Abstract
Targeted gene replacement is a powerful tool in Leishmania genetics that can be time-consuming to implement. One tedious aspect that delays progress is the multi-step construction of gene targeting vectors. To accelerate this process, we developed a streamlined method that allows the assembly of a complete targeting vector from all its constituent parts in a single-step multi-fragment ligation. The individual components to be assembled are flanked by sites for the restriction endonuclease SfiI that generates nonidentical, non-palindromic three base 3’-overhangs designed to allow annealing and ligation of the parts only in the proper order. The method was optimized by generating constructs for targeting the Leishmania donovani inosine monophosphate dehydrogenase gene (LdIMPDH) encoding six different drug resistance markers, and was found to be rapid and efficient. These constructs were successfully employed to generate heterozygous LdIMPDH gene replacement mutants. This method is adaptable for generating targeting vectors for a variety of species.
Keywords: Leishmania, gene targeting, gene knockout, homologous recombination, multi-fragment ligation, SfiI restriction endonuclease
Targeted gene replacement via homologous recombination has been an invaluable tool for the genetic dissection of important metabolic and virulence pathways in Leishmania species [1], as well as for many other protozoan parasites [2]. The general experimental approach for the genetic manipulation of model organisms is essentially the same: DNA sequences of sufficient length to direct homologous recombination flanking the gene to be targeted - referred to herein as 5’- and 3’-targeting sequences (TS) - are independently isolated and joined to an alternative gene (i.e., drug resistance gene) that allows selection of cells in which the appropriate integration event has occurred. The most common method for generating leishmanial and other parasite gene targeting constructs involves the sequential cloning of 5’-TS and 3’-TS DNAs into a vector encoding a drug resistance cassette flanked by restriction sites [3]. This multi-step process can be time consuming and is complicated by the fact that commonly used vectors have limited restriction sites for TS insertion, exchange of drug resistance markers, and excision of the targeting cassette from the vector backbone prior to transfection. The ability to assemble the complete gene targeting construct in a single step would greatly enhance the throughput of gene replacement studies. Numerous techniques have been described for the simultaneous assembly of multiple DNA fragments, including variations on ligation-independent cloning [4–6], overlap extension polymerase chain reaction (PCR) [7], site-specific recombination [8], and recombination in Escherichia coli, i.e., “recombineering”[9]. Some of these methods have been used to construct gene targeting constructs for various trypanosomatids [10,11]. However, these techniques can involve tedious, complicated, or unfamiliar technology [5,6,8,9,12], require expensive kits or reagents [4,8], and are complicated by inefficiency [13].
To overcome these many impediments to gene replacement in Leishmania, we have developed a simple, cost-effective, and efficient method (see Fig. 1) for construction of leishmanial targeting constructs that is based on the properties of the SfiI restriction endonuclease and requires only readily available and familiar technologies, (i.e., PCR, restriction digestion, and DNA ligation). The SfiI restriction endonuclease was chosen as the basis of this multi-fragment ligation strategy for several reasons. First, the recognition sequence for SfiI, GGCCNNNNNGGCC, is an interrupted palindrome that generates an asymmetric three base 3’-overhang (underlined and in boldface type in the sequence) upon DNA cleavage that can only ligate to complementary overhangs but not to itself nor to overhangs from non-identical SfiI sites. SfiI sites designed to produce non-identical 3’-overhangs incorporated at the ends of the individual targeting vector components allows the directional and ordered assembly of the targeting vector in a single ligation reaction (see Fig. 1C). Second, the eight base pair recognition sequence of SfiI should only rarely occur within any given TS, rendering this strategy applicable to the generation of targeting constructs for virtually any leishmanial gene. Indeed, examination of one megabase of L. infantum genome sequence identified SfiI sites with a frequency of approximately one in every 38 kb. If a TS encompasses an SfiI site, alternative restriction enzymes with six base pair recognition sequences that are also interrupted palindromes are potentially available for generating compatible 3’-overhangs (see Supplementary Protocol). Third, SfiI has been utilized by other groups for the ordered assembly of multiple DNA fragments and the method has proven to be quite efficient [13,14]. While the use of SfiI in other strategies for producing gene targeting constructs has been described [12,15], none of these schemes involve the simultaneous assembly of a complete targeting vector by single-step ligation of all of the constituent components.
