Abstract
Since their discovery in Drosophila, the heat shock proteins (Hsps) have been shown to regulate both stress resistance and life span. Aging is characterized by increased oxidative stress and the accumulation of abnormal (malfolded) proteins, and these stresses induce Hsp gene expression through the transcription factor HSF. In addition, a subset of Hsps is induced by oxidative stress through the JNK signaling pathway and the transcription factor Foxo. The Hsps counteract the toxicity of abnormal proteins by facilitating protein refolding and turnover, and through other mechanisms including inhibition of apoptosis. The Hsps are up-regulated in tissue-specific patterns during aging, and their expression correlates with, and sometimes predicts, life span, making them ideal biomarkers of aging. The tools available for experimentally manipulating gene function and assaying healthspan in Drosophila provides an unparalleled opportunity to further study the role of Hsps in aging.
Keywords: Aging, Hsps, proteotoxicity, ROS, HSF, p53
1. Introduction
The laboratory fruit-fly, Drosophila melanogaster, has been a leading model for the study of aging for almost a century because of its ease of culture, short life cycle, and highly developed genetics and molecular biology (Partridge and Tower, 2008). Moreover, the heat shock response and the Hsp genes were first discovered in Drosophila, as revealed by the “puffing” pattern of polytene chromosomes in response to heat and oxidative stress (Ritossa, 1962, 1996). Since then the Drosophila model has been key to understanding the regulation of the heat shock response, the function of the Hsps, and the intimate connection between the Hsps and the aging process.
At a genetic level, aging in Drosophila and other organisms is thought to result from antagonistic pleiotropy of gene function between developmental stages and the sexes (Hughes and Reynolds, 2005; Magwire et al., 2010; Tower, 2006; Tower and Arbeitman, 2009) . The force of natural selection declines with age, allowing for the accumulation of inherited alleles with late-acting deleterious effects (“mutation accumulation”). These same aging-promoting alleles may be maintained by positive selection if they provide a benefit earlier in life (“antagonistic plieotropy”). Because the sexes are subject to different and sometime opposing selective pressures, inherited alleles may accumulate that are relatively beneficial to one sex but relatively deleterious to the other sex, or detrimental to both sexes (“sexual antagonistic plieotropy”). Antagonistic pleiotropy is one possible explanation for why pathways that promote growth and differentiation, such as the insulin/IGF1-like signaling (IIS) pathway, the target-of-rapamycin (TOR) pathway, and the sex determination pathway (X/Y) are such important modulators of aging, as differentiation should be epistatic to the antagonist-pleiotropic activities of many genes (Figure 1). For example, these growth and differentiation-promoting pathways may act by up-regulating expression of life-span shortening genes, and down-regulating expression of longevity-promoting genes (such as Hsps), as was observed for IIS in C. elegans (Ayyadevara et al., 2008; Curran et al., 2009; Kenyon, 2010; Murphy et al., 2003). One of the first genes suspected of exhibiting antagonistic pleiotropy between developmental stages was mammalian p53, based upon its beneficial function in young animals to prevent cancer, but a possible detrimental effect in old animals by promoting counter-productive cell senescence or apoptosis (Campisi, 2003). Recently Drosophila p53 has been shown to exhibit sexual antagonistic pleiotropy, as indicated by opposing effects of p53 mutations and transgenes on life span in males versus females (Shen and Tower, 2010; Waskar et al., 2009). Interestingly the sexual-dimorphism of p53 transgene effects on life span were found to be regulated by the IIS target transcription factor Foxo, consistent with a role for IIS and Foxo in promoting sexual antagonistic pleiotropy (Shen and Tower, 2010). The activity of p53 is regulated by Hsps, making p53 one likely component of the mechanism(s) by which Hsps can affect aging and life span.
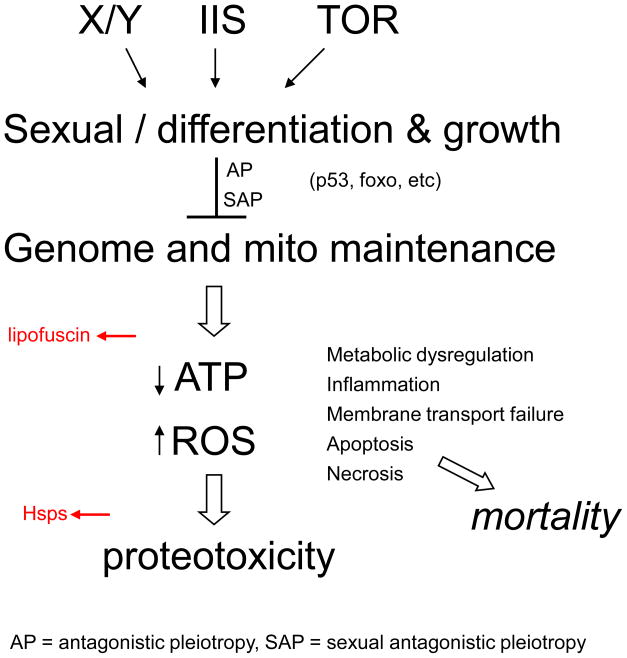
Figure 1.
Over-view of aging, proteotoxicity and Hsps. Three pathways, the sex-determination pathway (X/Y), the insulin/IGF1-like signaling pathway (IIS), and the target of rapamycin pathway (TOR) promote differentiation, sexual differentiation and growth. This differentiation and growth is epistatic to the antagonistic pleiotropic activities of numerous genes, thereby inhibiting optimal maintenance of the genome and mitochondria. Mitochondrial failure results in decreased production of ATP and increased production of ROS, resulting in the accumulation of abnormal proteins and proteotoxicity. Denatured proteins and ROS cause chronic induction of Hsp genes (indicated in red) providing one predictive biomarker of aging. Lipids, proteins and carbohydrates undergo complex cross-linking and biochemical changes to produce the autofluorescent age pigment called lipofuscin (indicated in red), another biomarker of aging. Normally regulated expression of Hsps can favor longevity by counteracting proteotoxicity, inhibiting apoptosis, and possibly other mechanisms.
Mechanistically, aging appears to involve an imbalance between damage and repair of macromolecules, including DNA damage and mutations, loss of epigenetic regulation, and telomere erosion (Vijg, 2008), as well as the response of the animal and its cells to such damage (Campisi and Vijg, 2009). Proteins are particularly subject to aging-related damages, including cleavage, covalent modifications, oxidative lesions, glycation, crosslinking, and denaturation (Semba et al., 2010; Stadtman, 2006). The correct synthesis, folding, and turnover of proteins is one of the most energetically costly functions of the cell, and proteotoxicity is a key component of aging and aging-related diseases (Morimoto and Cuervo, 2009). Mitochondrial malfunction is increasingly implicated in the aging process, and abnormal mitochondria have been found to accumulate with age in several Drosophila tissues (Calleja et al., 1993; Fleming et al., 1985; Schwarze et al., 1998; Sohal, 1975; Walker and Benzer, 2004). The resultant oxidative stress and reduction in ATP synthesis may play a central role in the creation and accumulation of abnormal proteins during aging (Bueler, 2009; Linford and Pletcher, 2009; Marzetti et al., 2009; Morrow and Tanguay, 2008).
