Abstract
Stellantchasmus falcatus is a minute intestinal fluke in the family Heterophyidae. Metacercariae, the infective stage, were reported in a marine fish, mullet Liza subviridis, and a fresh water fish, Dermogenus pusillus, in Thailand. Adults were found in chicks, rats, cats, and humans. Morphological studies were done for comparing Stellantchasmus sp. worms found in 2 different fish hosts; their shapes and organ arrangements were very similar except for the prepharynx length. Therefore, the present study aimed to compare their DNA fingerprints using the HAT-RAPD method for both types of Stellantchasmus and several other related species. Ten arbitrarily selected primers (OPA-04, OPA-09, OPN-02, OPN-03, OPN-09, OPN-12, OPP-11, OPR-15, OPX-13, and OPAD-01) were used. It was found that OPA-09, OPN-03, and OPAD-01 were able to generate S. falcatus specific fragments in mullets which consisted of 200, 760, and 280 bp, respectively. In addition, the results of morphologic, DNA fingerprinting, and phylogenetic analyses strongly suggest that the fresh water and marine specimens of Stellantchamus may be different species.
Keywords: Stellantchasmus falcatus, molecular marker, phylogenetic relationship, HAT-RAPD, mullet (Liza subviridis)
INTRODUCTION
Stellantchasmus falcatus is a minute intestinal fluke in the family Heterophyidae. Several mammals, including humans, rats, cats, dogs, and chicks, are its definitive hosts [1,2]. This intestinal fluke is found in many Asian countries. People become infected with this parasite by consuming undercooked food prepared from fish containing metacercariae. Even though, symptomatic features of S. falcatus infection are not yet clearly understood. The first clinical report in Thailand revealed that S. falcatus was found at autopsy and many embryonated eggs were found in cardiac blood vessels [1]. In this respect, S. falcatus should be considered as a food-borne zoonotic trematode which can affect human health.
Larval stages, including the miracidium, sporocyst, redia, and cercaria, of S. falcatus are found in the fresh water snails, Melanoides tuberculata and Tarebia granifera [3]. The metacercariae (the infective stage) are mainly found in brackish water or marine fish, especially the mullet group, including Mugil spp. [3] and Liza spp. [4]. However, in Thailand, several reports have also shown that S. falcatus metacercariae are found in fresh water fish, such as half-beaked fish (Dermogenus pusillus) and climbing perch (Anabas testudineus) [5,6] both of which have high infection rates of S. falcatus with endemics in northern Thailand. The 2 types of S. falcatus are morphologically similar except for the prepharynx length. However, they may be genetically distinct, and we propose that the marine and fresh water populations of S. falcatus may be different species.
Molecular approaches are the most effective and accurate means for the detection and identification of such organisms and for screening of genetic variation among populations. For example, PCR and filter-hybridization have been used to detect bird schistosomes cercariae in lakes [7], snail hosts, feces, water, and plankton samples [8-11]. PCR-RFLP has been developed to identify Schistosoma haematobium and Schistosoma bovis from Kenya [12], to detect the large liver fluke, Fasciola hepatica cercariae infected in the snail, Lymnaea collumella [13], and for the identification of schistosomiasis transmission sites [14]. PCR-based methods have also been used for coprodiagnosis to detect parasites, such as Echinococcus multilocularis in the definitive hosts [15], Opisthorchis viverrini in humans [16,17], and in experimental hosts [18]. A mitochondria-based PCR for identification and discrimination of Clonorchis sinensis and O. viverrini were also developed [19].
Another genetic technique, high annealing temperature-randomly amplified polymorphic DNA (HAT-RAPD), has been used for identification of lychee (Litchi chinensis Sonn., Sapindaceae) in Thailand [20]. This technique was also adopted for identification of S. falcatus [5], for the optimal DNA qualities and quantities of some trematodes for use in PCR reaction [21], and for intra-specific variation analysis of tree paramphistome flukes in Thailand [22]. Recently, a specific primer for detection of Haplorchis taichui infection has been successfully developed [23]. This method can yield more polymorphic DNA, has higher resolution, and reproducible results. Therefore, the HAT-RAPD method was chosen for use in this study to screen for genetic markers of S. falcatus among marine and fresh water populations.
