Abstract
Increasing evidence links genomic and epigenomic instability, including multiple fragile sites regions to neuropsychiatric diseases including schizophrenia and autism. Cancer is the only other disease associated with multiple fragile site regions, and genome and epigenomic instability is a characteristic of cancer. Research on cancer is far more advanced than research on neuropsychiatric disease; hence, insight into neuropsychiatric disease may be derived from cancer research results. Towards this end, this article will review the evidence linking schizophrenia and other neuropsychiatric diseases (especially autism) to genomic and epigenomic instability, and fragile sites. The results of studies on genetic, epigenetic and environmental components of schizophrenia and autism point to the importance of the folate-methionine-transulfuration metabolic hub that is diseases also perturbed in cancer. The idea that the folate-methionine-transulfuration hub is important in neuropsychiatric is exciting because this hub present novel targets for drug development, suggests some drugs used in cancer may be useful in neuropsychiatric disease, and raises the possibility that nutrition interventions may influence the severity, presentation, or dynamics of disease.
Keywords: Genomic and epigenomic instability, fragile sites, schizophrenia, autism, folate, methionine, transulfuration, s-adenosyl methionine, cancer.
INTRODUCTION
Genomic instability refers to an increased mutation rate that can take the form of chromosomal abnormalities, translocations, large or small insertions or deletions and base changes. Epigenomic instability refers to perturbed responses of gene regulation to environmental fluctuations. Fragile site regions of the genome have high levels of genetic and epigenetic instability.
In 2003, we reported a link between somatic mutations (genomic instability) and fragile sites and schizophrenia [1]. Later, we reported aberrant epigenetic regulation of genes involved in dopamine metabolism in the synaptic cleft in schizophrenia and bipolar disease brains [2, 3]. Today, there is increasing evidence for genome instability in neuropsychiatric diseases, including an association with fragile site regions. Cancer is the only other disease associated with multiple fragile site regions, and genome instability is a characteristic of cancer. This article will review the evidence linking schizophrenia and other neuropsychiatric diseases (especially autism) to genomic and epigenomic instability and fragile sites.
Schizophrenia and Autism
Schizophrenia and Autism are neuropsychiatric diseases linked to multiple genetic and environmental factors. Like many common illnesses these diseases remain an enigma because there is no single factor or small number of factors that accounts for a large number of patients.
The prevalence of schizophrenia is ~1% worldwide but varies between 0.3 to 2.7% [4]. Diagnosis is based on the appearance and duration of about 30 symptoms divided into positive (e. g. hallucinations (especially auditory are common)), negative (e.g. withdrawal, blunted affect etc), and cognitive (executive function). However, symptoms (endophenotypes) and outcome (Fig. 1) vary even in the same family, raising the possibility that several different diseases (i. e. the “schizophrenias”) presenting similar collections of symptoms have been grouped together [5, 6]. These and other observations suggest that a genetic predisposition is not sufficient by itself to cause disease. Further in some cases, the disease appears to be environmentally induced in the absence of detectable genetic predisposition (see below).
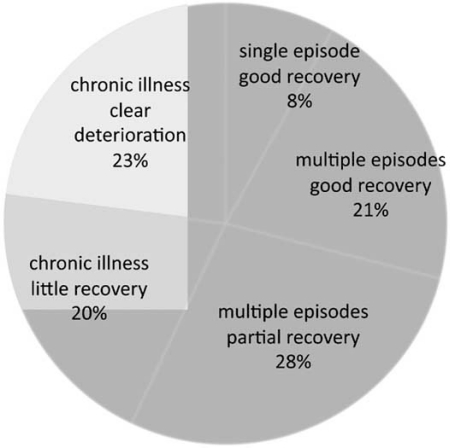
Fig. (1).
Classification of schizophrenia based on outcome. The outcome of schizophrenia disease is highly variable; suggesting different diseases may have been grouped together. (Adapted from Summary report of symposium “Schizophrenia and other Psychosis (http://www.science.org.au).
Autism is a complex, early onset (typically <5 years of age) lifelong illness that is difficult to diagnose and treat. Autism appears to be multiple diseases that make up autism spectrum disorder (ASD) defined by limits in three behaviors (1) social interactions, (2) communication and imaginative play, and (3) interests and activities. Other symptoms include impaired immunological responses, inflammation (especially in the gut), and oxidative stress [7]. Today, treatments include intensive educational and behavioral interventions with drugs to reduce remaining symptoms.
GENETICS
First-degree relatives of schizophrenia probands have a ~10% probability of becoming ill [8], while ~ 50% of cases of schizophrenia are spontaneous with no other affected family member [9]. Although variable [10-12], the general belief is that ~50% of monozygotic twins afflicted with schizophrenia are discordant for the disease, although progeny of both the well and ill discordant MZ twin have the elevated probably (~10%) typical of first degree relatives of ill individuals [13].
Genetic studies have linked many genes and chromosomal regions spread throughout the genome to schizophrenia in different families, but no single or small number of genes accounts for the majority of cases. Common alleles have small effects (e. g. ZNF804) while rare alleles (e. g. NRG1, DTNB1, DAOA and DISC1) have greater effects [14]. A summary of the genes linked to schizophrenia is shown in Table 1. Genes linked to schizophrenia do not affect a single neurobiological system, and include neurotrophic factors (e. g. BDNF, NRG), neuromodulatory receptors (DRD, HTR), members of the synaptic packaging and release machinery (SNAP25), and both inhibitory and excitatory neurotransmitter systems (GRIN, GRIK, GABR). Also, there are genes linked to folate processing (MTHFR) and methylation (e.g. DNMT, COMT) see below.
Table 1.
Fragile Sites in the Human Genome
| Chr | Locus | Location | R/C | Agent |
|---|---|---|---|---|
| 1 | FRA1E | 1p21.2 | C | Aph |
| 1 | FRA1M | 1p21.3 | R | FolA |
| 1 | FRA1D | 1p22 | C | Aph |
| 1 | FRA1L | 1p31 | C | Aph |
| 1 | FRA1C | 1p31.2 | C | Aph |
| 1 | FRA1B | 1p32 | C | Aph |
| 1 | FRA1A | 1p36 | C | Aph |
| 1 | FRA1J | 1q12 | C | 5-Aza |
| 1 | FRA1F | 1q21 | C | Aph |
| 1 | FRA1G | 1q25.1 | C | Aph |
| 1 | FRA1K | 1q31 | C | Aph |
| 1 | FRA1H | 1q42 | C | 5-Aza |
| 1 | FRA1I | 1q44 | C | Aph |
| 2 | FRA2L | 2p11.2 | R | FolA |
| 2 | FRA2E | 2p13 | C | Aph |
| 2 | FRA2D | 2p16.2 | C | Aph |
| 2 | FRA2C | 2p24.2 | C | Aph |
| 2 | FRA2A | 2q11.2 | R | FolA |
| 2 | FRA2B | 2q13 | R | FolA |
| 2 | FRA2F | 2q21.3 | C | Aph |
| 2 | FRA2K | 2q22.3 | C | Aph |
| 2 | FRA2G | 2q31 | C | Aph |
| 2 | FRA2H | 2q32.1 | C | Aph |
| 2 | FRA2I | 2q33 | C | Aph |
| 2 | FRA2J | 2q37.3 | C | Aph |
| 3 | FRA3B | 3p14.2 | C | Aph |
| 3 | FRA3A | 3p24.2 | C | Aph |
| 3 | FRA3D | 3q25 | C | Aph |
| 3 | FRA3C | 3q27 | C | Aph |
| 4 | FRA4D | 4p15 | C | Aph |
| 4 | FRA4A | 4p16.1 | C | Aph |
| 4 | FRA4B | 4q12 | C | BrdU |
| 4 | FRA4E | 4q27 | C | Unclas |
| 4 | FRA4C | 4q31.1 | C | Aph |
| 5 | FRA5A | 5p13 | C | BrdU |
| 5 | FRA5E | 5p14 | C | Aph |
| 5 | FRA5B | 5q15 | C | BrdU |
| 5 | FRA5D | 5q15 | C | Aph |
| 5 | FRA5F | 5q21 | C | Aph |
| 5 | FRA5C | 5q31.1 | C | Aph |
| 5 | FRA5G | 5q35 | R | FolA |
| 6 | FRA6C | 6p22.2 | C | Aph |
| 6 | FRA6A | 6p23 | R | FolA |
| 6 | FRA6B | 6p25.1 | C | Aph |
| 6 | FRA6D | 6q13 | C | BrdU |
| 6 | FRA6G | 6q15 | C | Aph |
| 6 | FRA6F | 6q21 | C | Aph |
| 6 | FRA6E | 6q26 | C | Aph |
| 7 | FRA7A | 7p11.2 | R | FolA |
| 7 | FRA7D | 7p13 | C | Aph |
| 7 | FRA7C | 7p14.2 | C | Aph |
| 7 | FRA7B | 7p22 | C | Aph |
| 7 | FRA7J | 7q11 | C | Aph |
| 7 | FRA7E | 7q21.2 | C | Aph |
| 7 | FRA7F | 7q22 | C | Aph |
| 7 | FRA7G | 7q31.2 | C | Aph |
| 7 | FRA7H | 7q32.3 | C | Aph |
| 7 | FRA7I | 7q36 | C | Aph |
| 8 | FRA8C | 8q24.1 | C | Aph |
| 8 | FRA8E | 8q24.1 | R | DistA |
| 8 | FRA8F | 8q13 | R | Unclass |
| 8 | FRA8B | 8q22.1 | C | Aph |
| 8 | FRA8A | 8q22.3 | R | FolA |
| 8 | FRA8D | 8q24.3 | C | Aph |
| 9 | FRA9A | 9p21 | R | FolA |
| 9 | FRA9C | 9p21 | C | BrdU |
| 9 | FRA9B | 9q32 | R | FolA |
| 9 | FRA9E | 9q32 | C | Aph |
| 9 | FRA9F | 9q12 | C | 5-Aza |
| 9 | FRA9D | 9q22.1 | C | Aph |
| 10 | FRA10B | 10q25.2 | R | BrdU |
| 10 | FRA10E | 10q25.2 | C | Aph |
| 10 | FRA10G | 10q11.2 | C | Aph |
| 10 | FRA10C | 10q21 | C | BrdU |
| 10 | FRA10D | 10q22.1 | C | Aph |
| 10 | FRA10A | 10q23.3 | R | FolA |
| 10 | FRA10F | 10q26.1 | C | Aph |
| 11 | FRA11C | 11p15.1 | C | Aph |
| 11 | FRA11I | 11p15.1 | R | DistA |
| 11 | FRA11E | 11p13 | C | Aph |
| 11 | FRA11D | 11p14.2 | C | Aph |
| 11 | FRA11H | 11q13 | C | Aph |
| 11 | FRA11A | 11q13.3 | R<R | FolA |
| 11 | FRA11F | 11q14.2 | C | Aph |
| 11 | FRA11B | 11q23.3 | R | FolA |
| 11 | FRA11G | 11q23.3 | C | Aph |
| 12 | FRA12A | 12q13.1 | R | FolA |
| 12 | FRA12B | 12q21.3 | C | Aph |
| 12 | FRA12C | 12q24 | R | BrdU |
| 12 | FRA12E | 12q24 | C | Aph |
| 12 | FRA12D | 12q24.13 | R | FolA |
| 13 | FRA13A | 13q13.2 | C | Aph |
| 13 | FRA13B | 13q21 | C | BrdU |
| 13 | FRA13C | 13q21.2 | C | Aph |
| 13 | FRA13D | 13q32 | C | Aph |
| 14 | FRA14B | 14q23 | C | Aph |
| 14 | FRA14C | 14q24.1 | C | Aph |
| 15 | FRA15A | 15q22 | C | Aph |
| 16 | FRA16B | 16q22.1 | R | DistA |
| 16 | FRA16C | 16q22.1 | C | Aph |
| 16 | FRA16E | 16p12.1 | R | Aph |
| 16 | FRA16A | 16p13.11 | R | FolA |
| 16 | FRA16D | 16q23.2 | C | Aph |
| 17 | FRA17A | 17p12 | R | DistA |
| 17 | FRA17B | 17q23.1 | C | Aph |
| 18 | FRA18A | 18q12.2 | C | Aph |
| 18 | FRA18B | 18q21.3 | C | Aph |
| 19 | FRA19B | 19p13 | R | FolA |
| 19 | FRA19A | 19q13 | C | 5-Aza |
| 20 | FRA20A | 20p11.23 | R | FolA |
| 20 | FRA20B | 20p12.2 | C | Aph |
| 22 | FRA22B | 22q12.2 | C | Aph |
| 22 | FRA22A | 22q13 | R | FolA |
| X | FRAXB | Xp22.31 | C | Aph |
| X | FRAXC | Xq22.1 | C | Aph |
| X | FRAXD | Xq27.1 | C | Aph |
| X | FRAXA | Xq27.3 | R | FolA |
| X | FRAXE | Xq28 | R | FolA |
| X | FRAXF | Xq28 | FolA |
Chr = chromosome number, R/C= Rare or common, Aph=amphidicolin, Fola= Folic acid, 5-Aza= Azacytidine, Data was compiled from [147, 148] and Genome Database. 1999. Chr = chromosome; R/C = rare/common, Aph = amphidicolin or folic acid, FolA = Folic Acid; 5-AzaC = 5-Azacytidine, BrdU –Bromo-uridine, Unclass = unclassified, DistA = Distamycin( http://ncbi.nlm.nih.gov).
