Abstract
Background
A simple and rapid IsoAmp® HSV assay has been developed for qualitative detection of herpes simplex virus (HSV) types 1 and 2 from genital lesions. Sample preparation involved a simple dilution step and the diluted specimens were directly add to the device and amplified by isothermal helicase-dependent amplification (HDA). Amplification products were then detected by a DNA strip embedded in a disposable cassette without any instrument. The total test turn-around time is less than 1.5 hours from specimen processing to result reporting.
Objectives
To evaluate the analytical and clinical performance of the IsoAmp® HSV assay as well as the robustness and reproducibility of the assay.
Study Design
The analytical sensitivity of the IsoAmp® HSV assay were determined using both HSV-1 and HSV-2. Clinical performance was evaluated using 135 frozen specimens collected from patients with suspected HSV infection in genital area.
Results
The analytical sensitivity of the assays was 5.5 and 34.1 copies/reaction for HSV-1 and HSV-2 respectively with a 95% confidence interval. When the herpes viral culture was used as the reference standard, the clinical sensitivity and specificity of the IsoAmp® HSV assay were 100.0% and 96.3% respectively. The inter-laboratory reproducibility achieved an overall 97.5% agreement by testing a total of 80 blinded HSV-1 samples among five laboratories.
Conclusion
Adequate analytical and clinical performance of the IsoAmp® HSV assay was demonstrated. This assay is simple to perform and has acceptable inter-laboratory reproducibility.
Keywords: herpes simplex viruses, isothermal helicase-dependent amplification, disposable detection device
1. Background
Herpes simplex viruses (HSV) are a common human pathogen found worldwide that cause a variety of diseases, including viral meningitis, skin infection, genital herpes, and neonatal herpes 1. Neonatal herpes virus infection is often severe, if not fatal 2. HSV transmission can result from direct contact with infectious secretions from either symptomatic or asymptomatic individuals. HSV have two different types: HSV-1 is generally associated with infections of the oro-pharynx; whereas HSV-2 is primarily associated with genital and neonatal infections 1. However, data from Europe and Canada showed that more cases of neonatal herpes are associated with HSV-1 infection than HSV-2 3.In the U.S., about one out of six individuals at 14 to 49 years of age have had genital HSV-2 infection (http://www.cdc.gov/std/herpes/stdfact-herpes.htm). The proportion of primary genital infections due to HSV-1 is increasing, from 10% in 1983 to 32% in 1995 4, 5.
Conventional and modified cell cultures remain the test of choice to detect HSV in mucocutaneous, genital and ocular lesions. The Enzyme-Linked Virus Inducible System (ELVIS®, Diagnostics Hybrids Inc., Athens, OH) has been widely used in clinical laboratories to detect the presence of HSV-1 or HSV-2 6–8. The transgenic shell vial cell culture-based test takes a minimum of 16 hours for results and it can only be performed in a laboratory setting with cell culture facility. Numerous molecular assays have been reported to detect and differentiate HSV-1 and HSV-2 with high sensitivities and specificities 7–9. Recently, a PCR-based test for the direct detection of HSV-1 and HSV-2 from vaginal lesion specimens w as cleared by the Food and Drug Administration 8. However, the MutiCode® HSV-1&2 assay requires extraction of nucleic acid from the specimen and expensive real-time PCR instrumentation.
Helicase-dependent amplification (HDA) is an isothermal nucleic acid amplification method that relies on the use of a DNA helicase to unwind double-stranded nucleic acids 10, 11. The amplified DNA can then be applied on to a vertical-flow strip embedded in a disposable cassette for rapid detection of the organism. This method has been used for the rapid amplification and detection of methicillin-resistant Staphylococcus aureus in positive blood cultures 12, Clostridium difficile in stool specimens from hospitalized patients with diarrhoea 13 and HIV-1 in plasma in low-resource settings 14.
2. Objectives
Develop a user friendly, rapid isothermal nucleic acid amplification test (IsoAmp® HSV assay) for the detection of HSV-1 and HSV-2 on samples collected from genital lesions. Evaluate the performance characteristics of IsoAmp® HSV assay including analytical sensitivity and specificity, clinical sensitivity and specificity as well as intra and inter-laboratory reproducibility.
