Abstract
A growing number of human diseases are linked to abnormal gene expression which is largely controlled at the level of transcriptional initiation. The gene-specific activator promotes the initiation of transcription through its interaction with one or more components of the transcriptional initiation machinery, hence leading to stimulated transcriptional initiation or activation. However, all activator proteins do not target the same component(s) of the transcriptional initiation machinery. Rather, they can have different target specificities, and thus, can lead to distinct mechanisms of transcriptional activation. Two such distinct mechanisms of transcriptional activation in yeast are mediated by the SAGA (Spt-Ada-Gcn5-Acetyltransferase) and TFIID (Transcription factor IID) complexes, and are termed as “SAGA-dependent” and “TFIID-dependent” transcriptional activation, respectively. SAGA is the target of the activator in case of SAGA-dependent transcriptional activation, while the targeting of TFIID by the activator leads to TFIID-dependent transcriptional activation. Both the SAGA and TFIID complexes are highly conserved from yeast to human, and play crucial roles in gene activation among eukaryotes. The regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID are discussed here.
Keywords: Transcriptional activation, SAGA, TFIID, Activator, TBP, Proteasome, RNA polymerase II
1. Introduction
An altered pattern of gene expression is directly correlated with various types of human diseases. In eukaryotes, gene expression is largely controlled at the level of transcription that is mechanistically composed of different steps such as initiation, elongation, and termination. During transcriptional initiation, several general transcription factors (GTFs) such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, as well as RNA polymerase II (RNAPII) holoenzyme are assembled at the promoter, thus resulting in the formation of preinitiation complex (PIC) to start transcription. Subsequently, transcriptional elongation follows, and finally termination occurs [1–4]. Among these, transcriptional initiation is an important step which is stimulated by gene-specific activator proteins (activators). A typical activator contains a promoter-targeting region, and a distinct activation domain that interacts with transcription factor(s) to promote transcription. Activation domains are generally acidic in nature (e.g., Gal4, Gcn4, VP16 and p53). However, several other activation domains of different types such as glutamine-rich (e.g., SP1), proline-rich (e.g., CTF/NF1), and zinc finger-containing (e.g., AdE1a) activation domains also exist in eukaryotes. An activator can stimulate multiple genes in the genome, thus generating a highly co-ordinated regulation of gene expression. However, a gene can also be controlled by the action of multiple activators, thereby leading to a combinatorial regulation.
A variety of studies indicate that activators work by increasing the PIC formation [3, 5–7]. Such stimulation of the PIC formation is mediated by the interaction of activator with one or more components of the transcription machinery, termed as “target” (Figure 1A). A number of transcription factors have been identified as the direct targets of activators based upon in vitro protein-protein interaction experiments [8–24]. For example, at least ten different proteins interact with the yeast activator, Gcn4, in vitro [13, 15–21, 23]. Similarly, another well-characterized yeast activator, Gal4, has been shown to have a large number of targets based on in vitro studies [8, 10, 11, 13, 14, 22]. However, whether any of these are bona fide targets in vivo was not known. Using the chromatin immunoprecipitation (ChIP) assay, we [25] and others [26] have identified SAGA (Spt-Ada-Gcn5-Acetyltransferase; a multi-subunit protein complex as described below) as an essential in vivo target of the yeast activator, Gal4. Further, our FRET (Fluorescence resonance energy transfer) studies have revealed that the Tra1 subunit of SAGA interacts directly with Gal4 in vivo [27]. Consistent with these results, Workman and colleagues [17] have also demonstrated that Tra1 interacts with Gal4, Gcn4, and several other yeast activators in vitro. Intriguingly, unlike Gal4, the activator Rap1 does not target the SAGA complex during transcriptional activation in yeast [25, 28–34]. Rather, it targets TFIID (Transcription factor IID; a multi-subunit protein complex as described below) to stimulate transcription of the ribosomal protein genes [28, 31, 33–35]. And, these genes do not require the SAGA complex for their transcriptional activation [25, 36–39]. Thus, there appears to be two distinct mechanisms of transcriptional activation, which are dictated by the target specificities of the activators. Consistently, the genome-wide DNA microarray analysis revealed the differential requirement of SAGA as well as TFIID in the regulation of transcription in yeast [36–38]. These results were further corroborated by the ChIP analysis which revealed the recruitment of the TFIID components to the core promoters of the TFIID-dependent, but not SAGA-dependent genes [25, 28, 35]. On the other hand, the SAGA components are recruited to the promoters of the SAGA-dependent genes [25, 26, 40]. Together, these studies have revealed two distinct mechanisms of transcriptional activation mediated by SAGA and TFIID. Intriguingly, the GTFs such as Mediator and TFIIB are dispensable for the recruitment of TBP/TFIID at the TFIID-dependent genes [28]. However, they are required for transcriptional initiation of the TFIID-dependent genes [28, 41–44]. In contrast, these GTFs are essential for the recruitment of TBP at the SAGA-dependent promoters [27, 28]. Thus, the transcriptional initiation complexes are differentially formed at the promoters of the TFIID and SAGA-dependent genes, hence leading to the complex regulation of gene activation. These two distinct mechanisms of eukaryotic transcriptional activation are discussed below.
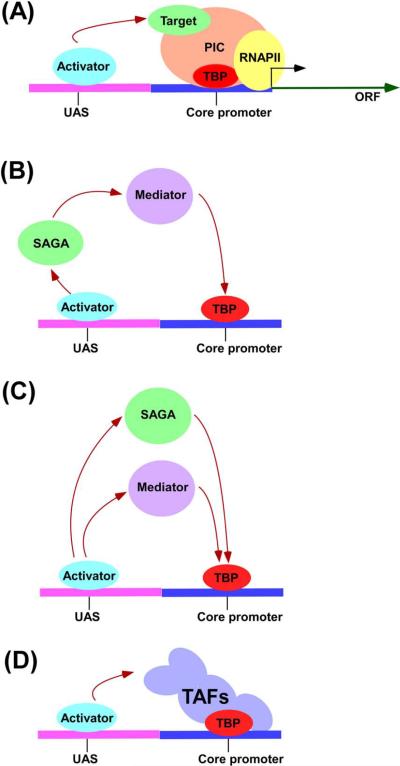
Figure 1.
Schematic representations of SAGA and TFIID-dependent transcriptional activation. (A) The transcriptional activator that binds to the UAS of the promoter stimulates the formation of the PIC assembly at the core promoter through interaction with one or more components of the transcriptional initiation machinery, termed as “target”. The schematic representation of how different activator–target interactions stimulate TBP recruitment at the (B and C) SAGA and (D) TFIID-dependent promoters. Subsequent to TBP recruitment, formation of the PIC continues to initiate transcription [1–4].
2. Mechanisms of transcriptional activation by SAGA
SAGA was first identified as a histone acetyltransferase (HAT) for histone H3 [45]. It is a large multi-subunit protein complex, and is highly conserved among eukaryotes through evolution [46]. In yeast, SAGA has fifteen non-essential and six essential components [46–48]. The non-essential components include Ada1, Ada2, Ada3, Gcn5, Ada5, Spt3, Spt7, Spt8, Spt20, Sgf11, Sgf29, Sgf73, Ubp8, Sus1 and Chd1, while the essential components are ATM/PI-3-kinase-related protein Tra1 as well as a subset of TAFs (TBP associated factors) such as TAF5, TAF6, TAF9, TAF10 and TAF12. The integrity of the SAGA complex is maintained by Ada1, Spt7 and Spt20 [49–51]. The Gcn5 and Ubp8 components possess HAT and histone deubiquitylase activities, respectively [47]. Both components are shown to regulate gene expression through modulation of the chromatin structure [47]. The other non-essential components play important roles in SAGA function and gene expression [47, 48]. The essential component, Tra1, has been implicated as the target of several activators [17, 24, 27]. TAFs have been shown to play important roles for SAGA function and hence transcription activation [16, 52]. Many conserved homologues of yeast SAGA components are found in human SAGA complex [46]. However, the human homologues of yeast Spt8 and Chd1 are not yet identified in human SAGA [46]. The human homologue of yeast Tra1 is TRRAP (transformation/transcription domain-associated protein). Like yeast Tra1, human TRRAP has been proposed to be the target of several activators, such as c-Myc and E2F [53] which regulate cell proliferation and apoptosis. Thus, Tra1/TRRAP functions as a direct target of many transcriptional activators such as Gal4, Gcn4, VP16, E1A, E2F, c-Myc, p53 and nuclear receptors in yeast and human [17, 24, 27, 53–63]. The ATM/PI-3-kinase-like domain near the C terminus of Tra1/TRRAP is important for interaction with activators [17, 63], and thus, plays a crucial role in targeting the HAT activity to the promoters. Like in yeast and human, the SAGA complex has also been identified and characterized in Drosophila [46]. The Drosophila SAGA shows remarkable similarity with yeast SAGA complex. However, several homologues of yeast SAGA components such as Spt8, Spt20, TAF6, Sgf73 and Chd1 are not yet identified in Drosophila [46]. Like yeast SAGA, the Drosophila SAGA complex also interacts with acidic activator VP16 and Drosophila p53 [64]. Thus, the targeting of the SAGA complex to the active genes appears to be evolutionary conserved.
