-evolved to increase the processivity of synthesis.
Processivity of the holoenzyme
The ability to catalyze processive synthesis is essential for replisomal complexes. However, most replicases have little processivity by themselves, generally synthesizing 15 nt or less per primer-binding event. Pol γA is somewhat exceptional in that it can synthesize ≥100 nts (Graves et al., 1998; Johnson et al., 2000). However, when bound to their accessory proteins all replicases exhibit high processivity, synthesizing thousands of nucleotides without dissociation (Hori et al., 1979; McHenry and Kornberg, 1977).
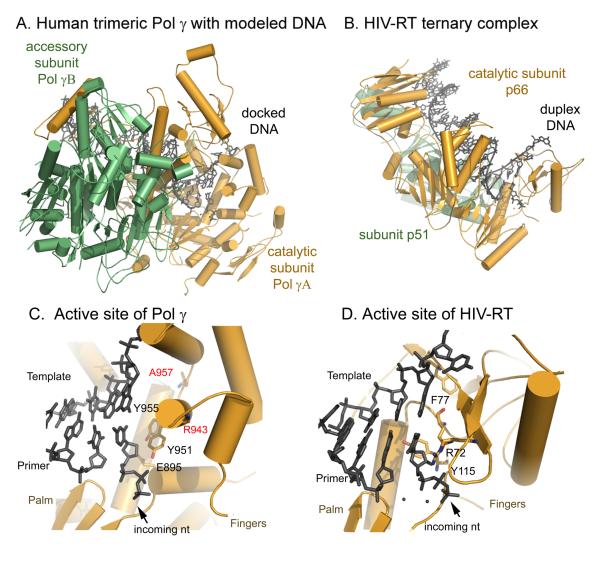
To begin to understand the mechanism of Pol γ processivity, we modeled a Pol γ-DNA complex by docking the primer-template DNA from the T7 DNAP-DNA complex (Brieba et al., 2004) onto the Pol γ holoenzyme after superimposing the active site domains of the two polymerases. Despite strong circumstantial evidence for a bacterial origin of mitochondria, the catalytic subunit Pol γA is more closely related to bacteriophage T7 DNAP than to any bacterial replicase. The two active site domains show high similarity and superimpose with an rmsr of 2.3 Å (Fig. S5), which enables modeling an enzyme-DNA complex with confidence.
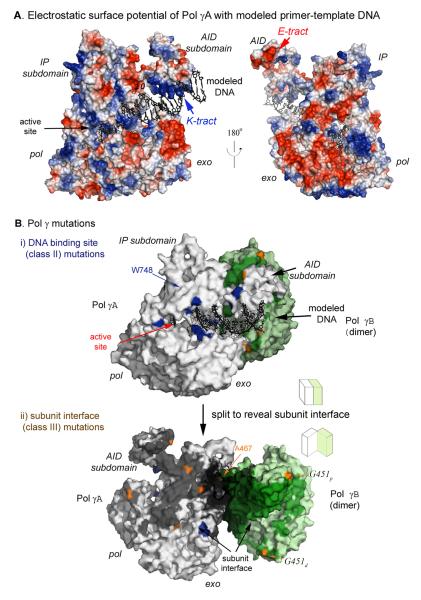
The docked DNA is cradled by a positively charged channel formed by the thumb, palm, and fingers of Pol γA (Fig. 4, 5); this pol domain makes contact with ~10 bp of template DNA that includes the primer terminus. In Pol γA holoenzyme, the hydrophobic interaction between Pol γB and the L-helix of the AID subdomain exposes a surface on Pol γA containing a high density of positively charged residues (496KQKKAKKVKK505, termed the K-tract, Figs. 4B, 5A). The K-tract interacts with the negatively charged phosphodiester backbone of DNA upstream to that bound in the pol domain, thus increasing the contact of holoenzyme to DNA to ~25 bp (Fig. 5A). The modeled complex reveals no direct contact between Pol γB and primer-template DNA, suggesting that the increased DNA-binding affinity of holoenzyme by Pol γB is mediated entirely through Pol γA. In support of this conclusion, weakening the hydrophobic interaction between the AID subdomain and Pol γB, which likely causes additional flexibility of the K-tract, reduces activity and processivity of holoenzyme (Fig. 3). This model now provides a structural basis for the known increased affinity of holoenzyme, relative to Pol γA, to DNA.