3. Study Design
3.1. IsoAmp® HSV assay development
A pair of primers, GBF1 and Bio-GBR1, was used to amplify the HSV glycoprotein B (gB) gene. Primer Bio-GBR1 was labeled with biotin at its 5’ end. To generate biotin-labeled single-stranded amplicon for probe hybridization, asymmetric HDA was performed with a primer ratio of 90 nM for primer Bio-GBR1 to 30 nM for primer GBF1. Probes for the HSV target, HSV-FI and HSV-Dig, were labeled with 6-carboxyfluorescien (FAM) and digoxigenin NHS ester (digoxN) at their 3’ ends respectively. A competitive internal control (IC) was developed to monitor specimen inhibition and reaction failure. The same primer pair (GBF1 and Bio-GBR1) was used to amplify the IC sequence and the IC amplicon which was detected by probe HSVIC-Dig labeled with digoxN at its 3’ end. All oligonucleotides were purchased from Integrated DNA Technology, Inc. (Coralville, IA). The HDA primers and probes used in this study are listed in Table 1.
Table 1.
HDA Primers and Probes
| Primers and Probes | Sequences and modifications * |
|---|---|
| GBF1 | 5'-TTCAAGGAGAACATCGCCCCGTACAA-3' |
| Bio-GBR1 | 5'-biotin-TAAACTGGGAGTAGCGGTGGCCGAAC-3' |
| HSV-FI | 5'-ATGTACTACAAAGACGT-FAM-3' |
| HSV-Dig | 5'-ATGTACTACAAAGACGT-digoxN-3' |
| HSVIC-Dig | 5'-CAAAGAGTCCGCAC-digoxN-3' |
FAM, 6-carboxyfluorescien; digoxN, digoxigenin NHS ester.
3.2. IsoAmp® HSV assay procedure
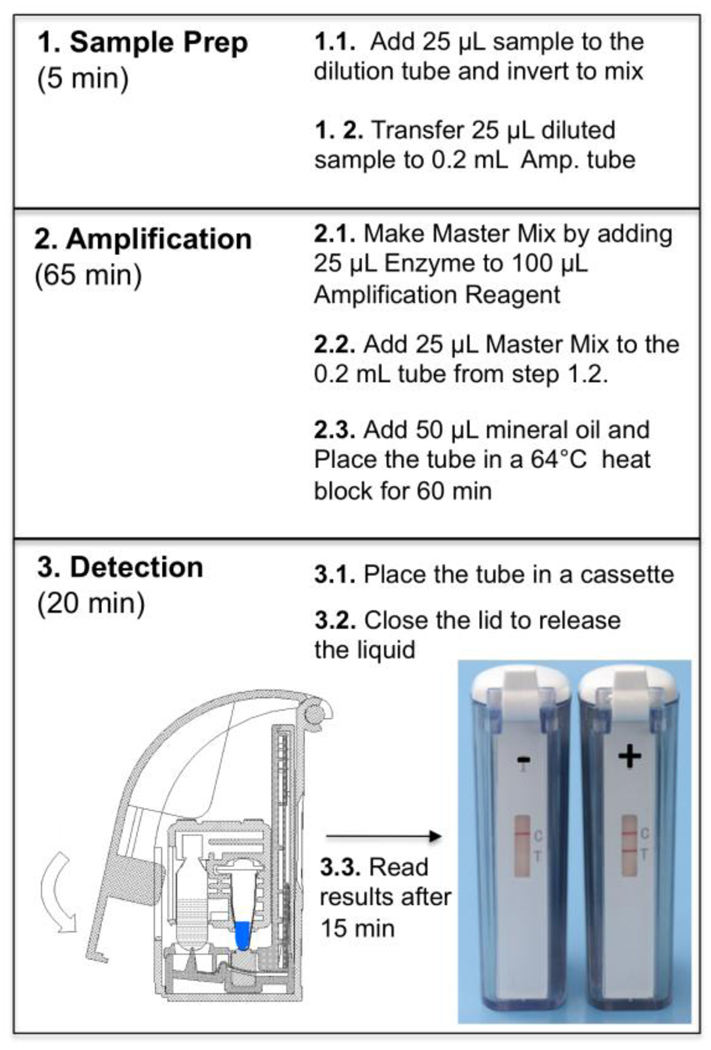
HDA reactions were performed using an IsoAmp® HSV assay (BioHelix Corporation, Beverly, MA) including three steps. (1) sample-preparation – 25 µl of viral transport medium containing genital lesion swab specimens was diluted with 1 mL dilution buffer. After mixing, 25 µl of each diluted specimens was transferred to a 0.2 mL thin-wall Amplification tube (Fig.1). (2) Isothermal amplification - The master mix was prepared by mixing 25 µl of Enzyme Reagent with 100 µl of Amplification Reagent prior use. 25 µl of master mix was then added into 25 µl diluted sample in each Amplification tube followed by adding 50 µl of mineral oil. The Amplification tube was placed in a 64°C heat block for 60 minutes to amplify the target DNA. (3) Instrument-free detection – After the amplification the 0.2 mL Amplification tube was placed in an amplicon cartridge of a Type II BESt™ Cassette (BioHelix Corporation, Beverly, MA) for amplicon detection 12–14. Closing the cassette-lid forced the tube to release amplicon into a vertical-flow DNA strip for colormatric detection without cross-contamination. The assay result can be visibly read by viewing through the detection window after 10 to 15 minutes (Fig. 1).
