Abstract
Background. Membranous glomerulonephropathy (MN) is the most prevalent cause of nephrotic syndrome in adult humans. However, the specific biomarkers of MN have not been fully elucidated. We examined the alterations in gene expression associated with the development of MN. Methods. Murine MN was induced by cationic bovine serum albumin (cBSA). After full-blown MN, cDNA microarray analysis was performed to identify gene expression changes, and highly expressed genes were evaluated as markers both in mice and human kidney samples. Results. MN mice revealed clinical proteinuria and the characteristic diffuse thickening of the glomerular basement membrane. There were 175 genes with significantly different expressions in the MN kidneys compared with the normal kidneys. Four genes, metallothionein-1 (Mt1), cathepsin D (CtsD), lymphocyte 6 antigen complex (Ly6), and laminin receptor-1 (Lamr1), were chosen and quantified. Mt1 was detected mainly in tubules, Lamr1 was highly expressed in glomeruli, and CtsD was detected both in tubules and glomeruli. The high expressions of Lamr1 and CtsD were also confirmed in human kidney biopsies. Conclusion. The murine MN model resembled the clinical and pathological features of human MN and may provide a tool for investigating MN. Applying cDNA microarray analysis may help to identify biomarkers for human MN.
1. Introduction
Membranous glomerulonephropathy (MN), characterized by the presence of diffuse thickening of the GBM and subepithelial in situ immune-complex disposition, is the most common cause of idiopathic nephrotic syndrome in adults [1]. The clinical course in the majority of patients is slow and indolent. Spontaneous remissions of proteinuria occur in approximately one quarter of patients; approximately half will have stable renal function with or without continued proteinuria and approximately 30%–40% of patients with MN develop progressive renal impairment, which results in end-stage renal failure after 10–15 years [2–4]. Effector cells, immunoglobulins, inflammatory cytokines, complements, and oxidative stress all participate in the pathogenesis of MN, although the definite pathomechanism of MN has not yet been fully elucidated [5–8]. Persistence of marked proteinuria, impaired estimated glomerular filtration rate at the time of discovery, increased urinary excretion rates for immunoglobulin G, α1-microglobulin, β-2 microglobulin, or complement proteins, increased interstitial fibrosis, and/or advanced stages of glomerular structural abnormalities are among the risk factors for a higher propensity of progression [9, 10]. However, no definite biomarkers or prognostic factors have been clearly documented.
Heymann nephritis (HN), a rat model of autoimmune-mediated glomerulonephritis, is induced by immunization with rat renal tubular antigen Fx1A or transfusion with anti-Fx1A Igs [11]. Alternatively, murine models are induced by repeated doses of cationic bovine serum albumin (cBSA), which has a similar clinical course and histopathology to human MN [12, 13]. Due to a limited source of human tissue being available and the low cost and advantages of using transgenic mice, this murine MN model may be suitable for sample experimental applications to investigate MN. Improved understanding of MN may create new opportunities for therapeutic intervention which may benefit patients with MN in the future. Microarray high throughput techniques have become increasingly important in basic and applied biomedical research due to the efficiency of analyzing large scale gene expressions simultaneously to investigate the complex molecular basis of pathological processes on a genomic scale [14–16].
In this study, we used cDNA microarrays to analyze global gene expressions of cortical renal tissue in this cBSA-induced MN model. The upregulated genes and their relevance to the evolution of MN were examined further by immunohistochemical staining of kidneys from the rats and humans.
2. Materials and Methods
2.1. Experimental Animals
This study was performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health and was approved by the Animal Care and Ethics Committee of the National Defense Medical Center (Taipei, Taiwan). BALB/c mice (4–6 weeks old, about 20 g body weight) were purchased from the National Laboratory Animal Center (Taipei, Taiwan). The mice were kept in the Laboratory Animal Center of the National Defense Medical Center under specific-pathogen-free conditions.
2.2. MN Animal Model: Induction Method
Six-week-old BALB/C mice were divided into an experimental group (group A) and a control group (group B). Both groups A and B were immunized with 1 mg of C-BSA and Freund's complete adjuvant. Two weeks later, group A was injected intravenously with 3 mg/kg of c-BSA 3 times weekly every other day for 4 weeks, and group B received saline with the same schedule. Preparation of cationic-bovine serum albumin (C-BSA) was performed as previously described [12, 13].
2.3. Clinical and Pathological Evaluation
Blood and urine sample are obtained from the mice then microfuged and stored at −70°C until assayed. Serum and urine biochemistries were measured as described previously [17]. Proteinuria was calculated as the ratio of urinary protein (mg/mL) to urinary creatinine (mg/dL) (Up/Ucr).
Renal tissues were snap frozen or fixed in either Carson-Millonig's solution for electron microscopy (EM) or 10% formalin fixative solution for immunohistochemistry (IHC) and hematoxylin and eosin staining. Frozen renal specimens were cut with a cryostat into 4-μm sections for the detection of immunofluorescence (IF).