Except for mitochondrial defects in a subset of patients, no other common genetic or environmental factor, nor is an effective intervention linked to a majority of patients [15]. Clearly, there a genetic component with multiple genes linked to the disease (for reviews see [16] and [17]). Many genes linked to autism are similar to those linked to schizophrenia and bipolar disorder ([18, 19], http://neuropsych.bu. edu).
EPIGENETICS
Epigenetic programming refers to factors that are “epi”, or "on top of" genetic (DNA) sequences and was coined by Waddington in the 1940s to link genes and development [20] (Fig. 2). Epigenetic regulation allows a single genome to code for functionally different cell types and short-term adaptation (for reviews see references [21-25]). In contrast, DNA sequence changes are responsible for long-term adaptation and evolution.
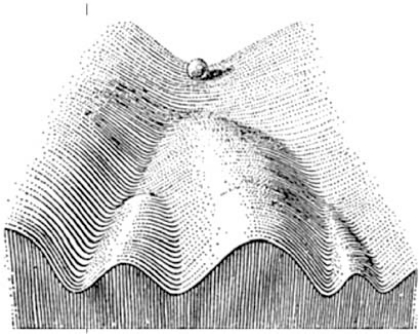
Fig. (2).
Genetics, epigenetics, and development. Waddington [20] coined the term epigenetics linking heritable factors to development. He likened development to a ball rolling down a valley, with epigenetic changes to DNA (DNA was proven to be the genetic material during this same period of time) directing a single genome towards different developmental outcomes, i.e. cell types. Epigenetic changes to DNA in a mature cell make development into another type of cell difficult (the ball cannot move into another valley).
The term “epigenetic programming” is evolving, and today refers to reversible molecular changes to DNA, RNA or proteins (e. g. histones) that regulate gene function but do not involve DNA base changes. Epigenetic changes include DNA methylation, RNA modification (e.g. editing (addition/deletion/change to base sequence), RNA interference) and both histone and non-histone proteins modifications (e. g. methylation, acetylation, phosphorylation, sumoylation, ubiquitination).
Epigenetic programming of chromatin begins shortly after DNA synthesis, although subsequent alterations may occur in response to variety of ordinary or pathological environmental or biological factors. Epigenetic changes occur globally early in development, and at specific loci throughout life and in disease states [26-28]. In cancer, the impact of epigenetic modification on gene expression has been studied for some time [29-35].
DNA Methylation
DNA methylation is the best-characterized epigenetic factor controlling gene expression (Fig. 3; for reviews see [24, 25, 36-38]). In vertebrates, 4-8% of all cytosines, and 70% of cytosines within the 5'CpG3' dinucleotide sequence, are methylated. In contrast, 70% of the cytosines at 5'CpG3' dinucleotide sequences within promoter regions of active genes are unmethylated. There are ~29,000 "CpG islands" (regions rich in 5'CpGs3') in the human genome 2 sequence. The methylation state of half of these islands regulates mRNA expression. About half of these islands are highly methylated [39]. DNA methyltransferase (DNMT) enzymes are responsible for methylation of CpG sequences [40], with the rate of methylation determined by the availability of DNMTs and their relative affinity for a given CpG site on DNA [41], and other co-factors (see below). Today, no DNA demethylase has been identified.
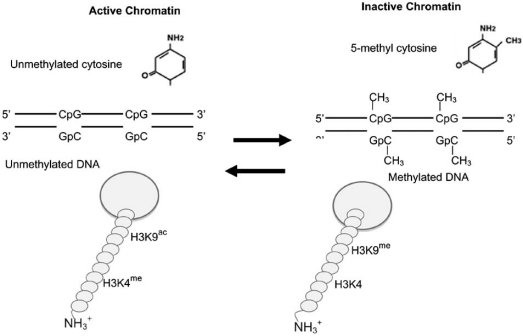
Fig. (3).
Epigenetic programming to chromatin. DNA methyl transferases (DNMTs) add methyl groups to the cytosines in CpG dinucleotide sequences. Histone 3 lysine 9 methylation (H3K9me) is concurrent with local DNA methylation in promoters. In the absence of promoter DNA methylation, histone 3 lysine 4 methylation (H3K4me) and histone 3 lysine 9 acetylation (H3K9ac) modification are found. Although both the DNA and histone modifications are reversible, only histone de-acetylases (HDAC) and de-methylases are known, no DNA de-methylase enzyme has been identified. Adapted from [25].
The number and location of methylated CpG sites in promoter regions usually, but not always, correlates with gene expression in vivo [24, 25, 36, 37, 38, 42]. Usually, dense DNA methylation is associated with irreversible silencing of gene expression, while a strong activator can overcome partial methylation. Partial promoter DNA methylation marks genes that may become unmethylated and expressed, allowing for re-adaptation to a changing micro- or macro- environment (e.g. season, ecological conditions, nutritional habits and demands of different developmental periods (see below)). More complexity in DNA methylation is introduced when the state of CpG sites within genes (i.e. outside the promoter regions) are compared to promoter dinucleotides. Ball et al. [39] show that methylation of CpG sites within genes is correlated with light promoter methylation; hence, gene body methylation appears to correlate with expression.
DNA methylation in promoter regions occurring at 5’CpG3’ dinucleotides within transcription factors recognition sites (e.g. GGGCGG and TGACGTCA for factors stimulatory protein 1 (SP1) and cAMP response element protein (CREB), respectively) may decrease expression of genes driven by these factors [25]. Gene activation itself may impact local DNA methylation. For instance, transcription factor (e.g. SP1) binding may interfere with DNA promoter methylation directly [43].
Transcription can be inhibited by proteins that bind directly or indirectly to methylated DNA (see referenced reviews above). One methylated DNA binding family, consisting of the MeCP2, MBD1, MBD2, MBD3, and MBD4 proteins, has a conserved methyl-binding domain (MBD) and binds singly methylated CpG dinucleotides [44]. Another repressor family, all containing a zinc-finger motif, consists of Kaiso protein, which binds CGCGs, the Kaiso binding sequence (KBS; recognition sequence = TCCTGCNA) protein, and the ZBTB4 and ZBTB38 proteins that bind lone methylated CpGs dinucleotides [45].
Epigenetic changes in DNA are correlated with amino terminal histone 3 modifications (methylation and acetylation)(for reviews see [46, 47, 25]; Fig. 3). Promoter regions of expressed genes (i.e., unmethylated regions) have histone 3 lysine-4 methylation (H3K4me) and histone 3 lysine-9 acetylation (H3K9ac) modifications. Promoter regions of unexpressed genes, (i.e. highly methylated regions) have no modification at histone 3 lysine 4 (H3K4) but have histone 4 lysine 9 methylation (H3K9me).
Generally, chromatin codes (DNA and histone) are preserved through mitosis, although reprogramming may occur [48]. During meiosis and early development, complex differential global chromatin reprogramming occurs, some specific for male or female germline and others for development. Some germline epigenetic patterns are inherited [48].
Epigenetic programming imprints some genes to be expression in a parental origin dependent manner [47]. Gene imprinting is proven for ~80 genes, and predicted for ~200 genes (http://www.geneimprint.com). Most imprinted genes are associated with growth and development. In female cells, epigenetic changes turn off all gene expression from one X chromosome randomly in each cell during early embryogenesis [49]. This insures that chromosome X gene expression levels are similar for female (XX) and male (XY) cells.
Although, epigenetic contributions to cancer phenotypes have been studied for some time, only recently has this area of research begun to impact neurological diseases. We and others have previously reviewed [24, 25, 50, 51] the connection between epigenetic modifications and neurological disease, including the effect of folic acid (a source of methyl groups for epigenetic modifications) metabolism on psychotic symptoms, and the co-morbidity of psychosis with diseases clearly linked to epigenetic changes (e. g. schizophrenia, bipolar disease, autism, Rett's and Angelmen's /Prader-Willi disease, mental retardation and degeneration (see below)).
GENETIC AND EPIGENETIC REGULATION OF DOPAMINE METABOLISM
The dopamine hypothesis of schizophrenia arose because many anti-psychotic medications used in the treatment of schizophrenia are dopamine receptor antagonists. Oxygen methylation of dopamine by Catechol-O-Methyl Transferase (COMT) appears to be the prominent means of dopamine catabolism after synaptic release in brain regions such as the prefrontal cortex (reviewed in [52]). The 5’ region of the COMT gene contains methylation sites that are actively regulated. Our experiments [2, 3] studied promoter methylation and gene expression levels in Brodmann Area 46 (DL-PFC) of normal versus neuropsychiatric (schizophrenia and bipolar) individuals (Fig. 4). The results revealed a significant correlation between membrane-bound COMT (MB-COMT) promoter hypo-methylation (especially at SP1 binding sites) and over-expression of the MB-COMT gene product in schizophrenia and bipolar disorder.
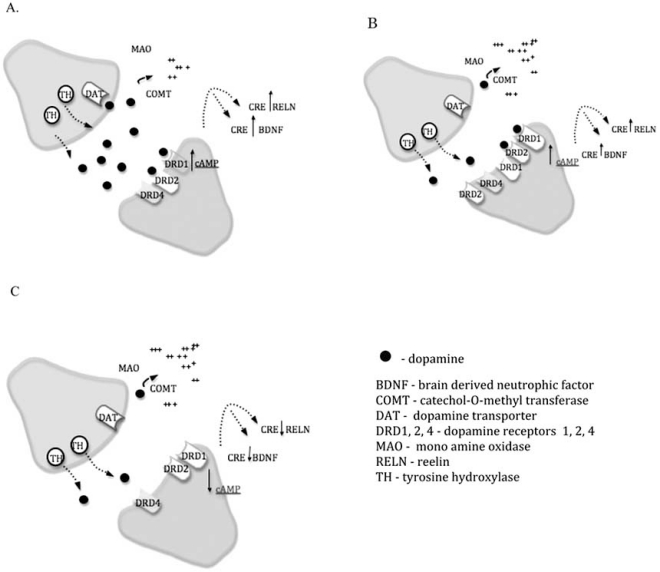
Fig. (4).
Genetic and epigenetic regulation of dopamine metabolism in schizophrenia. (A) Dopamine released by the pre-synaptic neuron into the synaptic cleft may dock with dopamine receptors on the post-synaptic neuron for downstream signaling; be degraded by MAO or COMT; or be taken back up into the pre-synaptic neuron by binding to DAT. (B) When dopamine degradation is high, for instance, by an increase in COMT activity, dopamine receptors expression is elevated to compensate for low amounts of dopamine in the synaptic cleft. (C) In schizophrenia, the coordinated up-regulation of the dopamine receptors does not exist, or exists at a greatly reduced level.
The same samples used above were genotyped for a common COMT allele (Val158Met single nucleotide polymorphism (SNP)). The results showed that schizophrenia samples were more likely to have a VAL allele, and less likely to be homozygous for the MET allele than controls. Bipolar patients were more likely to be homozygous for the VAL allele than controls.
