Abstract
In mammals, there are three neurofilament (NF) subunits (NF-L, NF-M, and NF-H), but it was thought that only a single NF, NF180, exists in lamprey. However, NF180 lacked the ability to self-assemble, suggesting that like mammalian NFs, lamprey NFs are heteropolymers, and that additional NF subunits may exist. The present study provides evidence for the existence of a lamprey NF-L homolog (L-NFL). Genes encoding two new NF-M isoforms (NF132 and NF95) also have been isolated and characterized. With NF180, this makes three NF-M-like isoforms. In situ hybridization showed that all three newly cloned NFs are expressed in spinal cord neurons and in spinal-projecting neurons of the brainstem. Like NF180, there were no KSP multiphosphorylation repeat motifs in the tail regions of NF132 or NF95. NF95 was highly identical to homologous parts of NF180, sharing 2 common pieces of DNA with it. Northern blots suggested that NF95 may be expressed at very low levels in older larvae. The presence of L-NFL in lamprey CNS may support the hypothesis that as in mammals, NFs in lamprey are obligate heteropolymers, in which NF-L is a required subunit.
Index terms: neurofilament, molecular biology, NF180, NF132, NF95, L-NFL, cytoskeleton, axonal regeneration
INTRODUCTION
The tips of regenerating giant reticulospinal axons in the lamprey differ from embryonic growth cones because they lack filopodia and lamellipodia, and are filled with densely packed neurofilaments (NFs) [20,21,32,40,47]. This and other findings [19,31] have suggested that NFs may be involved in the mechanism of axon regeneration, and have spurred interest in the structure of NFs in the lamprey. NFs belong to the intermediate filament (IF) family of cytoskeletal proteins [25,55]. Three subunits (NF-L, NF-M, and NF-H) are found in mammalian nervous systems. None of the NFs can self-assemble in cell lines lacking a preexisting IF network [8,38], although NF-L can form short homopolymeric filaments in vitro. NF-M and NF-H need the presence of NF-L to form normal filaments [16,39] and cannot self-assemble or form filaments with each other. All NFs consist of a head domain at the amino terminus, a highly conserved rod region (~10 kDa), and a sidearm of varying length at the carboxy terminus. The latter accounts for the differences in the sizes of NF subunits.
Bodian-stained gel electrophoresis suggested that only one NF exists in lamprey [36]. This was supported later by immunoblotting with anti-NF antibodies [48] and cDNA cloning [30]. The cloned peptide, NF180 had rod and sidearm epitopes characteristic of each of the three mammalian subunits [48], although its amino acid sequence was most similar to that of human NF-M [30]. However, in tests performed in vitro and in transfected cultured cells, NF180 lacked the ability to form typical filaments [67]. This suggested that lamprey neurons might contain a previously unsuspected factor required for NF assembly, e.g., a second IF protein. Although a 50kDa IF, nIF50, has been characterized immunochemically and localized to neurons [33], its nucleotide and amino acid sequences are unknown, and there is no evidence that it is a member of the NF family. The present study provides evidence for a lamprey homolog of NF-L and for two additional NF-M homologs, none of which is nIF50.
EXPERIMENTAL PROCEDURES
Molecular cloning of a lamprey NF-L gene
The NCBI trace archive - Petromyzon marinus-WGS (Whole Genome Shotgun, http://www.-ncbi.nlm.nih.gov/Traces/trace.cgi?) was searched by MegaBlast against the rat NF-L nucleotide sequence (Accession number: AF031880, 2032 bp) [7]. This led to discovery of one 843 bp of DNA fragment (TI number: 1229593736) encoding for a 273 amino acid peptide that showed a high homology to NF-L of other species. Several primers were designed to clone the full length of this neurofilament by PCR, using the Uni-ZAP XR cDNA library as template. Sequences in upstream regions were determined using the T3 primer (5’-AATTAACCCTCACTAAAGGG-3’, sense) and the primer 5’-GGAGTGCGTGCGTCTGAACGAGC-3’ (antisense), which covers a small region (86 bp) of the pBluescript II vector and 211 bp of a known sequence at the 5’-end. Primers 5’-GAAGGTGCAGCAGGAGGAGATCG-3’ and 5’-AACGCCAGGGTTTTCCCAG-TCACGACGTTG-3’ were used to amplify the 3’ end of known regions (1306 bp) of the gene and small regions (99 bp) of the vector. Sequencing this 1.4 kb cDNA fragment revealed that a poly-A sequence was included. The original 843 bp region of the gene was confirmed by sequencing PCR products generated using primers 5’-ATGAGCTCCTACACATCCGACTCTGCTGT-3’ and 5’-CCTGTACGCTGCAATTTCTATGTCCAATG-3’. Each primer was analyzed to avoid a high degree of homology to the determined sequences such as NF180, NF132, and NF95.
Screening of cDNA library and nucleotide sequence analysis
An oligo(dT) primed Uni-ZAP cDNA library constructed from polyadenylated RNA isolated from the brain and spinal cord of larval wild-type Petromyzon marinus lampreys was screened with a prelabeled cDNA fragment of cloned lamprey neurofilament NF180. The library was prepared by Stratagene Corp. with cloning sites EcoRI and XhoI. The cDNA fragment was generated by PCR using the primers 5’-AACGACCGCTTCGC-3’/5’-TTGAGCAGGTCCTGGTA-3’ and an NF180 [30] cDNA template, covering the most conserved rod regions of neurofilaments. PCR amplification was performed using the Expand™ Long Template PCR System (Boehringer Mannheim). The temperature profile for the PCR consisted of an initial denaturation step for 5 minutes at 94°C, followed by 34 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 55°C, extension for 1 minute at 72°C, with a final 7-minute extension at 72°C. Following amplification, PCR fragments of expected size (443–1293 of NF180, 851 bp) were purified on 1.5% agarose gels and ligated into pGEM®-T Easy vector (Promega) for subsequent characterization and preparation. Sequencing was carried out at the Cell Center, University of Pennsylvania. The insert was isolated by restriction enzymes and used as templates for probe preparations [Ready-To-Go DNA Labeling Beads (−dCTP), GE Healthcare, UK]. Synthesized 32P-probes were purified by passage through G-50 micro-columns (ProbeQuant™ G-50 Micro Columns, Pharmacia Biotech). Duplicate lifts of one million plaques yielded 33 positive phage clones. All of them were processed by in vivo excision as described by the manufacturer (Stratagene manual BN#937111). Sequencing was performed and the final sequences were determined for both cDNA strands.
In situ hybridization with digoxigenin-labeled riboprobes
Hybridization of digoxigenin-labeled riboprobes to sectioned and wholemounted lamprey brainstem was performed using techniques reported previously [58]. For wholemount preparations, brain or spinal cord was removed, stripped from the overlying meninx primitiva and choroid plexus, pinned flat on Sylgard strips, fixed in 2% paraformaldehyde, washed in phosphate-buffered saline (PBS) and stored in 70% ethanol at 4°C. For histological sectioning, tissue was cut into 1–2 cm lengths, fixed in 4% paraformaldehyde, washed in PBS, dehydrated in serial ethanols, cleared in toluene, infiltrated with Paraplast, and then embedded in paraffin. Ten µm transverse paraffin sections were collected, deparaffinized, and rehydrated in xylene and serial ethanols.
Digoxigenin-labeled riboprobes were constructed based on alignment analysis so that the most specific regions of the gene were amplified by PCR. Because NF95 shares almost all its DNA sequences with NF180, the probe for NF95 could not distinguish mRNA message of NF95 from NF180. Therefore we named this probe “NF95/NF180.” Four pairs of primers were used in PCR to amplify the specific regions of each NF: primers 5’-TCCCGCCTGACCATCAGCTCCTCCT-3’/5’-GACGTTAACACGTTCA-ACATGCACT-3’ for L-NFL, primers 5’-GCAAGGAGCCCAAGAAAGCCGTCAA-3’ /5’- TTTTTTTCTTTAACATTCTGTTTTC-3’ for NF95/NF180, primers 5’-AGGCCGGAGCTGAGGAAGAGGACGA-3’/5’-CTGCTTCTGTGTCGTCTTGTTCAGC-3’ for NF132, and primers 5’- GCCCCAGAGCCAAAAGCAGCTCCCA-3’/5’-TCTTCCTCCTCTTTGGCCGGGGCGG-3’ for NF180. The PCR products were ligated into pGEM®-T Easy vector (Promega) for subsequent sequencing. They have been proved to be 100% identical to nucleotides 1232–1620 of L-NFL, 1829–2245 of NF95, 1975–2261 of NF132, and 2510–2967 of NF180 by sequencing. The vectors were then digested with the NotI or NcoI restriction enzyme to form the templates for sense/antisense probe synthesis. In vitro transcription was performed with a RNA transcription kit (Roche, Nutley, NJ) as recommended by the manufacturer. The T7 or SP6 RNA polymerase was used for the sense or antisense probes, respectively.
Both wholemounted and sectioned tissues were pre-hybridized at 65°C in hybridization solution (50% deionized formamide, 5 × SSC, 100 pg/ml Torula yeast RNA, 100 pg/ml wheat germ tRNA, 50 pg/ml heparin, 0.1% Tween-20) followed by hybridization overnight at 65°C in the same solution plus 400 ng/ml digoxigenin-labeled cRNA. Specimens were washed in hybridization solution at 65°C followed by room temperature washes in PTw (0.1% Tween-20 in PBS), and PBT (0.1% bovine serum albumin, 0.2% Triton X-100 in PBS). Alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (0.75 U/ml, Roche, Nutley, NJ) were diluted (1:1,000) and applied to tissue overnight at 4°C. Tissue was washed sequentially in PBT and SMT (100 mM NaCl, 50 mM MgCl2, 100 mM Tris, pH 9.5, 0.1% Tween-20). The chromogenic reaction was carried out in ice-cold SMT containing 175 pg/ml 5-Bromo-4-chloro-3-indolylphosphate and 350 pg/ml 4-Nitro blue tetrazolium chloride for 30 minutes on ice in the dark. Finally, specimens were washed in PBS, dehydrated in serial ethanols, cleared with xylene and mounted under Permount (Fisher scientific).
Protein expression and Western blotting
To test if cloned L-NFL represents the same protein as nIF50, the neuronal intermediate filament protein previously characterized immunochemically [33], the L-NFL cDNA was ligated into HindIII/XbaI sites of pRC/CMV, an eukaryotic expression vector (Invitrogen) after PCR amplification with primers 5’- TTTAAGCTTCACCATGAGCTCCTACACATCCGACTCTGC -3’/5’-ATTTCTAGATATCAATGTGACTGGTGGGATTCGTC-3’ to form L-NFL-pRC/CMV. To identify L-NFL expression, a c-Myc epitope (10 amino acids, EQKLISEEDL) was added to the amino-terminal by including the c-myc sequence in the PCR primers. Human adrenal carcinoma cells (SW13cl.2Vim−) were cultured and transiently transfected with the myc-tagged L-NFL construct. The transfected cells were cultured at 37 °C for 48 hrs and harvested by centrifugation. The cell pellet was suspended and lysed by sonication in a lysis buffer for 2 minutes. Thirty µg of protein was separated in a 10% SDS mini-gel and was transferred electrophoretically onto nitrocellulose using a Bio-Rad transblot apparatus. An equal amount of cytoskeletal protein prepared from lamprey spinal cord [48] was used as a positive control. The membranes then were pre-incubated in 10% non-fat dry milk in PBS buffer containing 0.1% Tween-20. The blots were incubated with either a rabbit polyclonal antibody to c-Myc (sc-789) or LCM40 monoclonal antibody, which was previously shown to bind specifically to nIF50 [33]. The membranes were then washed and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit IgG secondary antibody (Santa Cruz Biotech., Santa Cruz, CA) for 1 hour. After washing with PBS containing 0.3% Tween-20 for 30 min, immunoreactive proteins were visualized using the enhanced chemiluminescence (ECL) kit.
To estimate molecular weights of the other newly cloned NFs in SDS gels, the full length NF132 or NF95 sequence was inserted into the pET 11b (Novagen) expression vector, which was transformed into BL21(DE3)pLysS cells (Novagen). The culture was incubated with ampicillin in a shaking incubator at 225 rpm at 37°C overnight. Expression was induced by the addition of 5 ml of IPTG (0.2 M; Fisher, Fair Lawn, NJ) per L of culture. The pre- and post-induction aliquots (2 ml) were collected by spinning down for 10 minutes at 5000 rpm. Twenty µl of 2 × SDS sample buffer (0.125 M Tris-chloride, pH 6.8, 4% SDS, 20% glycerol and 10% 2-mercaptoethanol) was added to the pellets. The samples were then boiled for 5 minutes and loaded on gels. The gels were stained with Coomassie Blue R-250.