Figure 4.
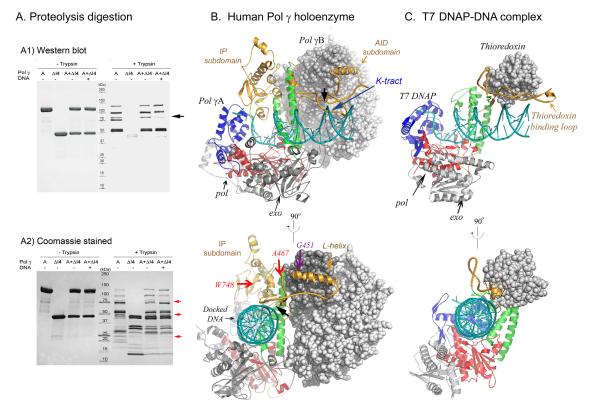
Comparison of a modeled Pol γ-DNA complex with that of the T7 DNAP-DNA complex. A) Limited proteolysis of Pol γ visualized after SDS-PAGE by Western blot or by Coomassie blue staining. B) Modeled Pol γ-DNA complex containing Pol γA (shown in ribbons), Pol γB (grey CPK) and a docked DNA (blue ribbons) shows that IP and AID subdomains enhance DNA-binding. Mutations and the region protected by DNA from proteolytic digestion (black arrow) are indicated. C) Crystal structure of T7 DNAP-DNA complex containing gp5 (ribbons), thioredoxin (grey CPK) and a primer-template DNA (blue ribbons).
Figure 5.
Pol γA charge distribution and mutational analysis. A) The electrostatic surface potential of Pol γA is shown in two views with positively charged regions highlighted in blue and negatively charged regions in red. A primer-template DNA duplex is modeled using black sticks. B) Locations of Pol γ mutants. i) Pol γA mutants in the DNA-binding channel (class II) are highlighted in blue. ii) A split open view of the subunit interface showing class III mutants (orange) affecting Pol γA and Pol γB (subscripts denote the proximal and distal monomer) interaction.
Further evidence supporting the model includes limited proteolysis of the holoenzyme with and without primer-template DNA. Comparison of protease digestion patterns suggests several regions of Pol γ are protected by DNA (Fig. 4A). Taking advantage that both Pol γA and Pol γB are His-tagged at their C-termini, Western blot analyses using anti-His antibody aided identification of proteolytic fragments. Digestion of the catalytic subunit Pol γA generated three C-terminal major fragments with apparent molecular masses of 105 kDa, 77 kDa, and 56 kDa, and a minor fragment of 84 kDa. The 77 kDa C-terminal fragment is absent when Pol γB is present, suggesting that the region around residue 560, corresponding to AID domain L-helix of Pol γA, is involved in subunit interactions. When holoenzyme is bound to DNA, the intensity of the 84 kDa fragment is significantly reduced, suggesting that it contains a DNA-binding site. The DNA-protected region lies near residue 500, corresponding to the K-tract, which is in good agreement with the modeled Pol γ-DNA complex. Other differentially protease-sensitive bands are apparent in Fig. 4A, but the protected regions cannot be unequivocally identified.
Although Pol γA and T7 DNAP’s catalytic subunit, gene 5 protein (gp5), have similar active site domains (Fig. 4 and S5), simply referring to Pol γA as a T7-like DNAP is only partially correct. Pol γA is more processive than T7 gp5 in the absence of an accessory protein (Graves et al., 1998; Tabor et al., 1987), and the corresponding processivity factors are different in structure and function. E. coli thioredoxin, the processivity factor for gp5, is a 104 aa monomeric protein whereas the dimeric Pol γB contains 970 aa. Functionally, the two accessory proteins use different mechanisms for processivity enhancement: thioredoxin increases T7 DNAP affinity for primer/template DNA by decreasing koff without affecting the rate of nucleotide incorporation, therefore, effectively prolonging the time of each binding event (Huber et al., 1987). In contrast, Pol γB does not alter koff but accelerates the polymerization rate, thereby increasing the number of nucleotides incorporated per binding event (Johnson et al., 2000).