The Val158Met polymorphism is known to directly affect the thermostability of the MB-COMT protein. The Met alleles is thermolabile, causing COMT enzyme activity in Met homozygotes to drop to approximately 1/3 the level of Val homozygotes at physiological temperature [53]. COMT hyperactivity (from the Val allele) has been linked to poor working memory as well as disturbed executive function and attention [54-58]. Genetic epigenetic gene expression results showed that dopamine degradation in the synaptic cleft is increased in individuals with schizophrenia because of increased COMT activity or expression.
Additional studies examined the expression and regulation of other genes involved in dopamine metabolism. The results revealed that expression of the dopamine receptor 1 (DRD1) was inversely correlated with MB-COMT expression in all groups, although to a lower level in the patient groups. DRD2 showed the reverse pattern: hypo-methylation of the MB-COMT promoter was nearly always associated with hypo-methylation of the DRD2 promoter and higher DRD2 gene expression levels. However, schizophrenia and bipolar patients show a significantly less severe decrease in methylation of their DRD2 promoters in response to MB-COMT hypo-methylation.
Also, the promoter methylation state of the RELN gene was significantly linked to Val158Met genotype. All schizophrenics and control subjects possessing a Val/Val genotype had a hyper-methylated RELN promoter and a decrease in RELN gene expression. This is consistent with results [59, 60] that hyper-methylation of the RELN promoter and subsequent low expression of the reelin gene in the frontal lobes is correlated with schizophrenia.
The fact that control subjects more strongly downregulate DRD1 expression and upregulate DRD2 expression when they possess a hypo-methylated MB-COMT promoter suggests that a mechanism exists for regulation of synaptic dopamine at the transcriptional level. Coordinated regulation was absent or decreased in neuropsychiatric patients. More recent unpublished data has detected aberrant methylation of the DAT1 and DRD4 promoters, but not the NRG1, HTR2A or NOS1 promoters, in samples from schizophrenic brains versus control subjects. The results suggest that aberrant synaptic dopamine metabolism in the schizophrenia/bipolar brain through genetic or epigenetic causes may contribute to disease pathogenesis.
Other groups have also examined methylation deficits in schizophrenia. For example, the methyltransferase DNMT1is up-regulated in the inhibitory inter-neurons of schizophrenia patients (reviewed in [61]). DNMT1 up-regulation is suggested to induce hyper-methylation and down-regulation of RELN and the GABA synthesizing enzyme GAD67 in prefrontal inter-neurons of schizophrenia patients. Woo et al. [62] and Costa et al. [61] speculated that down-regulation of the NMDA receptor subunit NR2A in these neurons may stem from hyper-methylation after DNMT1 up-regulation. RELN controls the surface expression of two other NMDA receptor subunits (NR2B and NR1, [63]) suggesting a possible deficit in NMDA receptors in the inter-neurons of schizophrenics. This supports the “NMDA hypofunction theory of schizophrenia” developed from observations that NMDA receptor antagonists, PCP and ketamine, both induce schizophrenia-like symptoms. In addition, the SOX10 (sex-determining region Y-box containing gene 10) gene, an oligodendrocyte specific transciption factor with a large CpG promoter island, is hyper-methylated and down-regulated in the prefrontal cortex (BA10) of schizophrenia patients [64].
GENOMIC INSTABILITY
Our initial research on schizophrenia focused on monozygotic twins. The goal was to understand disease discordance: how does one monozygotic twin avoid illness, and how do both the ill and well twin passed the same elevated genetic predisposition to progeny [1]. The specific aim was to identify, clone and sequence the expected small number of somatic changes present in monozygotic twins discordant for disease, and then do further studies to determine whether any differences were related to disease occurrence/presentation. The research targeted anonymous (CAG)n because these sequences are unstable and located within a number of genes linked to schizophrenia (e.g. [65-67], Fig. 5). The experiments examined anonymous restriction length polymorphism (RFLPs) of PCR amplicons containing (CAG)n repeating and adjacent sequences in lymphocytes using a method developed by us called Targeted Genomic Differential Display (TGDD) [68]. TGDD is similar to differential display [69], but examines subsets of DNA sequences sharing a targeted sequence.
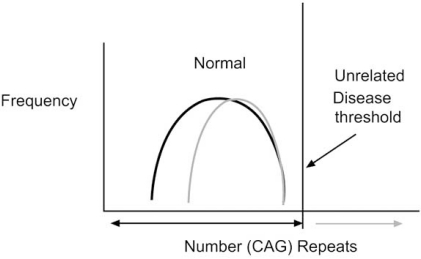
Fig. (5).
Trinucleotide repeat distribution in individuals with schizophrenia. Genes having (CAG)n and (CCG)n repeating sequences have been linked to specific diseases and to schizophrenia. The specific disease mutations are typical of repeat diseases where a repeat number over a threshold value (~50 repeats) leads to disease. Black = Distribution in unaffected individuals. Grey = In schizophrenia individuals, the repeat distribution is skewed towards larger sizes but not greater than the threshold value linked to specific disease.
Unexpectedly, a statistically significant high level of RFLP variability around (CAG)n was detected in monozygotic twins discordant for schizophrenia (Fig. 6). Twin pairs concordant for the disease had greater variability than controls, but for this small sample size this variability did not reach statistical significance. Assuming all the twin pairs were monozygotic (i. e., began life with identical DNA), RFLP variability must reflect somatic mutation rates after twinning. Hence, the results showed that a high somatic mutation rate was associated with schizophrenia, especially in monozygotic twins discordant for disease.
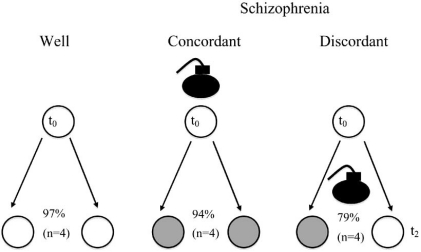
Fig. (6).
Somatic genomic instability in twins affected by schizophrenia. TGDD was used for RFLP analysis of genomic fragments containing (CAG)n repeats and adjacent sequence in 12 pairs of monozygotic twins. The results showed that twins concordantly well or concordantly affected by schizophrenia had fewer differences than twins discordantly affected by schizophrenia. Assuming these twins began life as with identical DNA (i.e. are monozygotic), the observed differences represent somatic mutations, and the results show a higher somatic mutation rate in twins discordantly affected by schizophrenia.
Evidence supporting the idea include that schizophrenia is linked to genome instability. Cytogenetic observations of increased chromosome aneuploidy in brain cells from individuals with schizophrenia [70, 71] and other neurological diseases including autism, ataxia-telangiectasia [72, 73], Alzheimer's disease [72], Down syndrome, Edwards syndrome, Patau syndrome, Parkinson's disease, spinal muscular atrophy, mental retardation, Turner syndrome, psychiatric disorders associated with trisome X and Klinefleter syndrome, and 47,XYY karyotype (reviewed in [74-77]). Other evidence (reviewed in [1]) is the skewed (CAG)n repeat distribution in schizophrenia (Fig. 5), and the inverse correlation of disease with some cancer (reviewed in [78]).
More recently, genome wide scanning of SNPs in association studies revealed an elevated rate of copy number variation (CNV) in schizophrenia [79-82], and a number of other neuropsychiatric diseases such as autism, mental retardation, bipolar disease, Rett syndrome, Tourette’s syndrome, Prader-Willi/Angelman syndrome etc. (e.g. [18, 83], for review see [84]). Clearly, genomic instability is linked to neurological disease.
FRAGILE SITES
Fragile sites are regions of the genome that are prone to mutation and epigenetic changes; hence, hot spots for genomic instability. A fragile site is defined as unstable DNA stretch that appears as a gap or break on metaphase chromosomes (Fig. 7A) when DNA replication of dividing cells is partially inhibited by incubation in culture medium deficient in folic acid or containing Bromodeoxyuridine (BrdU), distamycin, 5 azacytidine, or aphidicolin [85, 86].
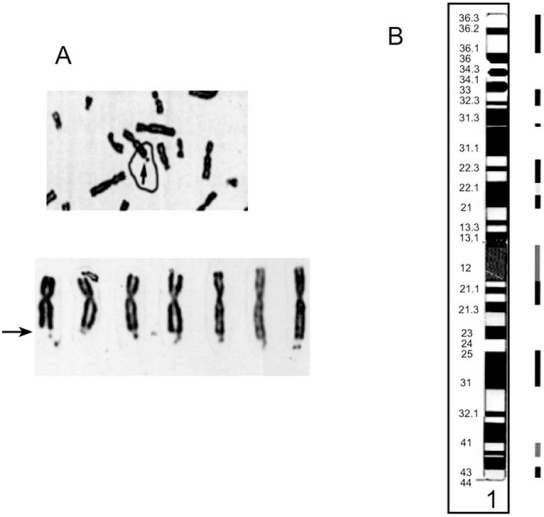
Fig. (7).
Fragile site appearance and distribution. A. Cytogenetic appearance of fragile X. Arrows point to fragile sites. B. Distributation of fragile sites along chromosome 1. The bars beside the cytogenetic bands represent the fragile site locations (see Table 1). Dark to light bars represent inducing agents. Amphidicolin, 5-Azacytidine, and Folic acid, respectively. Taken from [150].
Fragile sites are unusual chromosomal abnormalities because, although heritable, they appear only in a subset of cells, and usually only occur when induced. There are 119 known fragile sites (Tables 1 and 2), spread throughout the genome classified as common or rare based on frequency in the population (greater or less than 5%, respectively).
Table 2.
Summary of Fragile Sites within the Human Genome
| Inducer | Common | Rare | Total |
|---|---|---|---|
| Folic | 78 | 22 | 100 |
| Amphidicolin | 78 | 0 | 78 |
| BrdU | 7 | 2 | 9 |
| 5-AzaC | 4 | 0 | 4 |
| Distamycin | 0 | 5 | 5 |
| Unclassified | 1 | 0 | 1 |
The first identified and best studied example of the association between fragile sites and mental illness is Fragile X syndrome. Fragile X syndrome is associated with transcriptional silencing of either FMR1 or FMR2 (Fragile X mental retardation genes 1 and 2) on chromosome X (for review see [87]). Silencing of FMR1 or FMR2 is accompanied by hyper-methylation of the (CGG)n expansion within fragile sites FRAXA at Xq27.3 or FRAXE at Xq28, respectively. The number and methylation status of the (CCG)n repeating sequences influences the expression of the fragile X mental retardation genes. The FRAXA and FRAXE promoter sites behave similarly. For FRAXA sites, well individuals have 7 to 50 (CCG)n repeating sequences (with a mode of 30). Mental retardation occurs, and the fragile site becomes visible under folate deficient conditions, when the repeat number exceeds 230 and becomes hyper-methylated. Repeat numbers can reach up to 2000. Numbers between 50 and 200 are un-methylated and considered “pre-mutations”, but carriers may have symptoms other than mental retardation [88]. Schizophrenia is linked to several fragile sites (Table 4), some of which are unique (e.g. [89]). Neurological diseases and cancers [90, 91] are linked to specific sites as well (Table 3).
Table 4.