The Hsps are defined by their ability to bind to denatured proteins, their ability to alter the folded structure of other proteins, and by the induction of their expression in response to stresses that cause protein denaturation, such as heat and oxidative stress (Morimoto, 2008; Morrow et al., 2006). By mediating either protein refolding or degradation, the Hsps counteract proteotoxicity and favor stress resistance. These functions of the Hsps may underly their ability to favor life span and counteract aging-related malfunction in several systems (Lithgow and Walker, 2002; Morrow and Tanguay, 2003). In addition to their role in response to global protein damage, the Hsps also have more specific targets, and are involved in regulating pathways that are central to aging phenotypes such as protein turnover, cellular senescence, cancer, and apoptosis/programmed cell death (Morimoto and Cuervo, 2009; Tower, 2009), and the specific modulation of such pathways may also underly the effects of Hsps on aging and life span. Finally, the expression of Hsps correlates with, and sometimes predicts, life span, making Hsps among the best-known biomarkers of aging in Drosophila and other animals (Johnson, 2006; Tower, 2009).
2. Hsp classes and functions
The heat shock proteins are molecular chaperones a class of proteins characterized by their ability to modulate the structure and folding of other proteins (client proteins)(Kim et al., 2007). The expression of the Hsps is induced through the HSF (heat shock transcription factor) pathway, by stresses that cause protein denaturation (unfolding), such as heat stress, oxidative stress and other stresses (Figure 2). Activated HSF binds to heat shock response elements (HSEs) in the promoters of the Hsp genes, and activates high-level transcription (Fuda et al., 2009; Voellmy, 2004). Certain Hsps, including Hsp90, Hsp70 and Hsp40, feed-back inhibit HSF, thereby limiting the response (Voellmy and Boellmann, 2007)(Figure 2a). A subset of Hsp genes are also induced by oxidative stress through the JNK pathway and the transcription factor Foxo (Wang et al., 2005)(Figure 2b), and the Menin protein has been found to be required for sustained expression of Hsp genes during stress (Papaconstantinou et al., 2005).
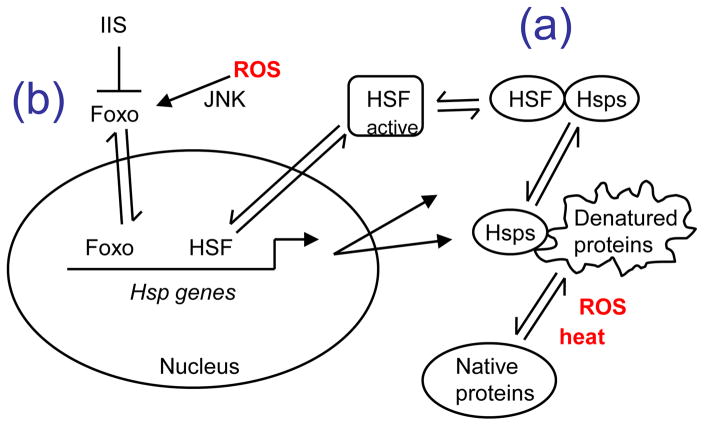
Figure 2.
Hsp gene induction by ROS and denatured proteins.
Reactive oxygen species (ROS, shown in red) and denatured proteins induce Hsp gene expression through at least two pathways, involving the transcription factors HSF (a) and Foxo (b). (a) Denatured proteins activate all Hsp genes through the transcription factor HSF. HSF is constitutively expressed, and under normal conditions HSF is present in the cytoplasm in a complex with several Hsps (including Hsp90, Hsp70 and Hsp40), that sequester the HSF protein in an inactive, monomeric form. The induction of Hsp genes by denatured proteins involves activation of HSF, and a negative feed-back loop that limits the response. Heat stress, ROS damage and other protein-denaturing stresses cause the unfolding of proteins and the exposure of hydrophobic residues in the denatured proteins. The exposed hydrophobic residues bind to Hsps, and these interactions inhibit aggregation, and facilitate re-folding or turnover, thereby reducing toxic effects on the cell. The binding of Hsps to denatured proteins titrates the Hsps away from HSF transcription factor, allowing HSF to assume an active trimeric form and translocate to the nucleus. In the nucleus, HSF binds to heat shock response elements (HSEs) in the promoters of the Hsp genes, and activates high-level Hsp gene expression. Once Hsp protein levels increase sufficiently they saturate the denatured proteins and titrate HSF back to the inactive Hsp-bound form, thereby limiting the response. The Hsps bind to a variety of clients, including inhibitory interactions with the tumor-suppressor (and senescence-regulatory protein) p53. In this way, stresses such as heat or oxidative stress that cause the denaturation of proteins can coordinately affect a variety of pathways by sequestering the Hsps away from their multiple regulatory-protein clients. (b) ROS also activates a subset of Hsp genes through the JNK signaling pathway and the transcription factor Foxo. JNK signaling activates Foxo in the cytoplasm, allowing Foxo to translocate to the nucleus where it binds to motifs in the promoters of a subset of Hsp genes and activates their expression, for example, the sHsp gene l(2)efl. The insulin/IGF1-like signaling (IIS) pathway negatively regulates Foxo activity by phosphorylating sites on Foxo that favor its retention in the cytoplasm. Therefore, decreased IIS can favor the expression of specific genes, such as l(2)efl, through increased activity of Foxo.
Key: HSF, heat shock factor; IIS, insulin/IGF1-like signaling pathway; JNK, Jun N-terminal kinase. Solid arrowheads indicate activation or production. T-bar indicates inhibition. Single-side arrowheads indicate partitioning between two subcellular compartments or states.
The Hsps are traditionally divided into families based upon their molecular weight and sequence conservation (Chang et al., 2007; Tang et al., 2007). These are the Hsp90/100 family, the Hsp70 family, the Hsp60 family, the Hsp40 family, and the small Hsps (sHsps). The constitutively expressed members of the Hsp70 family are referred to as Hsc70s. The Hsps often function with cofactors (co-chaperones) that determine specificity for interactions and functions. In Drosophila there is one member of the Hsp90/100 family, called Hsp83 (or Hsp90), that sometimes functions in complexes with members of the Hsp70 and Hsp40 families. Studies in Drosophila and mammals reveal that these chaperone complexes regulate the activity of transcription factors, such as p53 and HSF, as well as signaling pathway components, including phosphatases, protein kinases, and steroid hormone receptors, and other enzyme complexes such as telomerase (Morimoto, 2002; Morrow and Tanguay, 2003; Tower, 2009). Hsp90 is also involved in assembly of complex structures such as the centrosome, and in developmental processes such as sperm individualization.
The Hsp70 family in Drosophila includes five genes encoding the canonical stress-inducible protein Hsp70, another stress-inducible gene encoding a related protein Hsp68, and several genes encoding the constitutively expressed family members, the Hsc70s, several of which also exhibit up-regulation upon stress. The Hsc70 family members are involved in nascent protein folding at the ribosome, and trafficking and transport of proteins, including import into the mitochondria and endocytosis (Hartl and Hayer-Hartl, 2002; Neupert, 1997). While expression and function of the Hsc70s is conducive to cell growth, expression of Hsp70 has been shown to be inhibitory for cell division in certain circumstances, such as in dividing Drosophila cells (Feder et al., 1992). In contrast, many mammalian cancer cell lines are “addicted” to Hsps, in that the function of HSF and the elevated expression of Hsp70 or other Hsps is required for continued proliferation and tumorigenesis (Dai et al., 2007); one likely mechanisms is that the Hsps inhibit p53, thereby inhibiting cell senescence and apoptosis. In mammals, both Hsp70 and Hsp90 can bind to and modulate the activity of p53 (Blagosklonny et al., 1996; Muller et al., 2008; Walerych et al., 2010; Walerych et al., 2009), and in turn p53 has been implicated in the regulation of human HSP70 gene expression (Agoff et al., 1993; Tsutsumi-Ishii et al., 1995).