For IF, the frozen sections were air-dried, fixed in acetone for 10 min at room temperature, and incubated with fluorescein isothiocyanate-conjugated goat antimouse IgG (Cappel; Organon Teknika, Durham, NC). For IHC, a microwave heating procedure was used as described previously [18], followed by incubating with goat anti-Lamin receptor 1 (R&D, Minneapolis, MN) and Cathepsin D (Santa Cruz, Santa Cruz, CA) at 4°C overnight. Horseradish peroxidase-conjugated protein-G (Pierce, Rockford, IL) was then applied to the sections for 1 h. Reaction products were visualized using a colour solution consisting of AEC (DAKO, Carpinteria, CA) for 2-3 min, and the slides were counterstained lightly with haematoxylin.
2.4. Microarray Analysis
The cDNA microarray containing 15000 different mouse cDNA clones (http://lgsun.grc.nia.nih.gov/cDNA/15k.html) was provided by Biochip R&D Center, Tri-Service General Hospital, Taipei, Taiwan. Total RNA was extracted from the renal cortices of the normal control and MN mice and was then annealed to oligo(dT) and reverse transcribed in the presence of Cy5- and Cy3-labelled dUTP, respectively. After concentration, the two cDNA probes were further processed and applied to the slides for cDNA microarray analysis as described previously [18]. The slides were scanned with a GenePix 4000A scanner (Axon Instruments, Union City, CA). Data normalization and analysis were performed as described previously [18]. The average of median ratios from replicates was calculated for each spot. Spots representing housekeeping genes were used to normalize the entire slide so that all slides could be compared directly. Finally, the ratios were taken as log2 transformation, and the SD of the mean was then calculated from these log2 ratios for the determination of expression outliers.
2.5. RNA Extraction and Real-Time Quantitative PCR
Total RNA was extracted from the renal cortices with TRIzol reagent (Life Technologies, Gaithersburg, MD, USA). First-strand complementary DNA (cDNA) was synthesized using a SuperScript III Reverse Transcriptase kit (Life Technologies) following the standard protocol. Real-time polymerase chain reaction (PCR) analyses were performed using a SYBR Green Master Mix Kit (Bio-Rad) and an Opticon PCR thermal cycler (MJ Research, Waltham, MA, USA). The relative expression of each cytokine mRNA was determined and normalized to the expression of the internal housekeeping gene GAPDH. All samples were measured in triplicate three times. Primer and probe sequences are listed in Table 1.
Table 1.
The 175 genes with significantly different expressions in the MN kidneys compared with the normal kidneys.
| Increased expression | |
|---|---|
| Adamts1: a disintegrin and metalloproteinase with thrombospondin motif type 1 | Ly6a: lymphocyte antigen 6 complex, locus A |
| Akr1b3: aldo-keto reductase family 1, B3 | Ly6e: lymphocyte antigen 6 complex,, locus E |
| Aldo: aldolase 1, A isoform | Lyzs: lysozyme |
| Anxa2: annexin A2 | Macs: myristoylated alanine rich protein |
| Anxa5: annexin A5 | Mglap: matrix gamma-carboxyglutamate (gla) protein |
| Arbp: acidic ribosomal phosphoprotein | Morc3: MORC family CW-type zinc finger 3 |
| Arpclb: actin-related protein | Mpz: myelin protein zero |
| Axl: AXL receptor tyrosine kinase | Mrgpre: MAS-related GPR, member E |
| Bgn: biglycan | Mrp63: mitochondrial ribosomal protein 63 |
| C1ga: complement component C1ga | Mrps6: mitochondrial ribosomal protein S6 |
| C1gc: complement component C1gc | Mt1: metallothionein 1 |
| C3: complement component 3 | Mt2: metallothionein 2 |
| Calbl: calbindin-28K | Muc1: mucin 1, cell surface associated |
| Cap 1: adenylyl cyclase-associated protein 1 | Muc1: mucin 1, transmembrane |
| Capg: capping protein (actin filament) | Nes: nestin |
| Cd24a: CD24a antigen | Nfkbia: nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| Cd9: CD9 antigen | Nid1: nidogen 1 |
| Cldn7: claudin 7 | Npnt: nephronectin |
| Clic1: chloride intracellular channel | Pcolce: procollagen C-proteinase enhancer |
| Clu: clusterin | Pctp1: phosphatidylcholine transfer protein |
| Cmkbr2: chemokine (C-C) receptor 2 | Plat: plasminogen activator, tissue |
| Col 3a1: procollagen, type III, alpha 1 | Prdx6: peroxiredoxin 6 |
| Col4a1: procollagen, type IV, alpha 1 | Prkab1: protein kinase, AMP-activated, beta 1 noncatalytic subunit |
| Col4a2: procollagen, type IV, alpha 2 | Ptma: prothymosin alpha |
| Colla2: procollagen, type I, alpha 2 | Rab31: member RAS oncogene family |
| Collal: procollagen, type I, alpha 1 | Raph1: Ras association and pleckstrin homology domains 1 |
| Cox7a3: cytochrome c oxidase, subunit 7a3 | Rbm3: RNA binding motif protein 3 |
| Csnkld: casein kinase 1 delta | Rbm5: RNA binding motif protein 5 |
| Csrp: cysteine rich protein | Rhoc: ras homolog gene family, member C |