Summary of Genes Linked to Schizophrenia and Fragile Sites
| GENE | FRAGILE | |||||
|---|---|---|---|---|---|---|
| NAME | ALIAS | FUNCTION | ADDRESS | SITE | ADDRESS | |
| CHROMOSOME 1 | ||||||
| GSTM1 | glutathione S-transferase M1 | 1p13.3 | ||||
| GRIK3 | glutamate receptor ionotropic | 1p34-p33 | ||||
| HTR6 | 5-hydroxytryptamine (serotonin receptor type 6) | 1p36-p35 | FRA1A | 1p36 | ||
| RHD | Rhesus blood group D antigen | 1p36.11 | FRA1A | 1p36 | ||
| MTHFR | 5 10-methylenetetrahydrofolate | 1p36.3 | FRA1A | 1p36 | ||
| SCZD9 | schizophrenia disorer 9 | 1q21-q22 | FRA1F | 1q21 | ||
| SYT11 | Synaptotagamin X1 | 1q21.2 | FRA1F | 1q21 | ||
| KCNN3 | hSKCa3 | potassium intermediate/small c | 1q21.3 | FRA1F | 1q21 | |
| RGS4 | regulator: g-protein signaling 4 | 1q23.2 | ||||
| IL10 | interleukin 10 | 1q31-q32 | FRA1K | 1q31 | ||
| DISC2 | disrupted in schizophrenia 2 | 1q32.1 | ||||
| DISC1 | disrupted in schizophrenia 1 | 1q42.1 | FRA1H | 1q42 | ||
| CHROMOSOME 2 | ||||||
| NOGO | RTN4 | reticulon 4 | 2p13-p14 | FRA2E | 2p13 | |
| IL1B | interleukin 1 beta | 2q14 | ||||
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 2q22-23 | FRA2K | 2q22.3 | ||
| CTLA4 | cytotoxic T-lymphocyte-associative protein | 2q33 | FRA2I | 2q33 | ||
| CHROMOSOME 3 | ||||||
| GRM2 | GRM2 | glutamate receptor metabotropic 2 | 3p21.31 | |||
| CCK | cholecystokinin | 3p22-p21.3 | ||||
| GRM7 | GRM7 | glutamate receptor metabotropic 7 | 3p26.1-p25.1 | |||
| CHL1 CALL | cell-adhesion molecule with homology to L1CAM | 3p26.1 | ||||
| DRD3 | dopamine receptor D3 | 3q13.3 | ||||
| CHROMOSOME 4 | ||||||
| GABRB1 | GABRB1 | gamma-aminobutyric acid (GABA) receptor, beta 1 | 4p12 | |||
| CCKAR | cholecystokinin A receptor | 4p15.1-p15.2 | FRA4D | 4p15 | ||
| DRD5 | dopamine receptor D5 | 4p16.1 | FRA4A | 4p16.1 | ||
| CHROMOSOME 5 | ||||||
| GDNF | glial cell derived neurotrophic factor | 5p13.1-p12 | FRA5A | 5p13 | ||
| SCZD1 | schizophrenia disorder 1 | 5q11.2-q13.3 | ||||
| Homer 1 | homer homolog 1 (Drosoph) | 5q14.2 | ||||
| HTR4 | 5-hydroxytryptamine (serotonin) receptor 4 | 5q31-q33.2 | FRA5C | 5q31.1 | ||
| GABRB2 | GABA A receptor, beta 2 | 5q34 | ||||
| H2 rec | HRH2 | histamine H2 receptor | 5q35.3 | FRA5G | 5q35 | |
| DRD1 | dopamine receptor D1 | 5q35.1 | FRA5G | 5q35 | ||
| CHROMOSOME 6 | ||||||
| NQO2 | NADPH hydrogenase quinone 2 | 6pter-q12 | FRA6C/A/B | 6p22.2/23/25.1 | ||
| NOTCH4 | Notch homolog 4 (Drosophila) | 6p21.3 | ||||
| TNFA | Tumor necrosis factor alpha | 6p21.31 | ||||
| HLA | HLA-A | major histocompatability complex , class I, A | 6p21.3 | |||
| TNXB | tenascin XB | 6p21.3 | ||||
| DTNBP1 | dystrobrevin binding protein 1 | 6p22.3 | ||||
| SCZD3 | schizophrenia disorder 3 | 6p23 | FRA6A | 6p23 | ||
| SCA1 | spinocerebellar ataxia 1 (oliv) | 6p23 | FRA6A | 6p23 | ||
| CB1 | CNR1 | Cannabinoid receptor 1 | 6q14-q15 | FRA6G | 6q15 | |
| SCZD5 | schizophrenia disorder 5 | 6q13-q26 | FRA6D/E | 6q13,q26 | ||
| HTR1B | 5-hydroxytryptamine (serotonin) receptor 1B | 6q13 | FRA6D | 6q13 | ||
| Fyn kinase | FYN | FYN oncogene related to SRC, FGR, YES | 6q21 | FRA6F | 6q21 | |
| CHROMOSOME 7 | ||||||
| DDC | DDC | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 7p11 | FRA7A | 7p11.2 | |
| NPY | Neuropeptide Y | 7p15.1 | ||||
| GRM3 | glutamate receptor metabotropi fact. 3 | 7q21.1-q21.2 | FRA7E | 7q21.2 | ||
| RELN | reelin | 7q22 | FRA7F | 7q22 | ||
| CHROMOSOME 8 | ||||||
| NRG1 | neuregulin 1 | 8p21-p12 | ||||
| SCZD6 | schizophrenia disorder 6 | 8p21 | ||||
| PPP3CC | protein phosphotase 3 | 8p21.2 | ||||
| FDZ3 | frizzled homolog 3 | 8p21 | ||||
| DPYSL2 | human dihydroppyrimidinase-related protein 2 | 8p21-p22 | ||||
| CHROMOSOME 9 | ||||||
| OPRS1 | OPRS1 | opioid receptor, sigma 1 | 9p13.2 | |||
| DBH | dopamine beta-hydroxylase (dop) | 9q34 | ||||
| GRIN1 | NMDA | glutamate receptor ionotropic | 9q34.3 | |||
| CHROMOSOME 10 | ||||||
| SCA8 | spinocerebellar axia protein 8 | 10q23.3-24.1 | FRA10A | 10q23.3 | ||
| VMAT2 | SVMT | solute carrier family 18 (vesicular monoamine), member 2 | 10q25 | FRA10B/E | 10q25.2 | |
| CHROMOSOME 11 | ||||||
| PAX6 | paired box gene 6 (aniridia k) | 11p13 | FRA11E | 11p13 | ||
| BDNF | brain-derived neurotrophic fac | 11p13 | FRA11E | 11p13 | ||
| TPH1 | tryptophan hydroxylase | 11p15.3-p14 | FRA11D | 11p14.2 | ||
| TH | tyrosine hydroxylase | 11p15.5 | ||||
| cPLA2 | HTATIP2 | HIV-1 Tat Interactive Protein 60kDa | 11q13 | FRA11A/H | 11q13.3/ 13 | |
| GRIA4 | glutamate receptor ionotrophi | 11q22 | ||||
| DRD2 | Dopamine receptor D2 | 11q23 | FRA11B/G | 11q23.3 | ||
| HMBS | hydroxymethylbilane synthase | 11q23.3 | FRA11B/G | 11q23.3 | ||
| B3GAT | beta-1, 3-Glucronyltransferase-1 | 11q25 | ||||
| CHROMOSOME 12 | ||||||
| NR2B | GRIN2B | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | 12p12 | |||
| NTF3 | NT3 | neurotrophin 3 | 12p13 | |||
| B37 | DRPLA | dentatarubral-pallidoluysian atrophy (atrophin- 1) | 12p13.31 | |||
| PAH | phenylalanine hydroxlase | 12q22-24.2 | FRA12C/E/D | 12q24/24.13 | ||
| PLA2 | phospholipase A2. group IB | 12q23-q24.1 | FRA12C/E/D | 12q24/24.13 | ||
| NOS1 | nitric oxide synthase 1 (neuro) | 12q24.2-q24.31 | FRA12C/E | 12q24 | ||
| DAO | DAOA | d-amino acid oxidase | 12q24 | FRA12C/E/D | 12q24/24.13 | |
| CHROMOSOME 13 | ||||||
| CAGR1 *** | mab21-like 1 (c. elegans) | 13q13 | FRA13A | 13q13.2 | ||
| HTR2 | HTR2/ HTR2a | 5-hydorxytryptamine (serotonin) receptor | 13q14-q21 | FRA13B/C | 13q21-q21.2 | |
| SCZD7 | schizophrenia disorder 7 | 13q32 | FRA13D | 13q32 | ||
| G7G72 | DAOA | d-amino acid oxidase activator | 13q34 | |||
| CHROMOSOME 14 | ||||||
| NPAS3 | neuronal pas domain protein 3 | 14q12-q13 | ||||
| CHROMOSOME 15 | ||||||
| HERC2 | hect doman and RLD2 | 15q13 | ||||
| CHRNA7 | cholinergic receptor nicotini | 15q14 | ||||
| SCZD10 | schizophrenia disorder 10 | 15q15 | ||||
| CHROMOSOME 16 | ||||||
| GRIN2A | glutamate receptor, ionotropic 2A | 16p13.2 | ||||
| CHROMOSOME 17 | ||||||
| SLC6A4 | SLC6A4 | serotonin transporter | 17q11.2-q12 | |||
| ACE | angiotensin I converting enzym | 17q23 | FRA17B | 17q23.1 | ||
| CHROMOSOME 18 | ||||||
| IMPA2 | inositol(myo)-1(or 4)-monophos | 18p11.2 | ||||
| CHROMOSOME 19 | ||||||
| SCA6 | CACNA1A | calcium channel, voltage dependent, P/Q type, alpha 1A subunit | 19p13.2-p13.1 | FRA19B | 19p13 | |
| APOE | apolipoprotein E | 19q13.2 | FRA19A | 19q13 | ||
| DNMT | DNA methyltrasnferase 1 | 19q13.2 | FRA19A | 19q13 | ||
| CHROMOSOME 20 | ||||||
| PRNP | prion protein (p27-30) (Creutz) | 20pter-p12 | FRA20B | 20p12.2 | ||
| SNAP-25 | synaptosomal-associated protein 25kDa | 20p12-p11.2 | FRA20B/A | 20p12.2/11.23 | ||
| CHGB | chromogranin B ( secretogranin 1) | 20pter-p12 | FRA20B | 20p12.2 | ||
| CHROMOSOME 22 | ||||||
| COMT | catechol-O-methyltransferase | 22q11.21 | ||||
| SNAP29 | synaptosomal-associated protein | 22q11.21 | ||||
| PCQAP | PC2 (positive cofactor 2 mult | 22q11.2 | ||||
| PRODH/DGCR6 | DiGeorge Syndrome critical region, gene 6 | 22q11.21 | ||||
| UFD1L | ubiquitin fusion degradation 1 | 22q11.21 | ||||
| ZNF74 | zinc finger protein 74 (Cos52) | 22q11.21 | ||||
| APOL-4 | apolipoprotien L-4 | 22q11.2-13.2 | FRA22A/B | 22q12.2/13 | ||
| APOL-2 | apolipoprotien L2 | 22q12 | FRA22B | 22q12.2 | ||
| SYN3 | synaptin 3 | 22q12.3 | ||||
| TIMP3 | tissue inhibitor of metalloprot.3 | 22q12.3 | ||||
| YWHAH | tyrosine 3-monooxygenase/trypt | 22q12.3 | ||||
| APOL-1 | apolipoprotein L1 | 22q13.1 | FRA22A | 22q13 | ||
| SYNGR1 | synaptogyrin 1 | 22q13.1 | FRA22A | 22q13 | ||
| CYP2D6 | cytochrome P450 family 2 sub | 22q13.1 | FRA22A | 22q13 | ||
| IL2RB | interleukin 2 receptor beta | 22q13/13.1 | FRA22A | 2222q13 | ||
| BZRP | BZRP | benzodiazapine receptor (peripheral) | 22q13.31 | FRA22A | 22q13 | |
| WKL1 | MLC1 | megalencephalic leukoencephalopathy with subcortical cysts 1 | 22q13.33 | FRA22A | 22q13 | |
| X CHROMOSOME | ||||||
| HTR2C | 5-hydorxytryptamine (serotonin) receptor 2C | Xq24 | ||||
| L1CAM | L1 cell adhesion molecule | Xq28 | FRAXE/F | Xq28 | ||
Studies were obtained from the National Institute of Health’s database linking specific genes to schizophrenia at http://www.geneticassociationdb.com. In addition, a Pubmed search using the keywords "gene AND schizophrenia" yielded more unique studies. The genes found using these two methods were then searched more exclusively using the keywords “ gene name” AND schizophrenia” in order to more thoroughly assess whether at least one positive association was found between a gene and schizophrenia. Genes are organized by chromosomal locations, and appear in bold when co-localizing with a chromosomal fragile sites. The co-localizing fragile site name and address is shown. More information can be found at http://schizogad.bu.edu.
Table 3.