Besides the roles of activators in targeting SAGA, histone covalent modifications also play important roles in stabilizing the interaction of SAGA with the promoters. Several studies have indicated that SAGA interacts with acetylated-histone H3 through the bromodomain of its Gcn5 subunit [47, 65–72]. Thus, SAGA appears to write the acetylation mark on histone H3, and subsequently, reads it during the establishment of a transcriptionally competent chromatin structure. Further, the chromodomain of the Chd1 subunit of yeast SAGA has been shown to interact with di- and trimethylated-K4 (lysine 4) of histone H3 [73–75]. Such interaction of SAGA with methylated-K4 of histone H3 may play important role in recruitment and/or stabilization of SAGA onto chromatin. This interaction may also play a direct role to regulate the Gcn5's HAT activity, thus contributing to the establishment or maintenance of active chromatin. Intriguingly, several studies have also implicated that Gcn5 preferentially acetylate histone H3 that is phosphorylated at S10 [76–81], suggesting the role of histone H3 phosphorylation on SAGA's HAT activity. Together, these studies demonstrate that the interaction of SAGA with activator drives its recruitment, and subsequently, histone covalent modification leads to its stabilization onto the promoter.
How does activator-SAGA interaction stimulate PIC formation, and hence transcription? Genetic studies have implicated direct interaction between the Spt3 component of SAGA and TBP [26, 82, 83]. Further, biochemical experiments have demonstrated that the Spt8 component of SAGA interacts directly with TBP [49]. Consistent with the genetic and biochemical interactions, the ChIP experiments have revealed that Spt3 and Spt8 are required for recruitment of TBP at the promoters of several SAGA-dependent genes such as ADH1, PHO84 and VTC3 [51]. However, these components are not essential for global structural integrity of SAGA [25, 49]. Together, these observations support that SAGA stimulates recruitment of TBP (and hence PIC formation) via its Spt3 and Spt8 components. Intriguingly, the Spt3 and Spt8 components of SAGA have also been shown to inhibit TBP recruitment at HIS3, TRP3 and HO [84, 85]. Thus, these two components of SAGA perform differential functions in recruiting TBP at the two different sets of promoters. Additional factors such as Swi/Snf and FACT may play a role in such differential regulation of TBP recruitment. However, the factors associated with positive vs. negative regulation of TBP recruitment by Spt3 and Spt8 are not yet known. In addition to its role in recruiting TBP, SAGA also regulates transcriptional activation through its HAT activity [47, 51, 86–91]. When present within the SAGA complex, Gcn5 acetylates nucleosomal histone H3 at K9, K14, K18, and K23 [92–93]. The Gcn5-HAT activity is provided by other SAGA subunits [93, 94]. It has been demonstrated that Gcn5 with Ada2 and Ada3 forms a catalytic core which is minimally capable of nucleosomal acetylation [51, 95]. Ada2 has been implicated to potentiate the HAT activity of Gcn5, while Ada3 promotes acetylation and lysine specificity [95]. Although Gcn5, Ada2 and Ada3 form the core catalytic domain within SAGA, other SAGA subunits have also been implicated to regulate the HAT activity of Gcn5. For example, temperature-sensitive (ts) inactivation of TAF12 significantly impairs the SAGA's ability to acetylate nucleosomes, and subsequently, reduces transcriptional activation in vitro [52]. However, the molecular basis for such regulation of the Gcn5's HAT activity remains unclear. Nonetheless, SAGA-mediated acetylation of histone H3 is required to stimulate transcription [86–88]. Further, Yu et al [85] have demonstrated that such acetylation promotes the formation of the TFIIA-TBP-DNA complex. Consistently, the ChIP studies have demonstrated the requirement of Gcn5 for histone H3 acetylation as well as TBP recruitment (and hence transcriptional activation) at the SAGA-regulated promoters [90, 91]. Intriguingly, the HAT activity of SAGA has also been implicated in transcriptional repression at a subset of genes [96]. For example, Gcn5-mediated acetylation reduces the binding of TBP with the ARG1 promoter, and impairs significantly the expression of ARG1 in the rich medium. Thus, Gcn5 plays a differential role in the regulation of transcriptional activation, hence further increasing the complexity of gene activation. However, SAGA can also regulate transcription independently of its HAT activity. For example, SAGA acts as a “physical adaptor” at several genes to stimulate the recruitment of TBP (and hence formation of the PIC) via its Spt3 and Spt8 components, but not Gcn5 [25, 26, 51, 91]. Consistently, the DNA microarray analysis has demonstrated that a set of SAGA-dependent genes does not require Gcn5 for their expression [38]. Intriguingly, Qiu et al [97] have further demonstrated that SAGA can also exert its function by recruiting RNAPII independently of its stimulatory role for TBP recruitment. Together, these studies reveal that SAGA differentially controls transcriptional activation of a subset of RNAPII genes, thus leading to the complex regulation of gene activation.
In addition to its above roles in gene activation, SAGA also regulates transcription through histone H2B deubiquitylase activity of its Ubp8 subunit [40, 98–102]. Monoubiquitylation of lysine on histone H2B occurs by ubiquitin-activating, conjugating and ligase enzymes [101–106]. It is a reversible process. Ubiquitin moiety can be enzymatically removed from the ubiquitylated-H2B by Ubp8 deubiquitylating enzyme. Ubp8 hydrolyzes isopeptide bond at glycine 76 of ubiquitin, leading to deubiquitylated-H2B. Thus, the enzymatic activity of Ubp8 appears to provide another level of regulation. Further, monoubiquitylation of histone H2B is essential for di- and trimethylation of K4 of histone H3 [106–107] which is associated with active transcription. Therefore, it is anticipated that the loss of Ubp8 would increase histone H2B ubiquitylation, and subsequently, would promote histone H3 K4 di- and trimethylation, leading to the additional level of transcriptional regulation. Indeed, it has been demonstrated that the SAGA-associated Ubp8 is required for deubiquitylation of ubiquitinated-K123 of histone H2B at several SAGA-dependent genes [40, 99, 100]. Such alteration in the level of histone H2B ubiquitylation is correlated with histone H3 K4 methylation at several genes [40, 99, 100]. Intriguingly, Shukla et al [40] have demonstrated that the histone H2B deubiquitylation activity of Ubp8 can also differentially regulate H3 K4 methylation at the SAGA-dependent promoters. However, the molecular basis for such differential regulation of histone H3 K4 methylation by Ubp8 remains unclear. Nonetheless, the modulations of covalent modifications of histones by Ubp8 have been shown to promote the transcription of SAGA-dependent genes [99, 100]. However, Ubp8 has also been shown to repress transcription [102]. Such transcriptional repression has been correlated with the level of histone H2B ubiquitylation [102]. Interestingly, Shukla et al [40] have further demonstrated the dispensability of Ubp8 in formation of the PIC (and hence transcription) at the promoters of several SAGA-dependent genes. Thus, Ubp8 differentially controls transcription of the SAGA-regulated genes, revealing a complexity of gene activation in vivo. However, the factors associated with such differential regulation of transcriptional activation are not currently understood.
Recently, Ubp8 has been shown to form a distinct structural module with Sgf11 and Sus1 within the SAGA complex [46, 108], and this module is termed as “deubiquitylation module”. Each component of the deubiquitylation module plays important role in histone H2B-K123 deubiquitylation as well as histone H3 K4 methylation [109]. Interestingly, Sus1 has also been found to be an integral component of the TREX-2 complex that is attached with nuclear pore complex for nuclear export of mRNA [46, 110]. Thus, the deubiquitylation module appears to link the SAGA-dependent transcription with nuclear export of mRNA. Indeed, the sites of active transcription of the SAGA-dependent genes are found at the nuclear pore complexes [110–113]. Such localization of the SAGA-dependent genes during active transcription is dependent on Sus1 [110]. Consistent with these observations, the live-cell imaging studies at GAL genes have also revealed the presence of SAGA component at the nuclear envelope during transcriptional activation [112]. Such translocation of the SAGA-dependent genes by Sus1 during active transcription might be leading to an efficient mRNA export. However, it remains to be further elucidated how universal this link is. Nonetheless, these results point to the fact that Sus1 links SAGA with mRNA export machinery [46, 110]. Intriguingly, the recent studies have also implicated the role of another non-essential component of SAGA, Sgf73, in the regulation of mRNA export [108, 114, 115]. These studies have demonstrated that the loss of Sgf73 impairs partly the association between Sus1 and TREX-2 as well as partial mislocalization of Sus1 to cytoplasm [108, 114]. However, how Sgf73 is linked to Sus1 and TREX-2 remains to be further investigated. In addition to its role in mRNA export, Sgf73 has also been implicated in maintaining SAGA integrity (and hence PIC formation) at the SAGA-dependent genes [91, 116]. In human, it is involved in neurodegenerative diseases through modulation of chromatin structure and gene activation [117–120].