Figure 1.
IsoAmp® HSV assay workflow
3.3. Viral cultures
Viral cultures for HSV were performed using Enzyme-Linked Virus Inducible System (ELVIS®) shell vial assay (Diagnostic Hybrid Inc. Athens, OH). ELVIS® HSV uses a genetically engineered host cell line (transgenic baby hamster kidney cells) that was provided as fresh confluent monolayer cultures. The BHK cells contain the promoter sequence for HSV which can be activated only by viral proteins produced by HSV type 1 or 2 15. This viral promoter controls LacZ, the E coli gene for β-galactosidase which serves as a reporter molecule in the ELVIS HSV assay system. When an ELVIS Cell is infected with HSV present in a patient sample, the reporter enzyme quickly accumulates inside of the infected cells and catalyzes the reporter to produce intense blue color after cells are fixed and the ELVIS staining buffer is added. Results can be read with an inverted microscope per the manufacturer’s protocol.
3.3. Analytical sensitivity analysis
HSV-1 (9.0 × 1010 viral-particle/mL) and HSV-2 (1.6 × 1011 viral-particle/mL) viral stocks (Advanced Biotechnologies Inc, Columbia, MD 21046) were first diluted 1,000-fold to 9.0 × 107 viral-particle/mL and 1.6 × 108 viral-particle/mL. The viral concentrations of the primary dilutions were determined as 5.9 × 107 copies/mL for HSV-1 and 1.3 × 108 copies/mL for HSV-2 by real-time qPCR using HSV-1 or HSV-2 specific primers and quantified HSV-1 or HSV-2 viral DNA (Advanced Biotechnologies Inc, Columbia, MD 21046) as a standard. Serial dilutions of each HSV type, which were prepared from the primary dilutions in pooled HSV-negative clinical samples, were repeatedly tested in IsoAmp® HSV assay to generate hit rates at each concentration. The limit of detection (LoD) was calculated using Probit analysis (Minitab Inc, State College, PA).
3.4. Analytical specificity analysis
The potential cross-reactivity of the IsoAmp® HSV assay was evaluated in the presence of genomic DNA or microorganisms. Genomic DNAs were purchased from Advanced Biotechnologies Inc. (Columbia, MD) including varicella-zoster virus (VZV), Chlamydia trachomatis, Toxoplasma gondii, human papillomavirus (HPV)-16, and HPV-18. Microorganisms were purchased from American Type Culture Collection (ATCC) (Manassas, VA) including Trichomonas vaginalis (ATCC 30001), Mobiluncus mulieris (ATCC 35240), Candida albicans (ATCC 14053), Neisseria gonorrheae (ATCC 21823) and Gardnerella vaginalis (ATCC 14018 cells). The analytical specificity was tested at concentrations ranging from 1.2×103 to 2.5×106 copies/reaction (genomic DNA) or CFU)/reaction (microorganisms).
3.4. Intra- and inter-laboratory reproducibility
Five laboratories participated in the inter-laboratory reproducibility study, including four clinical laboratories plus BioHelix. A panel of blinded samples was prepared at BioHelix by diluting inactivated HSV-1 virus (Zeptometrix, Buffalo, NY) with M4 viral transport medium. Four different viral concentrations were made: 1) High (450 copies/reaction); 2) Medium (150 copies/reaction); 3) Low (50 copies/reaction); and 4) Blank (0 copy/reaction test). Each laboratory tested each concentration four times in a five-day period plus positive and negative controls.