The model suggests that the relatively high processivity of Pol γA in the absence of Pol γB can be attributed to the IP subdomain of the spacer, which provides a binding site for the upstream primer-template DNA duplex (Fig. 4B). An IP subdomain is not found in T7 gp5 (Fig. 4C), precluding significant DNA synthesis in the absence of thioredoxin. The modeled DNA-holoenzyme complex can also fully explain the remarkable ability of Pol γB to increase polymerase and decrease exonuclease activity simultaneously. Binding of Pol γB to Pol γA causes the primer terminus to be preferentially bound in the pol rather than the exo site, probably because less DNA bending is required.
Furthermore, Pol γB may function beyond processivity enhancement and play a role in replisome assembly. Despite it not contacting DNA directly in the modeled Pol γ-DNA complex, Pol γB is able to bind DNA. This activity, however, appears important only for DNA synthesis on duplex templates. Changing the positively charged residues 363RKK365 and, separately, 328RK329 to alanines abolishes DNA-binding activity. 363RKK365 is part of the I7 loop (residues 356-369) that contains several positively charged residues; the corresponding region in threonyl-tRNA synthetase (structurally homologous to Pol γB) is involved in tRNA binding. Deletion of I7 abolishes Pol γB DNA-binding (Carrodeguas et al., 2001). Nonetheless, mutant Pol γB-containing holoenzyme retains normal activity in copying single-stranded DNA (Farge et al., 2007), but is defective in replicating duplex DNA in the presence of SSB and helicase. These data are consistent with the substitutions lying distant from the primer-template channel (Fig. S3). Other Pol γB mutants are analyzed in supplementary material.
Structurally, the I7 region in both monomers is disordered in the apo Pol γB structure, but that in the proximal monomer becomes ordered in the holoenzyme structure. To assess whether the distal monomer remains disordered is because of asymmetrical interaction between dimer Pol γB with Pol γA or whether it is due to the loss of the four-helical bundle in the Pol γB—ΔI4 mutant, we crystallized and determined its structure to 3.3 Å resolution. Deletion of the helical bundle changed the crystal packing from that of the wild-type but the structure still remains a perfect dimer, being formed by two monomers related by a 2-fold crystallographic axis (Fig. S6). Aside from the deleted helical bundle, the structure of Pol γB—ΔI4 is essentially identical to that of the wild-type protein. As in the wild-type protein, the two I7 regions are disordered in Pol γB—ΔI4, indicating that the differential folding of the Pol γB loops in the holoenzyme is not a function of the deletion. Most likely, the ordering of I7 in holoenzyme is a direct consequence of the Pol γB proximal monomer interacting with Pol γA.
Although we do not have sufficient data to model the replisome, the electrostatic surface potential of Pol γA is informative. As expected, the putative DNA-binding channel is lined with positively charged residues but the opposite surface of the protein presents a large negatively charged region near the exo domain and the tip of the AID subdomain also contains four sequential glutamates (535EEEE538, E-tract; Fig. 5A). The human mitochondrial helicase, Twinkle, has a highly positively charged C-terminal region that could contact one of these regions. If the interaction is through the negatively charged E-tract in the replisome, Twinkle would be positioned in a location close to that of the 363RKK365 and 328RK329, residues important in Twinkle-dependent replication of duplex DNA.