Homologues of yeast Ubp8, Sgf11 and Sus1 have been identified in higher eukaryotes. Nonstop and USP22 are the homologues of yeast Ubp8 in Drosophila and human, respectively. Nonstop associates with Drosophila SAGA, and participates in an accurate axon guidance in the optic lobe in neural development [121]. Further, Nonstop appears to function as an enhancer of position-effect variegation [122]. Like in Drosophila, USP22 associates with human SAGA, and is recruited to specific genes by activators such as the MYC oncoprotein for transcription [123]. The enhanced transcription of MYC target genes is critical for malignant transformation in mammals. Further, MYC is the most potent activator involved in cell cycle progression, and its role in cancer is based on its ability to drive cell proliferation. Thus, it is expected to observe proliferative defect upon depletion of USP22. Indeed, proliferative defect and the G1 cell cycle arrest were observed upon depletion of USP22. Thus, a specific requirement of USP22 in cell cycle progression is linked to the etiology of cancer, and hence, can be used as a predictor of tumor growth [123]. Sus1 homologues in Drosophila and human are e(y)2 and ENY2, respectively [122, 124, 125]. Both e(y)2 and ENY2 are part of the Drosophila and human SAGA complexes [122, 125]. ENY2 has been shown to function as a cofactor for the full transcriptional activity of nuclear receptors, while e(y)2 is required for the barrier activity of Su(Hw)-dependent insulators [122, 125]. Sgf11 and ATXN7L3 are yeast Sgf11 homologues in Drosophila and human, respectively, and are part of the SAGA complex [46, 122]. Drosophila Sgf11 is proposed to function in position-effect variegation [122]. Together, these observations support that deubiquitylation module and its components are evolutionary conserved among eukaryotes. Indeed, like in yeast, the deubiquitylation module containing USP22, ENY2 and ATXN7L3 has been characterized in human SAGA [122].
In addition to its above roles in gene activation, SAGA also regulates transcriptional activation via the Mediator complex. The Mediator is an evolutionarily conserved-complex that contains at least 22 proteins [41]. However, its precise mechanism-of-action in transcriptional activation is not clearly understood. Our recent ChIP studies [27] have revealed that the Mediator complex is recruited to the GAL1 promoter subsequently to SAGA to stimulate the PIC formation. Consistently, Roberts and Winston [126] have demonstrated the functional interaction between SAGA and Mediator. Further, systematic genetic analysis has revealed that Δspt20 and Δspt7 show lethality with the null mutations of the two components (GAL11 and SIN4) of the Mediator complex [126]. In addition, the suppressor mutant of the Srb9 component of the Mediator complex has been shown to rescue partially the transcription defect of GAL1 in Δspt20 [127]. Thus, SAGA interacts functionally and genetically with Mediator, and acts as an “adaptor” that recruits the Mediator complex to the activation domain of Gal4 (Figure 1B). Intriguingly, Mediator and SAGA have also been shown to be independently recruited to the SAGA-dependent promoters by other activators such as Gcn4 and Met4 for the PIC formation (Figure 1C) [21, 128, 129]. These observations fit well with the model that have implicated Mediator as a target of several transcriptional activators in yeast and mammalian cells [41], and demonstrates how different activator–target interactions stimulates the PIC formation (and hence transcriptional activation). Despite these advances, our understanding of a complete picture of the protein–protein and protein–DNA interactions during transcription activation remains elusive in vivo. Thus, further investigation is essential to delineate the specific protein interaction network within eukaryotic transcriptional activation machinery in vivo.
Besides its role in transcriptional activation, SAGA also participates in other transcriptional regulatory steps such as transcriptional repression at telomere, transcriptional elongation, mRNA export, active gene translocation, and genome repair [46–48, 110, 130–133]. Thus, SAGA appears to act as a master regulator of gene expression. In yeast, SAGA is required for normal transcription of approximately 10 percent of the genes [36, 38]. These genes tend to be inducible with consensus TATA box [36]. Thus, SAGA appears to promote the transcription of genes that respond to environmental stress such as heat, DNA damage and metabolic starvation [36]. However, majority of genes are regulated by TFIID [36–38], and they appear to be house keeping genes without consensus TATA box [36]. The mechanisms of transcriptional activation by TFIID are discussed below.
3. Mechanisms of transcriptional activation by TFIID
TFIID is a multi-subunit protein complex, and was first characterized in Drosophila and mammals [134–136]. Subsequently, it was biochemically characterized in yeast Saccharomyces cerevisae [137–139]. TFIID is composed of TBP and a set of TAFs. TAFs are highly conserved from yeast to human [140, 141], and are ubiquitously distributed. However, some TAFs are expressed in a tissue-specific fashion in higher eukaryotes [140, 142–145]. In yeast, there are 14 TAFs, namely TAF1, TAF2, TAF3, TAF4, TAF5, TAF6, TAF7, TAF8, TAF9, TAF10, TAF11, TAF12, TAF13 and TAF14 [140, 141]. The orthologues of each of these TAFs except TAF14 have been identified as the components of the TFIID complex in Drosophila and mammals [140, 141]. In addition to the above TAFs, Drosophila genome encodes five additional TAFs. These are nht (no hitter) (TAF4L), can (cannonball) (TAF5L), mia (meiosis I arrest) (TAF6L), sa (spermatocyte arrest) (TAF8L), and rye (ryan express) (TAF12L), and are paralogues of TAF4, TAF5, TAF6, TAF8 and TAF12, respectively [146, 147]. These TAFs are specifically expressed in a co-ordinated fashion in spermatocytes, and are required for normal transcription of genes required for spermatid differentiation [146, 147]. Further, these testis-specific TAFs have histone fold domain, and are, therefore, likely to form stable TAF complex(es) to regulate the testis-specific gene expression in primary spermatocytes for terminal differentiation of male germ cells. However, unlike in Drosophila, the mouse genome encodes two TAF paralogues which are involved in spermatogenesis [148, 149]. Thus, the roles of the testis-specific TAFs do not appear to be evolutionary conserved.
Despite its significant importance in transcriptional initiation, it is not clearly understood how the canonical TFIID complex is assembled by TAFs. The primary structure analysis has revealed that there is a striking similarity in the amino acid sequences of TAF6, TAF9 and TAF12 with the core histones H4, H3 and H2B, respectively [150]. Such similarity suggests the existence of a histone octamer-like structure within the TFIID complex [150]. Further, nine TAFs have the histone fold domains, thus indicating the presence of distinct heterodimers within TFIID [151, 152]. Indeed, these heterodimers were found in native TFIID [151]. Thus, the histone fold domain appears to function as a building block of the TFIID complex. However, very little is known about the supramolecular assembly of the TFIID complex, even though structural information is available at the atomic level for few TAF subdomains or TAFs [153–155]. The electron microscopic analysis has provided an overall shape of the TFIID complex in yeast and human at low resolution [156–160]. The TFIID complexes in both yeast and human have three lobes which are linked by connecting regions to form a “horseshoe”-shaped molecular clamp [156–160]. Further, the electron microscopic analysis with immunolabelling of TFIID revealed the localization of individual TAFs within this horseshoe-shaped structure, thus defining the composition of the lobes as reviewed by Cler et al. [161]. However, these structural informations were obtained at a very low resolution. Future studies at high resolution will provide accurate structural information of the TFIID complex, and thus, will provide the molecular insight of its mode of action.