| Cst3: cystatin C | Rpia: ribose 5-phosphate isomerase A |
| Ctsc: cathepsin C | Rpl12: ribosomal protein L12 |
| Ctsd: cathepsin D | Rpl29: ribosomal protein L29 |
| Dbh: dopamine beta-hydroxylase | Rpl3: ribosomal protein L3 |
| Dgcr: DiGeorge syndrome critical region | Rpl31: ribosomal protein L31 |
| Dgk1: diacylglycerol kinase 1 | Rpl36: ribosomal protein L36 |
| Dok7: docking protein 7 | Rpl37a: ribosomal protein L37a |
| Dppa2: developmental pluripotency associated 2 | Rpl41: ribosomal protein L41 |
| Dtx1: deltex homolog 1 (Drosophila) | Rpl5: ribosomal protein L5 |
| Eef1a1: eukaryotic translation elongation factor 1 alpha 1 | Rpl7: ribosomal protein L7 |
| Eif3d: eukaryotic translation initiation factor 3, subunit D | Rplp1: ribosomal protein, large, P1 |
| Elk1: member of ETS oncogene family | Rps15: ribosomal protein S15 |
| Eras: ES cell expressed Ras | Rps15: ribosomal protein S15 |
| F2r: coagulation factor II (thrombin) | Rps16: ribosomal protein S16 |
| F9: coagulation factor IX | Rps20: ribosomal protein S20 |
| Fau: Finkel-Biskis-Reilly murine sarcoma | Rps3a: ribosomal protein S3a |
| Fcerlg: Fc receptor, IgE high affinity | Rps4x: ribosomal protein S4, X-linked |
| Fkbp5: FK506 binding protein 5 | Rps6: ribosomal protein S6 |
| Fn1: fibronectin 1 | Rrm2: ribonucleotide reductase M2 |
| Fn14: fibroblast growth factor-inducible 14 | Rsbn1: round spermatid basic protein 1 |
| Fstl: follistatin-like protein | Rundc3a: RUN domain containing 3A |
| Ft11: ferritin light chain 1 | S100a6: S100 calcium binding protein A |
| Gnb211: guanine nucleotide binding protein | Sam68: Src-associated in mitosis, 68kDa |
| Gpr56: G protein-coupled receptor 56 | Sdc4: syndecan 4 |
| H2-Ebl: histocompatibility 2, class 1E beta | Serpinb6: serpin peptidase inhibitor, B6 |
| Hcfc1: host cell factor C1 | Serping1: serpin peptidase inhibitor, G1 |
| Hdc-c: histidine decarboxylase | Serpinh1: serpin peptidase inhibitor, H1 |
| Hmga1: high mobility group A1 | Sh3bgrl3: SH3 domain binding glutamic acid-rich protein like 3 |
| Hmgb2: high mobility group box 2 | Sparc: secreted protein acidic and rich in cysteine |
| Hmgn2: high mobility group nucleosomal binding domain 2 | Spp1: secreted phosphoprotein 1 |
| Hn1: hematological and neurological expressed protein 1 | Star: steroidogenic acute regulatory protein |
| Hnrp1: heterogeneous nuclear ribonucleoprotein A1 | Syn1: synapsin 1 |
| Hsp25: heat shock protein, 25 kDa | Tbst1: protein-tyrosine sulfotransferase 1 |
| Hsp84-1: heat shock protein, 84 kDa 1 | Tgfb1i4: transforming growth factor beta 1 induced transcript 4 |
| Idb2: inhibitor of DNA binding 2 | Tip39: tuftelin-interacting protein 33 |
| Klhl2: kelch-like 2, Mayven (Drosophila) | Tmsb10: thymosin, beta 10 |
| Krt2-8: keratin complex 2, basic, gene 8 | Tmsb4x: thymosin, beta 4, X chromosome |
| Lamr1: laminin receptor 1 | Tpi: triose phosphate isomerase |
| Laptm5: lysosomal associated protein multispanning transmembrane 5 | Tspan2: tetraspanin 2 |
| Lcn2: lipocalin 2 | Tuba2: tubulin, alpha 2 |
| Lcn7: lipocalin 7 | Tubb5: tubulin, beta 5 |
| Ldh1: lactate dehydroaenase 1 | Ubc: ubiquitin C |
| Lgals3: lectin, galactoside-binding, soluble, 3 | Uchrb: Ubiquitin c-terminal hydrolase |
| Litaf: LPS-induced TNF-alpha factor | Ucp2: uncoupling protein 2 |
| Lrat: lecithin retinol acyltransferase | Wisp2: WNT1 inducible signaling pathway protein 2 |
| Lu: Lutheran blood group glycoprotein | |
| Decreased expression | |
| Ak4: adenylate kinase 4 | Lrp2: low density lipoprotein receptor-related protein 2 |
| Bhmt2: betaine-homocysteine methyltransferase 2 | Mad1l1: mitotic arrest deficient-like 1 |
| Bid: BH3 interacting domain death agonist | Map4: microtubule-associated protein 4 |
| Ccnc: cyclin C | Med28: mediator complex subunit 28 |
| Cdc2a: cell division cycle 2 homolog A | Megf10: multiple EGF-like-domains 10 |
| Degs2: degenerative spermatocyte homolog 2 | Narg1: NMDA receptor regulated 1 |
| Emp3: epithelial membrane protein 3 | Prlr: prolactin receptor |
| Ensa: endosulfine alpha | Rac GAP: Rac GTPase activating protein 1 |
| Epha2: Eph receptor A2 | Rhd: Rh blood group, D antigen |
| Evi5: ecotropic viral integration site 5 | Rhov: ras homolog gene family, member V |
| Fignl1: fidgetin-like 1 | Slc15a2: solute carrier family 15, member 2 |
| Hmgcs2: 3-hydroxy-3-methylglutaryl CoA synthase 2 | Ttr: Transthyretin |
| Hsd3b: 3-hydroxysteroid dehydrogenase type 2 | Xnp: X-linked nuclear protein |
2.6. Statistical Analyses
All data were expressed as mean ± SD. Statistical analysis was performed by the t-test for two groups or by analysis of variance (ANOVA) for multiple groups with Tukey's post-hoc test. Correlation analysis was examined by tests of linear regression. Significance was defined as P < .05.