Distinct mode of substrate binding
Polymerases are classic examples of enzymes using the induced-fit mechanism to achieve substrate specificity. The apo form of most DNA Pol I members adopt an ‘open’ conformation, where catalytically important residues on the fingers domain lie some distance away from the palm active site residues. After DNA-binding, the fingers domain undergoes structural changes to the ‘closed’ conformation, enabling the enzyme to align the important residues for catalysis and substrate selection. The desolvation effect generated by the conformational changes further enhances substrate specificity (Fersht, 1985; Petruska et al., 1986). This mechanism is utilized by all high-fidelity polymerases and is apparently absent in error-prone polymerases. Interestingly, the fingers domain of apo Pol γA directly abuts where the primer terminus will be positioned and apo Pol γ thus presents a partially ‘closed’ conformation. Nevertheless, the catalytically important residues on the fingers domain are still too far for catalysis, a rotational conformational change is still necessary after DNA-binding to position the catalytic residues correctly. The configuration of the Pol γA active site suggests that the conformational change in the fingers domain is coaxial with the duplex DNA, whereas it is perpendicular in other DNAPs.
Apo Pol γA further differs from other DNAPs in the active site by containing a small subdomain (residues 1050-1095) that partially blocks the DNA-binding channel. This type of subdomain has not been described before in other DNAPs but the apo form of phage N4 virion miniRNAP contains a similar subdomain that rotates out of the channel following DNA-binding (Gleghorn et al., 2008). If Pol γ indeed undergoes different conformational changes than other DNAPs when binding DNA, it may then use different mechanisms to ensure replication fidelity. These differences could reflect the high susceptibility of Pol γ to NRTIs.
Pol γ mutations and human diseases
The critical functions of Pol γ in mtDNA synthesis may, in part, rationalize the diversity and progressive effects of Pol γ mutations in degenerative human disorders. Many severe human diseases have been correlated with mutations affecting Pol γ (http://tools.niehs.nih.gov/polg/), and several mutant proteins have been characterized. Using the Pol γ structure we can now begin to rationalize the effects of some of these substitutions. Previous mutational analyses using the T7 DNAP structure as a model have successfully explained mutations predicted to affect the active site (Graziewicz et al., 2004), and as we have shown here the active sites of the two proteins are homologous. However, mutations located in the spacer region — as originally defined by sequence alignment with E. coli Pol I - show diverse biochemical behaviors. The structure of Pol γ now enables us to use a structure-based domain definition and distinct subdomain functions to re-analyze the effects of these mutations.
We divided all reported disease-associated Pol γ mutations into three classes (Table S1). Class I contains active site mutations that all result in reduced enzyme catalysis; class II includes substitutions located in the putative DNA-binding channel, thus reducing DNA-binding affinity directly. Class III contains subunit interface substitutions that disrupt the subunit interaction between Pol γA and Pol γB and thus naturally have a reduced processivity. It should be noted that, although clinically discovered, not all substitutions shown in Table S1 have been shown biochemically to adversely affect enzyme activity.
A cluster of substitutions found in PEO patients (Lamantea et al., 2002; Van Goethem et al., 2001): R943H/C, Y955C, and A957P/S fall into class I. A modeled Pol γ-DNA complex with an incoming dNTP (Fig. 6C) suggests that R943 may form a charged interaction with the triphosphate moiety. Substitution of the positively charged arginine should drastically reduce affinity for incoming nucleotides. Y955 abuts the templating base, in a position that is consistent with its known multi-functional roles in other Pol I family enzymes, including primer-template alignment and substrate selection (Joyce and Benkovic, 2004). A957 is adjacent to a critical glycine (G958); the equivalent residue in T7 RNAP (another Pol I family member) serves as a fulcrum during enzyme translocation and coordinates substrate binding (Yin and Steitz, 2004). Substitutions of A957 with bulkier residues likely interfere with both enzyme translocation and binding to an incoming dNTP. In general, the predicted consequences of all these mutations are a decreased affinity for dNTPs, increased error rates and/or reduced catalysis, in good agreement with solution studies (Graziewicz et al., 2004). Mutations giving rise to these defective proteins should tend to confer a dominant phenotype, likely because the mutant enzymes compete effectively with the wild-type enzyme for binding to the template DNA and cause error-prone DNA synthesis.
Figure 6.
Structural differences between human Pol γ and HIV reverse transcriptase. Panel A and B: overall structures of the two enzymes illustrate differences in the interaction between the catalytic and acces α-helical fingers domain (C), differs significantly from that of HIV RT (D), where the incoming dNTP binding site is comprised of β-sheet fingers.