Neurological Diseases Associated with Specific Fragile Sites. Gene Names for Abbreviations are Shown in Table 4
| Fragile Site | Associated Gene(s) | Neurological Disease |
|---|---|---|
| FRA2A | Mental retardation/schizophrenia | |
| FRA2B | Autism | |
| FRA4F | GRID2 | Tremor/Ataxia |
| FRA6A | Autism | |
| FRA6E | PARK2 | Autosomal Juvenile Parkinsonism |
| FRA6F | LAMA4 | Schizophrenia |
| FRA7I | CNTAP2 | Tourette's |
| FRA9F | Schizophrenia | |
| FRA11B | CBL2 | Jacobsen's Syndrome |
| FRA12A | DIP2B | Autism / Mental retardation |
| FRA13A | NBEA | Sporadic Autism |
| FRA15A | RORA | Tremor/Ataxia, Imbalance |
| FRAXA | FMR1 | Fragile X Mental Retardation / FRAXA Tremor Ataxia |
| FRAXC | IL1RAPL1, DMD | Mental Retardation associated with complex glycerol kinase deficiency |
| FRAXE | FMR2 | Fragile X Mental Retardation (mild) |
| Global FS Expression | ATR | Seckel syndrome |
Cells from schizophrenia patients grown in the absence of folate present a greater overall number of fragile sites per metaphase than controls [92, 93]. These results may indicate that schizophrenia patients may have a greater sensitivity to folic acid deficiency, or a higher number of fragile sites with borderline expansion (e. g. see (CAG)n repeats in schizophrenia above).
Most fragile sites are mapped only to the low-resolution chromosomal cytogenetic band level; ~15 fragile sites are characterized at the sequence level. One site appears to be ~3 million base pairs (bp) in size and contains 10 genes and multiple repeat sequences. Rare folate sensitive sites like FRAXA are composed of the expanded simple trinucleotide repeat (CCG)n while some contain other interspersed repeats (e.g. LINE) or AT-rich sequences (e. g. common fragile sites are linked to AT-rich sequences). Replication of repeating sequences, or any sequence that deviates from the mean G+C level, can stress metabolism because the DNA replication machinery requires a different ratio of deoxynucleoside triphosphates (i. e. the ratio of G+C vs A+T).
We calculated that ~70% of the human genome was devoid of fragile sites by determining what percent of the genome, at the cytogenetic band level, was linked to one or more fragile sites (Fig. 7B). Our preliminary analysis [1] using chromosome abnormalities and genes linked to schizophrenia (reported in [94] and [95], respectively) found that ~70%, rather than the expected ~30% (X^2, p = 0.001), co-localize to regions of the having fragile sites.
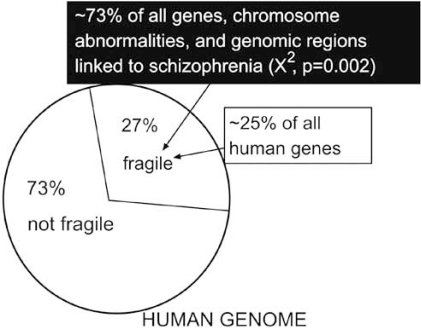
More recent studies by us reviewed 387 genetic studies from the literature that identified 111 unique genes linked to schizophrenia (Fig. 8). Of the 111 genes, 58 co-localized with at least one fragile site at the Giemsa band level (df = 1, χ2=14.227, p <0 .0001; Odds Ratio = 2.92). Moreover, a significant number of rare (CCG)n containing fragile sites co-localized with the sample of genes (df = 1, χ2=5.67, p < .025; Odds Ratio = 2.285). More detailed and updated information will be provided elsewhere.
Fig. (8).
Genomic distribution of genes, chromosomal regions, and chromosomal abnormalities linked to schizophrenia vs fragile sites. These results were obtained by cataloguing genes linked to schizophrenia from a Pubmed search (http://www.ncbi.nlm.nih.gov) using the words "schizophrenia" AND "genes", "genetic studies", or "chromosomal abnormalities". The genomic regions that contain a fragile site was determined from a Pubmed search using the words "fragile sites". The genome "real-estate" of each locus and all the fragile sites was taken as the highest known chromosome banding resolution. Negative controls consisting of (a) all human genes and (b) genes tested but not found to be associated with schizophrenia did not have any preferential association with fragile sites.
Expansion of repeating sequences within fragile sites is accompanied by local hyper-methylation (i.e. FRAXA and FRAXE) and the appearance of fragile sites in vitro.
Certainly, genes in fragile sites regions in the brain may be impacted in vivo when individuals are folate malnourished during development. In adults, DNA replication in the brain occurs in the dentate gyrus and olfactory bulb, hence folate deprivation could impact neurogenesis during all periods of life, perhaps transiently increasing the severity of disease.
In summary, fragile sites are more frequent in schizophrenia and co-localize with schizophrenia-linked genes. Fragile sites are sensitive to conditions that interfere with DNA replication, including folate deficiencies. Schizophrenia is linked to folate metabolism genetically (e. g. through hypoactive polymorphisms in genes that directly affect folate processing (e. g. MTHFR, MTR – see meta-analysis in [96])) and through epigenetic studies (see above) and environmental studies (see below).
ENVIRONMENTAL FACTORS AND SCHIZOPHRENIA
Some environmental factors linked to schizophrenia during early development are listed in Table 5. No factor is sufficient by itself to induce disease. Family history, CNS damage, bereavement, and rubella infection increase the odds ratio most for disease. Paternal age and nutrition, well-documented factors linked to schizophrenia, provide important clues for understanding the biochemistry of schizophrenia. Further, the metabolic links can be used to postulate a role for other environmental components in disease (see below).
Table 5.
Odds Ratio of Genetics and Environmental Factors Linked to Schizophrenia. Adapted from [149]
| Factor | Odds Ratio | |
|---|---|---|
| Place/time of birth | Winter | 1.2 |
| Urban | 1.5 | |
| Infection | Influenza | 2.0 |
| Respiratory | 2.2 | |
| Rubella | 5.2 | |
| Poliovirus | 1.1 | |
| CNS | 4.0 | |
| Prenatal | Famine | 2.0 |
| Bereavement | 6.2 | |
| Flood | 1.8 | |
| Unwantedness | 2.4 | |
| Maternal depr | 1.8 | |
| Obstetric | Rh incompatibility | 2.8 |
| Hypoxia | 3.0 | |
| CNS damage | 7.0 | |
| Low birth weight | 1.6 | |
| Pre-eclampsia | 2.5 | |
| Genetics | Family history | 9.7 |
Paternal Age
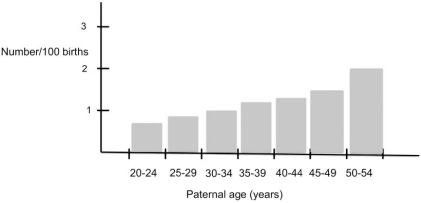
Since 1958, many studies have implicated paternal age as an environmental factor influencing the occurrence of schizophrenia (e. g. [97-99]). For instance, Malespina et al. [97] reported a three-fold increase in the incidence of schizophrenia in progeny of fathers over the age of 50 years (Fig. 9). Today, the association with maternal age is unclear. Paternal and maternal age are linked to autism [100].
Fig. (9).
The effect of paternal age on schizophrenia. The data shows a linear increase in the incidence of schizophrenia and paternal age, and a three-fold increase for children of fathers over the age of 50. Figure is adapted from [97].
The paternal age connection implicates changes to paternal germline DNA in some cases of schizophrenia because DNA is the sole paternal biological contribution to progeny. Paternal aging is linked to diminished semen quality [101] and fertility [102], increases in sperm DNA damage (e.g. [103-105]) spontaneous abortions [105, 106], birth defects [106, 107] and singe base changes in rare autosomal dominant diseases [108-110]. For instance, mutations in DF1 fibroblast growth factor receptor (FGFR3) are linked to Achondroplasia. Mutations in FGF2 are linked to Apert, Crouzon, and Pfeiffer syndrome (PS), although some PS mutations may occur in FGFR2. Mutations in the lamina A (LMNA) gene are linked to Progeria, while mutations in REarranged during transfection (RET) are linked to multiple endocrine neoplasia (MEN2A MEN2B) and medullary thyroid carcinoma (MTC).
Base substitutions account for all but progeria mutations in LMNA. The majority of mutations are transitions, (C to T) although some transversions (C to G) occur in a single dinucleotide CpG sequence. However, neither the number of replication cycles nor the observed mutation rates [110-113] accounts for the exponential rather than linear increase in disease as a function age; hence, it was suggested that these mutations confer a selection growth advantage to sperm. Lower and more linear-like increases as a function of paternal age are observed for a number of other rare autosomal dominant diseases such as neurobromatosis, bilateral retinoblastoma, Treacher Collins syndrome, multiple extostoses, and Sotos syndrome [108, 112, 113], as well as Down syndrome, neural tube defects, congenital cataracts, and reduction defects of the upper limb [105, 107].
Nutrition
Under-nutrition (general caloric or protein deficiency) and malnutrition (deficiencies in specific elements, e. g. folic acid, zinc, copper, etc.) occur worldwide and are the most common diseases of childhood and prenatal life. Moderate to severe under-nutrition occurring prior to 2 years of age is associated with persistent behavioral and cognitive deficits that resist nutritional rehabilitation [114]. Pregnant mothers exposed to famine [115, 116] or malnourished (e.g. for folate deficiencies [117]) have an increased risk for children with schizophrenia. Maternal exposure to nutritional insults leads to persistent physiological and biochemical effects on the offspring [118-121]. Nutritional, factors that have been linked to schizophrenia and autism, like folate deficiency, can impact both genetics (DNA damage and fragile site expression) and epigenetics (DNA methylation via folate deficiency) in affected individuals. Generally, the specific mechanism(s) by which nutritional deficiencies produce these birth defects are unknown.
Folic Acid
The importance of folic acid in preventing birth defects (e.g. neural tube defects including spina bifida) is well known, although the mechanism of disease induction is not understood [122]. Less well known is that fact that folic acid deficiencies are associated with a number of neurological diseases (e. g. [123, 124]) including schizophrenia and mood disorders [125-129], and are common in patients with psychopathology [130]. Furthermore, genes specifying proteins involved in folate metabolism are associated with schizophrenia and mood disorders as well as autism and other neuropsychiatric diseases [131]. Folic acid provides methyl groups to form S-adenosyl-methionine (SAM, see below), the universal intracellular methyl donor during methylation reactions such as those important in epigenetics.
Folic Acid Metabolism
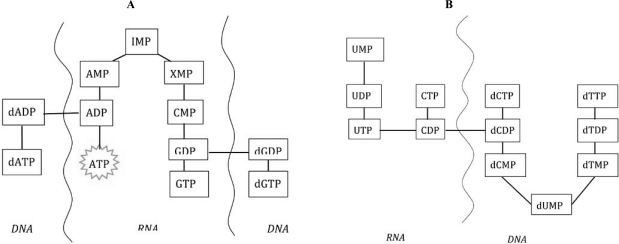
At the molecular level, folic acid deficiencies have the potential to disrupt nucleic acid metabolism, processes that require energy (i.e. ATP or NAD, GTP), activated nucleotide precursors (ribo - and deoxyribo- nucleoside triphosphates, e. g. DNA replication and RNA transcription), or SAM (or folate directly) for methylation (Fig. 10). Abbreviated schemes of de novo synthetic pathways for ribo- and deoxyribo- nucleoside triphosphate synthesis are shown in Fig. (11). Folate derivatives are required by thymidine synthase that converts dUMP to dTMP, and for two steps in the purine biosynthetic pathway to make IMP; hence impacting ribo and deoxyribo purine synthesis.
Fig. (10).
Folic Acid Cycle. Folate is an essential nutrient that is required in the synthesis of nucleic acid, s-adenosyl methionine (SAM) and amino acids. Further, synthesis of these monomers and their incorporation into polymeric molecules most times requires activated nucleosides like ATP, NAD and GTP whose synthesis depends on folic acid intermediates. Hence, the synthesis of DNA/RNA and SAM is heavily dependent on folic acid. (Figure adapted from http://www.tcd.ied/ IUBMB-Nicholson/pdf/29.pdf).
Fig. (11).
Abbreviated schematic of metabolic pathways leading to the de novo biosynthesis of RNA and DNA precursors. Purines are synthesized from a branchpoint intermediate, inosine monophosphate (IMP). In the primidine pathway, deoxyuridine and deoxythymidine intermedates are made from deoxycytidine diposphate. ATP, is predominantly synthesized from ADP in the mitochondria, and is the most used cofactor in the cell. Deoxynucleotides are made from ribonucleotides.