3. Results
3.1. Clinical and Pathological Evaluation of the MN Model
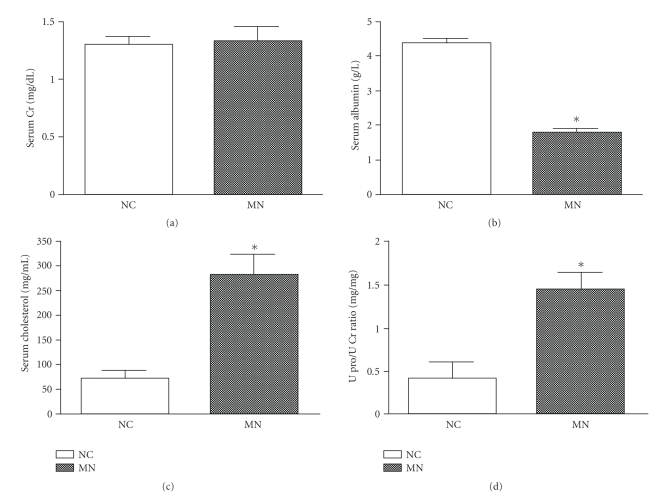
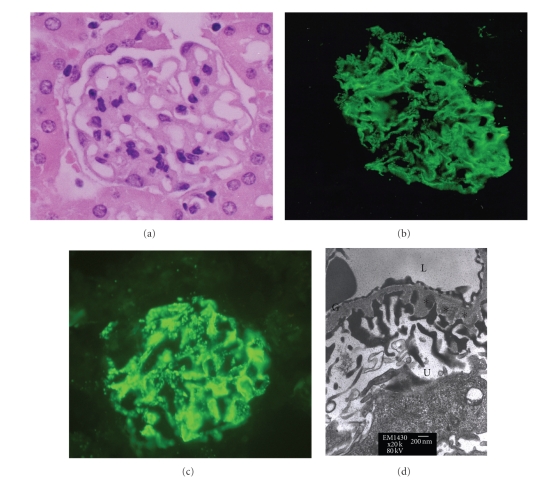
Mice with experimental MN developed normal renal function (Figure 1(a)) and the characteristic clinical symptoms of hypoalbuminemia (Figure 1(b)), hypercholesterolemia (Figure 1(c)), and overt proteinuria (Figure 1(d)). Renal histopathology revealed typical findings of diffuse basement thickening over the whole glomerular basement membrane (GBM) in hematoxylin and eosin staining (Figure 2(a)). Immunofluorescence of IgG and C3 showed a granular pattern of deposition along the GBM (Figures 2(b) and 2(c)). Electronic microscopy revealed a granular pattern of deposition over the subepithelial space but no mesangial electron dense deposits (Figure 2(d)).
Figure 1.
Clinical manifestations in mice with MN. The biochemical findings in MN mice revealed normal renal function (a), hypoalbuminemia (b), hypercholesterolemia (c), and overt proteinuria (d). *P < .05.
Figure 2.
Renal histopathology in mice with MN. Histopathology revealed findings characteristic of diffuse basement membrane thickening, as observed in the (a) hematoxylin and eosin staining, (b) positive granular immunofluorescent staining for IgG, (c) positive granular immunofluorescent staining for C3, and (d) subepithelial deposition (asterisk). NC: normal control; MN: membranous nephropathy; G: glomerular basement membrane; L: lumen of capillary; U: urinary space.
3.2. Gene Expression in the Renal Cortex
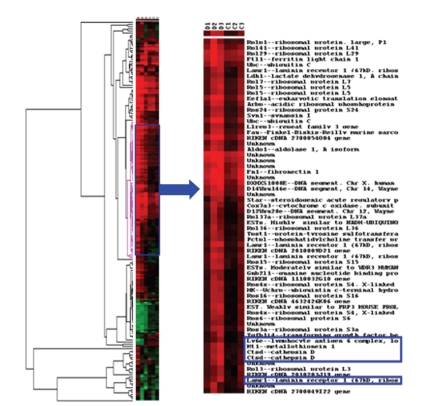
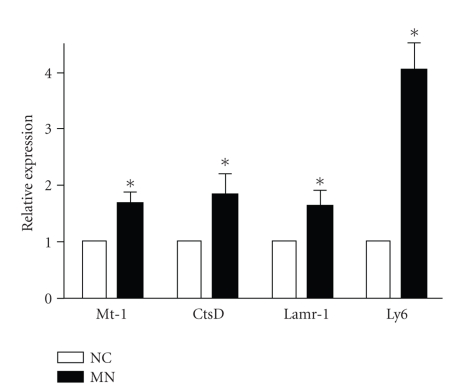
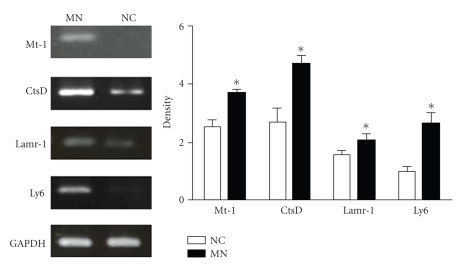
To determine the profile of altered gene expression associated with MN, cDNA microarray chip analysis was performed on cortical renal tissue from the NC and MN mice. There were 175 genes with significantly different expressions in the MN kidneys compared with the normal kidneys (Table 1). Four enhanced genes related to injury, inflammation, and cell-matrix interaction were chosen: Mt-1, CtsD, Lamr-1, and Ly6 (Figure 3). To further confirm the upregulated gene profiles in the cortical tissue, we designed primers and performed quantitative real-time PCR (Table 2). All four chosen genes revealed significant increases in expression as shown using the microarray chip (Figure 4). These protein expressions were also confirmed using Western blot (Figure 5).