The sHsps have in common a small size ranging from 10 to 40 kDa, a C-terminal alpha-crystallin domain and oligomeric structure, and the ability to function as molecular chaperones (de Jong et al., 1998; Morrow et al., 2006). Drosophila has eleven predicted open reading frames containing alpha-crystallin domains, several of which have been well -characterized for their developmental and tissue-specific expression patterns, including Hsp27, Hsp26, Hsp23 and Hsp22 (Michaud et al., 1997; Michaud et al., 2002; Michaud and Tanguay, 2003). These Drosophila sHsps differ in subcellular localization, with Hsp22 in the mitochondrial matrix (Morrow et al., 2000), Hsp23 and Hsp26 in the cytosol, and Hsp27 in the nucleus (Beaulieu et al., 1989; Marin and Tanguay, 1996). The sHsp gene on the X chromosome, called hspB8 (CG14207) appears to be the ancestral family member based on phylogenetic analysis (Li et al., 2009), followed closely by l(2)efl (“essential-for-life”) on chromosome 2. While unpublished data has been cited to suggest a lethal phenotype for l(2)efl, this has not yet been confirmed (Kurzik-Dumke and Lohmann, 1995). Both the mammalian sHsp HSP27 and Drosophila Hsp27 have been shown to confer heat resistance when expressed in mammalian cells (Landry et al., 1989; Rollet et al., 1992), and mammalian sHsps have been shown to interact with cytoskeletal proteins (Mounier and Arrigo, 2002). Finally, both mammalian and Drosophila sHsps can inhibit apoptosis and inhibit the activity of p53 in mammalian cells (Concannon et al., 2003; Mehlen et al., 1995; Mehlen et al., 1996; Wadhwa et al., 2010). Notably, mutations in human sHsp genes have been associated with cardiomyopathy and neurodegenerative phenotypes (Hu et al., 2007; Rajasekaran et al., 2007; Shemetov et al., 2008; Vicart et al., 1998).
3. Hsps and protein turnover
The Hsps can facilitate the refolding of denatured client proteins, and/or facilitate their entry into several degradation pathways, including the lysosome, autophagic vesicles, and the ubiquitin/proteosome pathway. By facilitating the clearance of damaged proteins, the Hsps are key components of the cells response to proteotoxicity, and can confer upon cells increased resistance to heat and other stresses.
Chaperone-mediated autophagy is a process where specific cytosolic proteins are targeted for degradation by the lysosome. CMA is stimulated by several types of stress, including fasting and oxidative stress, and CMA activity declines during aging in several organisms and cell types (Cuervo, 2008; Mizushima et al., 2008). A complex comprised of Hsp90, Hsc70, Hsp40 and cofactors unfolds client proteins and facilitates their entry into the lysosome (autophagosome) where they are degraded. In Drosophila, autophagy is required for normal stress resistance, longevity and maintenance of the nervous system (Juhasz et al., 2007). Surprisingly, conditional inhibition of autophagy in adult flies did not affect life span, indicating that autophagy is not rate-limiting for adult fly life span under typical laboratory conditions (Bjedov et al., 2010; Ren et al., 2009). However, over-expression of the autophagy gene Atg8a in fly nervous system was reported to increase life span (Simonsen et al., 2008), and autophagy gene function was required for the drug rapamycin to increase adult fly life span (Bjedov et al., 2010), suggesting that autophagy may be limiting for adult life span under certain physiological conditions but not others. Consistent with these results, in C. elegans autophagy was not rate-limiting for life span under normal conditions, but was required for the increased life span produced by dietary restriction (DR) (Hansen et al., 2008). An additional mechanism for chaperone-assisted autophagy has been characterized in Drosophila that is particularly important for muscle maintenance, and involves the sHsp HspB8, the co-chaperone Starvin, and the chaperone-associated ubiquitin ligase CHIP (Arndt et al., 2010). The role of the Hsps in regulating autophagy and lysosomal protein degradation during aging should be a particularly interesting area for future research.
The Ubiquitin/proteosome pathway is one of the key mechanisms for protein turnover in cells. In yeast Hsp90 has been shown to promote assembly of the proteosome (Imai et al., 2003), and Hsp70 and Hsp90 family members and co-chaperones have been shown in several systems to facilitate entry of client proteins into the Ubiquitin/proteosome degradation pathway. For example, CHIP associates with Hsp90, Hsp70 and Hsc70 protein complexes and marks their client proteins for destruction by the proteosome by attaching ubiquitin (Dickey et al., 2007). Ubiquitin itself is often considered an Hsp, as it is induced by heat stress and can regulate the conformation, activity and turnover of other proteins. Notably, over-expression of the Drosophila JNK scaffold protein and Ubiquitin ligase POSH (plenty of SH3s) in the nervous system can increase life span (Seong et al., 2001a; Tsuda et al., 2006). During fly aging, the assembly and activity of the proteosome declines, and ubiquitinated proteins accumulate in various tissues (Tonoki et al., 2009; Vernace et al., 2007a; Vernace et al., 2007b). Proteosome assembly and function is dependent upon normal ATP levels, and therefore mitochondrial malfunction and decreased ATP production during aging may be causally related to Ubiquitin/proteosome system decline (Schwarze et al., 1998; Vernace et al., 2007a). Increased protein damage due to increased ROS production along with decline of the protein turnover system(s) likely underlies the accumulation of abnormal proteins and the aging-related up-regulation of Hsp genes (Figures 1, 2).
Accumulation of unfolded proteins in the ER lumen induces the unfolded-protein response (UPR), involving the function and increased expression of several chaperones including BiP (an Hsp70 family member), GRP94 (an Hsp90 family member), and co-chaperones (Rasheva and Domingos, 2009). The function of the UPR declines with age in several systems (Naidoo, 2009; Salminen and Kaarniranta, 2010), suggesting another mechanism for aging-related accumulation of abnormal proteins, and one that may be particularly relevant to aging-related neurodegeneration. Decreased expression of BiP correlates with increased life span due to DR in mouse, and altered expression of UPR factors can increase life span in Drosophila and C. elegans (Liu et al., 2009), making the UPR a promising area for future research on aging-related proteotoxicity.
4. Hsps as modulators of stress resistance and proteotoxicity
Hsps have been demonstrated to be key players in conferring resistance to heat and other stresses in Drosophila (Morrow and Tanguay, 2003; Vermeulen and Loeschcke, 2007). For example, mutation of hsp22 decreases survival upon heat stress (Morrow et al., 2004a), as does mutation of all six copies of the hsp70 gene (Gong and Golic, 2006). Mutation of hsp83 sensitizes flies to the toxic effects of sleep deprivation (Shaw et al., 2002). Over-expression of certain Hsps can increase resistance to heat and other stresses. For example, over-expression of hsp70 can increase heat tolerance of Drosophila larvae (Welte et al., 1993), and over-expression of several Hsps can increase stress resistance of adult flies (Table 1). Hsps have been shown to be involved in acquired stress resistance (hormesis), which is the ability of a moderate stress to protect the animal from a subsequent and more severe stress; notably such hormesis correlates with increased Hsp expression and can produce small increases in life span (Hercus et al., 2003; Le Bourg et al., 2001; Moskalev et al., 2009; Scannapieco et al., 2007; Sorensen et al., 2007)(see also the article by Le Bourg, this issue).
Table 1.