In yeast, TFIID has been shown to be involved in transcription of the majority of genes [36, 38]. How is the TFIID complex targeted to the active genes? Early studies have revealed that TAFs are required for transcription in response to activators [140]. Consistently, these factors have been shown to interact directly with activators in yeast and higher eukaryotes [33, 34, 161–165]. Thus, TAFs appear to be required for the flow of genetic information from the activators to the core transcription machinery at the TFIID-dependent genes. However, whether activators work at the TFIID-dependent genes by recruiting TFIID in vivo was not clearly known. We [28] and others [35] have demonstrated that TAFs are co-recruited with TBP in a manner consistent with direct activator-TAF interactions. Consistently, we have demonstrated that TAFs are essential for recruitment of TBP at the TFIID-dependent genes, while TBP is dispensable for recruitment of TAFs [28, 39]. In addition, we show that TAF11 and TAF13 appear to mediate the interaction between TBP and other TAFs within the TFIID complex in vivo [39]. In support of this model, our studies have revealed that TAF11 and TAF13 are essential for recruitment of TBP, but not other TAF subunits of the TFIID complex [39]. Further, the physical and genetic interactions between TAF11 and TAF13 have been observed [39, 153, 166]. Together, these results provide some information on the structural organization of the TFIID complex in vivo. Further, these studies demonstrate that TAFs play crucial role in relaying the information from activator to TBP (Figure 1D). However, unlike the SAGA-dependent genes, the Mediator complex is dispensable for TBP recruitment at the TFIID-dependent genes, but essential for transcriptional activation [28]. Thus, the Mediator complex appears to be recruited following TFIID to the TFIID-dependent genes for promoting transcription. Like the Mediator complex, another GTF, namely TFIIB, is also dispensable for the recruitment of TBP at the TFIID-dependent genes, while TFIIB is essential for recruitment of TBP at the SAGA-dependent genes [25, 27, 28, 51]. Therefore, the Mediator complex and TFIIB play differential roles in regulation of transcriptional activation of the SAGA and TFIID-dependent genes. However, the molecular basis for such differential roles of the Mediator complex and TFIIB during transcriptional activation of the SAGA and TFIID-dependent genes is not clearly understood.
What determines whether a promoter would be TFIID-dependent or independent? Is TFIID-dependency specified by the core promoter elements or upstream activating sequences (UAS)? Several studies have attributed the core promoter elements for the TFIID-dependency [140]. For example, Drosophila and human TAFs have been shown to interact with initiator or downstream promoter element in a sequence-specific DNA binding manner [167]. Similarly, TFIID-dependency of some yeast genes has been mapped to the core promoter elements [168]. Besides core promoter elements, histone H3 K4 trimethylation has also been implicated to stimulate the binding of TFIID [169]. Such stimulation is mediated by the interaction between trimethylated-K4 of histone H3 and the plant homeodomain of TAF3 (absent in yeast TAF3, but present in higher eukaryotes) [169]. Further, acetylated-K9 and K14 of histone H3 interact with the bromodomains of TAF1 (absent in yeast TAF1, but present in higher eukaryotes) [161, 170]. The combination of histone H3 K4 trimethylation with histone H3 K9/14 acetylation significantly enhances the interaction of TFIID with promoter. However, such interaction is reduced when histone H3 K4 methylation is coupled with histone H3 R2 (arginine 2) dimethylation that is associated with transcriptional repression [161]. Together, these studies indicate that the core promoter elements as well as histone covalent modifications play important roles to regulate the stabilization of the interaction of TFIID with promoter. Interestingly, our studies [28, 29] have demonstrated that UAS is also responsible for recruitment of TFIID to the core promoter. When the UAS of the TFIID-dependent promoter is fused with the SAGA-dependent or TFIID-independent core promoter, TFIID is surprisingly recruited to the TFIID-independent or SAGA-dependent core promoters [29]. Conversely, TFIID is not recruited to the TFIID-dependent core promoter when it is fused with the UAS of the TFIID-independent or SAGA-dependent promoter [29]. These results clearly demonstrate that UAS drives the recruitment of the TFIID complex to the core promoter. Consistent with these results, Struhl and colleagues [31] have shown that the Rap1 activator which is associated with the UASs of the TFIID-dependent ribosomal protein genes is sufficient to recruit TAFs/TFIID to both TFIID-dependent and independent core promoters. Further, TAFs/TFIID are recruited to the core promoter in a Rap1-dependent manner, but independently of GTFs such as TBP, TFIIB and Mediator [28, 31], indicating that TAFs are the direct targets of Rap1. Indeed, the recent studies from the Weil laboratory [33, 34] have biochemically demonstrated the direct interaction between Rap1 and TAFs. Similarly, TAFs have been shown to be the direct targets of activators in mammalian cells [162–165]. The well-characterized examples are the interactions of the transcriptional activators SP1 and CREB (cyclic AMP response element binding) with TAF4. Together, these results demonstrate that TAF-dependency maps to both the UAS and core promoter. Thus, the functional interaction between the activator and core promoter is required for efficient recruitment of TAFs or TFIID at the TFIID-dependent promoters, and hence, TAFs can be direct targets of activators as well as core promoter recognition factors to activate transcription.
In higher eukaryotes, there are multiple variant TAFs and TBP-related factors (TRFs) [4, 171, 172]. These factors play important roles in tissue-specific gene expression [4]. For example, several muscle-specific genes are regulated by a complex containing TAF3 and TRF3, but not TFIID complex [173, 174]. It has been demonstrated that a very low level of canonical TFIID is present in differentiated muscle cells [173, 174]. In addition to its role in muscle-specific gene expression, TRF3 is also essential for development of the hematopoietic lineage in zebrafish through activation of the key differentiation genes [175]. Thus, it is likely to have different TFIID variants in higher eukaryotes for differential gene expression in a tissue-specific fashion. Further, it would be interesting to determine how TFIID variants are targeted to the specific genes. Does specific activators or specific DNA sequence in the promoter or histone covalent modifications drive the recruitment of the TFIID variants to the tissues-specific genes? The answers to these questions will significantly advance our understanding of the tissue-specific regulation of gene activation.
In addition to the above two distinct regulatory mechanisms for stimulation of the PIC assembly, activators also work through interaction with other histone modification factors and ATP-dependent chromatin remodeling complexes [3, 60, 176–178]. These chromatin remodeling/modification factors promote transcription by modulating the chromatin structures. Thus, besides SAGA and TFIID, the chromatin structures/chromatin remodeling factors also play crucial roles in the regulation of transcriptional activation. Intriguingly, several recent studies have also implicated a protein degradation machine, namely the 26S proteasome complex, in the regulation of eukaryotic transcriptional activation [179–182]. The proteasome regulates gene activation through diverse mechanisms as reviewed recently [182]. The proteasomal regulation of SAGA and TFIID-dependent transcriptional activation is discussed below.
4. Regulation of the SAGA and TFIID-dependent transcriptional activation by the proteasome complex
The 26S proteasome is a non-lysosomal proteolytic machinery in eukaryotes [183, 184], which consists of two subcomplexes, namely a 20S core particle (CP) and a 19S regulatory particle (RP). The 20S CP is responsible for the proteolytic activities, whereas the 19S RP shows ATP-dependence and specificity for ubiquitin protein conjugates. The 26S proteasome degrades polyubiquitylated proteins in an ATP-dependent manner. However, the 20S CP can also degrade non-ubiquitylated proteins without ATP [185, 186]. The 20S CP has a cylinder-like structure that is formed by two α rings and two β rings. Each ring consists of seven related proteins (α1–α7, or β1–β7), and are stacked in the order of α β β α. The cavity of the cylinder-like structure of the 20S CP has the protease activities [183, 184]. The binding of the 19S RP with the 20S CP opens the gate of the 20S CP in an ATP-dependent manner, and then allows the entry of the substrate into the 20S CP for degradation [183, 184, 187–189]. The 26S proteasome has the 19S RP attached on either end or both ends of the 20S CP. The 19S RP contains at least 17 different proteins, including six ATPases (Rpt1–Rpt6) belonging to the AAA (ATPases associated with a variety of cellular activities) family, as well as the non-ATPase proteins (Rpn1–Rpn3, and Rpn5–Rpn12) [182–184, 190]. The 19S RP is further divided into two subcomplexes, namely “base” and “lid” [182–184, 190]. The base has six AAA-ATPase proteins (Rpt1–Rpt6) and three non-ATPase proteins (Rpn1, Rpn2 and Rpn10), while the lid consists of eight non-ATPase proteins (Rpn3, Rpn5–Rpn9, Rpn11 and Rpn12) [183, 184]. The non-ATPase protein, Rpn10, exists at the interface between the base and the lid. The lid subcomplex associates with, and hydrolyzes polyubiquitin chains [183, 184]. The ATPases present in the base are required for the assembly of the 26S proteasome. Further, the base unfolds the substrate protein in an ATP-dependent manner, and then translocates it into the central chamber of the 20S CP for proteolysis [183, 184, 187].