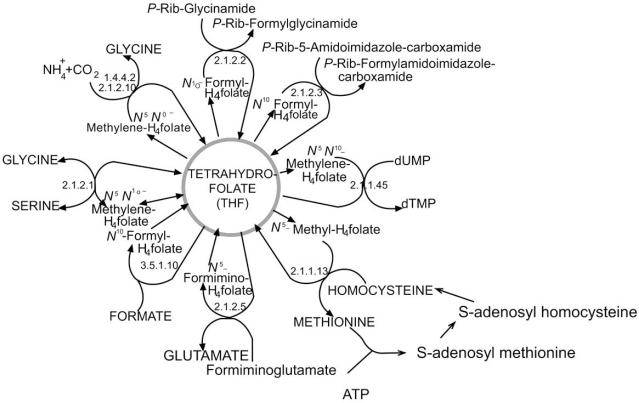
Folate participates in the methioine cycle to synthesize S-adenosyl methionine (SAM). SAM is the second most used cofactor in the cells after ATP (Fig. 12). SAM is used by >100 methyl transferases that act on DNA, RNA, proteins (e. g. DNA methyl transferase DNMT (for review see [40])), histone methyl transferases (HMT), and small molecules (e.g. COMT), and for the synthesis of polyamines that stabilize DNA.
Fig. (12).
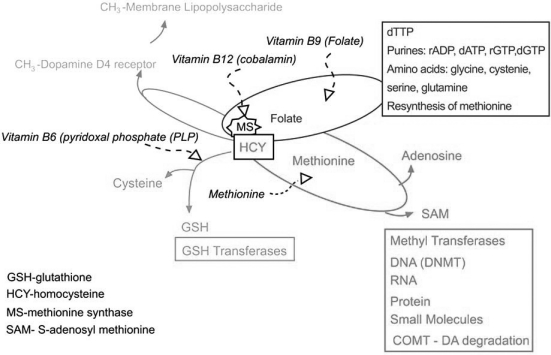
Confluence of the folate, methionine, trans-sulfuration, and dopamine D4 receptor methylation pathways. Folate is converted to derivatives that are utilized for the synthesis of dTMP, and IMP, and the amino acids serine, glycine, methionine and glutamate. SAM is formed from methionine and adenosine in the methionine cycle. Homocysteine (HCY), a degradation product of SAM, is converted to methionine by the enzyme methionine synthase (MS), utilizing a folate derivative, or by betaine homocysteine methyl transferase (BHMT) utilizing betaine (a choline derivative) as a methyl donor. In addition, HCY is a precursor for the biosynthesis of cysteine and the primary intracellular antioxidant, glutathione (GSH). The enzyme MS covalently adds a methyl group to the dopamine D4 receptor (DRD4), which transfers the methyl group to lipopolysaccharides. In mammals, folate, methionine, and vitamins B6, B9 and B12 required by these pathways, must be obtained from the diet or intestinal bacteria. Methionine may also be obtained from degradation of proteins.
In the methionine cycle, a methyl group from folate is use by the enzyme, Methionine Synthase (MS), to convert homocysteine (HCY) to methionine. Alternatively, Betaine Homocysteine Methyl Transferase (BHMT) regenerates methionine from HCY using a methyl group from betaine (choline). Dietary and regenerated methionine reacts with ATP to generate SAM, while HCY is the product of de-methylated (via methyl transferases) and de-adenylated SAM.
Besides being used to reform methionine, HCY may be directed towards the trans-sulfuration pathway to produce the amino acid cysteine, and the primary intracellular antioxidant, glutathionine (GSH) HCY is up-regulated in schizophrenia patients with a 5 microM plasma HCY level associated with a ~1.7 fold increase in schizophrenia risk [95].
MS, the enzyme that uses folate to reform methionine from HCY, covalently adds a folate derived methyl group to the dopamine D4 receptor. The dopamine D4 receptor acts like a methyl transferase when activated by dopamine and transfers the methyl group to membrane lipid polysaccharide, changing local membrane fluidity [131]. Dopamine function and metabolism is therefore tied to the folatemethionine-transulfuration metabolic hub in multiple ways: directly, through dopamine degradation by COMT, and indirectly through dopamine D4 receptor methyl transferase activity and promoter methylation of genes active in dopamine metabolism in the synaptic cleft. This metabolic hub (Fig. 12) links DNA replication and epigenetic changes through folate and SAM metabolism, and because epigenetic marking closely follows DNA replication at the macromolecular level. HCY, a key intermediate used for SAM metabolism, is required for the synthesis of GSH; hence, dopamine metabolism, DNA replication and epigenetic marking are linked to oxidative stress.
The brain is especially sensitive to oxidative stress. Oxidative stress (hypoxia) is linked to schizophrenia directly (Fig. 9), is a common consequence of obstetric complications linked to schizophrenia [132], and a potent inducer of fragile sites and genomic rearrangements [133]. Hence, oxidative stress through the transulfuration pathway is linked to DNA metabolism, and epigenetic marking. For instance, increased oxidative stress can direct HCY toward GSH production rather than SAM production, impacting many processes in vivo.
Nutrition is critical for maintaining the folate-methionine-transulfuration hub because vitamines B6, B9 (folate) and B12, and the amino acid methionine must be obtained from the diet. Other factors listed in Fig. (9) can impact the folate-methionine-transulfuration hub. For instance, winter births are associated with times of food scarcity [134], and many times bereavement and depression are accompanied by reduced food intact. Infection or inflammation increases metabolites requirements such as those needed for DNA replication, or transcription.
Aberrant folate metabolism in schizophrenia has been demonstrated in a number of studies, for review see [135, 136, 2]. In fact, the Nobel Laureate (twice), chemist Linus Pauling, advocated for nutritional interventions in psychiatry in the 1960s [137].
Aberrant folate metabolism has been detected in autistic patients. In an impressive series of experiments, James et al. [138-141] detected aberrant levels of metabolic markers for the folate-methionine-transulfuration hub in patients and their mothers. For instance, decreased levels of methionine cycle (e.g. methionine, SAM, S-adenosylhomocysteine (SAH), adenosine, and HCY), and trans-sulfuration pathway (e. g. cystathionine, cysteine and total glutathione (oxidized (GSH) + reduced GSSG)), metabolites were detected. Also reported was an increase in other methionine cycle (e.g. SAM, adenosine) and transulfuration (e.g oxidized glutathione) pathway metabolites. In 2006, James et al. [138] linked SNPs in genes within the folate cycle (in the reduced folate carrer (RFC), methylenetetrahydrofolate reductase (MTHR), the methionine cycle (COMT), and the transsulfuration pathway (glutathionine-S-transferase (GST) to autism. In a preliminary study, James et al. [141] demonstrated that a nutritional treatment regime (supplementation with methylcobalamine (methylated vitamin B6), and folic acid) improved but did not normalize abnormal metabolite blood values. An analysis of the effect of nutritional supplementation on disease symptoms was not measured, although anecdotal improvements were reported.
CONCLUSION
In summary, genetic and environmental components of schizophrenia and other neuropsychiatric diseases point to the importance of the folate-methionine-transulfuration pathway. This idea is exciting because this hub presents novel targets for drug development, and may lend themselves to nutrition interventions.
Folate supplementation has been successful in the prevention of spina bifida and related abnormalities. Similar therapies may decrease risk and severity for neuropsychiatric disease. Faulty DNA replication and epigenetic marking during brain development and adult neurogenesis may impact occurrence, presentation and dynamics of neuropsychiatric disease. Simply providing excess folate may not be useful (see [142]).
Reed and colleagues [143-146] have developed a dynamic model of the interaction of the folate and methionine cycles at the protein level. The Reed model is consistent with published data but does not yet include the entire folate-methionine-transulfuration hub, nor has the model been tested experimentally. However, this model is a beginning, and reminds us that an understanding the complex, dynamic behaviors of metabolic pathways are required to developed individualized nutritional and/or medical interventions in patients.
REFERENCES
- 1.Nguyen GH, Bouchard J, Boselli MG, Tolstoi LG, Keith L, Baldwin C, Nguyen NC, Schultz M, Herrera VL, Smith CL. DNA stability and schizophrenia in twins. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;120B(1):1–10. doi: 10.1002/ajmg.b.20010. [DOI] [PubMed] [Google Scholar]
- 2.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005;134B(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 3.Abdolmaleky HM, Cheng KH, araone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum. Mol. Genet. 2006;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol. Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 5.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25(12):528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen MJ, Craddock N, Jablensky A. The genetic deconstruction of psychosis. Schizophr. Bull. 2007;33(4):905–911. doi: 10.1093/schbul/sbm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohly HH, Panja A. Immunological findings in autism. Int. Rev. Neurobiol. 2005;71:317–341. doi: 10.1016/s0074-7742(05)71013-8. [DOI] [PubMed] [Google Scholar]
- 8.Kety SS, Wender PH, Jacobsen B, Ingraham LJ, Jansson L, Faber B, Kinney DK. Mental illness in the biological and adoptive relatives of schizophrenic adoptees. Replication of the Copenhagen Study in the rest of Denmark. Arch. Gen. Psychiatry. 1994;51(6):442–455. doi: 10.1001/archpsyc.1994.03950060006001. [DOI] [PubMed] [Google Scholar]
- 9.Malaspina D, Corcoran C, Fahim C, Berman A, Harkavy-Friedman J, Yale S, Goetz D, Goetz R, Harlap S, Gorman J. Paternal age and sporadic schizophrenia: evidence for de novo mutations. Am. J. Med. Genet. 2002;114(3):299–303. doi: 10.1002/ajmg.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kendler KS. Psychiatric genetics: a methodological critique. Am. J. Psychiatry. 2005;162(1):3–11. doi: 10.1176/appi.ajp.162.1.3. [DOI] [PubMed] [Google Scholar]
- 11.Joseph J. The gene illusion: genetic research in psychiatry and psychology under the microscope, XIV. New York: Algora Pub; 2004. p. 407. [Google Scholar]
- 12.Leo J. The fallacy of the 50% concordance rate for schizophrenia in identical twins. Hum. Nat. Rev. 2003;3:406–415. [Google Scholar]
- 13.Gottesman I, Bertelsen A. Confirming unexpressed genotypes for schizophrenia. Risks in the offspring of Fischer's Danish identical and fraternal discordant twins. Arch. Gen. Psychiatry. 1989;46(10):867–872. doi: 10.1001/archpsyc.1989.01810100009002. [DOI] [PubMed] [Google Scholar]
- 14.Owen MJ, Williams HJ, O'Donovan MC. Schizophrenia genetics: advancing on two fronts. Curr. Opin. Genet. Dev. 2009;19(3):266–2670. doi: 10.1016/j.gde.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Smith M, Spence MA, Flodman P. Nuclear and mitochondrial genome defects in autisms. Ann. N. Y. Acad. Sci. 2009;1151:102–132. doi: 10.1111/j.1749-6632.2008.03571.x. [DOI] [PubMed] [Google Scholar]
- 16.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int. J. Dev. Neurosci. 2007;25(2):69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Crespi B, Stead P, Elliot M. Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia. Proc. Natl. Acad. Sci. USA. 2009;107 (Suppl 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilmatre A, Dubourg C, Mosca AL, egallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, SaugierVeber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry. 2009;66(9):947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waddington C. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 21.Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010 Apr 10; doi: 10.1016/j.brainres.2010.03.109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neurosci. Biobehav. Rev. 2010;34(6):882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 24.Abdolmaleky HM, Smith CL, araone SV, Shafa R, Stone W, Glatt SJ, Tsuang MT. Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;127B(1):51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 25.Abdolmaleky HM, Zhou JR, hiagalingam S, Smith CL. Epigenetic and pharmacoepigenomic studies of major psychoses and potentials for therapeutics. Pharmacogenomics. 2008;9(12):1809–1823. doi: 10.2217/14622416.9.12.1809. [DOI] [PubMed] [Google Scholar]
- 26.Monk BC, Mason AB, Kardos TB, Perlin DS. Targeting the fungal plasma membrane proton pump. Acta Biochim. Pol. 1995;42(4):481–96. [PubMed] [Google Scholar]
- 27.Thomassin H, Flavin M, Espinas ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20(8):1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am. J. Clin. Nutr. 1999;69(2):179–197. doi: 10.1093/ajcn/69.2.179. [DOI] [PubMed] [Google Scholar]