MATERIALS AND METHODS
Parasite preparation
Eight trematode species were used in this study. This consisted of 4 intestinal flukes, i.e., S. falcatus (both in mullet and half-beaked fish), H. taichui, Haplorchoides sp., and Centrocestus caninus; 1 liver fluke, O. viverrini; 3 rumen flukes, Fischoederius elongatus, Orthocoelium streptocoelium, and Paramphistomum epiclitum, respectively. All specimens were obtained from cattle slaughtered at an abattoir in Lam Phun province, Thailand.
Total genomic DNA extraction
Genomic DNA of all parasites was extracted and purified from adults using the Dneasy Tissue Kit (QIAGEN Inc, Valencia, California, USA) according to the manufacturer's instructions. In addition, fish hosts, Liza subviridis, were also subjected to DNA preparation which had been used in PCR as a comparative study. All extracted genomic DNAs were diluted to a working concentration of 30 ng/µl and stored at -20℃ until use.
HAT-RAPD PCR
Ten commercially available arbitrary 10-mer primers (Operon Biotechnology, Huntsville, Alabama, USA) were performed to use individually in HAT-RAPD PCR. The reaction was carried out in a final volume of 20 µl containing 1x PCR buffer, 2 mM MgCl2, 10 µM of each dNTP, 1 µM of each primer, and 1 U of Vivantis Tag DNA polymerase. The reactions were performed in a MyCycler™ Thermocycler (Bio-RAD, Hercules, California, USA), and PCR protocols were indicated as follows: 1 cycle of 95℃ for 5 min, 30 cycle of 95℃ for 45 sec, 48℃ for 45 sec, 72℃ for 1 min, and 1 cycle of final extension at 72℃ for 7 min. HAT-RAPD PCR products were separated on 1.4% TBE agarose gel electrophoresis stained with ethidium bromide and photographed with a Kodak digital camera, Gel Logic 100.
Similarity index and phylogenetic analysis
Amplified HAT-RAPD markers were scored as '1' for the presence of a band, and '0' for none. Ambiguous bands that could not be clearly distinguished were not scored. The similarity of S. falcatus samples was calculated as follows: similarity = 2 NAB/NA+NB, NAB is the number of bands shared by individuals A and B. NA and NB are the number of bands of individuals A and B, respectively [24-25]. Phylogenetic relationships among 9 parasite samples were analyzed using a Multi-Variates Statistical Package (MVSP) program, and a UPGMA dendrogram was constructed.
RESULTS
Screening for a S. falcatus specific fragment
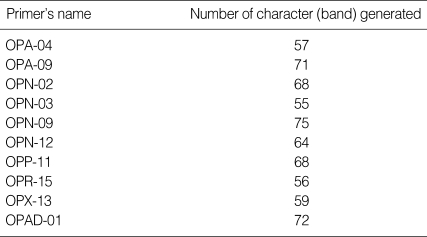
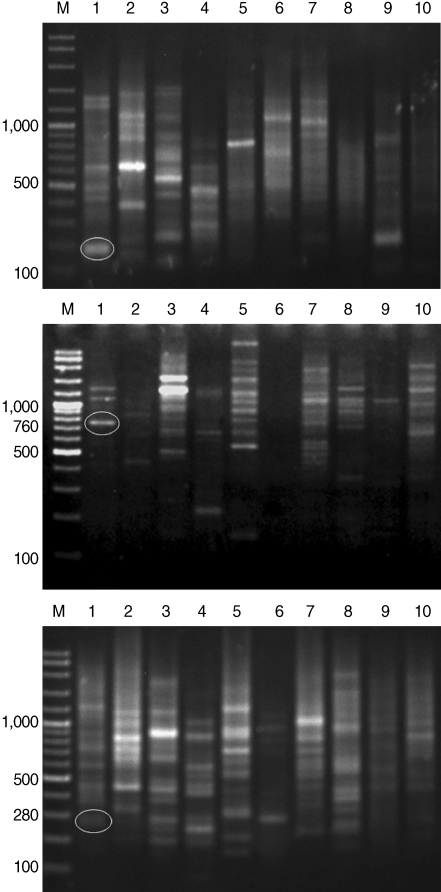
After genomic DNAs were amplified in PCR with 10 arbitrary primers, HAT-RAPD DNA profiles were generated and 456 polymorphic DNA markers were also scored. The number of characters (bands) generated in each primer is indicated in Table 1. Overall, 3 polymorphic markers (200, 760, and 280 bp) generated from OPA-09, OPN-03, and OPAD-01, respectively, were found to be a S. falcatus specific fragment (Fig. 1) which can be selected to construct a specific DNA marker (specific primer) for further detection and identification.