Northern blotting
Total RNA was isolated from brain and spinal cord of large larval (13–14 cm long) and adult (16–17 cm long) sea lampreys, Petromyzon marinus, as previously described [9]. Muscle also was collected for controls. RNA samples (15 µg/lane) were separated on a 1% agarose gel containing 2.2 M formaldehyde, 200 mM sodium acetate, and 5 mM EDTA. The gel was stained with ethidium bromide and visualized under UV light. After electrophoresis, RNA was blotted onto a nylon membrane overnight and baked for 2 hours at 80°C. The membrane was then pre-hybridized for 4 hr at 42 °C. Four cDNA fragments (423, 471, 321, and 492 bp) were cut from the pGEM®-T Easy vectors with inserts of PCR products for L-NFL, NF95/NF180, NF132, and NF180 with NotI and NcoI and used as templates for probe preparations [Ready-To-Go DNA Labeling Beads (-dCTP), GE Healthcare, UK]. Synthesized 32P-probes were purified by passage through G-50 micro-columns (ProbeQuant™ G-50 Micro Columns, Pharmacia Biotech). Following prehybridization, the buffer was replaced with 10 ml of hybridization buffer containing 2–4 × 106 dpm/ml of 32P-labeled cDNA probe. Membranes were incubated for 24 hours at 42°C in this solution, followed by washing twice in 100 ml of high-salt buffer [300 mM NaCl, 30 mM citric acid (pH 7.0), and 0.1% (wt/vol) SDS] for 20 minutes at room temperature and then twice in 100 ml of low-salt buffer [30 mM NaCl, 3 mM citric acid (pH 7.0), and 0.1% (wt/vol) SDS] at 65°C for 20 minutes. Membranes were exposed to X-OMAT AR film (Kodak) at −70°C with intensifying screens.
DNA sequencing, and DNA/protein sequence analysis
The reaction cocktail (approximately 1200 ng plasmid DNA and 8.0 pmol primer) was sent to a DNA Sequencing Facility (Cell Center, University of Pennsylvania, Philadelphia, PA) and analyzed on an ABI 3730XL DNA Analyzer (Applied Biosystems). Nucleotide or protein sequence alignments were carried out using a combination of Blastn, Blastx, and Discontiguous Mega Blast programs (NCBI, Bethesda, MD), and the programs Clustal X [62], GeneDoc [42] and BioEdit [22]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 [61].
RESULTS
Molecular cloning of lamprey NF-L
The DNA sequence of rat NF-L (AF031880) [7] was used for MegaBlast search of an NCBI trace archive (Petromyzon marinus-WGS), which led to the identification of a single 843 bp DNA fragment (TI number: 1229593736). Based on this DNA sequence, lamprey NF-L (L-NFL) was cloned using several primers and a Uni-ZAP XR cDNA library as template (Figure 1).
Figure 1. Nucleotide and predicted amino acid sequence of the lamprey neurofilament protein L-NFL.

Translation was begun at the first in-frame methionine of the longest open reading frame and terminated by the stop codon “TAA”. Nucleotide numbers are indicated at the beginning of each DNA sequence line. The protein sequence before the first bracket is the head region and that after the last bracket is the sidearm (tail) region. Protein sequence between the first and the last bracket is rod region, which consists of 3 coils (coil Ia, Ib, and II). In each coil, stars (*) mark hydrophobic residues at the first and fourth amino acid of putative heptad repeats, while exes (x) mark charged residues. The underlined region is the nucleotide sequence used for Northern blotting or in situ hybridization. The amino acids “GVPVG” in bold and underlined in the head region represents a sequence unique to L-NFL as compared with other NF-Ls listed in Table 1A.
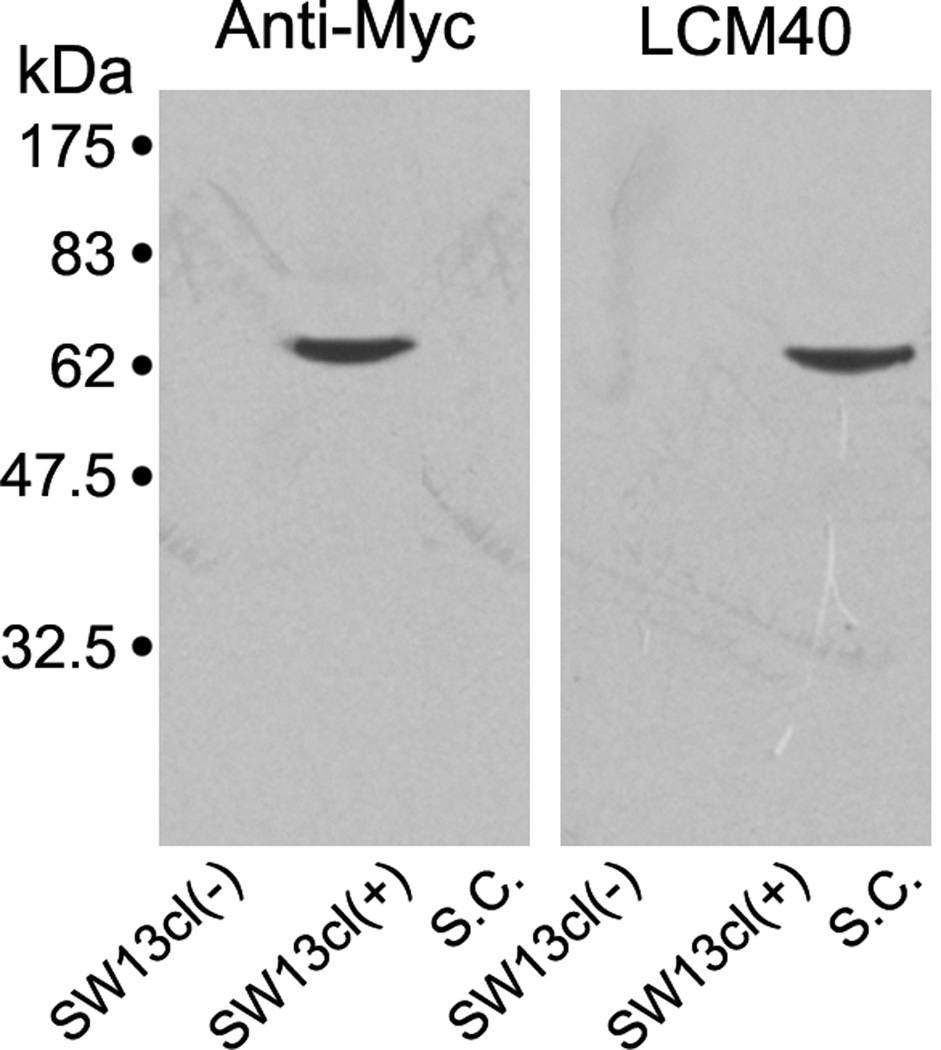
The L-NFL clone consisted of 1918 bp, with 22 bp 5’ untranslated region, and 1425 bp open reading frame encoding a 474 amino acid peptide with the putative TAA stop codon. It extended 471 bp further as a 3’ untranslated region, which contained a poly(A) tail (Figure 1). Estimation of the protein size is 52.7 KDa, and the apparent MW in SDS gel is around 64 kDa (Figure 2). To determine whether L-NFL is the same protein, nIF50, that we previously characterized immunochemically [33], we tagged L-NFL with c-Myc and expressed it in SW13cl.2Vim− cells, which had been used in a study of NF assembly [67]. Successful expression was confirmed by Western blot with anti-Myc antibody. However LCM40 did not bind to expressed L-NFL (Figure 2).
Figure 2. Recombinant L-NFL was not detected by LCM40 in Western blots.
The c-Myc-tagged L-NFL recombinant was expressed in SW13cl.2Vim− cells. Lysates of the cells with [SW13cl(+)] or without [SW13cl(−)] expression vector were separated on SDS-PAGE with cytoskeletal proteins prepared from lamprey spinal cord (S.C.), and transferred to nitrocellulose membrane. The membranes were then incubated with anti-Myc (Left) or LCM40 (Right) antibody. A band (~ 64 kDa) was labeled by anti-Myc but not by LCM40.
Molecular cloning of NF132 and NF95
Screening of a Petromyzon marinus brain Uni-ZAP XR cDNA library at low stringency with a fragment of NF180 cDNA probe led to the isolation of 33 positive phage clones, which hybridized strongly to the NF180 fragment. Partial sequencing indicated that one of them was 100% homologous with NF180 and another was a vimentin-like protein, which had 82% identity to European river lamprey vimentin (AJ278487) over a 250 amino acid sequence. Two others, designated as NF132 (DQ398933, Figure 3) and NF95 (DQ398934, Figure 4), contained a putative in-frame “ATG” start codon, a completed open reading frame and a stop codon.
Figure 3. Nucleotide and predicted amino acid sequence of the lamprey neurofilament protein NF132.
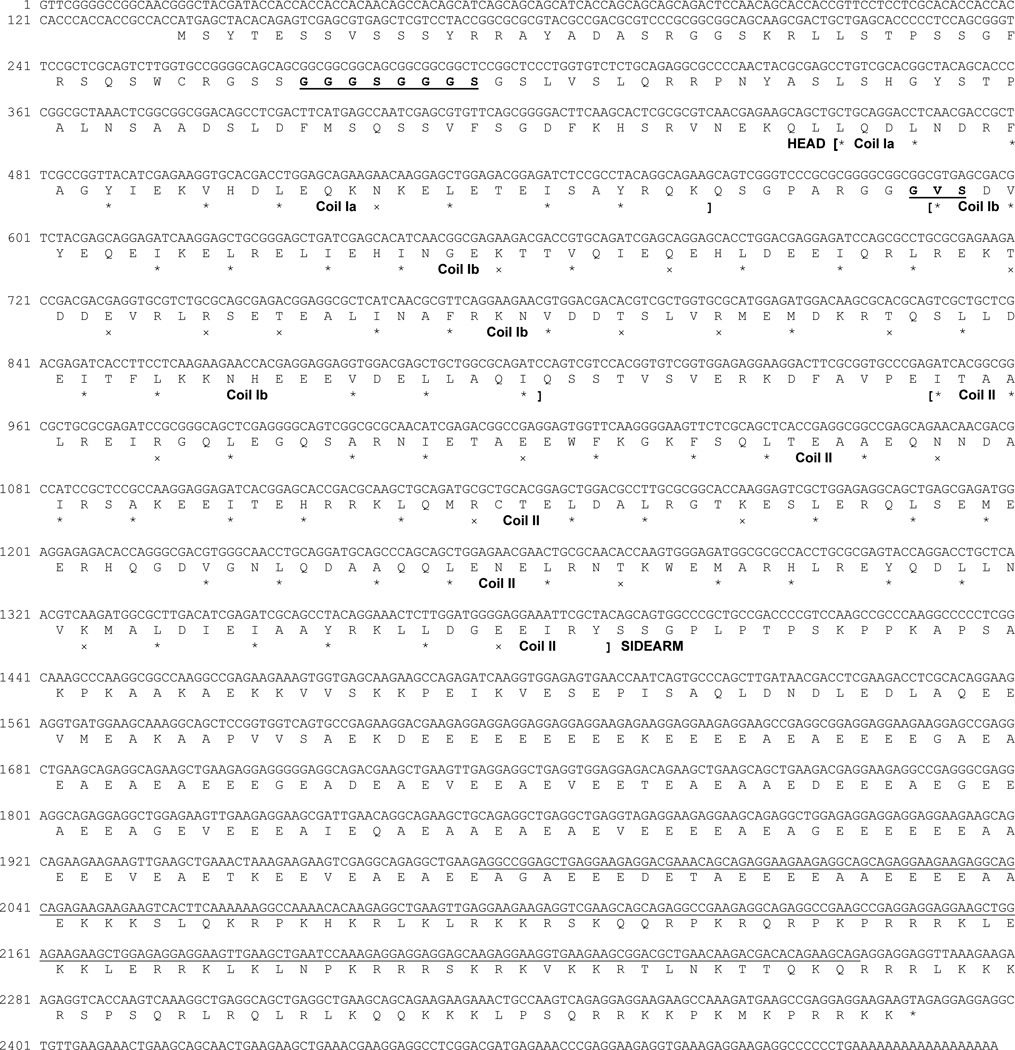
Translation was begun at the first in-frame methionine of the longest open reading frame and ended by a “TAG” stop codon. Nucleotide numbers are indicated at the beginning of each DNA sequence line. Amino acids “GGGSGGGS” and “GVS” in bold and underlined in the rod region indicates sequences unique to lamprey NF132 as compared with other NF-Ms listed in Table 1B, C.