- 29.Malik K, Brown KW. Epigenetic gene deregulation in cancer. Br. J. Cancer. 2000;83(12):1583–8. doi: 10.1054/bjoc.2000.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Volinia S, Bonome T, Calin GA, reshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johns-tone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA. 2008;105(19):7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 32.Fang JY, Lu J, Chen YX, Yang L. Effects of DNA methylation on expression of tumor suppressor genes and proto-oncogene in human colon cancer cell lines. World J. Gastroenterol. 2003;9(9):1976–1980. doi: 10.3748/wjg.v9.i9.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, aylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96(15):8681–9686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59(21):5438–5442. [PubMed] [Google Scholar]
- 35.Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin. Cancer Biol. 1999;9(5):349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 36.Illingworth RS, Bird AP. CpG islands--'a rough guide'. FEBS Lett. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 38.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010;44(1):71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- 39.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27(4):36136–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 41.McNamara AR, urd PJ, Smith AE, Ford KG. Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res. 2002;30(17):3818–3830. doi: 10.1093/nar/gkf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer. 2005;5(3):223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 43.Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, Rehli M. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70(4):1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 44.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell Biol. 2006;26(1):169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16(3):341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barlow DPBM. Genomic Imprinting in Mammals. In: CD A, editor. Epigenetics. Cold Spring Harbor: NY: Cold Spring Harbor Laboratory Press; 2007. pp. 357–375. [Google Scholar]
- 48.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 2002;132(8 Suppl):2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 49.Erwin JA, Lee JT. New twists in X-chromosome inactivation. Curr. Opin. Cell Biol. 2008;20(3):349–355. doi: 10.1016/j.ceb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr. Bull. 2009;35(6):1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schanen NC. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 2006;15(2):R138–150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 52.Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-o-methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol. Psychiatry. 2006;60(2):141–151. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 53.Chen J, Lipska BK, alim N, Ma QD, Matsumoto M, Mel-hem S, Kolachana BS, Hyde TM, Herman MM, Apud J, Egan MF, Kleinman JE, Weinberger DR. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am. J. Hum. Genet. 2004;75(5):807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Maz-zanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc. Natl. Acad. Sci. USA. 2001;98(12):6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blasi G, Mattay VS, ertolino A, Elvevag B, Callicott JH, Das S, Kolachana BS, Egan MF, Goldberg TE, Weinberger DR. Effect of catechol-O-methyltransferase val158met genotype on attentional control. J. Neurosci. 2005;25(20):5038–5045. doi: 10.1523/JNEUROSCI.0476-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruder GE, eilp JG, Xu H, Shikhman M, Schori E, Gor-man JM, Gilliam TC. Catechol-O-methyltransferase (COMT) genotypes and working memory: associations with differing cognitive operations. Biol. Psychiatry. 2005;58(11):901–907. doi: 10.1016/j.biopsych.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 57.deFrias CM, Annerbrink K, Westberg L, Eriksson E, Adolfsson R, Nilsson LG. Catechol O-methyltransferase Val158Met polymorphism is associated with cognitive performance in nondemented adults. J. Cogn. Neurosci. 2005;17(7):1018–1025. doi: 10.1162/0898929054475136. [DOI] [PubMed] [Google Scholar]
- 58.Galderisi S, Maj M, Kirkpatrick B, Piccardi P, Mucci A, Invernizzi G, Rossi A, Pini S, Vita A, Cassano P, Stratta P, Severino G, Del Zompo M. Catechol-O-methyltransferase Val158Met polymorphism in schizophrenia: associations with cognitive and motor impairment. Neuropsychobiology. 2005;52(2):83–89. doi: 10.1159/000087096. [DOI] [PubMed] [Google Scholar]
- 59.Abdolmaleky H M, Cheng K H, Russo A, Smith C L, Faraone S V, Wilcox M, Shafa R, Glatt S J, Nguyen G, Ponte J F, Thiagalingam S, Tsuang M T. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 60.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. USA. 2005;102(26):9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferaseoverexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2(1):29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- 62.Woo TU, Walsh JP, Benes FM. Density of glutamic acid decarboxylase 67 messenger RNA-containing neurons that express the N-methyl-D-aspartate receptor subunit NR2A in the anterior cingulate cortex in schizophrenia and bipolar disorder. Arch. Gen. Psychiatry. 2004;61(7):649–657. doi: 10.1001/archpsyc.61.7.649. [DOI] [PubMed] [Google Scholar]
- 63.Groc L, Choquet D, Stephenson FA, Verrier D, Manzoni OJ, Chavis P. NMDA receptor surface trafficking and synaptic subunit composition are developmentally regulated by the extracellular matrix protein Reelin. J. Neurosci. 2007;27(38):10165–10175. doi: 10.1523/JNEUROSCI.1772-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J. Neurosci. 2005;25(22):5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Culjkovic B, Stojkovic O, Savic D, Zamurovic N, Nesic M, Major T, Keckarevi D, Romac S, Zamurovi B, Vukosavic S. Comparison of the number of triplets in SCA1, MJD/SCA3, HD, SBMA, DRPLA, MD, FRAXA and FRDA genes in schizophrenic patients and a healthy population. Am. J. Med. Genet. 2000;96(6):884–887. doi: 10.1002/1096-8628(20001204)96:6<884::aid-ajmg41>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 66.Vincent JB, Yuan QP, Schalling M, Adolfsson R, Azevedo MH, Macedo A, Bauer A, DallaTorre C, Medeiros HM, Pato MT, Pato CN, Bowen T, Guy CA, Owen MJ, O'Donovan MC, Paterson AD, Petronis A, Kennedy JL. Long repeat tracts at SCA8 in major psychosis. Am. J. Med. Genet. 2000;96(6):873–876. doi: 10.1002/1096-8628(20001204)96:6<873::aid-ajmg37>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 67.Wei J, Hemmings GP. The NOTCH4 locus is associated with susceptibility to schizophrenia. Nat. Genet. 2000;25(4):376–377. doi: 10.1038/78044. [DOI] [PubMed] [Google Scholar]
- 68.Bouchard J, Foulon C, Storm N, Nguyen GH, Smith CL. Genomic discordance in monozygotic twins. In: Crusio WE G.R, editor. Techniques in behavioral and neural sciences. Amsterdam: Elsevier; 1999. pp. 237–259. [Google Scholar]
- 69.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257(5072):967–71. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 70.Yurov YB, Vostrikov VM, Vorsanova SG, Monakhov VV, Iourov IY. Multicolor fluorescent in situ hybridization on post-mortem brain in schizophrenia as an approach for identification of low-level chromosomal aneuploidy in neuropsychiatric diseases. Brain Dev. 2001;23(Suppl 1):S186–190. doi: 10.1016/s0387-7604(01)00363-1. [DOI] [PubMed] [Google Scholar]
- 71.Yurov YB, Iourov IY, Vorsanova SG, Demidova IA, Kravetz VS, Beresheva AK, Kolotii AD, Monakchov VV, Uranova NA, Vostrikov VM, Soloviev IV, Liehr T. The schizophrenia brain exhibits low-level aneuploidy involving chromosome 1. Schizophr. Res. 2008;98(1-3):139–147. doi: 10.1016/j.schres.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 72.Iourov IY, Vorsanova SG, Liehr T, Yurov YB. Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol. Dis. 2009;34(2):212–220. doi: 10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 73.Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum. Mol. Genet. 2009;18(14):2656–2669. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 74.Iourov IY, Vorsanova SG, Yurov YB. Chromosomal mosaicism goes global. Mol. Cytogenet. 2008;1:26. doi: 10.1186/1755-8166-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iourov IY, Vorsanova SG, Yurov YB. Molecular cytogenetics and cytogenomics of brain diseases. Curr. Genomics. 2008;9(7):452–465. doi: 10.2174/138920208786241216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, Monakhov VV, Soloviev IV. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2(6):e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yurov YB, Iourov IY, Vorsanova SG. Neurodegeneration mediated by chromosome instability suggests changes in strategy for therapy development in ataxia-telangiectasia. Med. Hypotheses. 2009;73(6):1075–6. doi: 10.1016/j.mehy.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 78.Catts VS, Catts SV. Apoptosis and schizophrenia: is the tumour suppressor gene, p53, a candidate susceptibility gene? Schizophr. Res. 2000;41(3):405–415. doi: 10.1016/s0920-9964(99)00077-8. [DOI] [PubMed] [Google Scholar]
- 79.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, ichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Ras-mussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Can-tor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460(7256):744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Consortium TIS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 82.Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, Cichon S, Hillmer A, Toncheva D, Owen MJ, O'Donovan MC. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol. Psychiatry. 2009;14(8):796–803. doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- 83.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, Gergel J, Hatchwell E, Gilliam TC, Gershon ES, Nowak NJ, Dobyns WB, Cook EH Jr. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol. Psychiatry. 2008;63(12):1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cook EH Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 85.Hecht F, Sutherland GR. Detection of the fragile X chromosome and other fragile sites. Clin. Genet. 1984;26(4):301–303. doi: 10.1111/j.1399-0004.1984.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 86.Hecht F, Hecht BK. Fragile sites and chromosome breakpoints in constitutional rearrangements I. Amniocentesis. Clin. Genet. 1984;26(3):169–173. doi: 10.1111/j.1399-0004.1984.tb04363.x. [DOI] [PubMed] [Google Scholar]
- 87.D'Hulst C, Kooy RF. Fragile X syndrome: from molecular genetics to therapy. J. Med. Genet. 2009;46(9):577–584. doi: 10.1136/jmg.2008.064667. [DOI] [PubMed] [Google Scholar]
- 88.Hagerman RJ, Leavitt BR, Farzin F, Jacquemont S, Greco CM, Brunberg JA, Tassone F, Hessl D, Harris SW, Zhang L, Jardini T, Gane LW, Ferranti J, Ruiz L, Leehey MA, Grigsby J, Hagerman PJ. Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation. Am J. Hum. Genet. 2004;74(5):1051–1056. doi: 10.1086/420700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen CH, Shih HH, Wang-Wuu S, Tai JJ, Wuu KD. Hum. Genet. 1998;103:702–706. doi: 10.1007/s004390050894. [DOI] [PubMed] [Google Scholar]
- 90.Hecht F, Sutherland GR. Fragile sites and cancer breakpoints. Cancer Genet. Cytogenet. 1984;12(2):179–181. doi: 10.1016/0165-4608(84)90132-8. [DOI] [PubMed] [Google Scholar]
- 91.Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17(6):339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 92.Demirhan O, Tastemir D, Sertdemir Y. Chromosomal fragile sites in schizophrenic patients. Genetika. 2006;42(7):985–992. [PubMed] [Google Scholar]
- 93.Tastemir D, Demirhan O, Sertdemir Y. Chromosomal fragile site expression in Turkish psychiatric patients. Psychiatry Res. 2006;144(2-3):197–203. doi: 10.1016/j.psychres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 94.Bassett AS, Chow EW, Weksberg R. Chromosomal abnormalities and schizophrenia. Am. J. Med. Genet. 2000;97(1):45–51. doi: 10.1002/(sici)1096-8628(200021)97:1<45::aid-ajmg6>3.0.co;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baron M. Genetics of schizophrenia and the new millennium: progress and pitfalls. Am. J. Hum. Genet. 2001;68(2):299–312. doi: 10.1086/318212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muntjewerff JW, Kahn RS, Blom HJ, den Heijer M. Homocysteine, methylenetetrahydrofolatereductase and risk of schizophrenia: a meta-analysis. Mol. Psychiatry. 2006;11(2):143–149. doi: 10.1038/sj.mp.4001746. [DOI] [PubMed] [Google Scholar]