Table 1.
Number of characters (bands) generated in each primer
Fig. 1.
HAT-RAPD markers of 200, 760, and 280 bp fragments generated by OPA-09 (upper), OPN-03 (middle), and OPAD-01 (lower), respectively. Lane M, DNA marker (VC ladder plus 100 bp); lane 1, S. falcatus (in mullet); lane 2, S. falcatus (in half-beaked fish); lane 3, mullet (Liza subviridis); lane 4, Haplorchis taichui; lane 5, Haplorchoides sp.; lane 6, Centrocestus caninus; lane 7, O. viverrini; lane 8, O. streptocoelium; lane 9, P. epiclitum; lane 10, F. elongates.
Similarity index and phylogenetic analysis
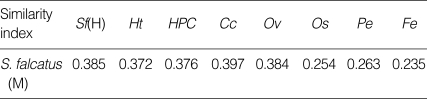
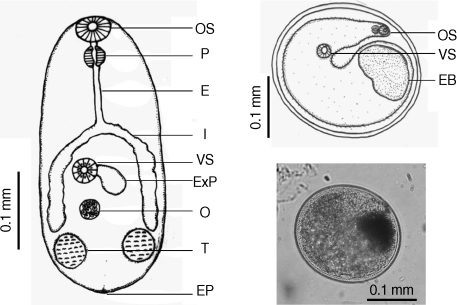
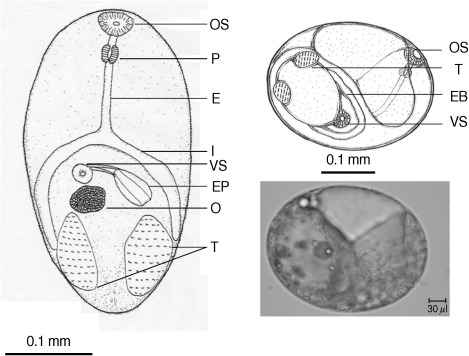
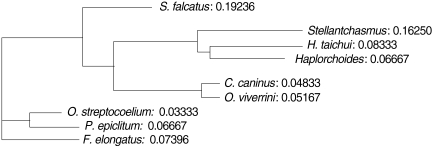
Similarity indices derived from HAT-RAPD markers of S. falcatus (from mullets) compared with those of other trematodes tested (Stellantchasmus sp. from half-beaked fish), H. taichui, Haplorchoides sp., C. caninus, O. viverrini, O. streptocoelium, P. epiclitum, and F. elongatus) were indicated as follows: 0.385, 0.372, 0.376, 0.397, 0.384, 0.254, 0.263, and 0.235, respectively (Table 2). There were a few morphological differences between the 2 types of S. falcatus; mostly in body size, organ arrangement, and the prepharynx length (Figs. 2, 3). An UPGMA dendrogram was constructed which showed that S. falcatus from mullets was separated from Stellantchasmus sp. from half-beaked fish which demonstrated divergent evolution along with other fresh water heterophyid species (Fig. 4). Based on DNA finger-printing and phylogenetic analysis, together with morphology, Stellantchasmus obtained from fresh water and marine habitats are distinct from each other.