Figure 3.
Dendrogram of microarray results from MN kidneys which revealed 175 genes with significantly different expressions compared with normal kidneys.
Table 2.
PCR gene sequences in four candidate genes and housekeeping genes.
| Name | Forward | Reverse | Product (bp) |
|---|---|---|---|
| Ly6e | 5′agtcttcctgcctgtgctgttg3′ | 5′cgccacaccgagattgagattg3′ | 253 |
| Lamr-1 | 5′ctcttatgtcaacctgcccacc3′ | 5′tgctcctccttctcaatctcctc3′ | 221 |
| CtsD | 5′agctgtcctacctgaacgtcac3′ | 5′tgtctttccaccctgcgatacc3′ | 287 |
| Mt-1 | 5′tcaacgtcctgagtaccttctcc3′ | 5′tgaagacctctgcttcctgtcc3′ | 397 |
| GAPDH | 5′tccgccccttctgccgatc3′ | 5′cacggaaggccatgccagtga3′ | 354 |
Ly6e: lymphocyte antigen 6 complex, Lamr-1: laminin receptor-1 (67kDa), CtsD: cathepsin D, Mt-1: metallothionein-1, GAPDH: housekeeping gene, bp: base pair.
Figure 4.
Candidate genes mRNA expression levels. Four candidate genes RNA were prepared from the kidney cortices of NC and MN mice, and the levels of mRNA expression were determined by RT-PCR (n = 3). *P < .05.
Figure 5.
Confirmation of candidate protein expressions based on the microarray results by Western blot of kidney protein extraction. NC: normal control; MN: membranous nephropathy; Mt-1: metallothionein-1; CtsD: cathepsin D; Lamr-1: laminin receptor-1; Ly6e: lymphocyte antigen 6 complex (n = 3). *P < .05.
3.3. Protein Expression and Localization in Kidney from Mice and Human
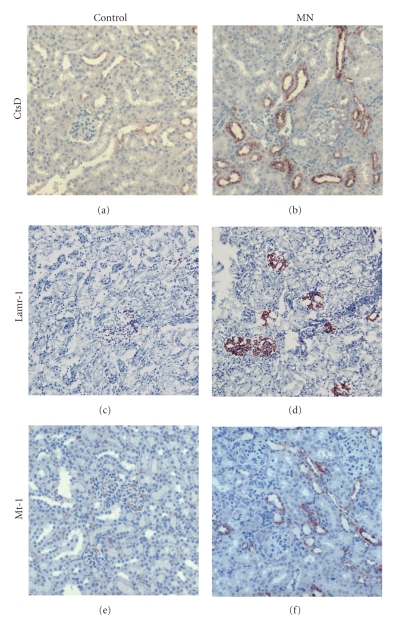
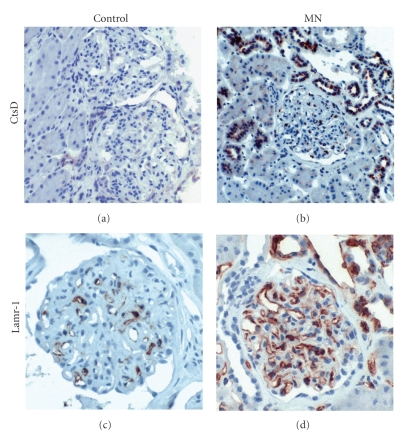
We further wanted to determine whether the gene-encoded protein expressions in the kidney cortices from the mice of the control and experimental groups correlated with gene expression. As the main source of Ly6e is from immune cells, we chose CtsD, Lamr-1, and Mt-1 gene-encoded proteins using IHC to identify the cellular source and the glomerular expression in the renal tissues. Compared with normal controls, MN mice showed enhanced expressions of all of these three proteins in the kidneys, which were similar to those shown by the microarray chip (Figure 6). The CtsD protein was expressed mainly in tubulointerstitium with a minority in glomeruli (Figures 6(a) and 6(b)). The expression of Lamr-1 protein was mainly in the glomeruli and Mt-1 was restricted to the tubulointerstitium (Figures 6(c)–6(f)). The major objective of our study was to test whether gene products identified from the experimental MN model induced by cBSA could be applied to human disease. We therefore chose human CtsD and Lamr-1 proteins, which are expressed in glomeruli, and performed IHC to check their expressions. As illustrated in Figures 7(a)–7(d), the enhanced expression pattern was similar to that in the murine model.