Figure 4. Nucleotide and predicted amino acid sequence of the lamprey NF protein NF95.

Translation was begun at the first in-frame methionine of the longest open reading frame and terminated by a “TGA” stop codon. A potential polyadenylation signal sequence “ATTAAA” in the 3’ unstranslated region is in bold. Note the amino acids “GGGS” and “GVS” in bold and underlined in the rod region. They represent inserted sequences unique to lamprey NF95 and NF180 as compared with other NF-Ms listed in Table 1B, C. Explanations for other marks are the same as in Fig. 1.
The NF132 sequence consisted of 2518 bp, with a 137 bp 5’ untranslated region, and a 2253 bp open reading frame coding for a 750 amino acid peptide and terminating with a “TAG” stop codon. It extended 128 bp further into a 3’ untranslated region which contained a poly(A) tail (Figure 3). NF95 had a full length of 2245 bp, with a 132 bp 5’ untranslated region, a 1842 bp open reading frame coding for a 613 amino acid peptide and terminating with a “TGA” stop codon. It extended 271 bp further into a 3’ untranslated region, which contained a potential polyadenylation signal “ATTAAA” (nucleotides 2199–2204) [3] (Figure 4).
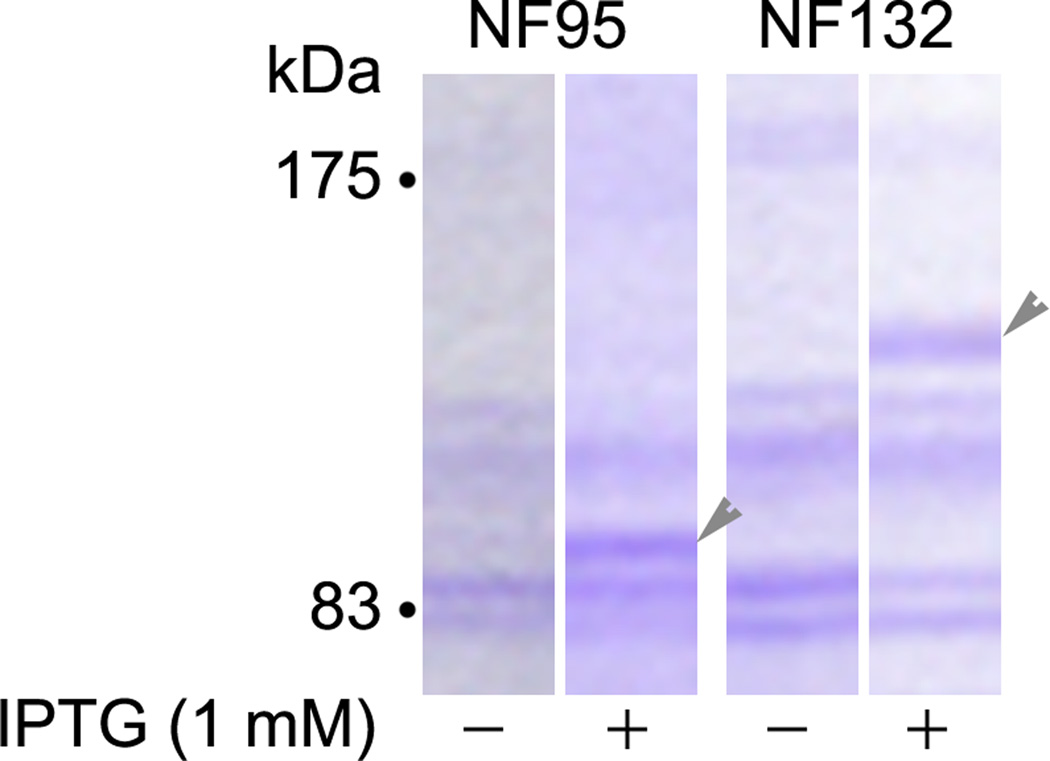
From the open reading frame, NF132 is predicted to be an 84 kDa peptide and NF95 a 68 kDa peptide. We have subcloned NF132 and NF95 into the pET 11b (Novagen) expression vector and transfected them into BL21(DE3)pLysS cells (Novagen). Expression was induced by IPTG and separated in SDS gels. Staining of the gels with Coomassie Blue R-250 revealed apparent MWs of approximately 132 and 95 kDa, respectively (Figure 5). A summary of the cloned lamprey NFs is shown in Table 2.
Figure 5. Estimation of molecular weights of recombinant NF95 and NF132 in Coomassie Blue-stained SDS gels.
The NF132 and NF95 genes were subcloned into the pET-11b (Novagen) expression vector and transfected into BL21(DE3)pLysS cells (Novagen). The cultures were harvested after incubation at 37°C overnight. Expression was induced by IPTG and controls were set without it. The cultures were allowed to grow for another 2 hours and lysed in homogenization buffer. The lysate proteins were separated on a 10% SDS-PAGE, stained with Coomassie Blue R-250 (0.1% in 50% methanol and 10% glacial acetic acid) and destained with 10% methanol and 7% acetic acid. A 95 kDa protein was expressed in NF95-transfected, IPTG-induced cells, while a 132 kDa protein was expressed in NF132-transfected, IPTG-induced cells (arrowheads).
Table 2. Amino acid numbers in the head, rod and sidearm of lamprey NFs.
The total number of amino acids in each NF subunit is in parentheses.
| Lamprey NF | Head | Core | Sidearm |
|---|---|---|---|
| NF180 (1110) | 103 | 311 | 696 |
| NF132 (750) | 107 | 311 | 332 |
| NF95 (613) | 103 | 311 | 199 |
| NFL (474) | 99 | 307 | 68 |
Positions of lamprey NFs in the phylogenetic tree
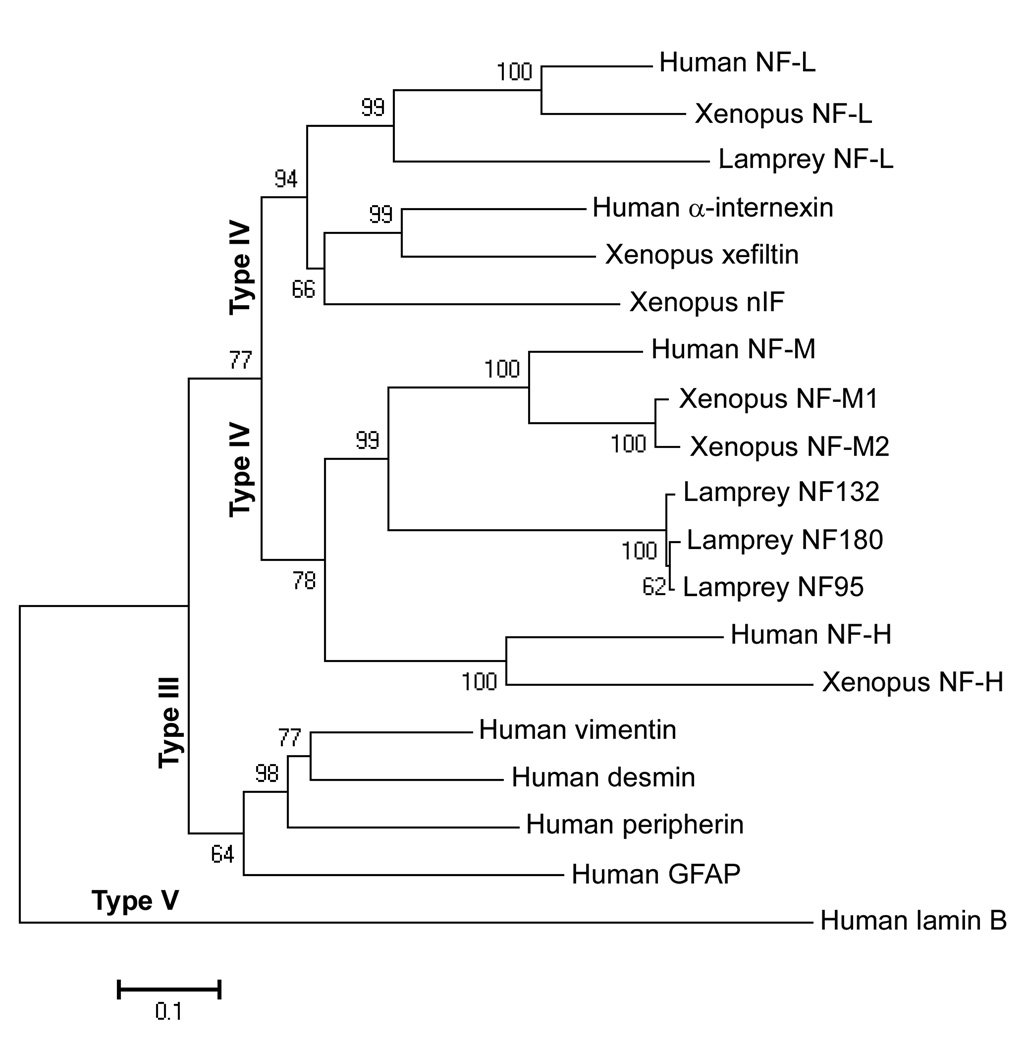
Different classes of IFs have characteristic amino acid sequences [46]. To confirm that the three newly cloned proteins are members of the type IV NF family, NF180 and the newly cloned NF sequences were aligned by Clustal X with human class III (vimentin, desmin, GFAP, and peripherin), class IV (α-internexin, NF-L, NF-M, NF-H), and class V (lamin B) IFs. Xenopus class III and IV IF protein sequences were used as a reference. Protein sequence alignments were conducted to construct an NJ tree (Figure 6) using MEGA version 4. The phylogenetic tree suggested that the newly cloned NF95 and NF132 and the previously cloned NF180 belong to the NF-M group, while the newly cloned L-NFL is related to the NF-L group. Within this clade, the branching order of human NF-L, Xenopus NF-L and lamprey NF-L mirrors the known phylogenetic relationships among mammals, frogs, and lampreys, respectively. It also is true for the subtree of NF-Ms. These relationships have high bootstrap support (99–100%, Figure 6). An additional conclusion is that in subtrees of the NF-L group, the relative branch lengths are comparable to those within the NF-M clades, which suggests that NF-L and NF-M are derived from different orthologs.
Figure 6. Positions of lamprey NFs in phylogenetic trees constructed with human and Xenopus IFs.
Amino acid sequences of 9 human IFs, 6 Xenopus IFs, 1 known lamprey NF (NF180), and 3 putative lamprey NFs (NF132, NF95, and L-NFL) were aligned using the Clustal X tool in BioEdit. The 19 nucleotide sequences obtained from GenBank (accession numbers in parentheses) were: Human desmin (NM_001927), Human GFAP (NM_001131019), Human lamin B (NM_032737), Human NF-H (NM_021076), Human NF-L (NM_006158), Human NF-M (NM_005382), Human peripherin (NM_006262), Human vimentin (NM_003380), Human α-internexin (NM_032727), Lamprey NF132 (DQ398933), Lamprey NF180 (U19361), Lamprey NF95 (DQ398934), Lamprey NF-L (DQ869188), Xenopus NF-H (BC136056), Xenopus NF-L (M86654), Xenopus NF-M1 (U85969), Xenopus NF-M2 (U85970), Xenopus nIF (M86653), Xenopus xefiltin (U63711). Amino acid sequences were inferred from the nucleotide sequences with the ORF finder program of NCBI. A bootstrap test was conducted to construct the NJ tree using MEGA version 4 {Tamura, 2007 #66} (Publication PDF at http://www.-kumarlab.net/publications). Numbers to the left of the branch points indicate the percent of 10,000 bootstrap replicates that support that branch. Lengths in the tree reflect distances between taxa (mean substitutions per residue). Human lamin B was chosen to provide an outgroup for rooting the IF tree because cytoplasmic IF proteins are believed to be derived from an ancestral nuclear lamin {Dodemont, 1990 #17}. Scale: 0.1 amino acid substitutions per site.
We have compared the similarity of amino acid sequences of L-NFL with that of other NF-Ls and the amino acid similarity of NF132 and NF95 with that of other NF-Ms. The highest identity (98–99%) was found in rod regions of lamprey NF-Ms (Table 1B, C). Identity across species was 53–64%. The same was true comparing L-NFL and NF-Ls of other species (Table 2A). Alignment of the rod regions of lamprey NFs with those in other species is shown in Figure 7. In the head region, identities among the lamprey NF-Ms were 94–98%, and between lamprey and other species 20–40%. Identities of L-NFL and other NF-Ls also were 20–40%. The sidearm region had the highest variability, with only 3–18% sequence identities.