Table 2.
Similarity index derived from HAT-RAPD markers of S. falcatus (in mullet) compared with those other trematodes tested
Fig. 2.
Drawing of excysted and encyst metacercariae of S. falcatus found in the half-beaked fish (Dermogenus pusillus).
Fig. 3.
Drawing of excysted and encyst metacercariae of S. falcatus found in the mullet (Liza subviridis).
Fig. 4.
UPGMA dendrogram (Phylogram) generated by Multi Variates Statistical Package (MVSP) program demonstrated the relationships calculated base on polymorphism of HAT-RAPD markers.
DISCUSSION
We report that the 3 expected fragments derived from HAT-RAPD markers seem to be S. falcatus (marine type) specific. They are 200, 760, and 280 bp fragments generated by OPA-09, OPN-03, and OPAD-01 primers and they could serve as S. falcatus specific fragments in mullets. Consequently, specific primers will be constructed based on the alignment of sequence data of S. falcatus specific fragments. Our results showed the advantages of sequence characterized amplified region (SCAR-marker) derived from HAT-RAPD. Since most reports designed specific primers based on known sequence data and were carried out in different locations of such parasite genomes, i.e. highly repeated sequences to detect Schistosoma mansoni [8,9], the use of tandem repeated DNA sequence to detect Trichobilharzia ocellata [10], mitochondrial 12S rRNA gene for the detection of Echinococcus multilocularis [15], complete mitochondrial sequence for Fasciola hepatica [13], and for Clonorchis sinensis and O. viverrini identification and discrimination [19]. Prior DNA sequence data or known location did not require SCAR-markers. The HAT-RAPD marker gives high resolution and reproducible data [21-23]. It was used in several studies due to an advantage of using SCAR-marker technique to construct specific means for detection of parasites and for keeping a lower cost and lower time consumption to carry out such work.
For the analysis of phylogenetic relationships, it was found that S. falcatus from mullets was separated from Stellantchasmus sp. from half-beaked fish. This demonstrated divergent evolution among fresh water heterophyid species. In contrast, 3 species of rumen flukes were clustered in the same clad to serve as an out-group control whereas other fresh water heterophyids were considered as closely related populations. Our results were similar to that of Sripalwit et al. [22] except in that the species closely related with the liver fluke O. viverrini was Haplorchoides sp., whereas in the present study it was C. caninus. It may have been due to different primers used and the number of parasite species used to generate DNA fingerprints.
The present study provided evidence that can demonstrate differences between fresh water and marine types of S. falcatus. The differences in morphology, habitat (fresh water or marine water), and DNA fingerprints and phylogenetic relationships strongly support that Stellantchasmus from fresh water and marine habitats may be different species.
The present study also provided valuable and useful information about the S. falcatus-specific DNA fragments which can be used for development of specific means for detection of either cercariae in snails and metacercariae in fish hosts, or eggs in fecal specimen of definitive hosts. More intensive studies to discover further useful evidences, such as specific DNA markers and DNA sequences, are required to discriminate the genetic and phylogenic characteristics of Stellantchasmus spp.
ACKNOWLEDGEMENTS
The authors wish to thank the Applied Technology in Biodiversity Research Unit, Institute for Science and Technology Research, Chiang Mai University, Thailand. Thanks are extended to the Parasitology Research Laboratory and the Economic Plant Genomes Research and Service Center, Department of Biology, Faculty of Science, Chiang Mai University, Thailand, for instrumental availability. Special thanks are also extended to Prof. Dr. Jong-Yil Chai, Seoul National University College of Medicine, Seoul, Korea, for valuable suggestions, Dr. J. F Maxwell for editing, and Mr. Suksan Chuboon for laboratory assistance.