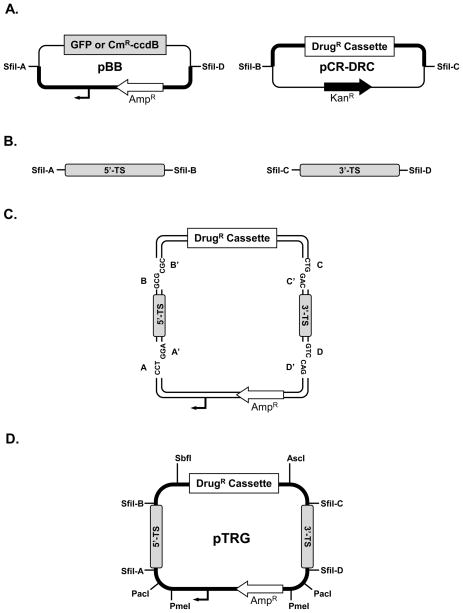
Fig. 1.
Strategy for assembly of gene targeting vectors via multi-fragment ligation. This method is modular in design with each component derived independently prior to simultaneous assembly in a single ligation reaction. (A) Plasmid pBB serves as the donor of a minimal (2165 bp) plasmid backbone (denoted by the thick black line) that encodes ampicillin resistance (AMPR- open arrow) and a plasmid replication origin (small black arrow). There are two versions of pBB, pBB-GFP (Genbank accession no. HQ416901) and pBB-CmR-ccdB (Genbank accession no. HQ416902), that encode either GFP (~800 bp) or CmR-ccdB (~1600 bp) stuffer fragments, respectively, flanked by non-identical SfiI restriction sites (SfiI-A and SfiI-D). Drug resistance cassettes (thick black line) are donated by the pCR-DRC plasmids that were constructed by cloning drug resistance cassettes (~3 kb) PCR amplified with primers encoding unique SfiI restriction sites (SfiI-B and SfiI-C) into pCR®-Blunt II-TOPO®. Six pCR-DRC plasmids were constructed that contain drug resistance cassettes conferring resistance to either neomycin/G418 (pCR-NEO), hygromycin (pCR-HYG), phleomycin (pCR-PHLEO), puromycin (pCR-PAC), blasticidin (pCR-BSD), or nourseothricin (pCR-SAT). A detailed description of pBB and pCR-DRC plasmid construction is provided in the supplementary materials and methods. For all of the drug resistance cassettes, the appropriate trans-splicing and polyadenylylation of the drug resistance gene is directed by 5’- and 3’-flanking sequences, respectively, derived from the L. major DHFR-TS gene as described [1] (B) The 5’- and 3’-targeting sequences (TS) are generated by PCR amplification using primers that add SfiI restriction sites SfiI-A and SfiI-B (5’-TS) or SfiI-C and SfiI-D (3’-TS) (see Supplementary Protocol). (C) Targeting vector assembly. The 5’- and 3’-TS PCR fragments, along with the pBB and pCR-DRC donor plasmids are digested with SfiI and the appropriate DNA fragments are agarose gel purified and combined in a ligation reaction. Each DNA fragment has unique three base 3’-overhangs on the sense (indicated by A, B, C, D) and antisense (indicated by A’, B’, C’, D’) strands that can only anneal to complementary overhangs. (D) The assembled targeting vector (pTRG) contains restriction sites for exchange of drug resistance cassettes (SbfI and AscI) and release of the targeting fragment from the plasmid backbone (PacI or PmeI). Note that the plasmids are not drawn to scale.