Figure 6.
Renal immunohistochemistry of candidate proteins in mice kidneys. Kidneys from mice of the normal control group ((a), (c), and (e)) and membranous nephropathy group ((b), (d), and (f)) were stained for CtsD ((a) and (b)), Lamr-1 ((c) and (d)), and Mt-1 ((e) and (f)). All images are at 400× magnification. CtsD: cathepsin D; Lamr-1: laminin receptor-1; Mt-1: metallothionein-1.
Figure 7.
Renal immunohistochemistry of candidate proteins in human kidneys. Kidneys from humans of the normal control group (a and c) and membranous nephropathy group (b and d) were stained for CtsD (a and b) and Lamr-1 (c and d). All images are at 400x magnification. CtsD: cathepsin D; Lamr-1: laminin receptor-1.
4. Discussion
The murine model of MN induced by cBSA resembles the clinical and pathological features of human MN and may provide a tool to investigate MN. In this study, we used a cursory approach of global gene expression, cDNA microarray, to analyze differential gene expression associated with the development of MN from a murine model. Using real-time mRNA analysis of kidney tissues, we observed that CtsD, Lamr-1, Mt-1, and Ly6e were differentially upregulated in affected kidneys and identified the expressions of these proteins in kidneys from mice and humans. These proteins may be viewed as biomarkers associated with the development of MN.
Although there was a lack of an evident mechanism in our findings, the identified genes that might contribute to the pathogenesis or the pathophysiological potential of each gene need to be further addressed. Normal GBM contains type IV collagen, noncollagenous glycoproteins, and heparan sulphate proteoglycans [19]. Laminin is the major glycoprotein binding to type IV collagen to form the backbone of basement membranes providing tensile strength and cell-adhesive properties. Previous studies have shown that there is an increased production of laminin with the appearance of abnormal laminin isoforms in MN. This has been proposed to play a role in the occurrence of proteinuria by modifying the functional properties of GBM and/or by modifying podocyte functions [20]. The presence of anti-LMN Ab has been reported in patients with various diseases involving basement membrane such as Goodpasture syndrome and poststreptococcal glomerulonephritis (GN) [21–23]. However, to the best of our knowledge, this is the first study to find an enhanced expression of laminin receptor-1 in MN. Laminin receptor-1 is a cell-surface receptor for the extracellular matrix (ECM) glycoprotein laminin with high affinity and specificity. It is well established that the receptor interacts directly with laminin to play important roles both in cell adhesion to the basement membrane and in signaling transduction. In addition to direct interactions with laminin, it also has been proposed to facilitate interactions between laminin and integrins as well as angiogenesis in vascular endothelial cells [24]. However, which cell type expresses Lamr-1, how the molecule is attributed to the process of MN, and its definite mechanism still require further research.
Cathepsin D, a 52-kDa protein and the major pepstatin A-sensitive aspartate protease within lysosomes, has been suggested to be involved in several biological activities including the regulation of apoptotic pathways, the activation of proteolytic enzymes involved in the degradation of extracellular matrix components and stimulatory effects towards cellular proliferation and angiogenesis [25]. Cathepsin D is also a renin-like enzyme that catalyzes angiotensinogen breakdown to AngI. In normal kidneys, this enzyme is preferably expressed in the distal tubular system and the collecting ducts, and its expression is upregulated in chronic renal disease [26]. Morphological, biochemical, and physiological heterogeneity of renal lysosomes has been confirmed in the kidney cortices of GN patients, such as Goodpasture disease and poststreptococcal GN [27, 28]. Changes in lysosome populations in rat kidney cortices induced by passive Heymann nephritis have been observed [29]. Furthermore, we confirmed the protein expression in human MN kidneys. Metallothionein is a heat stable, low molecular weight protein participating in chelating heavy metals and has a physiological role in the scavenging of free radicals [30]. Ly6e, which belongs to the Ly6 superfamily, is a small glycoprotein linked to the glycophosphatidylinositol (GPI) anchor participating in cell signaling and cell adhesion processes and is involved in lymphocyte activation [31]. Increased Ly6e gene expression in response to proteinuria, as well as being correlated to lupus activity and renal lesions, suggests a potential role of Ly6e in the pathophysiology of renal disease [32]. However, the definite mechanism is still unclear.
Microarray high throughput techniques are a very efficient method to analyze large-scale gene expressions simultaneously to investigate the complex molecular basis of pathological processes on a genomic scale. They have become increasingly important in basic and applied biomedical research including several renal diseases [14, 33]. However, a whole-genome gene expression analysis study of MN using a murine model and then being applied to human disease has not been reported. Cluster analysis of DNA microarrays can better provide an intuitive understanding of how to analyze microarray data, making it easier to interpret the meaning of the results in a biological framework. The main limitation of the present study is that it addressed gene expression in kidney cortex instead of glomeruli, which would have been more meaningful. Murine models of MN induced by cBSA not only resemble the clinical and pathological features of human MN, but also offer the advantages of being cheaper and easy to manipulate and having potential applications in gene-knockout or transgenic mouse studies on MN.