Through proteolysis, the 26S proteasome complex is involved in regulating the functions and fates of many transcription factors in a coordinated manner to control important cellular events [182, 191, 192]. In yeast (S. cerevisae), mutation of either the 19S RP or 20S CP alters the levels of ~70% of the genomic transcript at least by 2-fold [193]. The ts inactivation of the Rpt6 component of the 19S RP leads to the up-regulation of 1389 of 6400 yeast genes that encode proteasome subunits, ubiquitin-conjugating enzyme (Ubc5), and diverse proteins involved in mitochondrial function under stress conditions [193]. The ts inactivation of the Pre1 (β4) component of the 20S CP upregulates the expression of 1600 of 6400 yeast genes which encode proteins including protein folding factors, proteasomal subunits, mitochondrial proteins, stress sensors, and DNA repair factors (e.g., Rad7, Rad26, Rad59, and Hsh6). 60% of these genes were commonly up-regulated in both the ts mutants at the non-permissive temperature. However, 26% of genes were uniquely up-regulated in the ts mutant of Rpt6, while another 14% were up-regulated only in the pre1-ts mutant. Further, a large fraction of genes (2751 genes) were down-regulated in theses two ts mutants at the non-permissive temperature [193]. 63% of genes were commonly down-regulated in both mutants. However, 16% of genes were uniquely down-regulated in the ts mutant of Rpt6, while the expression of 21% of genes was decreased only in ts mutant of Pre1. Thus, a large number of genes in yeast are differentially regulated by the 26S proteasome in a 20S CP dependent and independent manner. Therefore, like in yeast, a large number of human genes might be dependent on the proteasome for their expression. Indeed, inhibition of the proteolytic function of the proteasome by Bortezomib (also known as Velcade) in retinoblastoma cell lines alters the expression of genes encoding stress-response proteins, heat shock proteins, pro-apoptotic proteins, cell-cycle regulators, cytokines, anti-apoptotic proteins, adhesion molecules, and a set of transcription factors [194]. Similarly, Bortezomib-treated pancreatic adenocarcinoma, classical Hodgkin lymphoma, multiple myeloma, and primary neuronal cells revealed altered-expression patterns of a variety of genes including those involved in cell cycle and apoptosis [195–198]. Thus, the proteasome appears to play crucial roles in human gene expression, and hence, has become an attractive target for treating human diseases including cancer. Indeed, Bortezomib is currently being used in the clinics to treat multiple myeloma and mantle cell lymphoma [199]. Further, several other proteasome inhibitors are in clinical trials [199–203].
The genome-wide DNA microarray analysis revealed that the expressions of both stress-response genes as well as housekeeping genes are regulated by the proteasome [182]. As previous studies [36, 38] have implicated SAGA and TFIID in the expression of stress-response and housekeeping genes, respectively, the proteasome thus appears to regulate the transcription of both SAGA and TFIID-dependent genes. However, whether the whole 26S proteasome or its subunits participate in the regulation of TFIID and SAGA-dependent transcriptional activation is not clear. Intriguingly, the genome-wide location analysis demonstrated that several hundred genes in yeast are associated with either the 19S RP or 20S CP [193, 204]. In support of this observation, Sulahian et al. [181] have shown that the transcription of several stress-responsive genes is regulated by the 19S RP independently of the proteolytic function of the 20S CP. These stress-responsive genes are dependent on SAGA for their transcriptional activation. On the other hand, the 20S CP or the proteolytic function of the proteasome regulates the transcription of the ribosomal protein genes [193, 205, 206]. As mentioned above, the ribosomal protein genes are dependent on the TFIID complex for their transcriptional activation. Thus, the 26S proteasome seems to have distinct functions in stimulation of transcription of SAGA and TFIID-dependent genes in the proteolysis-independent as well as -dependent manners, respectively.
How does the proteasome regulate transcriptional activation of the SAGA-dependent genes in a proteolysis-independent manner? Recently, Lee et al [207] have demonstrated that the 19S RP of the proteasome promotes the targeting of SAGA to the Gal4-driven promoter independently of the 20S CP. They show that the 19S RP uses the energy of ATP hydrolysis to stabilize the interaction between Gal4 and SAGA. In support of this observation, they demonstrate that the 19S RP interacts physically and genetically with SAGA, and enhances the HAT activity as well as DNA binding activity of SAGA. Further, they demonstrate that it is the base, but not the lid, of the 19S RP which stimulates SAGA targeting to the promoter. Thus, their data have provided a nice biochemical model as to why the 19S RP is required for GAL gene activation. However, the functional role of each 19S ATPase component in stabilization of the Gal4-SAGA interaction is not yet known. Further, it remains unclear whether the 19S RP regulates the transcriptional activation of other SAGA-dependent genes in a manner similar to the GAL genes. Moreover, it is yet to be elucidated whether the 19S RP functions similarly in the regulation of transcriptional activation of the SAGA-dependent genes in mammals. The answers to these questions will provide more functional insight on the 19S proteasomal regulation of the SAGA-dependent transcriptional activation.
In addition to its role in stimulation of activator-SAGA interaction, the 19S RP also plays a crucial role to regulate transcription by destabilizing the activator-promoter complex involved in the SAGA-dependent transcriptional activation [208]. For example, stability of the Gal4-promoter complex is regulated by the 19S RP [208]. Such regulation requires the ATPase activity of the 19S RP, but occurs independently of the proteolytic function of the 20S CP [208]. The monoubiquitylation of activator enhances the stability of the activator-promoter complex, while the 19S RP destabilizes activator-promoter interaction in the absence of monoubiquitylation [208]. Thus, the 19S RP and activator monoubiquitylation appear to function antagonistically in the regulation of activator-promoter interaction. In support of this model, Ferdous et. al. [208] have also demonstrated that the interaction of Gap71 (a Gal4 derivative that has a DNA binding domain, but is not monoubiquitylated) with promoter does not occur in the presence of the 19S RP. However, Gal4 forms a stable complex with promoter [208, 209]. Such stabilization of Gal4-promoter interaction is mediated by the monoubiquitylation of Gal4 [208, 209. However, how activator is monoubiquitylated remains to be elucidated.
Besides the regulation of activator-promoter/coactivator interaction, the 19S RP also promotes formation of the PIC at the SAGA-dependent genes, possibly through its molecular chaperonin activity. For example, using the ChIP assay in conjunction with mutational and transcriptional analyses, we [210] have recently demonstrated that the 19S proteasomal subcomplex establishes a specific protein interaction network of Gal4-SAGA-Mediator at the GAL1 UAS during transcriptional activation (Figure 2). Such an interaction network is essential for formation of the PIC at the core promoter to initiate transcription (Figure 2). Further, we [210] have demonstrated that formation of the transcriptional initiation complex at the GAL1 promoter is dependent on the 19S ATPase activity. Intriguingly, we [210] also find that the 19S ATPases function in a co-operative manner to facilitate the assembly of transcription factors at the GAL1 promoter. Thus, 19S RP is essential for formation of the PIC at the SAGA-dependent GAL1 promoter for transcriptional activation in vivo by establishing a specific protein interaction network at the UAS via its ATPase activity (Figure 2). However, it is not known whether the 19S RP directly participates in the PIC formation in vivo. Previous biochemical studies have implicated the interaction of the 19S RP with the components of the PIC assembly [211–213]. Thus, the 19S RP may directly regulate the PIC formation in vivo. Together, these studies have provided important insight on the regulatory mechanisms of transcriptional activation by the 19S RP. However, whether other SAGA-dependent genes in yeast are also regulated in a similar fashion by the 19S RP remains to be elucidated. Further, it will be interesting to verify whether 19S RP also regulates the PIC formation at the SAGA-regulated genes in mammals in a similar manner.
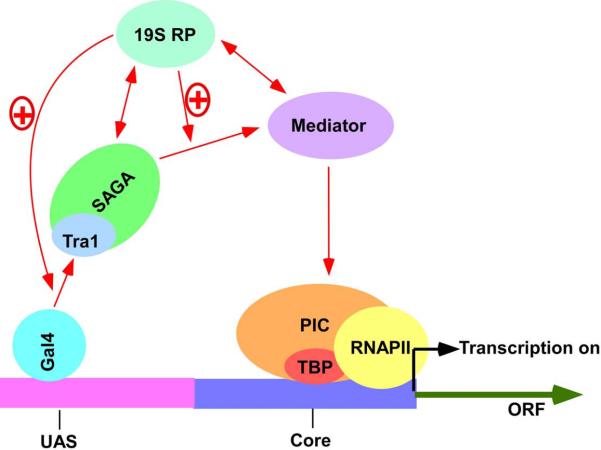
Figure 2.
A model showing the role of the 19S RP in regulation of transcriptional activation of the GAL1 genes in vivo. The activator, Gal4p recruits SAGA to the GAL1 UAS through its interaction with Tra1p [27]. However, the targeting of SAGA to the GAL1 UAS is less efficient in the absence of the 19S RP [210]. The ATPase activity of the 19S RP enhances SAGA targeting to the GAL1 UAS in a positive feedback manner [210]. Similar enhancement of SAGA targeting has also been demonstrated by biochemical studies [207]. The 19S ATPase activity is also essential for recruitment of the Mediator complex [210]. Mediator is required for formation of the PIC [27]. Further, in support of this model, several interactions have been demonstrated by biochemical and genetic studies [126, 127, 207, 211–213, 221]. For example, Lee et al. [207] have shown the physical and genetic interaction between SAGA and 19S RP. Sun et al. [221] have demonstrated the physical interaction between Mediator and the 19S RP. Functional and genetic interactions between SAGA and Mediator have also been reported [126, 127]. Further, several biochemical studies have shown the physical interaction of the 19S RP with the components of the PIC [211–213]. However, whether these interactions occur directly in vivo remains to be investigated. “+” referes “stimulation”.