3.5. Tolerance for variation
To test the tolerance for variation of the HSV assay, each critical component (i.e. dNTPs, Mg++, primers, etc.) was tested at three concentrations, the target concentration, 10% – 30% higher, and 10% – 30% lower. IsoAmp HSV assays were carried out with three concentrations of the testing component on three substrates, HSV-1 virus (3× LoD), HSV-2 virus (3× LoD), and Internal Control (IC) in 6 replicates for each condition. If all 6 replicates pass the variation tolerance test for a critical component, the tolerance range for this critical component is verified. If 1 (or more) reactions fail, the tolerance range will be reduced and the experiment will be repeated until it passes the narrowed test range.
4. Result
4.1. Robustness of the IsoAmp® HSV assay
The robustness of the IsoAmp® HSV RUO kit was evaluated by testing the HSV-1 assay positive control (a single-copy HSV-1 gB gene cloned in a plasmid), HSV-2 assay positive control (a single-copy HSV-2 gB gene cloned in a plasmid), and negative control (TE buffer) in 100 replicates. All 100 replicates per each HSV-1 (150 copies/reaction) and HSV-2 Assay Positive Controls (300 copies/reaction) showed positive test results. Out of 100 replicates tested for Negative Control, 99 replicates showed negative test results and one tested false-positive. Therefore, the invalid rates of IsoAmp® HSV Assay for positive and negative assay controls were 0% and 1%, respectively.
We also evaluated the IsoAmp® HSV assay on tolerance for variation in critical components and procedure. The variation tolerance study showed that the assay can tolerate minimal at 10% variation on dNTPs, MgSO4, primers, probes concentrations, 20% variation on enzymes, and 30% variation on IC control and positive controls based on the observations that the HSV assay was able detect both HSV-1 and HSV-2 at 3 times of LoD in the presence of these variations. The assay performed well with 10% variation in assay procedures including diluted sample input, enzyme reagents input and +/− 1°C incubation temperature variation.
4.2. Analytical sensitivity and specificity
For HSV-1, virus dilutions were tested in IsoAmp® HSV Assay at 50, 25, 12.5, 6 and 3 copies/assay. Each run tested the HSV-1 in 5 different concentrations in duplicates as well as one negative and positive control each. A total of 20 replicates were tested per each concentration. However, the positive rate was too high to estimate the LoD in the first series dilution experiment (Table 2). Therefore, the LoD study was repeated with lower concentrations of HSV-1 (6, 3, 1.5, 0.8 copies/reaction) and hit rates from 83% for the 6 copies/reaction to 17% for the 0.8 copies/reaction were observed. Probit analysis on combined data on HSV-1 showed that the analytical sensitivity for HSV-1 was estimated at 5.5 copies/test with a 95% confidence interval (CI) between 4.0 and 9.1 copies/test. Similar experiments conducted with IsoAmp® HSV assay on HSV-2 showed an estimation at 34.1 copies/test with a 95% CI between 21.9 and 60.8 copies/test (Table 2).
Table 2.
Limit of detection determination of IsoAmp® HSV for HSV-1/2 detection *
| HSV concentration | HSV-1 | HSV-2 | ||||
|---|---|---|---|---|---|---|
| (copies/assay) | Number tested |
Number positive |
Positive rate (%) |
Number tested |
Number positive |
Positive rate (%) |
| 50 | 20 | 20 | 100.0 | 20 | 19 | 95.0 |
| 25 | 20 | 20 | 100.0 | 20 | 17 | 85.0 |
| 12.5 | 20 | 20 | 100.0 | 20 | 17 | 85.0 |
| 6 | 20 | 20 | 100.0 | 20 | 10 | 50.0 |
| 3 | 20 | 15 | 75.0 | 20 | 8 | 40.0 |
| 0 | 10 | 0 | 0.0 | 10 | 0 | 0.0 |
The result of LoD studies was analyzed using Probit analysis. The LoD for HSV-1 was determined to be 5.5 copies/test. At this concentration, IsoAmp HSV Assay can detect 95% of the samples with a 95% confidence interval (CI) of 4.0 – 9.1 copies/test. The LoD for HSV-2 was determined to be 34.1 copies /test with a 95% CI of 21.9 – 60.8 copies/test.