Drosophila Hsps affecting life span
| Gene | Manipulation | Driver | Life span | Sex | Stress res.. | Refs |
|---|---|---|---|---|---|---|
| hsp70 | Increased copy #, mild heat stress | NA | Slight increase (~4%) | M | (Tatar et al., 1997) | |
| hsp68 | OE, tissue-general, development and adult | arm-GAL4 | Increase | M | ND | (Wang et al., 2003) |
| l(2)efl | OE, tissue-general, development and adult | arm-GAL4 | Increase | M | ND | (Wang et al., 2005) |
| OE, panneuronal, development and adult | elav-GAL4 | Increase | M | ND | (Wang et al., 2005) | |
| hsp27 | OE, tissue-general, development and adult | hsp70-GAL4 | Increase | M | Increase | (Wang et al., 2004) |
| OE, panneuronal, development and adult | elav-GAL4 Appl-GAL4 |
Increase | M | Increasea | (Liao et al., 2008) | |
| Mutation | NA | Decrease | M | Decreased starvation resistance | (Hao et al., 2007) | |
| hsp26 | OE, mild heat stress, tissue-general, preferentially in adult | hsp70-GAL4 | Increase | M | ND | (Seong et al., 2001b) |
| OE, tissue-general, development and adult | hsp70-GAL4 | Increase | M | Increase | (Wang et al., 2004) | |
| OE, panneuronal, development and adult | elav-GAL4 Appl-GAL4 |
Increase | M | Increasea | (Liao et al., 2008) | |
| hsp22 | OE, tissue-general, adult-specific | rtTA(3)E2 | Decrease | M | Decrease | (Bhole et al., 2004) |
| Mutation | NA | Decrease | M | Decrease | (Morrow et al., 2004a) | |
| OE, tissue-general, development and adult | actin-GAL4 | Increase | M | ND | (Kim et al., 2010; Morrow et al., 2004b) | |
| OE, panneuronal, development and adult | scabrous-GAL4 | Increase | M | ND | (Morrow et al., 2004b) | |
| OE, panneuronal, development and adult | elav-GAL4 | Slight decrease | M | ND | (Morrow et al., 2004b) | |
| OE, motorneurons, development and adult | D42-GAL4 | Increase | M | Increase | (Morrow et al., 2004b) |
stress resistance tested for Elav-GAL4 only. OE, over-expression. Res., resistance. M, male.
Several human neurodegenerative diseases involve the accumulation and aggregation of abnormal proteins, including Alzheimer’s Disease, Parkinson’s Disease and the polyglutamine repeat diseases, and these aggregates are typically associated with Hsps. When the human disease proteins are expressed in Drosophila they exhibit toxic and neurodegenerative phenotypes, and this toxicity can often be suppressed by over-expression and function of specific Drosophila Hsps including Hsp70, hsp68 and sHsps (Auluck et al., 2005; Bilen and Bonini, 2007; Ghosh and Feany, 2004; Gong and Golic, 2006; Latouche et al., 2007).
5. Regulation of Hsp expression in response to proteotoxic stress and aging
Denaturation of proteins and the resultant exposure of hydrophobic residues is the main mechanism for the induction of Hsp genes by the transcription factor HSF (Figure 2a). HSF is consitutively expressed, and normally resides in the cytoplasm in a complex with Hsps that hold HSF in an inactive state (Morimoto, 2002). When heat, ROS or other stress causes protein denaturation, this exposes hydrophobic residues that bind Hsps and titrate them away from HSF. This allows HSF to assume an active trimeric form and translocate to the nucleus where it binds to HSEs in the promoters of the Hsp genes, and activates their high-level expression. When Hsp levels rise sufficiently they inhibit HSF and limit the response.
Protein synthesis and turnover is one of the greatest energy demands in the cell, in part because of the inaccuracy of the cell’s machinery for producing proteins with the correct sequence and folded structure (Drummond and Wilke, 2009). Translation is inherently highly inaccurate, with an error rate of approximately one in every 1, 000 to 10,000 codons translated. At this error rate approximately 15% of average sized proteins will include at least one incorrect amino acid, and these substitutions are expected to inhibit the proper folding of many of these proteins. In addition, even with correctly translated proteins, folding often fails, and a significant fraction of newly synthesized proteins are rapidly degraded and turned-over (Schubert et al., 2000; Vabulas and Hartl, 2005). Because of the energetic costs involved in producing proteins, and the toxicity of abnormal proteins, protein translation and folding appears to be a dominant selective pressure in cells, and a major force shaping genome evolution (Drummond and Wilke, 2008). Hsps favor folding, and also likely function to suppress the consequences of amino acid mis-incorporation. During aging, mitochondrial malfunction and the consequent decrease in ATP and increase in ROS will exacerbate the production of abnormal proteins and inhibit their clearance, leading to the age-associated accumulation of abnormal protein species. Indeed age-associated accumulation of abnormal proteins was one of the first molecular changes characterized in metazoan aging (Gershon and Gershon, 1970). This proteotoxic stress and its increase with age may be one possible explanation for the recent observation that inhibiting translation using rapamycin or other interventions can favor longevity in several systems including Drosophila (Bjedov et al., 2010; Kennedy and Kaeberlein, 2009); conceivably reduced translation could reduce the production of one or more toxic proteins. However, other possibilities are that reduced global translation might produce increased autophagy and clearance of toxic proteins, or that reduced global translation might promote translation of specific longevity-promoting transcripts such as Hsps, or that reduced translation might inhibit differentiation and therefore reduce antagonistic pleiotropic gene functions.
Alterations in the expression of Hsps was among the first aging-related changes in gene expression to be characterized. Fleming and co-workers observed a lengthened period of Hsp expression in response to acute stress in old flies relative to young flies, suggesting a greater abundance of abnormal and readily-denatured proteins in the old animals (Fleming et al., 1988). Consistent with that conclusion, even in unstressed flies, the expression of Hsps has been found to be up-regulated during normal fly aging in tissue-specific patterns. For example, both hsp70 and hsp22 are up-regulated at the RNA and protein levels during normal Drosophila aging (King and Tower, 1999; Morrow and Tanguay, 2003; Wheeler et al., 1995; Wheeler et al., 1999). As expected, this age-dependent induction of Hsps requires functional HSEs in the gene promoters (King and Tower, 1999; Wheeler et al., 1999). Aging associated up-regulation of Hsps has also been observed in several genome-wide studies of gene expression changes during aging in the fly (Curtis et al., 2007; Landis et al., 2004; Pletcher et al., 2002; Zou et al., 2000). These studies confirmed up-regulation of a subset of Hsps, including Hsp70 and sHsp family members, and also revealed a dramatic up-regulation of the innate immune response, down-regulation of energy synthesis and electron transport chain genes, and an extensive over-lap between the aging response and the flies response to oxidative stress; each of these changes are consistent with a failure in mitochondrial maintenance during aging.
Finally, part of the induction of Hsps during aging appears to involve posttranscriptional regulation of Hsp expression. For example, up-regulated expression of transcripts for hsp22 appear several weeks before up-regulated expression of Hsp22 protein is detected, suggesting an age-related increase in Hsp translation or stability (King and Tower, 1999).
6. Regulation of Hsp expression in response to ROS and aging, a role for JNK and Foxo
A subset of Hsp genes are regulated by the transcription factor Foxo in both Drosophila and C. elegans. Foxo is particularly relevant to aging because in C. elegans the Foxo homolog Daf16, along with HSF, has been shown to be required for the life span extension caused by reductions in IIS (Kenyon, 2010). Notably, RNAi inhibition of the expression of sHsp genes has been shown to block part of this life span extension, suggesting that induction of sHsps by Foxo and HSF is a part of the mechanism by which reduced IIS increases C. elegans life span (Hsu et al., 2003; Morley and Morimoto, 2004; Murphy et al., 2003; Walker and Lithgow, 2003). Over-expression of Hsp16 in C. elegans could also increase life span, but interestingly, this life span extension was dependent upon Foxo, indicating a mechanism involving a feedback effect on the IIS signaling pathway (Walker and Lithgow, 2003).