The precise nature of the idiopathic MN-initiating antigen is unclear. A pathogenic antigen, megalin (gp 330), has been identified in Heymann nephritis but not in human or mice glomeruli [34]. Although the cBSA used in our model is an exogenous rather than an endogenous antigen, its immunopathological specificity has been proved during MN. Similar presentations were confirmed by IHC staining of the human specimens, also demonstrating the value of this murine MN model. We did not differentiate which component of the renal cortex contributed to the gene expression revealed by cDNA microarray analysis, but both glomerular and tubular responses composed the kidney response in MN. Although there was a lack of an evident functional assay of the genes in our findings, there is still the potential to identify biomarkers for human MN. However, whether these genes play an important role in the pathogenesis or pathophysiologies of glomerular injury needs to be further addressed. Our strategy of using cDNA microarrays to simultaneously monitor global gene expressions in renal tissue from the murine cBSA-induced MN model and then applying the findings to human disease seemed to work and to be practical.
In conclusion, the murine model of MN induced by cBSA resembled the clinical and pathological features of human MN and may provide a tool to investigate MN. Applying cDNA microarray analysis in this model may help to identify biomarkers for human MN, which may then broaden our understanding of the possible mechanisms of MN. This may provide further insight into the disease and generate new hypotheses for potential novel therapeutic targets in the future.
Acknowledgments
This work was supported by the National Science Council, Taiwan (NSC NSC98-2314-B-016-061-MY3) to Y-F Lin and Tri-Service General Hospital (TSGH-C100-031) to C-C. Wu.
References
- 1.Cattran DC. Idiopathic membranous glomerulonephritis. Kidney International. 2001;59(5):1983–1994. doi: 10.1046/j.1523-1755.2001.0590051983.x. [DOI] [PubMed] [Google Scholar]
- 2.Chadban SJ, Atkins RC. Glomerulonephritis. The Lancet. 2005;365(9473):1797–1806. doi: 10.1016/S0140-6736(05)66583-X. [DOI] [PubMed] [Google Scholar]
- 3.Glassock RJ. Diagnosis and natural course of membranous nephropathy. Seminars in Nephrology. 2003;23(4):324–332. doi: 10.1016/s0270-9295(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 4.Ponticelli C, Passerini P. Treatment of membranous nephropathy. Nephrology Dialysis Transplantation. 2001;16(5):8–10. doi: 10.1093/ndt/16.suppl_5.8. [DOI] [PubMed] [Google Scholar]
- 5.Cybulsky AV, Quigg RJ, Salant DJ. Experimental membranous nephropathy redux. American Journal of Physiology. 2005;289(4):F660–F671. doi: 10.1152/ajprenal.00437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nangaku M, Shankland SJ, Couser WG. Cellular response to injury in membranous nephropathy. Journal of the American Society of Nephrology. 2005;16(5):1195–1204. doi: 10.1681/ASN.2004121098. [DOI] [PubMed] [Google Scholar]
- 7.Ronco P, Debiec H. New insights into the pathogenesis of membranous glomerulonephritis. Current Opinion in Nephrology and Hypertension. 2006;15(3):258–263. doi: 10.1097/01.mnh.0000222692.99711.02. [DOI] [PubMed] [Google Scholar]
- 8.Wu CC, Chen JS, Chen SJ, et al. Kinetics of adaptive immunity to cationic bovine serum albumin-induced membranous nephropathy. Kidney International. 2007;72(7):831–840. doi: 10.1038/sj.ki.5002426. [DOI] [PubMed] [Google Scholar]
- 9.Ponticelli C. Prognosis and treatment of membranous nephropathy. Kidney International. 1986;29(4):927–940. doi: 10.1038/ki.1986.88. [DOI] [PubMed] [Google Scholar]
- 10.Branten AJW, Du Buf-Vereijken PW, Klasen IS, et al. Urinary excretion of β2-microglobulin and IgG predict prognosis in idiopathic membranous nephropathy: a validation study. Journal of the American Society of Nephrology. 2005;16(1):169–174. doi: 10.1681/ASN.2004040287. [DOI] [PubMed] [Google Scholar]
- 11.Heymann W, Hackel DB, Harwood S, Wilson SGF, Hunter JLP. Production of nephrotic syndrome in rats by Freund’s adjuvants and rat kidney suspensions. Journal of the American Society of Nephrology. 2000;11(1):183–188. [PubMed] [Google Scholar]
- 12.Chen JS, Chen A, Chang LC, et al. Mouse model of membranous nephropathy induced by cationic bovine serum albumin: antigen dose-response relations and strain differences. Nephrology Dialysis Transplantation. 2004;19(11):2721–2728. doi: 10.1093/ndt/gfh419. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, Chen JS, Lin SH, Chen A, Sytwu HK, Lin YF. Experimental model of membranous nephropathy in mice: sequence of histological and biochemical events. Laboratory Animals. 2008;42(3):350–359. doi: 10.1258/la.2007.06016e. [DOI] [PubMed] [Google Scholar]