Due to the multiple functions of the spacer, the phenotypes of what have traditionally been called spacer mutants vary with the spatial location of the substitution. Our structure suggests that the IP and AID subdomains of the spacer use distinct means to increase processivity: the IP subdomain functions independently of Pol γB, whereas AID acts only through its interaction with Pol γB. Accordingly, spacer mutations segregate into classes II and III, but in general both are likely to have reduced affinity for DNA.
A large number of class II mutations are arginine substitutions that are distributed along the modeled primer-template DNA binding channel. Substitution of positively charged arginine with neutral residues will decrease DNA binding and polymerase activity. Thus class II mutants tend to be recessive, as the mutant Pol γA is ineffective in competing with the wild-type enzyme for template DNA.
The class II mutation W748S is commonly associated with autosomal recessive ataxia and Alpers’ syndrome (Hakonen et al., 2005). W748 is located in the IP subdomain (Figs. 4B and 5B), away from the subunit interface, and is likely important in maintaining the local structure of the IP domain that contacts the downstream single-stranded DNA. W748 forms stacking-interaction with F750 and H733 in the local structure. Destabilizing this stack by the W748S substitution will undermine the enzyme’s interaction with template DNA, leading to lower polymerase activity. This interpretation is consistent with the biochemical observations of low DNA polymerase activity and processivity and a severe DNA-binding defect, but normal holoenzyme formation (Chan et al., 2006).
The most common substitution among all Pol γ mutations, A467T, is a representative of class III mutants and is associated with a wide range of mitochondrial disorders (Nguyen et al., 2005). Biochemical studies using the A467T mutant, which was thought to affect the spacer domain, unexpectedly was found to have reduced template binding, and the mutant-containing holoenzyme has lower processivity (Chan et al., 2005; Luoma et al., 2005), suggesting that the A467T mutant Pol γA has both reduced polymerase activity and subunit interaction. This observation can now be explained because the substitution actually lies in the thumb domain of Pol γA (Fig. 4A and 5B), which is well known to interact with template DNA. Although Pol γB interacts with the thumb containing A467, this residue faces away from the interaction surface. However, A467T may interrupt the local hydrophobic environment formed by L466 and L602, causing a slight spatial shift of the thumb domain that interferes with the interaction between the subunits.
The only Pol γB substitution that has been reported to be associated with disease is G451E, which was found in a single PEO patient with multiple mtDNA deletions (Longley et al., 2006). G451E is a class III mutant, as G451 is located near the interface formed between the C-terminal domain of the proximal Pol γB monomer and the AID subdomain of Pol γA (Fig. 5B). The G451E substitution may cause a steric clash with T556 on the AID subdomain; perhaps more importantly, it may disrupt the hydrophobic interaction that is essential for subunit interaction. This structural analysis agrees with the biochemical characterization of G451E-substituted Pol γB, which revealed a compromised subunit interaction and incomplete stimulation of catalytic subunit activity (Longley et al., 2006).
From their structural and biochemical properties, mutations in class III are expected to be autosomal recessive. A defective subunit-interface leads to reduced polymerase processivity and DNA-binding, defects that can be at least partially compensated in heterozygotes by the presence of wild-type enzyme. In addition, all class III mutations identified to date are located in the subunit hydrophilic interface that plays a secondary role in subunit interaction.
Structural dissimilarities between human Pol γ and HIV reverse transcriptase provides exploitable space for drug design
Anti-viral NRTIs present a unique opportunity for drug design, because both the target HIV RT and the adverse target human Pol γ are known. Although it has long been suspected that the two enzymes are dissimilar, for the first time we can make detailed structural comparisons of the human and viral polymerases and exploit differences in a rational design of antiviral drugs with higher selectivity.
There are several structural differences between human Pol γ and HIV RT that may be utilized in designing selective inhibitors. The distinct subunit interactions of the two enzymes result in substrate DNA being bound in the active site of Pol γ at an angle of 45° to that in HIV RT (Fig. 6 A, B). More importantly, while the catalytic aspartates of the HIV RT p66 subunit and Pol γA have a similar spatial arrangement, the incoming nucleotide-binding sites formed between the palm and fingers subdomains are structurally distinct. This portion of the fingers subdomain is α-helical in human Pol γ but β-sheet in HIV RT (Fig. 6C, D).