In Drosophila, over-expression of Foxo in fat body tissue has been found to increase life span, suggesting the possibility that Foxo function in life span regulation might be conserved between nematodes and flies (Giannakou et al., 2007; Giannakou et al., 2004; Hwangbo et al., 2004). Moreover, Drosophila Foxo regulates expression of the sHsp l(2)efl, and over-expression of l(2)efl during development and adulthood has been reported to increase fly life span (Wang et al., 2005), further suggesting a possible conservation of mechanism. Finally, in both Drosophila and C. elegans, activation of the JNK pathway can increase life span in a Foxo-dependant manner (Oh et al., 2005; Wang et al., 2005). Taken together, these studies suggest a possible conserved mechanism of life span regulation through Foxo activation of sHsp gene expression. However, Drosophila life span extension by JNK, reduced IIS, or Foxo over-expression has not yet been shown to require l(2)efl gene function in the fly.. Also, l(2)efl has not yet been shown to act in adult flies to affect life span, and the molecular mechanisms through which l(2)efl might act to regulate life span remain unknown. Each of these questions should be fruitful ones to address in future research.
7. Hsps as modulators of life span
Laboratory selection of Drosophila strains for increased life span produced flies with increased expression of sHsp genes as young adults, suggesting that increased sHsp expression might favor longevity (Kurapati et al., 2000). Consistent with this idea, mutation of hsp70 or hsp22 can reduce adult fly survival, and chemicals that increased life span, such as HDAC inhibitors, produced correlated increases in hsp70 and sHsp gene expression (Kang et al., 2002; Zhao et al., 2005). Direct testing of the effects of Hsps on life span has involved transgenic manipulations, and over-expression of several Hsps has been reported to increase fly life span (Table 1). Increased copy number of hsp70 genes resulted in reduced mortality rates upon mild stress, and a small increase in overall life span (Tatar et al., 1997). However, another study of hsp70 over-expression did not report increased life span (Minois et al., 2001), perhaps because it did not include a similar analysis of mortality rates. Over-expression of Hsp68 and four of the sHsps can produce moderate to large increases in life span when over-expression is produced in a tissue-general pattern (Table 1). This over-expression was accomplished using the GAL4/UAS system and various drivers whose expression pattern is known or expected to be tissue-general. In addition, over-expression of these four sHsps specifically in nervous tissue was found to be sufficient to increase fly life span, and for Hsp22, expression preferentially in motorneuron tissue using the D42-GAL4 driver was found to be sufficient (Morrow et al., 2004b). The Hsps may be acting through more than one pathway in different tissues to promote longevity, however, taken together, the studies suggest that nervous tissue is a particularly important site of action for the sHsps in Drosophila.
One technical limitation of the present information on the effect of Hsps on life span is that all GAL4 drivers produce some transgene expression during development as well as in the adult, so it is possible that some of the changes in life span might originate through alterations of development. In one study where hsp22 was specifically over-expressed in the adult fly using a conditional system, a negative effect on life span and stress resistance was observed, which was interpreted to suggest that increased Hsp22 levels late in adult life can be toxic to flies (Bhole et al., 2004). Conceivably, augmented Hsp expression early in life might be beneficial whereas increased accumulation of Hsps late in life could be toxic (Tower, 2009). However, negative effects (as well as positive effects) of transgene over-expression on life span must be interpreted with some caution, as it is conceivable that over-expression using heterologous promoters might produce unanticipated gain-of-function effects. Further studies using tissue-specific and developmental stage-specific alterations in Hsps should be particularly useful for identifying possible mechanisms for their effects on life span and the critical periods and tissues for their action.
Remarkably, no analysis of the effects of Hsps on female fly life span has yet been reported (Table 1). Because other genes and manipulations that affect Drosophila life span often have sex-specific effects (Burger and Promislow, 2004; Shen and Tower, 2010; Tower, 2006; Waskar et al., 2009), it is possible that results for the Hsps in females might differ from those obtained for males, and this should be a useful area for future research.
There are several possible non-exclusive mechanisms by which Hsps may be acting to increase fly life span. (i) First is by counteracting proteotoxicity. Denatured and aggregated proteins can be toxic to the cell by disrupting membrane structure, creating ROS, and by inducing apoptosis. By facilitating protein refolding, degradation or sequestration, the Hsps might decrease these toxic effects and favor cell and animal survival. (ii) Second is by suppressing mutations. By favoring the folding of proteins containing incorrect amino acids, the Hsps may facilitate the function of these abnormal proteins in supporting vital cellular processes (Blagosklonny et al., 1996). This mechanism may be related to the ability of hsp90 to act as a capacitor, and to modulate the expression of genetic variation (Levy and Siegal, 2008; Rutherford and Lindquist, 1998; Yeyati and van Heyningen, 2008). (iii) Third is by directly suppressing apoptosis. Hsp70 and sHsps can bind to and inhibit pro-apoptotic factors such as p53, thereby suppressing the apoptosis pathway (Garrido et al., 2006; Wadhwa et al., 2010). (iv) The fourth potential mechanism is by causing other favorable changes in gene expression, such as was observed in long-lived flies with hsp22 over-expression (Kim et al., 2010).
What specific mechanism(s) might be operating to produce the increases in life span observed upon over-expression of specific Hsps in Drosophila (Table 1) is not yet known. While nervous system appears to be one relevant target tissue, there may be additional tissues in which Hsps can act to affect fly life span, such as stem cells in the gut, malphighian tubule or gonads. In the future it will be of interest to determine if the life span increases observed upon over-expression of specific Hsps in Drosophila is dependent upon the presence of functional p53 or Foxo, which might indicate an effect on a specific signaling or apoptotic pathway as opposed to a mechanism involving a global suppression of proteotoxicity.
8. Hsps are predictive biomarkers of life span
Drosophila undergo a number of functional declines that correlate with increased age and increased mortality rate, including decreased spontaneous movement, decreased climbing speed, decreased memory, decreased heart function, and decreased reproductive capacity (Grotewiel et al., 2005; Iliadi and Boulianne, 2010; Piazza et al., 2009; Reenan and Rogina, 2008). Correlated molecular changes include decreases in protein turnover system activities (Ubiquitin/proteosome and autophagy/lysosomal), corresponding to the increase in abnormal proteins and the age-related up-regulation of Hsps. Abnormal mitochondria accumulate, and there is increased oxidative stress, accumulation of iron and decreased ATP production, all consistent with a failure in mitochondrial maintenance with age (Garesse and Kaguni, 2005; Massie et al., 1985; Morrow and Tanguay, 2008). Another correlated change is the accumulation of age pigment (lipofuscin) (Jacobson et al., 2010). However, predictive biomarkers of aging in Drosophila have only recently begun to be identified, and require longitudinal (repeated, non-lethal) assays. Both decreased movement and decreased egg laying have been shown to have some predictive power for remaining life span (Carey et al., 2006; Mueller et al., 2009). Fluorescent reporter transgenes, driving expression of proteins such as GFP, have proven to be particularly effective as predictive biomarkers. The first fluorescent reporter transgenes shown to act as predictive biomarkers of life span in any metazoan were anti-microbial peptide (AMP) gene promoters fused to GFP, which were found to be readily assayed in young Drosophila, and to negatively correlate with remaining life span (Landis et al., 2004); like the Hsps, the AMP genes are induced during normal aging and in response to oxidative stress. Since then, transgenic reporters for both hsp70 and hsp22 linked to GFP and DsRED fluorescent transgenes have been shown to be negatively correlated with remaining life span of the fly (Yang and Tower, 2009). Hsp gene expression shows promise as a biomarker of aging in other organisms as well, including C. elegans (Rea et al., 2005; Wu et al., 2006) and humans (Terry et al., 2006). Finally, longitudinal assay of individual Drosophila using real-time video tracking of GFP fluorescence revealed that the hsp22 and hsp70 transgenic reporters begin to spike in expression ~10 hours before death of the animal, providing an additional predictive biomarker for mortality (Grover et al., 2009; Grover et al., 2008). Video tracking holds great promise for future studies of Drosophila aging, as it allows for simultaneous real-time assay of movement, behavior and gene expression, including Hsp gene expression.