- 14.Hauser PV, Perco P, Mühlberger I, et al. Microarray and bioinformatics analysis of gene expression in experimental membranous nephropathy. Nephron Experimental Nephrology. 2009;112(2):e43–e58. doi: 10.1159/000213505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moch H, Schraml P, Bubendorf L, et al. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. American Journal of Pathology. 1999;154(4):981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudnicki M, Eder S, Perco P, et al. Gene expression profiles of human proximal tubular epithelial cells in proteinuric nephropathies. Kidney International. 2007;71(4):325–335. doi: 10.1038/sj.ki.5002043. [DOI] [PubMed] [Google Scholar]
- 17.Wu CC, Lu KC, Chen JS, et al. HO-1 induction ameliorates experimental murine membranous nephropathy: anti-oxidative, anti-apoptotic and immunomodulatory effects. Nephrology Dialysis Transplantation. 2008;23(10):3082–3090. doi: 10.1093/ndt/gfn247. [DOI] [PubMed] [Google Scholar]
- 18.Ka SM, Rifai A, Chen JH, et al. Glomerular crescent-related biomarkers in a murine model of chronic graft versus host disease. Nephrology Dialysis Transplantation. 2006;21(2):288–298. doi: 10.1093/ndt/gfi229. [DOI] [PubMed] [Google Scholar]
- 19.Haraldsson B, Nyström J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiological Reviews. 2008;88(2):451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 20.Fischer E, Mougenot B, Callard P, Ronco P, Rossert J. Abnormal expression of glomerular basement membrane laminins in membranous glomerulonephritis. Nephrology Dialysis Transplantation. 2000;15(12):1956–1964. doi: 10.1093/ndt/15.12.1956. [DOI] [PubMed] [Google Scholar]
- 21.Kefalides NA, Pegg MT, Ohno N. Antibodies to basement membrane collagen and to laminin are present in sera from patients with poststreptococcal glomerulonephritis. Journal of Experimental Medicine. 1986;163(3):588–602. doi: 10.1084/jem.163.3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foidart JM, Yaar M, Figueroa A. Abortion in mice induced by intravenous injections of antibodies to type IV collagen or laminin. American Journal of Pathology. 1983;110(3):346–357. [PMC free article] [PubMed] [Google Scholar]
- 23.Yaar M, Foidart JM, Brown KS. The Goodpasture-like syndrome in mice induced by intravenous injections of anti-type IV collagen and anti-laminin antibody. American Journal of Pathology. 1982;107(1):79–91. [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson J, McFerran NV, Pivato G, et al. The 67 kDa laminin receptor: structure, function and role in disease. Bioscience Reports. 2008;28(1):33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- 25.Press EM, Porter RR, Cebra J. The isolation and properties of a proteolytic enzyme, cathepsin D, from bovine spleen. The Biochemical Journal. 1960;74:501–514. doi: 10.1042/bj0740501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graciano ML, Cavaglieri RDC, Dellê H, et al. Intrarenal renin-angiotensin system is upregulated in experimental model of progressive renal disease induced by chronic inhibition of nitric oxide synthesis. Journal of the American Society of Nephrology. 2004;15(7):1805–1815. doi: 10.1097/01.asn.0000131528.00773.a9. [DOI] [PubMed] [Google Scholar]
- 27.Zou J, Hannier S, Cairns LS, et al. Healthy individuals have goodpasture autoantigen-reactive T cells. Journal of the American Society of Nephrology. 2008;19(2):396–404. doi: 10.1681/ASN.2007050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karan A, Saatci U, Bakkaloglu A. The role of cathepsin D in pathogenesis of acute post streptococcal glomerulonephritis. Acta Paediatrica Scandinavica. 1976;65(3):355–360. doi: 10.1111/j.1651-2227.1976.tb04897.x. [DOI] [PubMed] [Google Scholar]
- 29.Andonian S, Hermo L. Cell- and region-specific localization of lysosomal and secretory proteins and endocytic receptors in epithelial cells of the cauda epididymidis and vas deferens of the adult rat. Journal of Andrology. 1999;20(3):415–429. [PubMed] [Google Scholar]
- 30.Nakajima K, Kodaira T, Kato M, et al. Development of an enzyme-linked immunosorbent assay for metallothionein-I and -II in plasma of humans and experimental animals. Clinica Chimica Acta. 2010;411(9-10):758–761. doi: 10.1016/j.cca.2010.02.058. [DOI] [PubMed] [Google Scholar]
- 31.Pflugh DL, Maher SE, Bothwell ALM. Ly-6 superfamily members Ly-6A/E, Ly-6C, and Ly-6I recognize two potential ligands expressed by B lymphocytes. Journal of Immunology. 2002;169(9):5130–5136. doi: 10.4049/jimmunol.169.9.5130. [DOI] [PubMed] [Google Scholar]
- 32.Tang J, Gu Y, Zhang M, et al. Increased expression of the type I interferon-inducible gene, lymphocyte antigen 6 complex locus E, in peripheral blood cells is predictive of lupus activity in a large cohort of Chinese lupus patients. Lupus. 2008;17(9):805–813. doi: 10.1177/0961203308089694. [DOI] [PubMed] [Google Scholar]
- 33.Böttinger EP, Ju W, Zavadil J. Applications for microarrays in renal biology and medicine. Experimental Nephrology. 2002;10(2):93–101. doi: 10.1159/000049904. [DOI] [PubMed] [Google Scholar]
- 34.Kerjaschki D, Farquhar MG. The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(18):5557–5561. doi: 10.1073/pnas.79.18.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]