Both human Pol γ and HIV RT utilize electrostatic interactions of positively charged residues on the fingers domain to bind the negatively charged triphosphate of an incoming dNTP. However, their interaction with the nucleoside moiety is different. In Pol γA, the nucleoside binding site is likely bounded by E895, Y951, Y955 and Q1102; in HIV RT it is bounded by R72, F77, Y115 and Q151 (Fig. 6C, D) (Huang et al., 1998). Not only is a charge of a residue reversed (E895 in Pol γA vs. R72 in RT) but the positions of Y951 and Y115 are also altered. A highly conserved bulky residue (a Y or F) in members of the DNA Pol I family is known to play a major role in discriminating against incorporation of ddNMP. Pol γA Y951 is located on the O-helix of the fingers domain, and has been shown to be responsible for the lack of discrimination between dNTPs and ddNTPs (Lim et al., 2003). Y115 of the p66 subunit of HIV RT has been shown to be important in the discrimination of 3′-OH residues (Klarmann et al., 2007), and may be the equivalent residue to Pol γA Y951. However, Y115 in HIV RT p66 lies on a loop behind the ribose moiety of the incoming dNTP (Fig. 6C, D). The different angles at which Y951 and Y115 approach the sugar moiety of an incoming dNTP suggests that they differentially shape the active site and, further, that HIV RT and Pol γ may interact differently with nucleoside analogues. The HIV RT residue in the equivalent spatial position as Pol γA Y951 is R72, which actually functions in pyrophosphorolysis rather than discrimination against the 2′-OH of ribose (Sarafianos et al., 1995). The differences between the two enzymes suggest that it may therefore be possible to design small molecules that exploit these structural and functional dissimilarities.
The modeled complex of Pol γ and DNA also illustrate how the fingers domain may undergo conformational changes in order to accommodate the primer/template DNA duplex, changes that can be contrasted with those in the thumb domain of HIV RT (Ding et al., 1998; Huang et al., 1998). The differences may influence how the two enzymes maintain their different degrees of substrate specificity and their different responses to nucleoside inhibitors. However, a clearer picture of how correct and incorrect or analog nucleotides are differentially accommodated within the active sites of HIV RT and Pol γA must await co-crystal structures of the latter enzyme with DNA and dNTPs.
EXPERIMENTAL PROCEDURES
Preparation of Pol γA and Pol γB
The His-tagged, exonuclease deficient (exo-) catalytic subunit Pol γA (which also lacks the mitochondrial localization sequence (residues 1-29) and ten of 13 sequential glutamines (residues 43-52)), was prepared by substituting catalytic residues D198 and E200 to alanines. Other mutants were made using this construct as starting material. Details of these constructions and of the deletion mutant Pol γB—ΔI4 are described in supplementary material. Pol γA was expressed in insect Sf9 cells, Pol γB was expressed in E. coli.
Selenomethionine-substituted Pol γA was produced by a variation of the procedure of Bellizi et al., (1999). Briefly, Sf9 growth medium (5% FBS in SF-900 SFM; Invitrogen) was exchanged 12 hr post-infection for a methionine-deficient medium containing 5% dialyzed FBS; 7 hr later the medium was made 50 mg/L L-(+)-Se-Met (Acros). After 48 hr, cells were harvested, lysed, and proteins were purified by sequential application to Ni-NTA, SOURCE S and Superdex 200 columns (Yakubovskaya et al., 2006). Holoenzyme was formed by combining Pol γA and Pol γB monomer at a 1:2 molar ratio, the complex was then isolated by gel filtration through Superdex 200.