Standard targeting vectors are normally composed of four parts: a minimal plasmid backbone to permit selection and propagation in Escherichia coli, a drug resistance cassette to allow selection in Leishmania, and a 5’-TS and a 3’-TS to facilitate gene replacement via homologous recombination. In the multi-fragment ligation method, all four parts are digested with SfiI, gel purified, and combined in a single ligation reaction to generate the complete targeting construct. As depicted in Fig. 1A, the minimal plasmid backbone is donated by the pBB plasmid that contains two incompatible SfiI sites (SfiI-A and SfiI-D) flanking a stuffer fragment that allows the plasmid backbone fragment to be readily distinguished from uncut vector during gel purification. An expression cassette encoding one of six possible drug resistance genes currently available for Leishmania transfection [1,16–18] is donated by the corresponding pCR-DRC plasmid (Fig. 1A) and is also flanked by incompatible SfiI sites (SfiI-B and SfiI-C). Importantly, the SfiI overhangs encoded by the plasmid backbone fragment and the drug resistance cassettes cannot ligate to each other, but, rather, require the complementary SfiI overhangs provided by the 5’- and 3’-TS PCR fragments (Fig. 1B) to serve as a bridge between them (Fig. 1C). The complete targeting vector (Fig. 1D) encodes ampicillin resistance and the fact that the pCR-DRC plasmids are kanamycin-resistant eliminates the possible contribution of uncut pCR-DRC plasmid to the occurrence of background colonies following transformation of the ligation reaction. All the drug resistance cassettes are flanked by restriction sites for the rare cutting enzymes SbfI and AscI to facilitate the ready exchange of drug resistance cassettes between targeting constructs (i.e. pTRG in Fig. 1D) and the pCR-DRC plasmids (Fig. 1A). The targeting cassette can be conveniently excised from the plasmid backbone using either PacI or PmeI endonucleases, whose eight bp recognition sequences occur rarely.
To demonstrate the efficacy of the multi-fragment ligation method for targeting vector construction, we generated constructs for targeted replacement of the L. donovani inosine monophosphate dehydrogenase (LdIMPDH) gene [19]. As a first step, the conditions for the four-way ligations were optimized by varying the molar ratio of inserts (3’- and 5’-IMPDH TS fragments and puromycin (PAC) or phleomycin (PHLEO) drug resistance cassettes) to plasmid backbone fragment in trial ligation reactions. While the transformation efficiencies were uniformly high for all insert to vector ratios tested, the percentage of clones containing plasmids with the correct structure was consistently lower at the 0.5:1 molar ratio (Table 1). SfiI digestion of plasmid DNA from twenty colonies each from the 2:1 ratio ligations of both the PAC and PHLEO LdIMPDH targeting constructs revealed that 85% and 75%, respectively, had the correct structure (data not shown) confirming the efficiency of the method at this ratio. Therefore, a 2:1 insert to vector ratio was employed for construction of LdIMPDH targeting constructs encoding neomycin (NEO), hygromycin (HYG), blasticidin (BSD), and nourseothricin (SAT) expression cassettes, since it consistently allowed smaller ligation volumes to be used and yielded a large number of colonies containing a high percentage of targeting vectors with the expected structure in each case (data not shown).
Table 1.
Optimization of multi-fragment ligation conditions
| Inserts : Plasmid Backbone Ratio | Number of Colonies | Transformation efficiency (cfu/μg) | Positive clones (%) (N=4) | |
|---|---|---|---|---|
| PHLEO | 0.5:1 | 450 | 3.2 × 106 | 25 |
| 1:1 | 900 | 6.4 × 106 | 75 | |
| 2:1 | 1180 | 8.4 × 106 | 75 | |
| 3:1 | 1500 | 1.1 × 107 | 75 | |
| PAC | 0.5:1 | 524 | 3.7 × 106 | 25 |
| 1:1 | 1110 | 7.9 × 106 | 75 | |
| 2:1 | 1467 | 1.0 × 107 | 100 | |
| 3:1 | 1880 | 1.3 × 107 | 75 | |
| pUC19 | - | 2000 | 4.8 × 108 | - |
| Plasmid backbone fragment | - | 70 | 5.0 × 105 | - |
The molar ratio of inserts (5’- and 3’- IMPDH TS fragments and the drug resistance cassette) to plasmid backbone was varied with respect to 10 ng of the plasmid backbone. The Inserts: Plasmid Backbone Ratio denotes the molar ratio of each insert fragment relative to the plasmid backbone. The number of colonies from plating 100 μl (21% of total transformation volume) of E.coli transformed with 2 μl ligation reaction (representing 0.67 ng plasmid backbone fragment) is presented. Transformation efficiencies are given as colony forming units per μg of DNA and are calculated based on 0.67 ng of plasmid backbone fragment per transformation. Plasmid pUC19 (0.02 ng) was included to assess the transformation efficiency of the lot of commercially prepared competent E. coli used in this experiment. Transformation of a ligation reaction performed using only 10 ng of the purified plasmid backbone fragment (originating from the same batch used in inserts:vector ratio optimization) served to indicate the contribution of pBB-GFP to background. Further assessment of transformants derived from ligation reactions using 2:1 inserts to plasmid backbone ratio showed that the percentage of clones positive for the correct PHLEO- or PAC-containing targeting vector was 75% and 85%, respectively (N=20).