9. Summary
Altered expression of Hsps was among the first molecular changes characterized during aging in Drosophila. Since then the Hsps have been shown to directly regulate aging phenotypes, including life span, stress resistance and function. The tractability of the Drosophila system promises to allow continued discovery of the role of Hsps in aging, and facilitate our understanding of human aging and disease.
Acknowledgments
The author was supported by a grant from the Department of Health and Human Services (AG011833) and by Scientific Opportunity Funds from the Genetics of Longevity Consortium (U19 AG032122).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agoff SN, Hou J, Linzer DI, Wu B. Regulation of the human hsp70 promoter by p53. Science. 1993;259:84–7. doi: 10.1126/science.8418500. [DOI] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Hohfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–8. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Meulener MC, Bonini NM. Mechanisms of Suppression of {alpha}-Synuclein Neurotoxicity by Geldanamycin in Drosophila. J Biol Chem. 2005;280:2873–8. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu JF, Arrigo AP, Tanguay RM. Interaction of Drosophila 27,000 Mr heat-shock protein with the nucleus of heat-shocked and ecdysone-stimulated culture cells. J Cell Sci. 1989;92 ( Pt 1):29–36. doi: 10.1242/jcs.92.1.29. [DOI] [PubMed] [Google Scholar]
- Bhole D, Allikian MJ, Tower J. Doxycycline-regulated over-expression of hsp22 has negative effects on stress resistance and life span in adult Drosophila melanogaster. Mech Ageing Dev. 2004;125:651–63. doi: 10.1016/j.mad.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–64. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci U S A. 1996;93:8379–83. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson's disease. Exp Neurol. 2009;218:235–46. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Burger JM, Promislow DE. Sex-specific effects of interventions that extend fly life span. Sci Aging Knowledge Environ. 2004;2004:pe30. doi: 10.1126/sageke.2004.28.pe30. [DOI] [PubMed] [Google Scholar]
- Calleja M, Pena P, Ugalde C, Ferreiro C, Marco R, Garesse R. Mitochondrial DNA remains intact during Drosophila aging, but the levels of mitochondrial transcripts are significantly reduced. J Biol Chem. 1993;268:18891–7. [PubMed] [Google Scholar]
- Campisi J. Cancer and ageing: rival demons? Nat Rev Cancer. 2003;3:339–49. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- Campisi J, Vijg J. Does damage to DNA and other macromolecules play a role in aging? If so, how? J Gerontol A Biol Sci Med Sci. 2009;64:175–8. doi: 10.1093/gerona/gln065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey JR, Papadopoulos N, Kouloussis N, Katsoyannos B, Muller HG, Wang JL, Tseng YK. Age-specific and lifetime behavior patterns in Drosophila melanogaster and the Mediterranean fruit fly, Ceratitis capitata. Exp Gerontol. 2006;41:93–7. doi: 10.1016/j.exger.2005.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Tang YC, Hayer-Hartl M, Hartl FU. SnapShot: molecular chaperones, Part I. Cell. 2007;128:212. doi: 10.1016/j.cell.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Concannon CG, Gorman AM, Samali A. On the role of Hsp27 in regulating apoptosis. Apoptosis. 2003;8:61–70. doi: 10.1023/a:1021601103096. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–84. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavare S, Tower J. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin--small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–62. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Patterson C, Dickson D, Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med. 2007;13:32–8. doi: 10.1016/j.molmed.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–52. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–24. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–13. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- Fleming JE, Miquel J, Bensch KG. Age dependent changes in mitochondria. Basic Life Sci. 1985;35:143–56. doi: 10.1007/978-1-4899-2218-2_7. [DOI] [PubMed] [Google Scholar]
- Fleming JE, Walton JK, Dubitsky R, Bensch KG. Aging results in an unusual expression of Drosophila heat shock proteins. Proc Natl Acad Sci U S A. 1988;85:4099–103. doi: 10.1073/pnas.85.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garesse R, Kaguni LS. A Drosophila model of mitochondrial DNA replication: proteins, genes and regulation. IUBMB Life. 2005;57:555–61. doi: 10.1080/15216540500215572. [DOI] [PubMed] [Google Scholar]
- Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- Gershon H, Gershon D. Detection of inactive enzyme molecules in aging organisms. Nature. 1970;227:1214–1217. doi: 10.1038/2271214a0. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum Mol Genet. 2004;13:2011–8. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L. Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell. 2007;6:429–38. doi: 10.1111/j.1474-9726.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics. 2006;172:275–86. doi: 10.1534/genetics.105.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewiel MS, Martin I, Bhandari P, Cook-Wiens E. Functional senescence in Drosophila melanogaster. Ageing Res Rev. 2005;4:372–97. doi: 10.1016/j.arr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Grover D, Yang J, Tavare S, Tower J. Simultaneous tracking of fly movement and gene expression using GFP. BMC Biotechnol. 2008;8:93. doi: 10.1186/1472-6750-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover D, Yang J, Ford D, Tavare S, Tower J. Simultaneous tracking of movement and gene expression in multiple Drosophila melanogaster flies using GFP and DsRED fluorescent reporter transgenes. BMC Res Notes. 2009;2:58. doi: 10.1186/1756-0500-2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Zhang S, Timakov B, Zhang P. The Hsp27 gene is not required for Drosophila development but its activity is associated with starvation resistance. Cell Stress Chaperones. 2007;12:364–72. doi: 10.1379/CSC-308.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–8. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hercus MJ, Loeschcke V, Rattan SI. Lifespan extension of Drosophila melanogaster through hormesis by repeated mild heat stress. Biogerontology. 2003;4:149–56. doi: 10.1023/a:1024197806855. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–5. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen L, Zhang J, Li T, Tang J, Xu N, Wang X. Structure, function, property, and role in neurologic diseases and other diseases of the sHsp22. J Neurosci Res. 2007;85:2071–9. doi: 10.1002/jnr.21231. [DOI] [PubMed] [Google Scholar]
- Hughes KA, Reynolds RM. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 2005;50:421–45. doi: 10.1146/annurev.ento.50.071803.130409. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–6. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Iliadi KG, Boulianne GL. Age-related behavioral changes in Drosophila. Ann N Y Acad Sci. 2010;1197:9–18. doi: 10.1111/j.1749-6632.2009.05372.x. [DOI] [PubMed] [Google Scholar]
- Imai J, Yashiroda H, Maruya M, Yahara I, Tanaka K. Proteasomes and molecular chaperones: cellular machinery responsible for folding and destruction of unfolded proteins. Cell Cycle. 2003;2:585–90. [PubMed] [Google Scholar]