Polymerization assay
The substrate was single-stranded M13mp18 DNA annealed to a 26 nt primer. Reaction mixtures contained 50 nM Pol γA (wt, L549N, L552N, K553A or ΔL), 100 nM Pol γB (wt), and 50 nM primer/template DNA in 20 μL reaction buffer (10 mM HEPES, pH 7.5, 50 μg/ml BSA and 3 mM β-mercaptoethanol). Reactions were initiated by the addition of MgCl2 (10 mM) and dNTPs (50μM dGTP, dATP, dTTP, 5μM dCTP and 0.1 μM [α-32P]dCTP) and NaCl (90 or 190 mM), and incubated at 37°C for 10 min. Reactions were stopped by adding 1% SDS, 20 mM EDTA and 0.1 mg/ml Protease K, and incubating at 42°C for 30 min. Mixtures were then applied to Micro Bio-Spin 6 columns (Bio-Rad) to remove free nucleotides, heat-denatured at 95°C for 5 min in gel loading buffer (70% formamide, 1x TBE, 100 mM EDTA) and were analyzed on a 6% polyacrylamide / 7 M urea gel. Reaction products were visualized by autoradiography.
Crystallography
Crystals of Pol γ holoenzyme were grown using the hanging-drop method at 20°C at 2-3 mg/mL of the protein complex against a well solution containing 5.5-7% PEG 8000, 100 mM NaH2PO4, and 100 mM ACES (pH 7.0). Osmium derivatives were prepared by soaking crystals in mother liquor containing 3 mM K2OsO4 for 7 hr. Prior to freezing in liquid nitrogen, crystals were transferred into solutions with stepwise increasing concentrations of glycerol up to 20%. Single-wavelength anomalous diffraction data (SAD) for Se and Os derivatives were collected at Advanced Photon Source 19-ID. All data sets were processed using the program HKL (Otwinowski and Minor, 1997).
Se atoms (30 out of 34 total) and Os atoms (8) were located by the anomalous difference Fourier method using phases obtained from molecular replacement with apo Pol γB as a search model. Initial phases (with a figure-of-merit 0.56) were calculated using combined phases from Se-SAD, Os-SAD and molecular replacement using the program CNS (Brunger et al., 1998). Density modification using Solomon in the CCP4 suite (Collaborative Computational Project, 1994) was applied to the initial phases; this procedure and B-factor sharpening drastically improved the quality of electron density maps (Fig. S7). The diffraction data of the holoenzyme were initially indexed to a hexagonal space group P3221 containing one complex per asymmetric unit (asu). Although the electron density map was readily interpretable, the resulting atomic structure could only be refined to a high Rfree (~49%). After careful examination, we reassigned the diffraction data to space group P32 with two copies per asu. In the new space group, diffraction intensity analysis indicated that the diffraction data were partially twinned with a twinning operator (h,-h-k,-l) and a twinning factor of 0.46. Refinement was subsequently carried out in space group P32, utilizing the detwinning procedure in the program CNS.
Crystals of Pol γB—ΔI4 were grown by the hanging-drop method at 4°C using 10-15 mg/mL protein and a well solution containing 100 mM Tris-HCl, pH 7-7.5, 100 mM KCl, 6-8% PEG8000, and 30% glycerol. Crystals were directly flash-frozen in liquid nitrogen. Diffraction data were collected at APS 19-ID. The structure was determined by the molecular replacement method using wild-type human Pol γB as a search model with the program AMORE (Navaza, 2001) and was refined with Refmac (Winn et al., 2003).
Limited proteolysis
Experiments were conducted with 20 μg of purified Pol γA, Pol γB-ΔI4, and holoenzyme in 20 μl of reaction buffer (20 mM HEPES, pH 7.5, 140 mM KCl, 1 mM EDTA, 5 mM β-mercaptoethanol) with or without equal molar of 25/45mer DNA, to which trypsin (0.1 μg) was added. Samples were incubated on ice for 3 min and treated with an equal volume of 2 × SDS sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 100 mM DTT) to stop the digestion reaction. Reaction products were separated by SDS-PAGE; Western blots were performed used anti-His antibody following the manufacturer Abcam’s protocol.