Table 1. Comparisons of amino acid sequence identities between newly cloned lamprey NFs and previously identified NFs within different regions of the protein.
A. The deduced amino acid sequence of lamprey NF-L was compared to the NF-Ls of other species. Sequences were aligned first to identify the tripartite regions (Head-Rod-Sidearm). For each individual region, sequences were aligned and the identities were calculated using the tool “sequence identity matrix” in BioEdit. H-NFL: Human NF-L (BC039237); R-NFL: Rat NF-L (NM_031783); M-NFL: Mouse NF-L (DQ201635); C-NFL: Chicken NF-L (XM_417679); X-NFL: Xenopus NF-L (M86654); X-nIF: Xenopus nIF (M86653); Z-NFL: Zebrafish NF-L (BC114232); L-NFL: Lamprey NF-L (DQ869188). B & C: The deduced amino acid sequences of lamprey NF132 (B) or NF95 (C) proteins were compared to NF-Ms of other species. H-NFM: human NF-M (NM_005382); R-NFM: Rat NF-M (NM_017029), M-NFM: Mouse NF-M (NM_008691); C-NFM: Chicken NF-M (NM_001101730); NF180: Lamprey NF180 (U19361); X-NFM1: Xenopus NF-M1(BC074454); X-NFM2: Xenopus NF-M2 (BC078128); Z-NFM: Zebrafish NF-M (BC133105); NF132: Lamprey NF132 (DQ398933); NF95: Lamprey NF95 (DQ398934).
| A. L-NFL vs. other NF-Ls | Percentage Sequence Identity (%) | |||
|---|---|---|---|---|
| Head | Rod | Tail | Overall | |
| H-NFL | 35 | 63 | 9.0 | 44 |
| R-NFL | 35 | 63 | 9.8 | 45 |
| M-NFL | 36 | 63 | 9.7 | 45 |
| C-NFL | 34 | 64 | 8.3 | 43 |
| X-NFL | 30 | 59 | 10 | 41 |
| X-nIF | 27 | 54 | 11 | 41 |
| Z-NFL | 25 | 54 | 7.5 | 34 |
| B. NF132 vs. other NF-Ms | Percentage Sequence Identity (%) | |||
|---|---|---|---|---|
| Head | Rod | Tail | Overall | |
| H-NFM | 36 | 64 | 13 | 32 |
| R-NFM | 36 | 64 | 13 | 35 |
| M-MFM | 37 | 64 | 14 | 35 |
| C-NFM | 34 | 64 | 15 | 33 |
| NF180 | 94 | 98 | 30 | 55 |
| NF95 | 96 | 99 | 39 | 72 |
| X-NFM1 | 28 | 62 | 14 | 33 |
| X-NFM2 | 34 | 60 | 16 | 33 |
| Z-NFM | 22 | 53 | 13 | 31 |
| C. NF95 vs. other NF-Ms | Percentage Sequence Identity (%) | |||
|---|---|---|---|---|
| Head | Rod | Tail | Overall | |
| H-NFM | 38 | 64 | 11 | 31 |
| R-NFM | 37 | 64 | 12 | 33 |
| M-MFM | 38 | 64 | 12 | 33 |
| C-NFM | 35 | 64 | 13 | 32 |
| NF180 | 98 | 98 | 28 | 54 |
| X-NFM1 | 29 | 62 | 8.7 | 30 |
| X-NFM2 | 35 | 60 | 10 | 31 |
| Z-NFM | 23 | 53 | 9.6 | 29 |
Figure 7. Alignment of lamprey NFs with known vertebrate NF sequences.
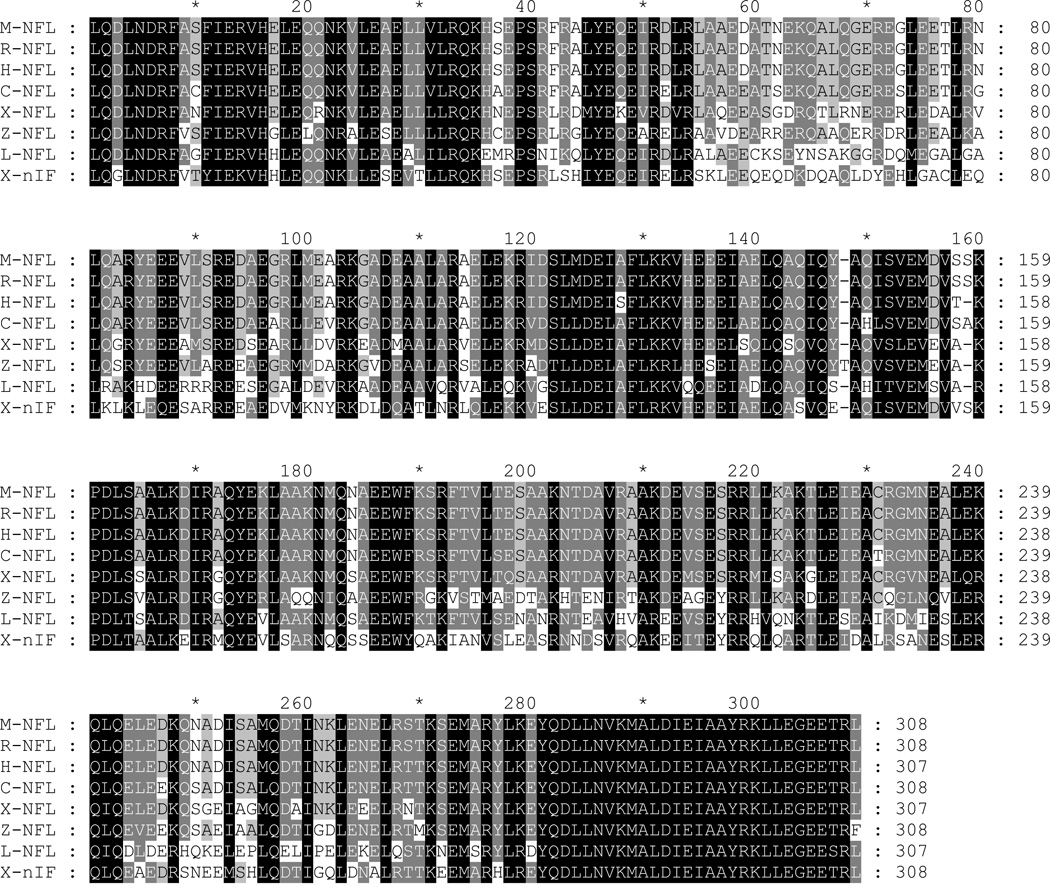
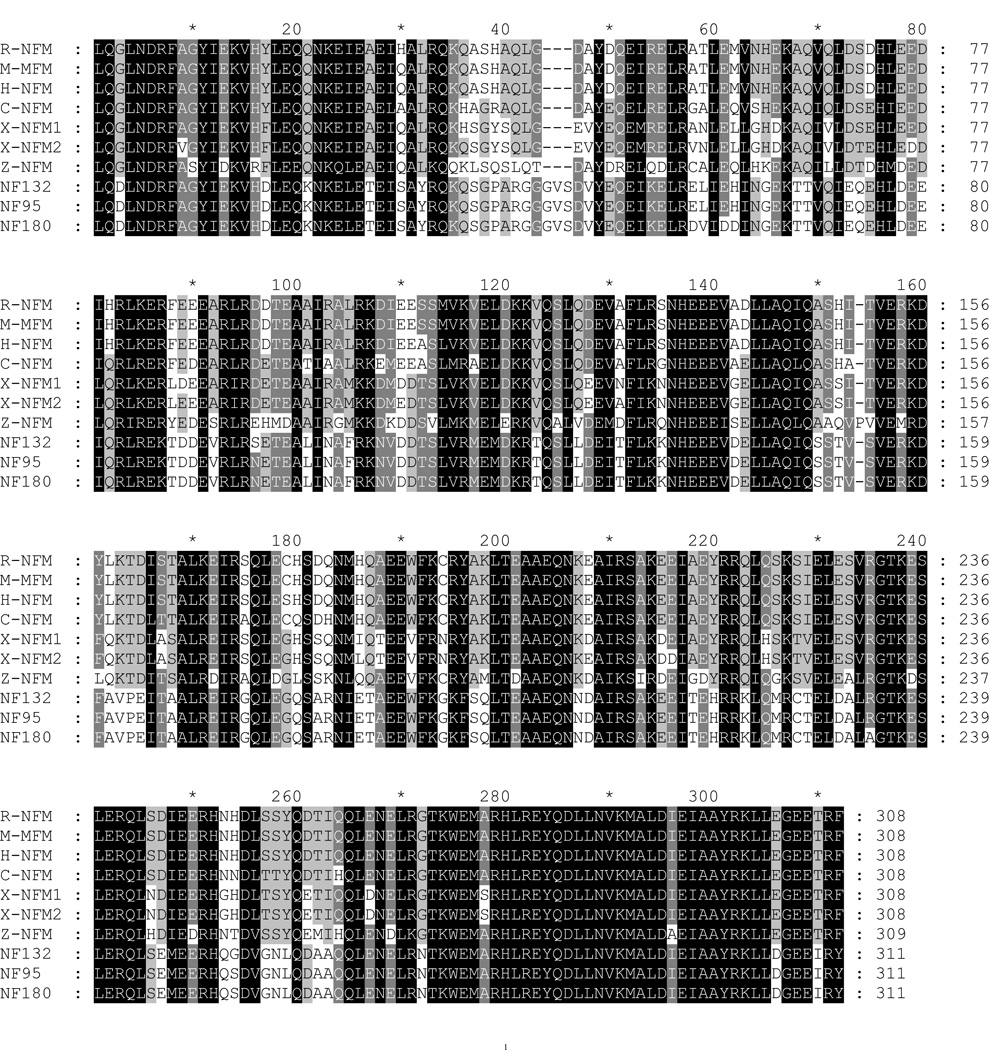
The rod regions of individual NFs were aligned with Clustal X. The generated MSF file was opened and edited with GeneDoc. Conserved amino acid residues are shaded black. Conservative substitutions are in gray. Abbreviations are as in Table 1. A, the amino acid sequence in the rod region of lamprey NF-L was aligned with NF-Ls of other species. B, the amino acid sequences in the rod regions of lamprey NF180, NF132, and NF95 were aligned with NF-Ms of other species. Note the unique sequence “GVS” inserted into the lamprey NF-Ms at position 44–46. At the carboxyl end, a conservative substitution of “D” for “E” occurs at the fourth position of “KLLEGEE” in the lamprey NF-Ms (306th position in the present sequence).
Like other IF protein sequences, the central rod domain of the all the newly cloned lamprey NFs contained a quartet of heptad repeats (a–b–c–d–e–f–g) (lA, lB, 2A, 2B), in which the first “a” and fourth “d” amino acids were non-polar (uncharged), while polar (charged) amino acids occupied the other positions (Figures 1, 3, and 4). In addition, the rod domain contained a highly conserved sequence “LNDRF” at the amino end (Figure 7A, 7B). The carboxyl end of the rod domain of NF-Ms was highly conserved among all species, except for a conservative substitution of aspartate (D) for glutamate (E) and isoleucine (I) for threonine (T) at positions 306 and 310, respectively (Figure 7B) in lamprey NF-Ms. Within the segment linking helices 1A and 1B (L1) there were three amino acids (glycine-valine-serine) not found in any of the mammalian, amphibian or fish NF-M sequences investigated thus far (Figure 7B). There were no such insertions in L-NFL, which maintained a highly conserved carboxy terminus sequence at the end of the rod domain (Figure 7A).
We also compared lamprey NFs at the DNA level and found that NF95 is highly homologous to NF180. If NF180 (1–3725, n.t.) was divided into three fragments I, II, and III as in Figure 8, NF95 was composed of fragment I and III, but lacked fragment II (1821–3308, n.t.). The identity between the Fragment I of NF180 (1–1820, 1820 bp) and that of NF95 (15–1828, 1814 bp) was 99.0%. They differed only in the deletion of 6 nucleotides in three loci within NF95, as indicated in Figure 8. Because the deletion of the first 3 nucleotides “ACC” occurred before the start codon (Figure 8A), there was no impact on the open reading frame (ORF). The second and third deletions occurred within the ORF. Deletion of an “A” caused a frameshift mutation that was corrected in the subsequent seven nucleotides because of the deletion of two more nucleotides “CG”. Two deletions resulted in a small change in amino acid sequence from “…EDRGRKE…” of NF180 to “…EAEAEE…” of NF95. Identity for fragment III (3309–3725 in NF180; 1829–2229 in NF95) was 99.7%. Fragment I codes the complete head domain, rod domain and partial sidearm, as indicated in Figure 8. NF132 has much lower homology to NF180 and NF95 (84% and 93%, respectively) because it has many gaps when aligned with them.