References
- 1.Kliks M, Tantachumrun T. Heterophyid (trematoda) parasites of cats in Northern Thailand, with notes on a human case found at necropsy. Southeast Asian J Trop Med Public Health. 1974;5:547–555. [PubMed] [Google Scholar]
- 2.Pearson JC, Ow-Yang CK. New species of Haplorchis from Southeast Asia, together with keys to the Haplorchis-group of heterophyid trematode of the region. Southeast Asian J Trop Med Public Health. 1982;13:35–60. [PubMed] [Google Scholar]
- 3.Chai JY, Lee SH. Food-borne intestinal trematode infections in the Republic of Korea. Parasitol Int. 2002;51:129–154. doi: 10.1016/s1383-5769(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 4.Pubua J, Wongsawad C. Redescription of the trematode metacercariae from the mullet (Liza subviridis) and half-beak (Dermogenus pusillus) Southeast Asian J Trop Med Public Health. 2007;38(suppl 1):106–109. [Google Scholar]
- 5.Sripalwit P, Wongsawad C, Chai JY, Rojanapaibul A, Anantalabhochai S. Development of HAT-RADP technique for the identification of Stellanchasmus falcatus; 4th Seminar on Food and Water borne Parasitic Zoonoses, 2nd International Meeting on Gnathostomiasis and Joint International Tropical Medicine Meeting; 2003. 289 pp. [Google Scholar]
- 6.Wongsawad C, Rojtinnakorn J, Wongsawad P, Rojanapaibul A. Helminths of vertebrates from Maesa stream. Southeast Asian J Trop Med Public Health. 2004;35(suppl 1):140–146. [Google Scholar]
- 7.Hertel J, Hamburger J, Harberl B, Hass W. Detection of bird schistosomes in lakes by PCR and filter-hybridization. Exp Parasitol. 2002;101:57–63. doi: 10.1016/s0014-4894(02)00036-x. [DOI] [PubMed] [Google Scholar]
- 8.Hamburger J, He-Na, Xin XY, Ramzy RM, Jourdane J, Ruppel A. A polymerase chain reaction assay for detecting snails infected with bilharzia parasites (Schistosoma mansoni) from very early prepatency. Am J Trop Med Hyg. 1998;59:872–876. doi: 10.4269/ajtmh.1998.59.872. [DOI] [PubMed] [Google Scholar]
- 9.Hamburger J, Xin XY, Ramzy RM, Jourdane J, Ruppel A. Development and labolatory evaluation of a polymerase chain reaction for mornitoring Schistosoma mansoni infestatin of water. Am J Trop Med Hyg. 1998;59:468–473. doi: 10.4269/ajtmh.1998.59.468. [DOI] [PubMed] [Google Scholar]
- 10.Hamburger J, He-Na, Abbasi I, Ramzy RM, Jourdane J, Ruppel A. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am J Trop Med Hyg. 2001;65:907–911. doi: 10.4269/ajtmh.2001.65.907. [DOI] [PubMed] [Google Scholar]
- 11.Pontes LA, Dias-Neto E, Rabello A. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am J Trop Med Hyg. 2002;66:157–162. doi: 10.4269/ajtmh.2002.66.157. [DOI] [PubMed] [Google Scholar]
- 12.Barber KE, Mkoji GM, Loker ES. PCR-RFLP analysis of the ITS2 region to identify Schistosoma haematobium and S. bovis from Kenya. Am J Trop Med Hyg. 2000;62:434–440. doi: 10.4269/ajtmh.2000.62.434. [DOI] [PubMed] [Google Scholar]
- 13.Magalhães KG, Jannotti-passos LK, Cavalho OS. Detection of Lymnea columella infection by Fasciola hepatica through multiplex PCR. Mem Inst Oswaldo Cruz. 2004;99:421–424. doi: 10.1590/s0074-02762004000400013. [DOI] [PubMed] [Google Scholar]