No cross-reactivities were observed when VZV (4×105 copies/reaction), C. trachomatis (3.6×105 copies/reaction), T. gondii (6.1×103 copies/reaction), HPV-16 (6.1×103 copies/reaction), HPV-18 (1.2×103 copies/reaction), T. vaginalis (2.5×106 CFU/reaction), M. mulieris (2.5×106 CFU/reaction), C. albicans (6.1×103 CFU/reaction), N. gonorrheae (6.1×103 CFU/reaction) and G. vaginalis (6.1×103 CFU/reaction) were tested by the IsoAmp® HSV assay.
4.3. Clinical sensitivity and specificity
One hundred and thirty-five un-linked frozen specimens collected by swbs (Microbiology lab, Cleveland Clinic) from lesions on patients with suspected HSV infection in genital area were tested with IsoAmp HSV assay. These samples consisted of 60 positives and 75 negative determined by HSV culture method (ELVIS® shell vial assay; DHI, Athens, OH). IsoAmp® HSV showed a 100% sensitivity and 96.3% specificity when compared to the reference method (Table 3).
Table 3.
Performance of IsoAmp® HSV assay on 135 clinical specimens in comparison to the ELVIS shell vial assay
| Reference Method | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
|
IsoAmp® HSV |
Positive | 60 | 5 | 65 |
| Negative | 0 | 70 | 70 | |
| Total | 60 | 75 | 135 | |
| Sensitivity | 100% | |||
| Specificity | 96.3% | |||
4.4. Inter-laboratory reproducibility
A total of 80 samples spiked with different concentrations of HSV-1 virions were tested in five sites (blinded). One site reported an indeterminate result on medium HSV-1 concentration specimen which probably was due to detection cassette failure. The same site also reported one false positive on a negative matrix. An overall 97.5% agreement was observed by testing a total of 80 blind-coded HSV-1 samples between five laboratories (Table 4).
Table 4.
Inter-laboratory reproducibility of HSV-1 detection
| HSV-1 concentrationa |
Number tested |
Site 1 | Site 2 | Site 3 | Site 4 | Site 5 |
|---|---|---|---|---|---|---|
| High | 20 | 4 (100.0) b | 4 (100.0) | 4 (100.0) | 4 (100.0) | 4 (100.0) |
| Medium | 20 | 4 (100.0) | 4 (100.0) | 4 (100.0) | 4 (100.0) | 3 (75.0)c |
| Low | 20 | 4 (100.0) | 4 (100.0) | 4 (100.0) | 4 (100.0) | 4 (100.0) |
| Blank (matrix only) | 20 | 4 (100.0) | 4 (100.0) | 4 (100.0) | 4 (100.0) | 3 (75.0)d |
High, medium, low, and matrix only indicated 450, 150, 50, and 0 copies/reaction of HSV-1 concentration respectively. Each run included 2 positive and 2 negative controls.
Number positive (%)
One was detected indeterminate.
One blank matrix was tested positive
5. Discussion
The IsoAmp® HSV assay for genital herpes combines a simple sample-prep (dilution-only), an isothermal HDA amplification, and a self-contained disposable amplicon detection device for rapid diagnosis of HSV-1 and HSV-2. As it does not require any special instrumentation, or complicated specimen handling, the assay enables more laboratories to perform rapid nucleic acid test for diagnosis of HSV infections without culture or expensive real-time PCR instrumentation. This study showed that the IsoAmp® HSV assay has 100% clinical sensitivity and 96.3% specificity for the detection of HSV from genital herpes when compared to the reference culture method. Good inter-laboratory reproducibility was observed in five sites with all positive and negative controls worked as expected. Based on the analytical sensitivity data, the IsoAmp® HSV assay demonstrated significantly lower LoD for HSV-1 (5.5 copies/test) than that of HSV-2 (34.1 copies/test). This LoD difference (HSV-1 from HSV-2) is probably caused by variation in the gB sequence since there are three single nucleotide polymorphisms (SNPs) between the two primers. Other factors such the accuracy of the quantification method may also contribute to the difference of the LoDs between HSV-1 and HSV-2.
The preliminary evaluation of the IsoAmp® HSV assay on clinical samples showed a sensitivity of 100%. The IsoAmp® assay also detected 5 culture-negative samples. Further investigation revealed that these 5 IsoAmp-positive/culture-negative samples were repeatedly tested positive by nucleic acid amplification test on another HSV target sequence (UL30, DNA polymerase). It is likely that IsoAmp® HSV assay has higher detection sensitivity than ELVIS® culture and therefore it was able to detect low-viral load and/or non-viable herpes simplex viruses. No cross-reactivities were observed when other related microorganisms including VZV, C. trachomatis, T. gondii, HPV-16, HPV-18, T. vaginalis, M. mulieris, C. albicans, N. gonorrheae and G. vaginalis were tested by the IsoAmp® assay.