Figure 8. Schematic representations of lamprey NF cDNAs.
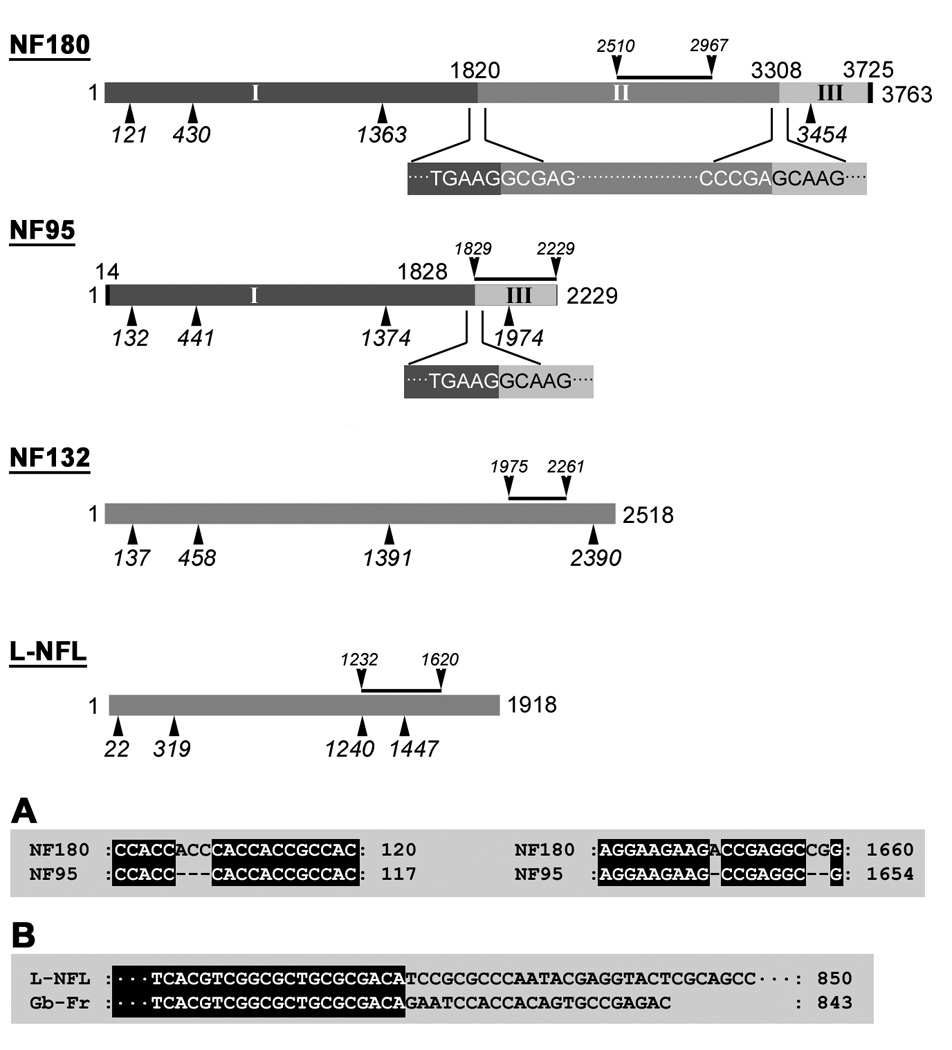
Four full-length lamprey NF cDNAs are drawn as rods in proportion to their length. The number on the right end represents the nucleotide number. The numbers below the rod represent the numbers of the nucleotides at the beginning of (from left to right) the head, rod, sidearm, and stop codon, respectively. The black line above each rod indicates the region of the individual probe used for Northern blot and in situ hybridizations. Numbers above NF180 and NF95 represent the last nucleotide of the previous fragment. Top 2 rods: NF180 was divided into 3 fragments: I, II, and III. NF95 shared two DNA fragments (I and III) with NF180 but lacked fragment II. Nucleotide sequences between fragments I & II or around fragment II and III in NF180, or the sequences between fragment I & III in NF95, were given below each rod. For fragment I: identity between NF180 and NF95 was 99.0%. For fragment III identity was 99.7%. A: Three gaps in NF95 as compared with NF180 in the fragment I region (ACC106–108, A1650, and CG1658–1659). B: Alignment analysis of L-NFL and an 843 bp sequence of a GenBank DNA fragment (Gb-Fr, TI number: 1229593736) obtained by MegaBlast search of the NCBI trace archive (Petromyzon marinus-WGS).
Northern blot and in situ hybridization analysis of lamprey NFs in the CNS
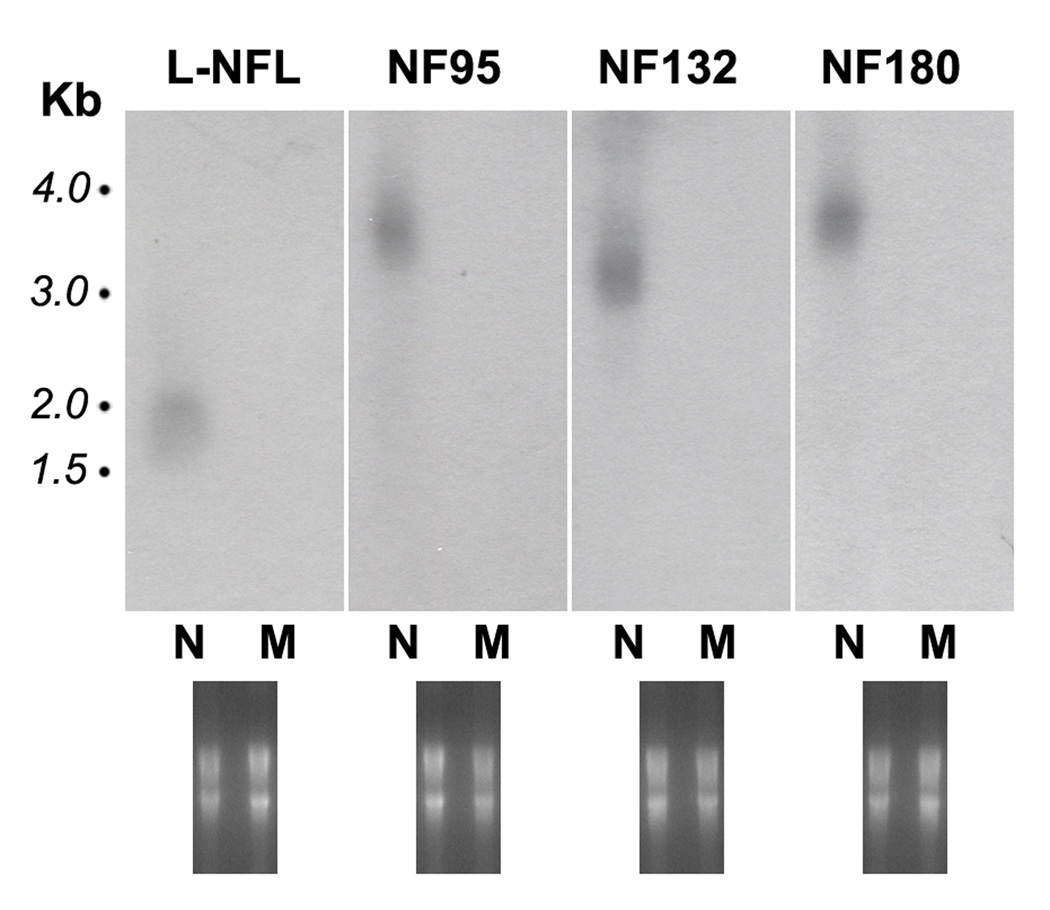
To examine the tissue expression of cloned lamprey NF mRNAs, we designed probes for NF132, NF95, L-NFL, and NF180. The sequences of the probes for the three newly cloned NFs are shown in Figures 1, 3, and 4 (regions underlined) respectively, and in Figure 8. The sequence of the NF180 probe was within the Fragment II area (see Figure 8). Specificities of each probe were analyzed with the BioEdit program, in order to avoid cross interactions. Probe identities among NF132, L-NFL and NF180 or NF132, L-NFL and NF95 were low (< 40%). The probe for NF180 was specific, but the probe for NF95 also had high identity to NF180 for the reason described above (Figure 8). Muscle mRNA was used as a control (Figure 9). Complementary DNA probes specific for each lamprey NF were hybridized to their corresponding targets. The L-NFL probe bound to a ~1.8 kb band, the NF95 probe bound to a band of the same size (~3.7 kb) as that detected by the NF180 probe, and the NF132 probe bound to a ~3.2 kb band. All probes bound to RNA prepared from the CNS, but not from muscle (Figure 9).
Figure 9. Northern blot analysis of tissue-specific distributions of lamprey NF mRNAs.
Aliquots of 15 µg of total RNA from pooled CNS or muscle from 20 wild-type P. marinus larvae were separated on 1% denaturing agarose gels, capillary blotted to nylon membranes and hybridized with cDNA probes specific to each individual lamprey NF. The blot was exposed to autoradiographic film overnight. Upper panel: For all 4 probes, only a single band was detected, and only in the CNS (N), not in muscle (M). The mRNA sizes for L-NFL, NF95, NF132 and NF180 are ~1.8 kb, 3.7 kb, 3.2 kb, and 3.7 kb. Note: NF95 labeled a band of the same size as NF180. No band showed in the area between L-NFL and NF132 where NF95 mRNA should be. Lower panel: Gels stained with ethidium bromide before transferring to the membrane to show the loading of RNA in each lane.
Cellular expression of cloned lamprey NF mRNAs was analyzed by in situ hybridization with digoxigenin-labeled cRNA probes constructed from the carboxyl-terminus of the same region as was used in Northern blotting. The lamprey CNS lacks myelin [5] and has no blood vessels entering spinal cord parenchyma. Thus it can be viewed in wholemount preparations and many “giant” neurons can be identified on the basis of their location, size and morphological features [52,54].
Brain
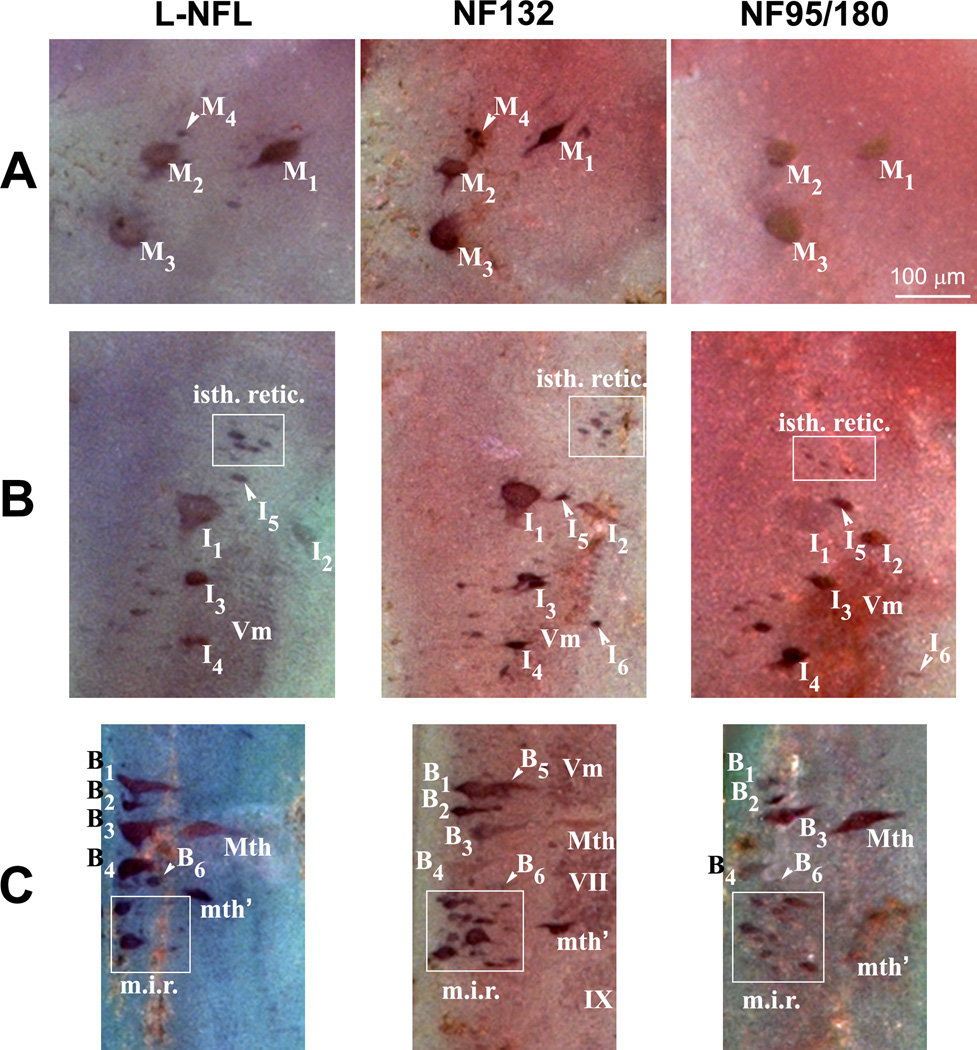
The neuronal cytoarchitecture of the lamprey brain has been well described [49,51,57,59]. A neuronal map of the lamprey brain is shown in Figure 10. The telencephalon and diencephalon contain no giant cells. The mesencephalon contains identified spinal-projecting neurons M1-4, the posterior commissure, and the optic tectum on each side (Figure 10). The brain extends to the isthmic region, which contains identified spinal-projecting neurons I1-6 and the rostral part of the trigeminal motor nucleus. The middle rhombencephalon has the bulbar identified spinal-projecting neurons B1-6, the Mauthner neuron, the auxiliary Mauthner neuron, and cranial nerve nuclei Vm, VII, IX and X (Figure 10, 11) on each side.