Although the non-proteolytic function of the proteasome has been implicated to regulate the transcriptional activation of the SAGA-dependent genes, the proteolytic function of the proteasome can also provide additional levels of regulation to the SAGA-dependent genes by controlling activator abundance, localization and destruction as reviewed recently [182]. Further, the destruction of the co-activator or co-repressor can also regulate transcriptional activation [182]. In addition, the proteasome has been implicated to regulate transcription by disassembling the PIC formation and modulating chromatin structures [182]. Possibly through these mechanisms, the TFIID-dependent genes are also regulated. Although the detailed mechanisms-of-action of the proteasome in regulation of the TFIID-dependent transcriptional activation has not been explored, the DNA microarray analysis in yeast has revealed that the TFIID-dependent ribosomal protein genes are regulated by the proteolytic function of the proteasome [193, 205, 206]. Similarly, several TFIID-dependent genes such as hormone-regulated genes [161, 214–220] in human are also controlled by the proteasome [182]. The activators or steroid receptors of hormone-regulated genes are polyubiquitylated, and then degraded by the 26S proteasome complex for tight regulation of gene activation [182]. The proteasome degrades estrogen receptor in response to estradiol during transcriptional activation, and subsequently, leads to the disassembly of the active transcriptional initiation complex from the promoter [182]. Such turnover of estrogen receptor in response to estradiol leads to periodic dissociation/association of estrogen receptor with the promoter for disassembly and reassembly of the active transcriptional initiation complex for gene activation as the inhibition of the proteolytic activity of the proteasome lowers transcription of the target genes of the estrogen receptor [182]. Similarly, several other steroid receptors of hormone-regulated genes are controlled by the proteasome [182]. The proteolytic turnover of steroid receptors is regulated by basal transcription factors. For example, the Cdk7 subunit of TFIIH phosphorylates estrogen receptor for recruitment of ubiquitin ligase and subsequent polyubiquitylation. Then polyubiquitylated estrogen receptor is degraded by the proteasome (Figure 3). Similarly, progesteron receptor and retinoic acid receptors are also phophorylated prior to proteasomal degradation. Thus, basal transcription machinery plays an important role in activator turnover or disassembly of transcriptional initiation complex in a feedback manner (Figure 3). Likewise, certain activators in yeast undergo proteasomal degradation by basal transcription machinery [182]. Together, these studies shed some light on the proteasomal regulation of transcriptional activation of a selective set of TFIID-dependent genes. However, it is not yet known whether other TFIID-dependent genes are also similarly regulated by the proteasome. Further, the complete regulatory mechanisms of transcriptional activation of the TFIID-dependent genes by the proteasome are not clearly understood. Thus, future investigations are crucial to delineate as to how the proteasome regulates the transcriptional activation of the TFIID-dependent genes. Further, the global genome-wide studies will advance our understanding on the proteasomal regulation of the SAGA and TFIID-dependent transcriptional activation.
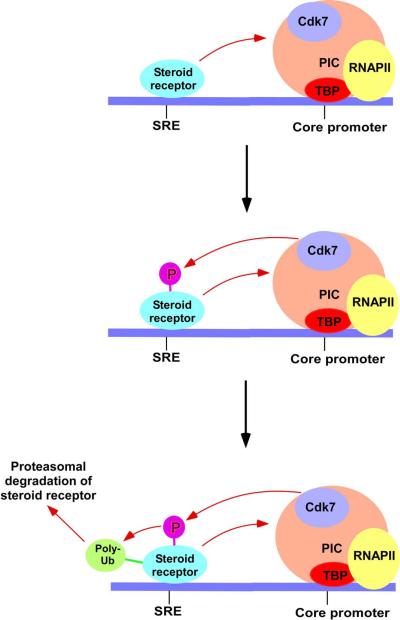
Figure 3.
Schematic representations showing the destruction of the steroid receptors in a transcription-dependent manner. In the first round of transcriptional activation, receptor stimulates the assembly of GTFs to form the PIC, and then the component of the basal transcription machinery (e.g. the Cdk7 component of TFIIH) facilitates phosphorylation of the steroid receptor. Such phosphorylation triggers polyubiquitination of steroid receptor which is subsequently recognized and degraded by the 26S proteasome. SRE, steroid response element; p, phosphorylation; and Ub, ubiquitylation.
5. Concluding remarks
Here, I have discussed two distinct mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. These distinct mechanisms of gene activation are dictated by the target specificity of the activator. In case of SAGA-dependent transcriptional activation, the activator targets SAGA which subsequently promotes the recruitment of TBP, and hence formation of the PIC to initiate transcription. On the other hand, TBP is recruited to the promoter of the TFIID-dependent gene via the interaction of activator with the TAF components of TFIID, subsequently promoting formation of the PIC. Intriguingly, the Mediator complex plays differential roles in these two distinct gene activation pathways. Mediator is essential for recruitment of TBP at the SAGA-dependent genes, but dispensable for TBP recruitment at the TFIID-dependent genes. Like the Mediator complex, another GTF, namely TFIIB, also differentially participates in the SAGA and TFIID-dependent transcriptional activation. TFIIB is essential for recruitment of TBP at the SAGA-dependent genes. However, the recruitment of TBP to the TFIID-dependent genes does not require TFIIB. Although both the Mediator complex and TFIIB are dispensable for recruitment of TBP at the TFIID-dependent genes, they are required for transcriptional initiation subsequent to TBP recruitment at the TFIID-dependent genes. Thus, GTFs function at the different steps of the transcriptional activation pathways of the SAGA and TFIID-dependent genes, implying a complex regulation of gene activation. Such a complex regulation of gene activation is further complicated by the involvement of the proteasome complex as discussed above and in a recent review article [182]. Briefly, the proteasome complex regulates transcriptional activation in a proteolysis dependent as well as independent manner. The proteolytic function of the proteasome regulates gene activation by controlling the abundance, localization and destruction of activators, destruction of co-activator or co-repressor, and disassembling the PIC. The non-proteolytic function of the proteasome has been implicated to control the interaction of activator with co-activator or promoter, and modulate chromatin structure for regulation of gene activation.
Although a large number of studies have significantly enriched our knowledge on the regulatory mechanisms of gene activation by TFIID and SAGA, these two distinct mechanisms of transcriptional activation have been deciphered from the studies of a selective set of RNAPII genes. However, how general these two distinct mechanisms of transcriptional activation are not fully explored. In addition, there is a small subset of RNAPII genes whose transcription is dependent on both the TFIID and SAGA complexes [38]. However, the mechanisms of transcriptional activation of these genes remain unclear. Further, how these gene regulatory mechanisms are controlled by the proteasome in a proteolysis-dependent or independent manner is yet to be deciphered. It would also be interesting to determine in mammals how conserved these mechanisms are. Investigations of these important questions will shed much light on the complex regulatory mechanisms of gene activation, and thus, will certainly provide crucial information for designing the therapeutic agents to maintain normal cellular function, since a large number of human diseases are correlated with the altered patterns of gene expression.