Acknowledgements
We thank Fran White and Bertrand Lemieux for discussion, and Haijing Li and Geetha Vanga for technical help.
Funding
This study was supported by National Institutes of Health grants 2R44AI066487-03 and 5R44AI066487-04.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethical approval
This study received approval from the institutional review boards in all participant sites
Conflict of interest
Hyun-Lin Kim, Yanhong Tong, Wen Tang, Xiaojing Pan, Aurelie Motre, Jian Hong, and Huimin Kong are employees of BioHelix Corporation, the commercial manufacturer of the IsoAmp® HSV assay. Yi-Wei Tang is a scientific advisory board member of BioHelix Corporation.
References
- 1.Yeung-Yue KA, Brentjens MH, Lee PC, Tyring SK. Herpes simplex viruses 1 and 2. Dermatol Clin. 2002 Apr;20(2):249–266. doi: 10.1016/s0733-8635(01)00003-1. [DOI] [PubMed] [Google Scholar]
- 2.Roberts S. Herpes simplex virus: incidence of neonatal herpes simplex virus, maternal screening, management during pregnancy, and HIV. Curr Opin Obstet Gynecol. 2009 Apr;21(2):124–130. doi: 10.1097/GCO.0b013e3283294840. [DOI] [PubMed] [Google Scholar]
- 3.Kropp RY, Wong T, Cormier L, et al. Neonatal herpes simplex virus infections in Canada: results of a 3-year national prospective study. Pediatrics. 2006 Jun;117(6):1955–1962. doi: 10.1542/peds.2005-1778. [DOI] [PubMed] [Google Scholar]
- 4.Corey L, Adams HG, Brown ZA, Holmes KK. Genital herpes simplex virus infections: clinical manifestations, course, and complications. Ann Intern Med. 1983 Jun;98(6):958–972. doi: 10.7326/0003-4819-98-6-958. [DOI] [PubMed] [Google Scholar]
- 5.Wald A, Zeh J, Selke S, Ashley RL, Corey L. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med. 1995 Sep 21;333(12):770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- 6.Crist GA, Langer JM, Woods GL, Procter M, Hillyard DR. Evaluation of the ELVIS plate method for the detection and typing of herpes simplex virus in clinical specimens. Diagn Microbiol Infect Dis. 2004 Jul;49(3):173–177. doi: 10.1016/j.diagmicrobio.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Patel N, Kauffmann L, Baniewicz G, Forman M, Evans M, Scholl D. Confirmation of low-titer, herpes simplex virus-positive specimen results by the enzyme-linked virus-inducible system (ELVIS) using PCR and repeat testing. J Clin Microbiol. 1999 Dec;37(12):3986–3989. doi: 10.1128/jcm.37.12.3986-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvaraju SB, Wurst M, Hor vat RT, Selvarangan R. Evaluation of three analyte-specific reagents for detection and typing of herpes simplex virus in cerebrospinal fluid. Diagn Microbiol Infect Dis. 2009 Mar;63(3):286–291. doi: 10.1016/j.diagmicrobio.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Wald A, Huang ML, Carrell D, Selke S, Corey L. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J Infect Dis. 2003 Nov 1;188(9):1345–1351. doi: 10.1086/379043. [DOI] [PubMed] [Google Scholar]
- 10.An L, Tang W, Ranalli TA, Kim HJ, Wytiaz J, Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem. 2005 Aug 12;280(32):28952–28958. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004 Aug;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldmeyer J, Li H, McCormac M, et al. Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol. 2008;46(4):1534–1536. doi: 10.1128/JCM.02234-07. Epub 2008 Jan 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow WH, McCloskey C, Tong Y, et al. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J Mol Diagn. 2008 Sep;10(5):452–458. doi: 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang W, Chow WH, Li Y, Kong H, Tang YW, Lemieux B. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J Infect Dis. 2010 Apr 15;201 Suppl 1:S46–S51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stabell EC, O'Rourke SR, Storch GA, Olivo PD. Evaluation of a genetically engineered cell line and a histochemical beta-galactosidase assay to detect herpes simplex virus in clinical specimens. J Clin Microbiol. 1993 Oct;31(10):2796–2798. doi: 10.1128/jcm.31.10.2796-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]