Figure 10. Expression patterns for lamprey NF mRNAs in the lamprey brain.
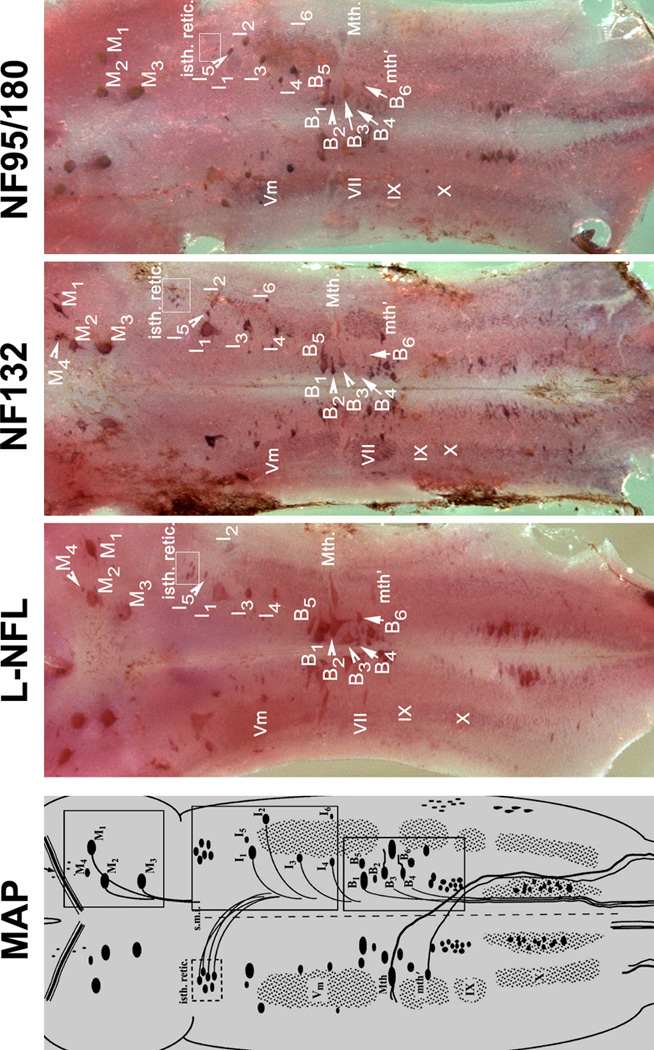
MAP: Schematic drawing of mature (5-year-old) larval sea lamprey brain stem, showing major anatomical features and the locations of identified neurons and the neuron groups. The view is from the dorsal surface after removal of the mesencephalic and rhombencephalic choroid plexus, transection of the cerebrotectal commissure and obex, and lateral reflection of the alar plate. The identified spinal-projecting neurons include M1-4, I1-6, B1-6, the Mauthner (Mth) and auxiliary Mauthner (Mth’) neurons, the isthmic reticulospinal (isth.retic.), and medial inferior reticulospinal (m.i.r.) cell groups, some cranial nerve nuclei [Vm (trigeminal motor nucleus), VII, IX and X] (Reprinted with permission from Jacobs et al., 1997, J Neurosci 17:5206–5220, ©1997 by the Society for Neuroscience). Three boxes on the right are the locations of 3 identified neuron groups (M-, I-, and B-groups), Mauthner (Mth) and auxiliary Mauthner (mth’) neurons. L-NFL, NF132, and NF95/180 are wholemount in situ hybridizations of lamprey brain stem with corresponding probes. Animals used were large lavae, approximately 4 years old with body lengths 130-, 135-, and 135-mm, respectively. Comparison of regions within boxes are given in Figure 11.
Figure 11. Anatomical distribution of L-NFL, NF132, and NF95/180 in lamprey brain.
Based on the nomenclature of Rovainen {Rovainen, 1967 #54} as modified by {Swain, 1993 #64} for identified neurons and neuron groups in mature larval sea lamprey brainstem. M, mesencephalic; I, isthmic; B, bulbar; Mth, Mauthner cell; mth’, auxiliary Mauthner cell. A: Labeled neurons in the mesencephalon. M4 was not detected with the NF95/180 probe. B: Labeled neurons in the isthmic region. Most I group neurons (I1-5), trigeminal nucleus (Vm), and the isthmic reticulospinal neuron group (isth.retic., within white box) were labeled by the probes for L-NFL, NF132, and NF95/180. I6 was not detected with the L-NFL probe. C: Labeled neurons in the middle rhombencephalon. The B1-4 neurons, the Mauthner neuron (Mth), the auxiliary Mauthner neuron (mth’), and the medial inferior reticulospinal (m.i.r., within white box) cell group were labeled by the probes for L-NFL, NF132, and NF95/180. B5 was not detected with the L-NFL and NF95/180 probes.
In situ hybridization to whole-mounted larval lamprey brains indicated that L-NFL, NF132, and NF95/180 mRNAs were abundant in all of the identifiable spinal-projecting neurons, and in neurons of the cranial motor nuclei (Figure 10). The mRNA for NF95/180 was expressed in mesencephalic neurons M1–3. The mRNAs for L-NFL and NF132 were expressed in M1-4 (Figure 10; Figure 11A). All three mRNAs were expressed in neurons I1-6 of the isthmic region, except that L-NFL expression was not apparent in I6 (Figure 10, 11B). All three mRNAs were expressed in B neurons (Figure 10, 11C) and in the Mauthner and auxiliary Mauthner neurons (Figure 11C). In situ hybridization with the probe specific for NF180 generated a similar staining pattern as that with the NF95/180 probe. Intensity of in situ labeling for NF132 and NF95/180 mRNA in the cranial motor nuclei was consistently darker than that for L-NFL (Figure 10). Expression was scarcely detected in the telencephalon, olfactory lobes, habenulopeduncular tract, and the small interneurons throughout the CNS. No cells were labeled by sense RNA probes.
Spinal cord
The lamprey spinal cord is a flattened, ribbon-like structure that extends the length of the vertebral canal. A transverse section through the spinal cord shows bilateral wing-like areas of gray matter surrounded by a zone of “white” matter. Based on their function, there are three types of neurons: 1) motor neurons situated at the lateral horn ventrally on either side; 2) dorsal cells, primary sensory neurons located in the dorsomedial part of the spinal gray; 3) interneurons, including giant interneurons, lateral cells, edge cells, and subependymal cells. A high level of NF95/180 mRNA expression was found along the entire length of the intact spinal cord (Figure 12). Label was found primarily in the medium-sized neurons of the lateral gray matter. By their sizes and locations, some of these may have been motor neurons (MN) and others interneurons. A low intensity of expression of L-NFL was detected in lateral gray matter (Figure 12, left column). Higher expression intensity was seen with the NF132 (Figure 12, middle column) probe, while the most intense labeling was seen with NF95/180 (Figure 12, right column). There did not appear to be specific neuronal types labeled selectively by one or another probe, except that the smallest cells, including the subependymal cells near the midline, were not labeled with NF132.
Figure 12. Cellular localization of NFs in the spinal cord.
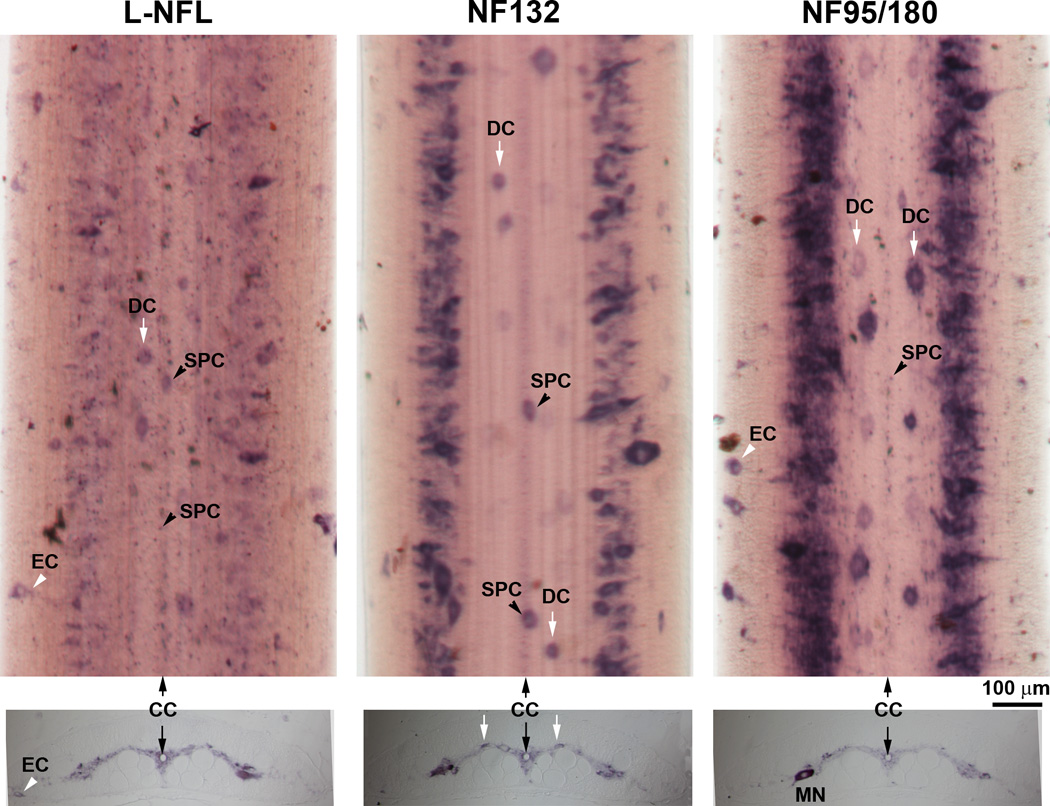
Upper panel: In situ hybridization in spinal cord wholemounts showed a high level of NF95/180 mRNA expression along the lateral gray matter. L-NFL was expressed relatively evenly and weakly along the entire spinal cord. Several neuron types can be identified by their morphology and location, including: dorsal cells (DC); subependymal cells (SPC) surrounding the central canal (CC), and edge cells (EC). Note: NF132 labeled only large and medium sized cells. Lower panel: In situ hybridization in transverse sections from a 12 cm long larva (approximately 3–4 yrs old). All neurons are localized in the gray matter except for ECs, which sit along the edge of the spinal cord. MN: possible motor neuron, based on size, location and the ventrally projecting axon hillock, which is faintly labeled.
DISCUSSION
It had been thought that lampreys have only one NF subunit, NF180 [30,36,48], and that therefore, lamprey NFs would have to be homopolymers. This was unusual because in other vertebrates, type IV IFs, including the NF triplet proteins (NF-L, NF-M and NF-H) and α-internexin are obligate heteropolymers [8]. Later it was found that NF180 by itself did not form NFs, either in cells or in vitro [67], prompting a search of additional NFs in the lamprey. Biochemical and immunohistochemical evidence uncovered another neuronal IF in lamprey, nIF50 [33]. There is no evidence that nIF50 is a NF. It could be another type IV IF found in neurons, such as α-internexin. The present study reports the characterization of cDNA clones encoding three additional lamprey neurofilaments, L-NFL, NF95, and NF132. Thus, like the heteropolymeric NFs found in mammalian neurons, lamprey NFs may be composed of several subunits, including an NF-L. While the sequence of nIF50 remains unknown, immunochemical data show that it is not any of the newly cloned NFs described in the present study.