While the overall number of colonies and the percentage of colonies containing the correct construct was high, it was noted that all of the incorrect plasmids could be readily identified as contaminating pBB-GFP plasmid (data not shown). Other systems for simultaneous assembly of multiple DNA fragments reduce the occurrence of background by including a conditionally lethal gene such as ccdB or sacB in donor vectors [8,12]. To reduce the potential for background colonies derived from the plasmid backbone donor plasmid, the GFP stuffer fragment of pBB-GFP was replaced with a chloramphenicol resistance/ccdB expression cassette to generate plasmid pBB-CmR-ccdB. This plasmid cannot be propagated in standard E. coli strains used for cloning but instead requires a specialized bacterial strain expressing the antitoxin to CcdB encoded by the ccdA gene [20]. When pBB-CmR-ccdB was used as the plasmid backbone donor in a four-way ligation to generate LdIMPDH targeting constructs containing a PAC cassette, no background was observed (data not shown). In fact, in eight additional targeting constructs have been produced in our laboratory using pBB-CmR-ccdB as the plasmid backbone donor and no background has been observed (A. Fulwiler and R. Soysa, unpublished observations).
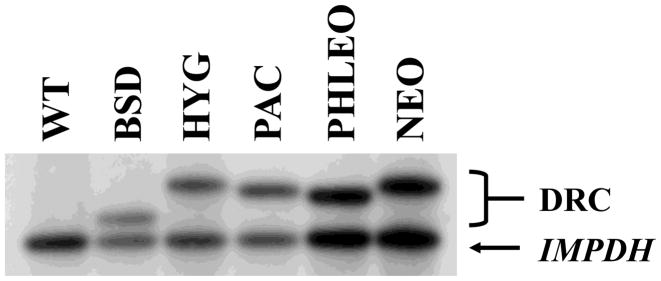
To establish the utility of constructs generated by multi-fragment ligation for targeted gene replacement, LdIMPDH targeting constructs encoding BSD-, HYG-, NEO-, PHLEO-, and PURO resistance cassettes were transfected into wild type L. donovani parasites and plated on the appropriate selective medium. Southern blot analysis indicated that each targeting construct was capable of generating IMPDH/impdh heterozygotes with the expected genomic structure (Fig. 2).
Fig. 2.
Southern blot analysis of Δimpdh/IMPDH parasites. Total genomic DNA (~2 μg) from wild type L. donovani (lane 1), or strains in which one IMPDH allele has been replaced by BSD (lane 2), HYG (lane 3), PAC (lane 4), PHLEO (lane 5), or NEO (lane 6) cassettes was digested with XhoI, fractionated on a 1% agarose gel, and blotted onto a nylon membrane. The blot was hybridized under high stringency conditions with a probe to the 3’-IMPDH TS. Bands above the IMPDH band correspond to the targeted IMPDH allele and vary in size in accordance with the sizes of the drug resistance cassettes (labeled DRC). Targeted replacement of LdIMPDH using a targeting construct containing a SAT cassette (pTRG-IMPDH-SAT) was not attempted.
The multi-fragment ligation strategy presented here for production of leishmanial gene targeting vectors is markedly faster than traditional approaches [3] and is significantly cheaper and more straightforward than alternatives [11]. Using this technique, we routinely generate gene targeting constructs in three to four days from the time of TS fragment PCR to confirmation of vector structure by restriction analysis. The general strategy is adaptable to the generation of targeting constructs for other parasites and genetically manipulable organisms by simply producing species-specific selectable markers flanked by the appropriate SfiI sites. All of the components of this system are available by request from the authors.
Supplementary Material
Acknowledgments
We gratefully acknowledge the generosity of Dr. Stephen Beverley for providing the pX63 and pXG vectors. We thank Dr. Nicola Carter for critical reading of the manuscript. This work was supported in part by grants AI023682 and AI041622 from the National Institute of Allergy and Infectious Disease.