- 97.Johanson E. A study of schizophrenia in the male: a psychiatric and social study based on 138 cases with follow up. Acta Psychiatr. Neurol. Scand. 1958;(Suppl., 125):1–132. [PubMed] [Google Scholar]
- 98.Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, Susser ES. Advancing paternal age and the risk of schizophrenia. Arch. Gen. Psychiatry. 2001;58(4):361–367. doi: 10.1001/archpsyc.58.4.361. [DOI] [PubMed] [Google Scholar]
- 99.Perrin MC, Brown AS, Malaspina D. Aberrant epigenetic regulation could explain the relationship of paternal age to schizophrenia. Schizophr. Bull. 2007;33(6):1270–1273. doi: 10.1093/schbul/sbm093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reichenberg A, Gross R, Sandin S, Susser ES. Advancing paternal and maternal age are both important for autism risk. Am. J. Public Health. 2010;100(5):772–773. doi: 10.2105/AJPH.2009.187708. author reply 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eskenazi B, Wyrobek AJ, loter E, Kidd SA, Moore L, Young S, Moore D. The association of age and semen quality in healthy men. Hum. Reprod. 2003;18(2):447–454. doi: 10.1093/humrep/deg107. [DOI] [PubMed] [Google Scholar]
- 102.Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertil. Steril. 2001;75(2):237–348. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- 103.Spano M, Kolstad AH, Larsen SB, Cordelli E, Leter G, Giwercman A, Bonde JP. The applicability of the flow cytometric sperm chromatin structure assay in epidemiological studies. Asclepios. Hum. Reprod. 1998;13(9):2495–2505. doi: 10.1093/humrep/13.9.2495. [DOI] [PubMed] [Google Scholar]
- 104.Bonde JP, Joffe M, Apostoli P, Dale A, Kiss P, Spano M, Caruso F, Giwercman A, Bisanti L, Porru S, Vanhoorne M, Comhaire F, Zschiesche W. Sperm count and chromatin structure in men exposed to inorganic lead: lowest adverse effect levels. Occup. Environ. Med. 2002;59(4):234–242. doi: 10.1136/oem.59.4.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fisch H, Hyun G, Golden R, Hensle TW, Olsson CA, Liberson GL. The influence of paternal age on Down syndrome. J. Urol. 2003;169(6):2275–2278. doi: 10.1097/01.ju.0000067958.36077.d8. [DOI] [PubMed] [Google Scholar]
- 106.McIntosh GC, Olshan AF, Baird PA. Paternal age and the risk of birth defects in offspring. Epidemiology. 1995;6(3):282–288. doi: 10.1097/00001648-199505000-00016. [DOI] [PubMed] [Google Scholar]
- 107.Wyrobek AJ, Eskenazi B, Young S, Arnheim N, Tiemann-Boege I, Jabs EW, Glaser RL, Pearson FS, Evenson D. Advancing age has differential effects on DNA damage, chromatin integrity, gene mutations, and aneuploidies in sperm. Proc. Natl. Acad. Sci USA. 2006;103(25):9601–9606. doi: 10.1073/pnas.0506468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Risch N, Reich EW, ishnick MM, McCarthy JG. Spontaneous mutation and parental age in humans. Am. J. Hum. Genet. 1987;41(2):218–248. [PMC free article] [PubMed] [Google Scholar]
- 109.Glaser RL, Broman KW, Schulman RL, Eskenazi B, Wy-robek AJ, Jabs EW. The paternal-age effect in Apert syndrome is due, in part, to the increased frequency of mutations in sperm. Am. J. Hum. Genet. 2003;73(4):939–947. doi: 10.1086/378419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1(1):40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 111.Tiemann-Boege I, Navidi W, Grewal R, Cohn D, Eskenazi B, Wyrobek AJ, Arnheim N. The observed human sperm mutation frequency cannot explain the achondroplasia paternal age effect. Proc. Natl. Acad. Sci. USA. 2002;99(23):14952–14957. doi: 10.1073/pnas.232568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Glaser RL, Jabs EW. Dear old dad. Sci. Aging Knowledge Environ. 2004;(3):re1. doi: 10.1126/sageke.2004.3.re1. [DOI] [PubMed] [Google Scholar]
- 113.Crow JF. Development. There's something curious about paternal-age effects. Science. 2003;301(5633):606–607. doi: 10.1126/science.1088552. [DOI] [PubMed] [Google Scholar]
- 114.Galler JR, Shumsky JS, Morgane PJ. Malnutrition and brain development. In: Walker W.A. W.J.B, editor. Nutrition in Pediatrics: Basic Science and Clinical Application. 2. Neuilly-sur-Seine, France: JB Decker Europe Inc; 1996. pp. 196–212. [Google Scholar]
- 115.Susser ES, Lin SP. Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944-1945. Arch. Gen. Psychiatry. 1992;49(12):983–988. doi: 10.1001/archpsyc.1992.01820120071010. [DOI] [PubMed] [Google Scholar]
- 116.Hoek HW, Brown AS, Susser E. The Dutch famine and schizophrenia spectrum disorders. Soc. Psychiatry Psychiatr. Epidemiol. 1998;33(8):373–379. doi: 10.1007/s001270050068. [DOI] [PubMed] [Google Scholar]
- 117.Brown AS, Susser ES. Prenatal nutritional deficiency and risk of adult schizophrenia. Schizophr. Bull. 2008;34(6):1054–1063. doi: 10.1093/schbul/sbn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hirschi KK, Keen CL. Nutrition in embryonic and fetal development. Nutrition. 2000;16(7-8):495–499. doi: 10.1016/s0899-9007(00)00362-2. [DOI] [PubMed] [Google Scholar]
- 119.Ramachandran P. Maternal nutrition--effect on fetal growth and outcome of pregnancy. Nutr. Rev. 2002;60(5 Pt 2):S26–34. doi: 10.1301/00296640260130704. [DOI] [PubMed] [Google Scholar]
- 120.Poledne R, Hajna J. Imprinting of high sensitivity to a high-cholesterol diet by nutrition in early life. Physiol. Res. 1998;47(2):95–101. [PubMed] [Google Scholar]
- 121.Tamura T, Goldenberg RL, reeberg LE, Cliver SP, Cutter GR, Hoffman HJ. Maternal serum folate and zinc concentrations and their relationships to pregnancy outcome. Am. J. Clin. Nutr. 1992;56(2):365–70. doi: 10.1093/ajcn/56.2.365. [DOI] [PubMed] [Google Scholar]
- 122.Litwack G. In: Folic Acid and Folates, Vitamins and Hormones. Litwack G, editor. Vol. 79. Amsterdam: Elsevier; 2008. p. 443. [DOI] [PubMed] [Google Scholar]
- 123.Kwik-Uribe CL, Golub MS, Keen CL. Chronic marginal iron intakes during early development in mice alter brain iron concentrations and behavior despite postnatal iron supplementation. J. Nutr. 2000;130(8):2040–2048. doi: 10.1093/jn/130.8.2040. [DOI] [PubMed] [Google Scholar]
- 124.Jankowski-Hennig MA, Clegg MS, Daston GP, Rogers JM, Keen CL. Zinc-deficient rat embryos have increased caspase 3-like activity and apoptosis. Biochem. Biophys. Res. Commun. 2000;271(1):250–256. doi: 10.1006/bbrc.2000.2608. [DOI] [PubMed] [Google Scholar]
- 125.Mischoulon D, Fava M. Role of S-adenosyl-L-methionine in the treatment of depression: a review of the evidence. Am. J. Clin. Nutr. 2002;76(5):1158S–1161S. doi: 10.1093/ajcn/76/5.1158S. [DOI] [PubMed] [Google Scholar]
- 126.Picker JD, Coyle JT. Do maternal folate and homocysteine levels play a role in neurodevelopmental processes that increase risk for schizophrenia? Harv. Rev. Psychiatry. 2005;13(4):197–205. doi: 10.1080/10673220500243372. [DOI] [PubMed] [Google Scholar]
- 127.Bottiglieri T. Homocysteine and folate metabolism in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2005;29(7):1103–1112. doi: 10.1016/j.pnpbp.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 128.Pancheri P, Scapicchio P, Chiaie RD. A double-blind, randomized parallel-group, efficacy and safety study of intramuscular S-adenosyl-L-methionine 1,4-butanedisulphonate (SAMe) versus imipramine in patients with major depressive disorder. Int. J. Neuropsychopharmacol. 2002;5(4):287–294. doi: 10.1017/S1461145702003085. [DOI] [PubMed] [Google Scholar]
- 129.Reif A, Pfuhlmann B, Lesch KP. Homocysteinemia as well as methylenetetrahydrofolatereductase polymorphism are associated with affective psychoses. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2005;29(7):1162–1168. doi: 10.1016/j.pnpbp.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 130.Young SN, Ghadirian AM. Folic acid and psychopathology. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1989;13(6):841–863. doi: 10.1016/0278-5846(89)90037-7. [DOI] [PubMed] [Google Scholar]
- 131.Deth R. Molecular origins of human attention: the dopaminefolate connection. Boston: Kluwer Academic Publishers; 2003. [Google Scholar]
- 132.McNeil TF. Handbook of schizophrenia. Vol. 3. Elsevier; 1988. Obstetric factors and perinatal injuries; pp. 319–343. [Google Scholar]
- 133.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell. 1998;2(2):259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- 134.Battle YL, Martin BC, Dorfman JH, Miller LS. Seasonality and infectious disease in schizophrenia: the birth hypothesis revisited. J. Psychiatr. Res. 1999;33(6):501–509. doi: 10.1016/s0022-3956(99)00022-9. [DOI] [PubMed] [Google Scholar]
- 135.Kale A, Naphade N, Sapkale S, Kamaraju M, Pillai A, Joshi S, Mahadik S. Reduced folic acid, vitamin B12 and docosahexaenoic acid and increased homocysteine and cortisol in never-medicated schizophrenia patients: implications for altered one-carbon metabolism. Psychiatry Res. 2010;175(1-2):47–53. doi: 10.1016/j.psychres.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 136.Krebs MO, Bellon A, Mainguy G, Jay TM, Frieling H. One-carbon metabolism and schizophrenia: current challenges and future directions. Trends Mol. Med. 2009;15(12):562–570. doi: 10.1016/j.molmed.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 137.Pauling L. Orthomolecular psychiatry. Varying the concentrations of substances normally present in the human body may control mental disease. Science. 1968;160(825):265–271. doi: 10.1126/science.160.3825.265. [DOI] [PubMed] [Google Scholar]
- 138.James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, Neubrander JA. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 2004;80(6):1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- 139.James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Cutler P, Bock K, Boris M, Bradstreet JJ, Baker SM, Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141B(8):947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.James SJ, elnyk S, Jernigan S, Hubanks A, Rose S, Gaylor DW. Abnormal transmethylation/transsulfuration metabolism and DNA hypomethylation among parents of children with autism. J. Autism Dev. Disord. 2008;38(10):1966–1975. doi: 10.1007/s10803-008-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.James SJ, Melnyk S, Fuchs G, Reid T, Jernigan S, Pavliv O, Hubanks A, Gaylor DW. Efficacy of methylcobalamin and folinic acid treatment on glutathione redox status in children with autism. Am. J. Clin. Nutr. 2009;89(1):425–430. doi: 10.3945/ajcn.2008.26615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ulrich CM, Potter JD. Folate supplementation: too much of a good thing? Cancer Epidemiol. Biovmarkers Prev. 2006;15(2):189–193. doi: 10.1158/1055-9965.EPI-152CO. [DOI] [PubMed] [Google Scholar]
- 143.Nijhout HF, Reed MC, Budu P, Ulrich CM. A mathematical model of the folate cycle: new insights into folate homeostasis. J. Biol. Chem. 2004;279(53):55008–55016. doi: 10.1074/jbc.M410818200. [DOI] [PubMed] [Google Scholar]
- 144.Nijhout HF, Reed MC, Anderson DF, Mattingly JC, James SJ, Ulrich CM. Long-range allosteric interactions between the folate and methionine cycles stabilize DNA methylation reaction rate. Epigenetics. 2006;1(2):81–87. doi: 10.4161/epi.1.2.2677. [DOI] [PubMed] [Google Scholar]
- 145.Nijhout HF, Reed MC, Lam SL, Shane B, Gregory JF 3rd, Ulrich CM. In silico experimentation with a model of hepatic mitochondrial folate metabolism. Theor. Biol. Med. Model. 2006;3:40. doi: 10.1186/1742-4682-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reed MC, Nijhout HF, Sparks R, Ulrich CM. A mathematical model of the methionine cycle. J. Theor. Biol. 2004;226(1):33–43. doi: 10.1016/j.jtbi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 147.Sutherland GR. Heritable fragile sites on human chromosomes. IX. Population cytogenetics and segregation analysis of the BrdU-requiring fragile site at 10q25. Am. J. Hum. Genet. 1982;34:753–756. [PMC free article] [PubMed] [Google Scholar]
- 148.Kooy RF, Oostra BA, Willems PJ. The fragile X syndrome and other fragile site disorders. Results Probl. Cell. Differ. 1998;21:1–46. doi: 10.1007/978-3-540-69680-3_1. [DOI] [PubMed] [Google Scholar]
- 149.Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e21. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Verma RS, Babu A. Human Chromosomes: Manual of Basic Techniques. New York City: Pergamon Press; 1989. [Google Scholar]