The three new clones are NFs
IF proteins have been grouped into five types [15,25,34,44]. The central (rod) region of the amino acid sequences of all IF protein chains contains four segments with heptad substructures of the form (a–b–c–d–e–f–g) where “a” and “d” positions are commonly occupied by non-polar residues such as leucine, valine and isoleucine [24,45,56]. These segments are known as 1A, 1B, 2A and 2B and are about 5–6, 14–15, 2–3 and 17–18 heptads long, respectively [10]. The heptad motif is characteristic of an α-helical coiled-coil in which right-handed α-helices coil around one another in a left-handed manner [45]. The coiled-coil segments are joined by linkers L1, L12, and L2, respectively. The sequences display the greatest degree of homology in regions located at both ends of the rod domain.
Sequence analyses of the deduced polypeptides demonstrated that the 3 newly cloned proteins conformed to the “head-rod-tail” tripartite structure exhibited by all IF proteins. Since it would be difficult to define a newly-cloned peptide based only on sequence identity studies, we used a phylogenetic tree to characterize the newly discovered proteins. Although it seems unrelated to phylogenetics or to evolution per se, such trees provide a powerful tool for understanding the biological functions of some unknown proteins [18]. The phylogenetic tree included most human class III and class IV IFs, including vimentin, desmin, GFAP, peripherin, α-internexin, and the neurofilament triplet, supplemented by Xenopus class III and IV IFs. The three newly discovered sequences and NF180 were assigned positions in the tree. The results indicated that all three newly cloned cDNAs encode NFs. Among these, N132 and NF95 belong to the NF-M group, and L-NFL to the NF-L group. The bootstrap values for the newly cloned lamprey NF nodes are 99–100, high enough to be confident that the topology is correct (Figure 6).
NFs may be heteropolymeric in the lamprey nervous system: Implications for NF evolution
The agnatha, i.e., lampreys and hagfishes, diverged from their sister vertebrate group, the gnathostomes, close to the root of the vertebrate phylogenetic tree about 460 million years ago [23,53]. NFs represent a hallmark of neuronal development in vertebrates [2]. It is unclear whether all three mammalian NF subunits evolved from a single precursor (either NFL-like or NFM-like). It has been hypothesized that an invertebrate progenitor IF gene closely related to the nuclear lamins may have developed into two early genes, one for the type IV gene lineage in vertebrates and the other for types I–III [36,55]. Because the lamprey occupies a phylogenetic position between cephalochordates (lancelets such as Amphioxus) and gnathostomes (jawed vertebrates), identification and characterization of new NF genes in P. marinus, especially NF-L-like genes, will help to test this hypothesis. In invertebrates, some IFs are simply derived from alternative RNA processing. In the gastropod Helix aspersa, two cytoplasmic IF proteins in non-neuronal cells originated from a single gene related to the nuclear lamins [11]. In squid, the 60, 70, and 220 kDa “NF” proteins also are derived from a single gene through alternate splicing [60,65]. Like the cytopolasmic non-neuronal IF proteins of Helix, the gene structures of squid NFs are more closely related to that of the nuclear lamins, which are classed as type III IF proteins in vertebrates. Interestingly, the largest of the squid NFs contains 21 KSP repeats in its sidearm, similar to the multiphosphorylation KSP repeats of most vertebrate NF-Ms and NF-Hs [65]. Although the tail regions of the lamprey NF-Ms contain several serine residues that can serve as phosphorylation sites, they lack these KSP sequences. Thus the presence of KSP repeats in squid and in gnathostomes is an apparent example of convergent evolution. In addition, the presence of both NF-M and NF-L homologues in lamprey suggests that more than one type of NF was probably present at the beginning of vertebrate evolution.
In vertebrate species other than lampreys, NFs are obligate heteropolymers, in which NF-L is the essential subunit, requiring either NF-M or NF-H to form filaments in cells [37,38]. During early neurite formation, NF-M precedes NF-H as a partner for NF-L [6]. The demonstration of an NF-L homolog in the lamprey provides a rational basis on which to postulate that lamprey NFs, like other vertebrate NFs, are obligate heteropolymers, and that NF-L is required to form filaments. Direct evidence for the heteropolymeric properties of lamprey NFs will be presented soon in another study (Zhang et al, unpublished).
Are there other NF-L-like proteins in lamprey?
In fish and Xenopus, multiple low molecular weight neuronal intermediate filaments have been identified. In zebrafish, two NF-L genes have been cloned (BC114232, BC096868). They differ only in the last 80 amino acids. A third small gene (NM_001144784) is more closely related to α-internexin than to NF-L. In Xenopus, one NF-L (M86654), one α-internexin-like (U63711) and one other neuronal IF (M86653) gene have been cloned. In lamprey, only 1 NF-L gene has been cloned so far. The present L-NFL is not the neuronal intermediate filament (nIF50) previously characterized biochemically [33] because the recombinant protein was not recognized by the same antibody (LCM40) in Western blot (Figure 2), and the size is slightly larger than nIF50. It is possible that nIF50 is another NF-L or a non-NF IF, such as an α-internexin-like protein. This is supported by analysis of DNA sequences obtained from GenBank. An 843 bp DNA fragment (TI number: 1229593736; Figure 8B) includes a tail (22 n.t.) that is not identical to the sequence cloned in L-NFL. The sequence just before unmatched region is unusual for the boundary of an Exon, neither “GT” in the GT-AG consensus sequence, nor “AT” in the AT-AC consensus sequence [4,66] (Figure 9B), suggesting the presence of another NF-L-like molecule. Moreover, three protein spots with sizes 50–65 kDa in 2-D gels of lamprey CNS cytoskeletal fragments have not been characterized (see spots 7, 8, 9 in Figure 4G of [33]. One or more could be a new NF-L. Finally, the low glutamic acid and high serine content in the sidearm of the newly cloned L-NFL is a feature usually seen in α-internexins but not in most NF-Ls. Therefore, there might be another NF-L with a sidearm amino acid sequence that is closer to that in mammalian NF-Ls.
Duplication of NF-M genes in sea lamprey
Analysis of nucleotide differences among lamprey NF-Ms suggests that the newly cloned NF-M homologs (NF132, NF95) and NF180 are the products of 3 separate genes. They may be derived from an ancestral gene by gene duplication. Gene duplication, followed by functional divergence of new genes has played an important role in vertebrate evolution [26,64]. It has been proposed that two rounds of duplication of the entire genome (polyploidization) occurred early in vertebrate history: one duplication was proposed to have occurred in the vertebrate lineage after the divergence of cephalochordates from the chordate evolutionary line, and the second after the divergence of jawless vertebrates (hagfish and lampreys) from the vertebrate line [27,43]. Because of its pivotal position in phylogeny, the lamprey has been used to study the timing and mechanism of gene duplication [12,14,29]. Our results for lamprey NF-Ms support the hypothesis of gene duplications, as occurred in Xenopus, which has multiple NF-M isoforms (BC135490, BC047248, BC078128, BC043878). Although we did not screen the lamprey cDNA library exhaustively in cloning lamprey NF-Ms, the available data indicate that two gene duplications took place to produce lamprey NF95/NF180 and NF132. It is not clear whether a paralogous form of NF132 exists, leading to a topology of the form (AB)(CD) in two rounds of duplication of the genome [28]. Isolation of additional lamprey NF-M genes might resolve this question. The significance of having more than one NF-M for NF structure and function is not clear but the stability of the rod domain in the evolution of NF-Ms is striking. By comparison, the lengths and sequences of NF tail domains vary extensively, both for orthologous forms of the same NF protein in different species and for different NF proteins within the same organism [17]. In sea lamprey, the lengths of the tail domains of the three NF-M forms differed by 497 (NF180 vs. NF95), 364 (NF180 vs. NF132), and 133 (NF132 vs. NF95) amino acids (Table 2), respectively.
NF95 is highly homologous to NF180 except for deletion of the nucleotide sequence from 1821 to 3308 (Figure 8). However, alternative splicing from a common gene cannot explain the sequence differences because the remaining sequence, while 99% homologous, is not identical to the comparable portion of NF180. On the other hand, alternative splicing has been described in the processing of several intermediate filament genes, such as peripherin in mice [35], lamins A and C in humans [13,41], and plasticin in zebrafish [1]. Alternative splicing is possible in lamprey NF95 processing when its sequence is compared with that of NF180. According to the GT-AG rule, a consensus sequence 5’-A/CAGgta/gagt-3’ representing a 5’ splice site (donor site), and 5’-yyyyyyxcagGG/T-3’ representing a 3’ splice site (acceptor site) are present at the boundary between exons and introns (nt in capitals are in Exons; nt in lower case are in introns; y = pyrimidine, x = a/c/g/t) [4]. Thus nucleotide sequences in the joint regions between fragments I and II in NF180 and fragments I and III in NF95 are consistent with the GT-AG rule (see Figure 8), and therefore support the hypothesis that NF95 might be alternatively spliced during RNA processing from an NF180-like pre-mRNA, but NF95 is not an alternate splice product of the NF180 gene itself.
Selective expression of lamprey NF mRNAs in projection neurons
The probe for NF95 detected only one band in Northern blotting, with a size identical to the band detected by the NF180 probe (~3.7 kb). This could be because: 1) NF95 message is present only at low levels in larval sea lampreys; or 2) messages of NF95 and NF180 both are expressed abundantly in larval lampreys, but mRNA of NF95 has longer (~1.5 kb) upstream and/or downstream untranslated regions than those of NF180, so that their total mRNA sizes are similar. A more exhaustive search of the available preassembled lamprey genomic libraries might help to determine this. In any case, because it was not possible to find a probe sequence that labeled NF95 but not NF180, the NF95 probe must be assumed to label both on in situ hybridizations. We do have probes that are specific for NF180 (because they bind to a region that is not present in NF95), but the labeling patterns for both the NF180 and NF95/180 probes were the same.
In the lamprey brainstem, NF messages were expressed primarily in neurons whose axons projected long distances outside the brainstem, including reticulospinal neurons and the cranial motor nuclei (trigeminal, gIossopharygeal, and vagal motor nuclei). The probes for all three new NFs (L-NFL, NF132 and NF95/180) showed in situ labeling similar to the pattern previously reported for NF180 [58]. Labeling was mainly independent of the diameters of cell bodies or axons. For example, the large Müller and Mauthner reticulospinal neurons, whose cell body diameters are 50–100 µm and whose axons in the spinal cord can exceed 25 µm, stained similarly to smaller reticulospinal and cranial motor neurons, some of whose axons have diameters less than 1 µm. However, reticulospinal neurons and cranial motor neurons of all sizes represent less than 10% of the total neurons in the lamprey brainstem [50]. Most local neurons were not labeled (some might have been too small to be identified above background staining). This is not unique to lamprey, but has been observed in the mammalian CNS. In rat, NF immunostaining was seen in only two thirds of neurons of the cerebral and cerebellar cortices, hypothalamus, spinal cord and retina [63]. The unstained neurons typically were small with minimal cytoplasm. The observation did not preclude the possibility that NFs are synthesized at a low level. In the squid, in situ hybridization also showed NF expression selectively in the large neurons of the stellate ganglion [60,65]. It would seem that in the squid, as in lampreys and mammals, neurons with longer-projecting axons have more NF content and need more de novo NF synthesis to maintain cytoskeletal dynamic balance.
Acknowledgements
The work was supported by NIH Grant R01 NS14837, R01-NS38537, and R24-HD050838; Grant sponsor: Craig H. Neilsen Foundation: Grant number: 2781.