Acknowledgements
I thank the members of my laboratory for critical reading of this manuscript. The work in my laboratory was supported by a National Institutes of Health grant (1R15GM088798-01), a National Scientist Development Grant (0635008N) from American Heart Association, a Research Scholar Grant (06-52) from American Cancer Society, a Mallinckrodt Foundation award, and several internal grants from Southern Illinois University. I apologize to the authors whose work could not be cited owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- [2].Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- [3].Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- [4].Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- [6].Ptashne M, Gann AA. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- [7].Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- [8].Melcher K, Johnston SA. GAL4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- [10].Wu Y, Reece RJ, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- [11].Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol. Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- [12].Zaman Z, Ansari AZ, Gaudreau L, Nevado J, Ptashne M. M. Gene transcription by recruitment. Cold Spring Harb. Symp. Quant. Biol. 1998;63:167–171. doi: 10.1101/sqb.1998.63.167. [DOI] [PubMed] [Google Scholar]
- [13].Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jeong CJ, Yang SH, Xie Y, Zhang L, Johnston SA, Kodadek T. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry. 2001;40:9421–9427. doi: 10.1021/bi010011k. [DOI] [PubMed] [Google Scholar]
- [15].Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2 with acidic activation domains and TATA binding protein. J. Biol. Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- [16].Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- [17].Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- [18].Ansari AZ, Koh SS, Zaman Z, Bongards C, Lehming N, Young RA, Ptashne M. Transcriptional activating regions target a cyclin-dependent kinase. Proc. Natl. Acad. Sci. USA. 2002;99:14706–14709. doi: 10.1073/pnas.232573899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator interaction domains. Mol. Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- [20].Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, Hinnebusch AG. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J. Biol. Chem. 2010;285:2438–2455. doi: 10.1074/jbc.M109.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- [24].Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- [25].Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Gene Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- [29].Li XY, Bhaumik SR, Zhu X, Li L, Shen W-C, Dixit B, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences: implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- [30].Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- [31].Mencía M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- [32].Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- [33].Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Layer JH, Miller SG, Weil PA. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J Biol Chem. 2010;285:15489–15499. doi: 10.1074/jbc.M110.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- [36].Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- [37].Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- [38].Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- [39].Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a Histone Deubiquitinase Whose Association with SAGA Is Mediated by Sgf11p, Differentially Regulates Lysine 4 Methylation of Histone H3 In Vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- [42].Struhl K. Transcriptional activation: mediator can act after preinitiation complex formation. Mol Cell. 2005;17:752–754. doi: 10.1016/j.molcel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- [43].Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- [44].Fan X, Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- [46].Rodríguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Daniel JA, Grant PA. P. A. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA- binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- [53].McMahon SB, Buskirk H. A. Van, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- [54].Grant PA, Schieltz D, Pray-Grant MG, Yates JR, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- [55].Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- [56].Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol. 2002;22:5650–5661. doi: 10.1128/MCB.22.16.5650-5661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- [58].Memedula S, Belmont AS. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 2003;13:241–246. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- [59].Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon SB, Cole MD, Tora L, Takahashi N, Kato S. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell. 2002;9:553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- [60].Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- [61].Gamper AM, Kim J, Roeder RG. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol. 2009;29:266–280. doi: 10.1128/MCB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kulesza CA, Buskirk H. A. Van, Cole MD, Reese JC, Smith MM, Engel DA. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene. 2002;21:1411–1422. doi: 10.1038/sj.onc.1205201. [DOI] [PubMed] [Google Scholar]
- [63].Park J, Kunjibettu S, McMahon SB, Cole MD. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 2001;15:1619–1624. doi: 10.1101/gad.900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Winston F, Allis CD. The bromodomain: a chromatintargeting module? Nat. Struct. Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- [66].Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- [68].Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- [69].Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- [70].Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- [71].Yoon S, Qiu H, Swanson MJ, Hinnebusch AG. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell Biol. 2003;23:8829–8845. doi: 10.1128/MCB.23.23.8829-9945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- [74].Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- [75].Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- [76].Oevelen C. J. van, Teeffelen H. A. van, Timmers HT. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell Biol. 2005;25:4863–4872. doi: 10.1128/MCB.25.12.4863-4872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- [78].Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- [79].Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu Y, Xu X, Singh-Rodriguez S, Zhao Y, Kuo MH. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5-mutant in HIS3 expression. Mol. Cell Biol. 2005;25:10566–10579. doi: 10.1128/MCB.25.23.10566-10579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- [82].Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- [83].Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yu Y, Eriksson P, Bhoite LT, Stillman DJ. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell Biol. 2003;23:1910–1921. doi: 10.1128/MCB.23.6.1910-1921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kuo MH, Baur E. vom, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- [90].Deckert J, Struhl K. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol Cell Biol. 2002;22:6458–6470. doi: 10.1128/MCB.22.18.6458-6470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- [93].Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- [94].Grant PA, Berger SL. Histone acetyltransferase complexes. Semin. Cell Dev. Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- [95].Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- [96].Ricci AR, Genereaux J, Brandl CJ. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol Cell Biol. 2002;22:4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. An array of coactivators is required for optimal recruitment of TATAbinding protein and RNApolymerase II by promoter-bound Gcn4p. Mol. Cell Biol. 2004;24:4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [99].Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao C,F,, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- [101].Weake VW, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- [102].Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- [104].Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief. Funct. Genomic Proteomic. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- [105].Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- [106].Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- [107].Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci. 2009;66:1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- [109].Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodríguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pascual-García P, Rodríguez-Navarro S. A tale of coupling, Sus1 function in transcription and mRNA export. RNA Biol. 2009;6:141–144. doi: 10.4161/rna.6.2.7793. [DOI] [PubMed] [Google Scholar]
- [111].Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- [112].Cabal GG, Genovesio A, rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- [113].Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- [114].Pascual-Garcia P, Govind CK, Queralt E, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGAand Slik(SALSA) HATcomplexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci U S A. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, Mandel JL, Tora L, Devys D. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- [118].Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, Spada A. R. La, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration, Proc. Natl Acad. Sci. USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, Belal S, Lebre AS, Abada-Bendib M, Grid D, Holmberg M, Yahyaoui M, Hentati F, Chkili T, Agid Y, Brice A. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Hum. Mol. Genet. 1998;7:165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- [120].Del-Favero J, Krols L, Michalik A, Theuns J, Lofgren A, Goossens D, Wehnert A, Van den Bossche D, Van Zand K, Backhovens H, van Regenmorter N, Martin JJ, Van Broeckhoven C. Molecular genetic analysis of autosomal dominant cerebellar ataxia with retinal degeneration (ADCA type II) caused by CAG triplet repeat expansion. Hum. Mol. Genet. 1998;7:177–186. doi: 10.1093/hmg/7.2.177. [DOI] [PubMed] [Google Scholar]
- [121].Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhao Y, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, schüle R, Takeyama K, Kato S, Tora L, Devys D. ATFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- [123].Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGAcomplex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A. The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol Cell Biol. 2001;21:5223–5231. doi: 10.1128/MCB.21.15.5223-5231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P, Krasnov A. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell. 2007;27:332–338. doi: 10.1016/j.molcel.2007.05.035. [DOI] [PubMed] [Google Scholar]
- [126].Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Larschan E, Winston F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol Cell Biol. 2005;25:114–123. doi: 10.1128/MCB.25.1.114-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Leroy C, Cormier L, Kuras L. Independent recruitment of mediator and SAGA by the activator Met4. Mol Cell Biol. 2006;26:3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- [131].Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Jacobson S, Pillus L. The SAGA subunit Ada2 functions in transcriptional silencing. Mol Cell Biol. 2009;29:6033–6045. doi: 10.1128/MCB.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].gonzález-Aguilera C, Tous C, Gómez-González B, Huertas P, Luna R, Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell. 2008;19:4310–4318. doi: 10.1091/mbc.E08-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- [135].Kokubo T, Takada R, Yamashita S, Gong DW, Roeder RG, Horikoshi M, Nakatani Y. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J Biol Chem. 1993;268:17554–17558. [PubMed] [Google Scholar]