Supplementary Material
Acknowledgements
We thank Ian Molineux for his insight, helpful discussions and critically commenting on the manuscript, Kenneth Johnson for his long-term support, help in initiating the crystallography of Pol γ and for his financial support through NIH (GM044613). We thank D. Bogenhagen, W. Copeland and all members of the Yin lab for helpful discussion. We are grateful for excellent technical support from Stephan Ginell and the staff at Argonne National Laboratory, SBC at the Advanced Photon Source, which is supported by DOE under contract DE-AC02-06CH11357. The research was supported by grants from the Welch Foundation (F-1592) and NIH (GM083703) to YWY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bellizzi JJ, Widom J, Kemp CW, Clardy J. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure. 1999;7:R263–267. doi: 10.1016/s0969-2126(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- Chan SS, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- Chan SS, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase gamma by the E1143G polymorphismin mitochondrial disorders. Hum Mol Genet. 2006;15:3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Zeviani M. 155th ENMC workshop: polymerase gamma and disorders of mitochondrial DNA synthesis, 21-23 September 2007, Naarden, The Netherlands. Neuromuscul Disord. 2008;18:259–267. doi: 10.1016/j.nmd.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002.
- Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jr., Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- Fan L, Kim S, Farr CL, Schaefer KT, Randolph KM, Tainer JA, Kaguni LS. A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase. J Mol Biol. 2006;358:1229–1243. doi: 10.1016/j.jmb.2006.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge G, Pham XH, Holmlund T, Khorostov I, Falkenberg M. The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function. Nucleic Acids Res. 2007;35:902–911. doi: 10.1093/nar/gkl1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G, Lamantea E, Donati A, Filosto M, Briem E, Carrara F, Parini R, Simonati A, Santer R, Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain. 2005;128:723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. 2nd edition W.H. Freeman & Company; 1985. [Google Scholar]
- Gleghorn ML, Davydova EK, Rothman-Denes LB, Murakami KS. Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Mol Cell. 2008;32:707–717. doi: 10.1016/j.molcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves SW, Johnson AA, Johnson KA. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry. 1998;37:6050–6058. doi: 10.1021/bi972685u. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am J Hum Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci U S A. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Hori K, Mark DF, Richardson CC. Deoxyribonucleic acid polymerase of bacteriophage T7. Purification and properties of the phage-encoded subunit, the gene 5 protein. J Biol Chem. 1979;254:11591–11597. [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- Klarmann GJ, Eisenhauer BM, Zhang Y, Gotte M, Pata JD, Chatterjee DK, Hecht SM, Le Grice SF. Investigating the “steric gate” of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase by targeted insertion of unnatural amino acids. Biochemistry. 2007;46:2118–2126. doi: 10.1021/bi061772w. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Lewis W. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ Mol Mutagen. 2007;48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, et al. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Lim SE, Ponamarev MV, Longley MJ, Copeland WC. Structural determinants in human DNA polymerase gamma account for mitochondrial toxicity from nucleoside analogs. J Mol Biol. 2003;329:45–57. doi: 10.1016/s0022-2836(03)00405-4. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Clark S, Man C. Yu Wai, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet. 2006;78:1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum Mol Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- McHenry C, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977;252:6478–6484. [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, McKiernan PJ, Gray G, Naviaux RK. POLG mutations in Alpers syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor M. In: Processing of X-ray Diffraction Data Collected in Oscillation Mode In Methods in Enzymology. C.W.C. Jr., Sweet RM, editors. Volume 276: Macromolecular Crystallography. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Petruska J, Sowers LC, Goodman MF. Comparison of nucleotide interactions in water, proteins, and vacuum: model for DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1986;83:1559–1562. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos SG, Pandey VN, Kaushik N, Modak MJ. Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. J Biol Chem. 1995;270:19729–19735. doi: 10.1074/jbc.270.34.19729. [DOI] [PubMed] [Google Scholar]
- Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernette CM, Kaguni LS. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster. Purification, subunit structure, and partial characterization. J Biol Chem. 1986;261:14764–14770. [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E, Lukin M, Chen Z, Berriman J, Wall JS, Kobayashi R, Kisker C, Bogenhagen DF. The EM structure of human DNA polymerase gamma reveals a localized contact between the catalytic and accessory subunits. Embo J. 2007;26:4283–4291. doi: 10.1038/sj.emboj.7601843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.