Abbreviations
- NFs
Neurofilaments
- NF-H
neurofilament high molecular subunit
- NF-M
neurofilament middle molecular subunit
- NF-L
neurofilament low molecular subunit
- IF
intermediate filament
- nIF
neuronal intermediate filament
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Asch WS, Leake D, Canger AK, Passini MA, Argenton F, Schechter N. Cloning of zebrafish neurofilament cDNAs for plasticin and gefiltin: increased mRNA expression in ganglion cells after optic nerve injury. J Neurochem. 1998;71:20–32. doi: 10.1046/j.1471-4159.1998.71010020.x. [DOI] [PubMed] [Google Scholar]
- 2.Bennett GS. Changes in intermediate filament composition during neurogenesis. Curr Top Dev Biol. 1987;21:151–183. doi: 10.1016/s0070-2153(08)60136-2. [DOI] [PubMed] [Google Scholar]
- 3.Bhat BM, Wold WS. ATTAAA as well as downstream sequences are required for RNA 3'-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985;5:3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 5.Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci Lett. 1984;48:145–148. doi: 10.1016/0304-3940(84)90010-7. [DOI] [PubMed] [Google Scholar]
- 6.Carden MJ, Trojanowski JQ, Schlaepfer WW, Lee VM. Two-stage expression of neurofilament polypeptides during rat neurogenesis with early establishment of adult phosphorylation patterns. J Neurosci. 1987;7:3489–3504. doi: 10.1523/JNEUROSCI.07-11-03489.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin SS, Liem RK. Expression of rat neurofilament proteins NF-L and NF-M in transfected non-neuronal cells. Eur J Cell Biol. 1989;50:475–490. [PubMed] [Google Scholar]
- 8.Ching GY, Liem RK. Assembly of type IV neuronal intermediate filaments in nonneuronal cells in the absence of preexisting cytoplasmic intermediate filaments. J Cell Biol. 1993;122:1323–1335. doi: 10.1083/jcb.122.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Crewther WG, Dowling LM, Steinert PM, Parry DAD. Structure of intermediate filaments. International Journal of Biological Macromolecules. 1983;5:267–274. [Google Scholar]
- 11.Dodemont H, Riemer D, Weber K. Structure of an invertebrate gene encoding cytoplasmic intermediate filament (IF) proteins: implications for the origin and the diversification of IF proteins. Embo J. 1990;9:4083–4094. doi: 10.1002/j.1460-2075.1990.tb07630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- 13.Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci U S A. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Force A, Amores A, Postlethwait JH. Hox cluster organization in the jawless vertebrate Petromyzon marinus. J Exp Zool. 2002;294:30–46. doi: 10.1002/jez.10091. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 16.Geisler N, Weber K. Self-assembly in Vitro of the 68,000 molecular weight component of the mammalian neurofilament triplet proteins into intermediatesized filaments. J Mol Biol. 1981;151:565–571. doi: 10.1016/0022-2836(81)90011-5. [DOI] [PubMed] [Google Scholar]
- 17.Gervasi C, Szaro BG. Sequence and expression patterns of two forms of the middle molecular weight neurofilament protein (NF-M) of Xenopus laevis. Brain Res Mol Brain Res. 1997;48:229–242. doi: 10.1016/s0169-328x(97)00096-x. [DOI] [PubMed] [Google Scholar]
- 18.Hall BG. Phylogenetic Trees Made Easy: A How-To Manual. Third Edition edn. Sinauer Associates Inc.; 2007. [Google Scholar]
- 19.Hall GF, Lee VM, Kosik KS. Microtubule destabilization and neurofilament phosphorylation precede dendritic sprouting after close axotomy of lamprey central neurons. Proc Natl Acad Sci U S A. 1991;88:5016–5020. doi: 10.1073/pnas.88.11.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall GF, Poulos A, Cohen MJ. Sprouts emerging from the dendrites of axotomized lamprey central neurons have axonlike ultrastructure. Journal of Neuroscience. 1989;9:588–599. doi: 10.1523/JNEUROSCI.09-02-00588.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall GF, Yao J, Selzer ME, Kosik KS. Cytoskeletal changes correlated with the loss of neuronal polarity in axotomized lamprey central neurons. Journal of Neurocytology. 1997;26:733–753. doi: 10.1023/a:1018562331003. [DOI] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; Nucleic Acids Symposium Series; 1999. pp. 95–88. [Google Scholar]
- 23.Halstead LB. Agnatha. In: Benton MJ, editor. The fossil record 2. London: Chapman & Hall; 1993. pp. 573–581. [Google Scholar]
- 24.Herrmann H, Aebi U. Intermediate filament assembly: fibrillogenesis is driven by decisive dimer-dimer interactions. Curr Opin Struct Biol. 1998;8:177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann H, Aebi U. Intermediate filaments and their associates: multitalented structural elements specifying cytoarchitecture and cytodynamics. Curr Opin Cell Biol. 2000;12:79–90. doi: 10.1016/s0955-0674(99)00060-5. [DOI] [PubMed] [Google Scholar]
- 26.Holland PW. Gene duplication: past, present and future. Semin Cell Dev Biol. 1999;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- 27.Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 28.Hughes AL. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J Mol Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- 29.Irvine SQ, Carr JL, Bailey WJ, Kawasaki K, Shimizu N, Amemiya CT, Ruddle FH. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J Exp Zool. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs AJ, Kamholz J, Selzer ME. The single lamprey neurofilament subunit (NF-180) lacks multiphosphorylation repeats and is expressed selectively in projection neurons. Brain Res Mol Brain Res. 1995;29:43–52. doi: 10.1016/0169-328x(94)00227-6. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs AJ, Swain GP, Snedeker JA, Pijak DS, Gladstone LJ, Selzer ME. Recovery of neurofilament expression selectively in regenerating reticulospinal neurons. J Neurosci. 1997;17:5206–5220. doi: 10.1523/JNEUROSCI.17-13-05206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin LQ, Zhang G, Jamison C, Jr, Takano H, Haydon PG, Selzer ME. Axon regeneration in the absence of growth cones: acceleration by cyclic AMP. J Comp Neurol. 2009;515:295–312. doi: 10.1002/cne.22057. [DOI] [PubMed] [Google Scholar]
- 33.Jin LQ, Zhang G, Selzer ME. Lamprey neurofilaments contain a previously unreported 50-kDa protein. J Comp Neurol. 2005;483:403–414. doi: 10.1002/cne.20459. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Coulombe PA. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007;21:1581–1597. doi: 10.1101/gad.1552107. [DOI] [PubMed] [Google Scholar]
- 35.Landon F, Lemonnier M, Benarous R, Huc C, Fiszman M, Gros F, Portier MM. Multiple mRNAs encode peripherin, a neuronal intermediate filament protein. Embo J. 1989;8:1719–1726. doi: 10.1002/j.1460-2075.1989.tb03564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lasek RJ, Phillips L, Katz MJ, Autilio-Gambetti L. Function and evolution of neurofilament proteins. Ann N Y Acad Sci. 1985;455:462–478. doi: 10.1111/j.1749-6632.1985.tb50429.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee MK, Cleveland DW. Neuronal intermediate filaments. Annu Rev Neurosci. 1996;19:187–217. doi: 10.1146/annurev.ne.19.030196.001155. [DOI] [PubMed] [Google Scholar]
- 38.Lee MK, Xu Z, Wong PC, Cleveland DW. Neurofilaments are obligate heteropolymers in vivo. J Cell Biol. 1993;122:1337–1350. doi: 10.1083/jcb.122.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liem RK, Hutchison SB. Purification of individual components of the neurofilament triplet: filament assembly from the 70 000-dalton subunit. Biochemistry. 1982;21:3221–3226. doi: 10.1021/bi00256a029. [DOI] [PubMed] [Google Scholar]
- 40.Lurie DI, Pijak DS, Selzer ME. Structure of reticulospinal axon growth cones and their cellular environment during regeneration in the lamprey spinal cord. J Comp Neurol. 1994;344:559–580. doi: 10.1002/cne.903440406. [DOI] [PubMed] [Google Scholar]
- 41.McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 42.Nicholas KB, Nicholas HB., Jr GeneDoc: a tool for editing and annotating multiple sequence alignments. 1997 Distributed by the author. [Google Scholar]
- 43.Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 44.Parry DA. Microdissection of the sequence and structure of intermediate filament chains. Adv Protein Chem. 2005;70:113–142. doi: 10.1016/S0065-3233(05)70005-X. [DOI] [PubMed] [Google Scholar]
- 45.Parry DA, Steinert PM. Intermediate filaments: molecular architecture, assembly, dynamics and polymorphism. Q Rev Biophys. 1999;32:99–187. doi: 10.1017/s0033583500003516. [DOI] [PubMed] [Google Scholar]
- 46.Parry DAD. Guidebook to the cytoskeletal and motor proteins. Second Edition edn. Oxford: Oxford University Press; 1999. pp. 285–291. [Google Scholar]
- 47.Pijak DS, Hall GF, Tenicki PJ, Boulos AS, Lurie DI, Selzer ME. Neurofilament spacing, phosphorylation, and axon diameter in regenerating and uninjured lamprey axons. J Comp Neurol. 1996;368:569–581. doi: 10.1002/(SICI)1096-9861(19960513)368:4<569::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Pleasure SJ, Selzer ME, Lee VM. Lamprey neurofilaments combine in one subunit the features of each mammalian NF triplet protein but are highly phosphorylated only in large axons. J Neurosci. 1989;9:698–709. doi: 10.1523/JNEUROSCI.09-02-00698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rovainen CM. Neurobiology of lampreys. Physiol Rev. 1979;59:1007–1077. doi: 10.1152/physrev.1979.59.4.1007. [DOI] [PubMed] [Google Scholar]
- 50.Rovainen CM. Neurophysiology. Vol. 4A. Academic Press; 1982. pp. 1–136. [Google Scholar]
- 51.Rovainen CM. Physiological and anatomical studies on large neurons of central nervous system of the sea lamprey (Petromyzon marinus). I. Muller and Mauthner cells. J Neurophysiol. 1967;30:1000–1023. doi: 10.1152/jn.1967.30.5.1000. [DOI] [PubMed] [Google Scholar]
- 52.Rovainen CM. Synaptic interactions of identified nerve cells in the spinal cord of the sea lamprey. J Comp Neurol. 1974;154:189–206. doi: 10.1002/cne.901540206. [DOI] [PubMed] [Google Scholar]
- 53.Salaneck E, Fredriksson R, Larson ET, Conlon JM, Larhammar D. A neuropeptide Y receptor Y1-subfamily gene from an agnathan, the European river lamprey. A potential ancestral gene. Eur J Biochem. 2001;268:6146–6154. doi: 10.1046/j.0014-2956.2001.02561.x. [DOI] [PubMed] [Google Scholar]
- 54.Selzer ME. Variability in maps of identified neurons in the sea lamprey spinal cord examined by a wholemount technique. Brain Res. 1979;163:181–193. doi: 10.1016/0006-8993(79)90348-2. [DOI] [PubMed] [Google Scholar]
- 55.Steinert PM, Roop DR. Molecular and cellular biology of intermediate filaments. Annu Rev Biochem. 1988;57:593–625. doi: 10.1146/annurev.bi.57.070188.003113. [DOI] [PubMed] [Google Scholar]
- 56.Steinert PM, Steven AC, Roop DR. The molecular biology of intermediate filaments. Cell. 1985;42:411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- 57.Swain GP, Ayers J, Selzer ME. Metamorphosis of spinal-projecting neurons in the brain of the sea lamprey during transformation of the larva to adult: normal anatomy and response to axotomy. J Comp Neurol. 1995;362:453–467. doi: 10.1002/cne.903620403. [DOI] [PubMed] [Google Scholar]
- 58.Swain GP, Jacobs AJ, Frei E, Selzer ME. A method for in situ hybridization in wholemounted lamprey brain: neurofilament expression in larvae and adults. Exp Neurol. 1994;126:256–269. doi: 10.1006/exnr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 59.Swain GP, Snedeker JA, Ayers J, Selzer ME. Cytoarchitecture of spinal-projecting neurons in the brain of the larval sea lamprey. J Comp Neurol. 1993;336:194–210. doi: 10.1002/cne.903360204. [DOI] [PubMed] [Google Scholar]
- 60.Szaro BG, Pant HC, Way J, Battey J. Squid low molecular weight neurofilament proteins are a novel class of neurofilament protein. A nuclear lamin-like core and multiple distinct proteins formed by alternative RNA processing. J Biol Chem. 1991;266:15035–15041. [PubMed] [Google Scholar]
- 61.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 62.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trojanowski JQ, Walkenstein N, Lee VM. Expression of neurofilament subunits in neurons of the central and peripheral nervous system: an immunohistochemical study with monoclonal antibodies. J Neurosci. 1986;6:650–660. doi: 10.1523/JNEUROSCI.06-03-00650.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 65.Way J, Hellmich MR, Jaffe H, Szaro B, Pant HC, Gainer H, Battey J. A high-molecular-weight squid neurofilament protein contains a lamin-like rod domain and a tail domain with Lys-Ser-Pro repeats. Proc Natl Acad Sci U S A. 1992;89:6963–6967. doi: 10.1073/pnas.89.15.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu Q, Krainer AR. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol. 1999;19:3225–3236. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang G, Spencer PH, Jin LQ, Cohlberg JA, Beaulieu JM, Julien JP, Selzer ME. The single neurofilament subunit of lamprey may need another element for filament assembly. J Comp Neurol. 2004;471:188–200. doi: 10.1002/cne.20026. [DOI] [PubMed] [Google Scholar]