- Jacobson J, Lambert AJ, Portero-Otin M, Pamplona R, Magwere T, Miwa S, Driege Y, Brand MD, Partridge L. Biomarkers of aging in Drosophila. Aging Cell. 2010 doi: 10.1111/j.1474-9726.2010.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE. Recent results: biomarkers of aging. Exp Gerontol. 2006;41:1243–6. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–6. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HL, Benzer S, Min KT. Life extension in Drosophila by feeding a drug. Proc Natl Acad Sci U S A. 2002;99:838–43. doi: 10.1073/pnas.022631999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Kaeberlein M. Hot topics in aging research: protein translation, 2009. Aging Cell. 2009;8:617–23. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation. Mol Cells. 2007;23:123–31. [PubMed] [Google Scholar]
- Kim HJ, Morrow G, Westwood JT, Michaud S, Tanguay RM. Gene expression profiling implicates OXPHOS complexes in lifespan extension of flies over-expressing a small mitochondrial chaperone, Hsp22. Exp Gerontol. 2010;45:611–20. doi: 10.1016/j.exger.2009.12.012. [DOI] [PubMed] [Google Scholar]
- King V, Tower J. Aging-specific expression of Drosophila hsp22. Dev Biol. 1999;207:107–18. doi: 10.1006/dbio.1998.9147. [DOI] [PubMed] [Google Scholar]
- Kurapati R, Passananti HB, Rose MR, Tower J. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J Gerontol A Biol Sci Med Sci. 2000;55:B552–9. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- Kurzik-Dumke U, Lohmann E. Sequence of the new Drosophila melanogaster small heat-shock-related gene, lethal(2) essential for life [l(2)efl], at locus 59F4,5. Gene. 1995;154:171–5. doi: 10.1016/0378-1119(94)00827-f. [DOI] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavaré S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–8. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Chretien P, Lambert H, Hickey E, Weber LA. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. Journal of Cellular Biology. 1989;109:7–15. doi: 10.1083/jcb.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latouche M, Lasbleiz C, Martin E, Monnier V, Debeir T, Mouatt-Prigent A, Muriel MP, Morel L, Ruberg M, Brice A, Stevanin G, Tricoire H. A conditional pan-neuronal Drosophila model of spinocerebellar ataxia 7 with a reversible adult phenotype suitable for identifying modifier genes. J Neurosci. 2007;27:2483–92. doi: 10.1523/JNEUROSCI.5453-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourg E, Valenti P, Lucchetta P, Payre F. Effects of mild heat shocks at young age on aging and longevity in Drosophila melanogaster. Biogerontology. 2001;2:155–64. doi: 10.1023/a:1011561107055. [DOI] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Li X, Yu QY, Xiang ZH, Kishino H, Zhang Z. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Lin HY, Yuh CH, Yu LK, Wang HD. The effect of neuronal expression of heat shock proteins 26 and 27 on lifespan, neurodegeneration, and apoptosis in Drosophila. Biochem Biophys Res Commun. 2008;376:637–41. doi: 10.1016/j.bbrc.2008.08.161. [DOI] [PubMed] [Google Scholar]
- Linford NJ, Pletcher SD. Aging: fruit flies break the chain to a longer life. Curr Biol. 2009;19:R895–8. doi: 10.1016/j.cub.2009.08.050. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, Walker GA. Stress resistance as a determinate of C. elegans lifespan. Mech Ageing Dev. 2002;123:765–71. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lu WC, Brummel TJ, Yuh CH, Lin PT, Kao TY, Li FY, Liao PC, Benzer S, Wang HD. Reduced expression of alpha-1,2-mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell. 2009;8:370–9. doi: 10.1111/j.1474-9726.2009.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Yamamoto A, Carbone MA, Roshina NV, Symonenko AV, Pasyukova EG, Morozova TV, Mackay TF. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 2010;6:e1001037. doi: 10.1371/journal.pgen.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin R, Tanguay RM. Stage-specific localization of the small heat shock protein Hsp27 during oogenesis in Drosophila melanogaster. Chromosoma. 1996;105:142–9. doi: 10.1007/BF02509495. [DOI] [PubMed] [Google Scholar]
- Marzetti E, Anne Lees H, Eva Wohlgemuth S, Leeuwenburgh C. Sarcopenia of aging: underlying cellular mechanisms and protection by calorie restriction. Biofactors. 2009;35:28–35. doi: 10.1002/biof.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie HR, Aiello VR, Williams TR. Iron accumulation during development and ageing of Drosophila. Mech Ageing Dev. 1985;29:215–20. doi: 10.1016/0047-6374(85)90020-x. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Preville X, Chareyron P, Briolay J, Klemenz R, Arrigo A-P. Constitutive expression of human hsp27, Drosophila hsp27, or human αB-crystallin confers resistance to TNF- and oxidative stress-induced cytotoxicity in stably transfected murine L929 fibroblasts. J Immunol. 1995;215:363–374. [PubMed] [Google Scholar]
- Mehlen P, Schulze-Osthoff K, Arrigo AP. Small stress proteins as novel regulators of apoptosis. Heat shock protein 27 blocks Fas/APO-1- and staurosporine-induced cell death. J Biol Chem. 1996;271:16510–4. doi: 10.1074/jbc.271.28.16510. [DOI] [PubMed] [Google Scholar]
- Michaud S, Marin R, Tanguay RM. Regulation of heat shock gene induction and expression during Drosophila development. Cellular and Molecular Life Sciences. 1997;53:104–113. doi: 10.1007/PL00000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Michaud S, Tanguay RM. Expression of the Hsp23 chaperone during Drosophila embryogenesis: association to distinct neural and glial lineages. BMC Dev Biol. 2003;3:9. doi: 10.1186/1471-213X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minois N, Khazaeli AA, Curtsinger JW. Locomotor activity as a function of age and life span in Drosophila melanogaster overexpressing hsp70. Exp Gerontol. 2001;36:1137–53. doi: 10.1016/s0531-5565(00)00263-1. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Dynamic remodeling of transcription complexes by molecular chaperones. Cell. 2002;110:281–4. doi: 10.1016/s0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–38. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64:167–70. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–64. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275:31204–10. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM. Heat shock proteins and aging in Drosophila melanogaster. Semin Cell Dev Biol. 2003;14:291–9. doi: 10.1016/j.semcdb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004a;279:43382–5. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. Faseb J. 2004b;18:598–9. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Morrow G, Heikkila JJ, Tanguay RM. Differences in the chaperone-like activities of the four main small heat shock proteins of Drosophila melanogaster. Cell Stress Chaperones. 2006;11:51–60. doi: 10.1379/CSC-166.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM. Mitochondria and ageing in Drosophila. Biotechnol J. 2008;3:728–39. doi: 10.1002/biot.200800015. [DOI] [PubMed] [Google Scholar]
- Moskalev A, Shaposhnikov M, Turysheva E. Life span alteration after irradiation in Drosophila melanogaster strains with mutations of Hsf and Hsps. Biogerontology. 2009;10:3–11. doi: 10.1007/s10522-008-9147-5. [DOI] [PubMed] [Google Scholar]
- Mounier N, Arrigo AP. Actin cytoskeleton and small heat shock proteins: how do they interact? Cell Stress Chaperones. 2002;7:167–76. doi: 10.1379/1466-1268(2002)007<0167:acashs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LD, Shahrestani P, Rauser CL. Predicting death in female Drosophila. Exp Gerontol. 2009;44:766–72. doi: 10.1016/j.exger.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–83. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Naidoo N. The endoplasmic reticulum stress response and aging. Rev Neurosci. 2009;20:23–37. doi: 10.1515/revneuro.2009.20.1.23. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–9. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaconstantinou M, Wu Y, Pretorius HN, Singh N, Gianfelice G, Tanguay RM, Campos AR, Bedard PA. Menin is a regulator of the stress response in Drosophila melanogaster. Mol Cell Biol. 2005;25:9960–72. doi: 10.1128/MCB.25.22.9960-9972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Tower J. Yeast, a Feast: The Fruit Fly Drosophila as a Model Organism for Research into Aging. In: Guarente LP, Partridge L, Wallace DC, editors. Molecular Biology of Aging. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2008. pp. 267–308. [Google Scholar]