- 14.Melo FL, do Vale Gomes AL, Barbosa CS, Werkhauser RP, Abath FGC. Development of molecular approaches for the identification of transmission site of schistosomiasis. Trans R Soc Trop Med Hyg. 2006;100:1049–1055. doi: 10.1016/j.trstmh.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Dinkel A, Nickisch-Rosenegk M, Bilger B, Merli M, Lucius R, Romig T. Detection of Echinococcus multilocularis in the definitive host: coprodiagnosis by PCR as an alternative to necropsy. J Clin Microbiol. 1998;36:1871–1876. doi: 10.1128/jcm.36.7.1871-1876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stensvold CR, Saijuntha V, Sithithaworn P, Wongratanacheewin S, Strandgaard H, Ornbjerg N, Johansen MV. Evaluation of PCR based coprodiagnosis of human opisthorchiasis. Acta Trop. 2006;97:26–30. doi: 10.1016/j.actatropica.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Wongratanacheewin S, Phumidonming W, Sermsawan RW, Pipitgool V, Maleewong W. Detection of Opisthorchis viverrini in human stool specimens by PCR. J Clin Microbiol. 2002;40:3879–3880. doi: 10.1128/JCM.40.10.3879-3880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maleewong W, Intapun PM, Wongkam C, Wongsaroj T, Komsuwan T, Phumidonming W, Pongsakulchoti P. Detection of Opisthorchis viverrini in experimentally infected bithynid snails and cyprinoid fishes by PCR-based method. Parasitology. 2003;126:63–67. doi: 10.1017/s0031182002002573. [DOI] [PubMed] [Google Scholar]
- 19.Le TH, De NV, Blair D, Sithithaworn P, McManus DP. Clonorchis sinensis and Opisthorchis viverrini: development of a mitochondrial-based PCR for their identification and discrimination. Exp Parasitol. 2006;112:109–114. doi: 10.1016/j.exppara.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Anuntalabhochai S, Chiangda J, Chandet R, Apawat P. Genetic diversity within lychee (Litchi chinensis Soonn.) based on RAPD analysis; Int Symp Trop Subtrop Fruit; 26th Nov-1st Dec; Cairhs, Australia. 2000. 45 pp. [Google Scholar]
- 21.Wongsawad C, Wongsawad P, Chai JY, Paratasilpin T, Anuntalabhochai S. DNA quantities and qualities from various stages of some trematodes using optical and HAT-RAPD methods. Southeast Asian J Trop Med Public Health. 2006;37(suppl 3):62–68. [PubMed] [Google Scholar]
- 22.Sripalwit P, Wongsawad C, Wongsawad P, Anantalabhochai S. High annealing temperature-random amplified polymorphic DNA (HAT-RAPD) analysis of tree paramphistome flukes from Thailand. Exp Parasitol. 2007;115:98–102. doi: 10.1016/j.exppara.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Wongsawad C, Wongsawad P, Chai JY, Anantalabhochai S. Haplorchis taichui Witenberg, 1930: development of HAT-RAPD marker for the detection of minute intestinal fluke infections. Exp Parasitol. 2009;123:158–161. doi: 10.1016/j.exppara.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Chapco W, Ashton NW, Martel RKB, Antonishyn N, Crosby WL. A feasibility study of the use of random ampliWed polymorphic-DNA in the population genetics and systematic of grasshoppers. Genome. 1992;35:569–574. doi: 10.1139/g92-085. [DOI] [PubMed] [Google Scholar]
- 25.Wilde J, Waugh R, Powell W. Genetic fingerprinting of Theobroma clones using randomly ampliWed polymorphic DNA markers. Theor Appl Genet. 1992;83:871–877. doi: 10.1007/BF00226710. [DOI] [PubMed] [Google Scholar]