- [136].Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly JM, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Reese JC, Apone L, Walker SS, Griffin LA, Green MR. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- [138].Moqtaderi Z, Yale JD, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Poon D, Weil PA. (1993) Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- [140].Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- [141].Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- [142].Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- [143].White-Cooper H, Schäfer MA, Alphey LS, Fuller MT. Transcriptional and posttranscriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- [144].Lin T-Y, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- [145].Deato MD, Tjian R. An unexpected role of TAFs and TRFs in skeletal muscle differentiation: switching core promoter complexes. Cold Spring Harb Symp Quant Biol. 2008;73:217–25. doi: 10.1101/sqb.2008.73.028. [DOI] [PubMed] [Google Scholar]
- [146].Hiller M, Lin T-Y, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissuespecific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- [148].Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localization of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- [149].Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- [150].Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- [151].Gangloff Y, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- [152].Gangloff YG, Pointud JC, Thuault S, Carre L, Romier C, Muratoglu S, Brand M, Tora L, Couderc JL, Davidson I. The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)155) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol Cell Biol. 2001;21:5109–5121. doi: 10.1128/MCB.21.15.5109-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. (1998) Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- [154].Werten S, Mitschler A, Romier C, Gangloff YG, Thuault S, Davidson I, Moras D. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAF(II)135) and hTAF12 (hTAF(II)20) J Biol Chem. 2002;277:45502–45509. doi: 10.1074/jbc.M206587200. [DOI] [PubMed] [Google Scholar]
- [155].Romier C, James N, Birck C, Cavarelli J, Vivares C, Collart MA, Moras D. Crystal structure, biochemical and genetic characterization of yeast and E. cuniculi TAF(II)5 N-terminal domain: implications for TFIID assembly. J Mol Biol. 2007;368:1292–1306. doi: 10.1016/j.jmb.2007.02.039. [DOI] [PubMed] [Google Scholar]
- [156].Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. EMBO J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Threedimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- [158].Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- [159].Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- [160].Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, Schultz P. Mapping the initiator binding TAF2 subunit in the structure of hydrated yeast TFIID. Structure. 2009;17:363–373. doi: 10.1016/j.str.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, Dailey G, Nogales E, Tjian R. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–21. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Rojo-Niersbach E, Furukawa T, Tanese N. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- [165].Asahara H, Santoso B, Guzman E, Du K, Cole PA, Davidson I, Montminy M. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol Cell Biol. 2001;21:7892–7900. doi: 10.1128/MCB.21.23.7892-7900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Komarnitsky KB, Michel B, Buratowski S. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II mediated transcription in vivo. Genes Dev. 1999;13:2484–2489. doi: 10.1101/gad.13.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes & Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- [168].Shen WC, Walker SS, Green MR. Yeast TAFII145 functions as a core promoter-selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- [169].Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- [170].Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- [171].Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- [172].Reina JH, Hernandez N. On a roll for new TRF targets. Genes Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- [173].Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- [177].Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:D1054–64. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- [178].Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- [179].Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- [180].Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;423:1009–1013. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- [181].Sulahian R, Sikder D, Johnston SA, Kodadek T. The proteasomalATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 2006;34:1351–1357. doi: 10.1093/nar/gkl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Bhaumik SR, Malik S. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol. 2008;43:419–33. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- [183].Voges D, Zwick IP, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- [184].Coux O. The 26S proteasome. Prog Mol Subcell Biol. 2002;29:85–107. doi: 10.1007/978-3-642-56373-7_6. [DOI] [PubMed] [Google Scholar]
- [185].Tarcsa E, Szymanska G, Lecker S, O'Connor CM, Goldberg AL. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26S proteasomes without ubiquitylation. J Biol Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- [186].Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- [187].Larsen CN, Finley D. Protein translocation channels in the proteasome and other proteases. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]
- [188].Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- [189].Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Glickman MH, Rubin DM, Fried VA, Finley D. The Regulatory Particle of the Saccharomyces cerevisiae Proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- [192].He H, Qi XM, Grossmann J, Distelhorst CW. C-Fos degradation by the proteasome. An early, Bcl-2 regulated step in apoptosis. J Biol Chem. 1998;273:25015–25019. doi: 10.1074/jbc.273.39.25015. [DOI] [PubMed] [Google Scholar]
- [193].Sikder D, Johnston SA, Kodadek T. Widespread, but nonidentical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem. 2006;281:27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- [194].Poulaki V, Mitsiades CS, Kotoula V, Negri J, McMillin D, Miller JW, Mitsiades N. The proteasome inhibitor bortezomib induces apoptosis in human retinoblastoma cell lines in vitro. Invest Ophthalmol Vis Sci. 2007;48:4706–4719. doi: 10.1167/iovs.06-1147. [DOI] [PubMed] [Google Scholar]
- [195].Yew EH, Cheung NS, Choy MS, Qi RZ, Lee AY, Peng ZF, Melendez AJ, Manikandan J, Koay ES, Chiu LL, Ng WL, Whiteman M, Kandiah J, Halliwell B. Proteasome inhibition by lactacystin in primary neuronal cells induces both potentially neuroprotective and pro-apoptotic transcriptional responses: a microarray analysis. J Neurochem. 2005;94:943–956. doi: 10.1111/j.1471-4159.2005.03220.x. [DOI] [PubMed] [Google Scholar]
- [196].Tang ZY, Wu YL, Gao SL, Shen HW. Effects of the proteasome inhibitor bortezomib on gene expression profiles of pancreatic cancer cells. J Surg Res. 2008;145:111–123. doi: 10.1016/j.jss.2007.03.061. [DOI] [PubMed] [Google Scholar]
- [197].Zhao X, Qiu W, Kung J, Zhao X, Peng X, Yegappan M, Yen-Lieberman B, His ED. Bortezomib induces caspasedependent apoptosis in Hodgkin lymphoma cell lines and is associated with reduced c-FLIP expression: a gene expression profiling study with implications for potential combination therapies. Leuk Res. 2008;32:275–285. doi: 10.1016/j.leukres.2007.05.024. [DOI] [PubMed] [Google Scholar]
- [198].Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, Koenig E, Fergus A, Huang Y, Richardson P, Trepicchio WL, Broyl A, Sonneveld P, Shaughnessy JD, Bergsagel PL, Schenkein D, Esseltine DL, Boral A, Anderson KC. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- [199].Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- [200].Montagut C, Rovira A, Albanell J. The proteasome: a novel target for anticancer therapy. Clin Transl Oncol. 2006;8:313–317. doi: 10.1007/s12094-006-0176-8. [DOI] [PubMed] [Google Scholar]
- [201].Voorhees PM, Orlowski RZ. The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- [202].Bennett MK, Kirk CJ. Development of proteasome inhibitors in oncology and autoimmune diseases. Curr Opin Drug Discov Devel. 2008;11:616–625. [PubMed] [Google Scholar]
- [203].Sterz J, von Metzler I, Hahne JC, Lamottke B, Rademacher J, Heider U, Terpos E, Sezer O. The potential of proteasome inhibitors in cancer therapy. Expert Opin Investig Drugs. 2008;17:879–895. doi: 10.1517/13543784.17.6.879. [DOI] [PubMed] [Google Scholar]
- [204].Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- [205].Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc Natl Acad Sci U S A. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Dembla-Rajpal N, Seipelt R, Wang Q, Rymond BC. Proteasome inhibition alters the transcription of multiple yeast genes. Biochim Biophys Acta. 2004;1680:34–45. doi: 10.1016/j.bbaexp.2004.08.008. [DOI] [PubMed] [Google Scholar]
- [207].Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- [208].Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, Johnston SA. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes & Dev. 2007;21:112–123. doi: 10.1101/gad.1493207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Archer CT, Burdine L, Liu B, Ferdous A, Johnston SA, Kodadek T. Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J Biol Chem. 2008;283:21789–21798. doi: 10.1074/jbc.M803075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Malik S, Shukla A, Sen P, Bhaumik SR. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J Biol Chem. 2009;284:35714–35724. doi: 10.1074/jbc.M109.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Weeda G, Rossignol M, Fraser RA, Winkler GS, Vermeulen W, van 't Veer LJ, Ma L, Hoeijmakers JH, Egly JM. The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res. 1997;25:2274–2283. doi: 10.1093/nar/25.12.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Makino Y, Yoshida T, Yogosawa S, Tanaka K, Muramatsu M, Tamura TA. Multiple mammalian proteasomal ATPases, but not proteasome itself, are associated with TATA-binding protein and a novel transcriptional activator, TIP120. Genes Cells. 1999;4:529–539. doi: 10.1046/j.1365-2443.1999.00277.x. [DOI] [PubMed] [Google Scholar]
- [213].Yanagi S, Shimbara N, Tamura TA. Tissue and cell distribution of a mammalian proteasomal ATPase, MSS1, and its complex formation with the basal transcription factors. Biochem. Biophys. Res. Commun. 2000;279:568–573. doi: 10.1006/bbrc.2000.3969. [DOI] [PubMed] [Google Scholar]
- [214].Wu SY, Thomas MC, Hou SY, Likhite V, Chiang CM. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J Biol Chem. 1999;274:23480–23490. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- [215].Lavigne AC, Mengus G, Gangloff YG, Wurtz JM, Davidson I. Human TAF(II)55 interacts with the vitamin D(3) and thyroid hormone receptors and with derivatives of the retinoid X receptor that have altered transactivation properties. Mol Cell Biol. 1999;19:5486–5494. doi: 10.1128/mcb.19.8.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Verrier CS, Roodi N, Yee CJ, Bailey LR, Jensen RA, Bustin M, Parl FF. High-mobility group (HMG) protein HMG-1 and TATA-binding protein-associated factor TAF(II)30 affect estrogen receptor-mediated transcriptional activation. Mol Endocrinol. 1997;11:1009–1019. doi: 10.1210/mend.11.8.9962. [DOI] [PubMed] [Google Scholar]
- [217].Petty KJ, Krimkevich YI, Thomas D. A TATA binding protein-associated factor functions as a coactivator for thyroid hormone receptors. Mol Endocrinol. 1996;10:1632–1645. doi: 10.1210/mend.10.12.8961272. [DOI] [PubMed] [Google Scholar]
- [218].Mengus G, May M, Carré L, Chambon P, Davidson I. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- [219].Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- [220].May M, Mengus G, Lavigne AC, Chambon P, Davidson I. Human TAF(II28) promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- [221].Sun L, Johnston SA, Kodadek T. Physical association of the APIS complex and general transcription factors. Biochem Biophys Res Commun. 2002;296:991–999. doi: 10.1016/s0006-291x(02)02026-0. [DOI] [PubMed] [Google Scholar]