Abbreviations
- AMPR
ampicillin resistance gene
- BSD
blasticidin resistance
- CmR
chloramphenicol resistance gene
- GFP
green fluorescent protein
- HYG
hygromycin resistance
- KANR
kanamycin resistance gene
- LdIMPDH
Leishmania donovani inosine monophosphate dehydrogenase gene
- NEO
neomycin resistance
- PAC
puromycin resistance
- PHLEO
phleomycin resistance
- SAT
nourseothricin resistance
- TS
targeting sequence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cruz A, Coburn CM, Beverley SM. Double targeted gene replacement for creating null mutants. Proc Natl Acad Sci U S A. 1991;88 (16):7170–4. doi: 10.1073/pnas.88.16.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997;89 (2):287–96. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 3.Cruz A, Beverley SM. Gene replacement in parasitic protozoa. Nature. 1990;348 (6297):171–3. doi: 10.1038/348171a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhu B, Cai G, Hall EO, Freeman GJ. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques. 2007;43 (3):354–9. doi: 10.2144/000112536. [DOI] [PubMed] [Google Scholar]
- 5.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4 (3):251–6. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 6.Rashtchian A, Buchman GW, Schuster DM, Berninger MS. Uracil DNA glycosylase-mediated cloning of polymerase chain reaction-amplified DNA: application to genomic and cDNA cloning. Anal Biochem. 1992;206 (1):91–7. doi: 10.1016/s0003-2697(05)80015-6. [DOI] [PubMed] [Google Scholar]
- 7.Darveau A, Pelletier A, Perreault J, Gobinda S. Methods in Neurosciences. Academic Press; 1995. [6] PCR-mediated synthesis of chimeric molecules; pp. 77–85. [Google Scholar]
- 8.Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14 (10B):2111–20. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2 (10):769–79. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee A, Roy G, Guimond C, Ouellette M. The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol Microbiol. 2009;74 (4):914–27. doi: 10.1111/j.1365-2958.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu D, Brandan CP, Basombrio MA, Tarleton RL. Evaluation of high efficiency gene knockout strategies for Trypanosoma cruzi. BMC Microbiol. 2009;9:90. doi: 10.1186/1471-2180-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtsuka M, Mizutani A, Kikuti YY, Kulski JK, Sato M, Kimura M, Tanaka M, Inoko H. One-step generation of recombineering constructs by asymmetric-end ligation and negative selection. Anal Biochem. 2007;360 (2):306–8. doi: 10.1016/j.ab.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, Mansoorabadi K, Jeffries T. Comparison of multiple gene assembly methods for metabolic engineering. Appl Biochem Biotechnol. 2007;137–140(1–12):703–10. doi: 10.1007/s12010-007-9090-y. [DOI] [PubMed] [Google Scholar]
- 14.Tsuge K, Matsui K, Itaya M. One step assembly of multiple DNA fragments with a designed order and orientation in Bacillus subtilis plasmid. Nucleic Acids Res. 2003;31 (21):e133. doi: 10.1093/nar/gng133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamper J. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Genet Genomics. 2004;271 (1):103–10. doi: 10.1007/s00438-003-0962-8. [DOI] [PubMed] [Google Scholar]
- 16.Goyard S, Beverley SM. Blasticidin resistance: a new independent marker for stable transfection of Leishmania. Mol Biochem Parasitol. 2000;108 (2):249–52. doi: 10.1016/s0166-6851(00)00210-3. [DOI] [PubMed] [Google Scholar]
- 17.Joshi PB, Webb JR, Davies JE, McMaster WR. The gene encoding streptothricin acetyltransferase (sat) as a selectable marker for Leishmania expression vectors. Gene. 1995;156 (1):145–9. doi: 10.1016/0378-1119(95)00042-5. [DOI] [PubMed] [Google Scholar]
- 18.Freedman DJ, Beverley SM. Two more independent selectable markers for stable transfection of Leishmania. Mol Biochem Parasitol. 1993;62 (1):37–44. doi: 10.1016/0166-6851(93)90175-w. [DOI] [PubMed] [Google Scholar]
- 19.Wilson K, Collart FR, Huberman E, Stringer JR, Ullman B. Amplification and molecular cloning of the IMP dehydrogenase gene of Leishmania donovani. J Biol Chem. 1991;266 (3):1665–71. [PubMed] [Google Scholar]
- 20.Salmon MA, Van Melderen L, Bernard P, Couturier M. The antidote and autoregulatory functions of the F plasmid CcdA protein: a genetic and biochemical survey. Mol Gen Genet. 1994;244 (5):530–8. doi: 10.1007/BF00583904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.