- Piazza N, Hayes M, Martin I, Duttaroy A, Grotewiel M, Wessells R. Multiple measures of functionality exhibit progressive decline in a parallel, stochastic fashion in Drosophila Sod2 null mutants. Biogerontology. 2009 doi: 10.1007/s10522-008-9210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Current Biology. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–39. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheva VI, Domingos PM. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis. 2009;14:996–1007. doi: 10.1007/s10495-009-0341-y. [DOI] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–8. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA, Rogina B. Acquired temperature-sensitive paralysis as a biomarker of declining neuronal function in aging Drosophila. Aging Cell. 2008;7:179–86. doi: 10.1111/j.1474-9726.2008.00368.x. [DOI] [PubMed] [Google Scholar]
- Ren C, Finkel SE, Tower J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2009;44:228–35. doi: 10.1016/j.exger.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- Ritossa F. Discovery of the heat shock response. Cell Stress Chaperones. 1996;1:97–8. doi: 10.1379/1466-1268(1996)001<0097:dothsr>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollet E, Lavoie JN, Landry J, Tanguay RM. Expression of Drosophila's 27 kDa heat shock protein into rodent cells confers thermal resistance. Biochem Biophys Res Commun. 1992;185:116–20. doi: 10.1016/s0006-291x(05)80963-5. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–42. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. ER stress and hormetic regulation of the aging process. Ageing Res Rev. 2010;9:211–7. doi: 10.1016/j.arr.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Scannapieco AC, Sorensen JG, Loeschcke V, Norry FM. Heat-induced hormesis in longevity of two sibling Drosophila species. Biogerontology. 2007;8:315–25. doi: 10.1007/s10522-006-9075-1. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Weindruch R, Aiken JM. Oxidative stress and aging reduce COX I RNA and cytochrome oxidase activity in Drosophila. Free Radic Biol Med. 1998;25:740–7. doi: 10.1016/s0891-5849(98)00153-1. [DOI] [PubMed] [Google Scholar]
- Semba RD, Nicklett EJ, Ferrucci L. Does Accumulation of Advanced Glycation End Products Contribute to the Aging Phenotype? J Gerontol A Biol Sci Med Sci. 2010;65:963–75. doi: 10.1093/gerona/glq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong KH, Matsuo T, Fuyama Y, Aigaki T. Neural-specific overexpression of drosophila plenty of SH3s (DPOSH) extends the longevity of adult flies. Biogerontology. 2001a;2:271–81. doi: 10.1023/a:1013249326285. [DOI] [PubMed] [Google Scholar]
- Seong KH, Ogashiwa T, Matsuo T, Fuyama Y, Aigaki T. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology. 2001b;2:209–17. doi: 10.1023/a:1011517325711. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Shemetov AA, Seit-Nebi AS, Gusev NB. Structure, properties, and functions of the human small heat-shock protein HSP22 (HspB8, H11, E2IG1): a critical review. J Neurosci Res. 2008;86:264–9. doi: 10.1002/jnr.21441. [DOI] [PubMed] [Google Scholar]
- Shen J, Tower J. Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects. Exp Gerontol. 2010;45:97–105. doi: 10.1016/j.exger.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Sohal RD. Mitochondrial changes in flight muscles of normal and flightless Drosophila melanogaster with age. J Morphol. 1975;145:337–53. doi: 10.1002/jmor.1051450307. [DOI] [PubMed] [Google Scholar]
- Sorensen JG, Kristensen TN, Kristensen KV, Loeschcke V. Sex specific effects of heat induced hormesis in Hsf-deficient Drosophila melanogaster. Exp Gerontol. 2007;42:1123–9. doi: 10.1016/j.exger.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–8. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Hayer-Hartl M, Hartl FU. SnapShot: molecular chaperones, Part II. Cell. 2007;128:412. doi: 10.1016/j.cell.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Terry DF, Wyszynski DF, Nolan VG, Atzmon G, Schoenhofen EA, Pennington JY, Andersen SL, Wilcox MA, Farrer LA, Barzilai N, Baldwin CT, Asea A. Serum heat shock protein 70 level as a biomarker of exceptional longevity. Mech Ageing Dev. 2006;127:862–8. doi: 10.1016/j.mad.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–18. doi: 10.1016/j.mad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Tower J. Hsps and aging. Trends Endocrinol Metab. 2009;20:216–22. doi: 10.1016/j.tem.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Arbeitman M. The genetics of gender and life span. J Biol. 2009;8:38. doi: 10.1186/jbiol141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Seong KH, Aigaki T. POSH, a scaffold protein for JNK signaling, binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 2006;580:3296–300. doi: 10.1016/j.febslet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Tsutsumi-Ishii Y, Tadokoro K, Hanaoka F, Tsuchida N. Response of heat shock element within the human HSP70 promoter to mutated p53 genes. Cell Growth Differ. 1995;6:1–8. [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–3. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Vermeulen CJ, Loeschcke V. Longevity and the stress response in Drosophila. Exp Gerontol. 2007;42:153–9. doi: 10.1016/j.exger.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. Faseb J. 2007a doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernace VA, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging and regulated protein degradation: who has the UPPer hand? Aging Cell. 2007b;6:599–606. doi: 10.1111/j.1474-9726.2007.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prevost M-C, Faure A, Chateau D, Chapon F, Tome F, Dupret J-M, Paulin D, Fardeau M. A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nature Genetics. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Vijg J. The role of DNA damage and repair in aging: new approaches to an old problem. Mech Ageing Dev. 2008;129:498–502. doi: 10.1016/j.mad.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–33. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Ryu J, Gao R, Choi IK, Morrow G, Kaur K, Kim I, Kaul SC, Yun CO, Tanguay RM. Proproliferative functions of Drosophila small mitochondrial heat shock protein 22 in human cells. J Biol Chem. 2010;285:3833–9. doi: 10.1074/jbc.M109.080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D, Olszewski MB, Gutkowska M, Helwak A, Zylicz M, Zylicz A. Hsp70 molecular chaperones are required to support p53 tumor suppressor activity under stress conditions. Oncogene. 2009;28:4284–94. doi: 10.1038/onc.2009.281. [DOI] [PubMed] [Google Scholar]
- Walerych D, Gutkowska M, Klejman MP, Wawrzynow B, Tracz Z, Wiech M, Zylicz M, Zylicz A. ATP binding to Hsp90 is sufficient for effective chaperoning of p53 protein. J Biol Chem. 2010 doi: 10.1074/jbc.M110.112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DW, Benzer S. Mitochondrial "swirls" induced by oxygen stress and in the Drosophila mutant hyperswirl. Proc Natl Acad Sci U S A. 2004;101:10290–5. doi: 10.1073/pnas.0403767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–9. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Wang HD, Kazemi-Esfarjani P, Benzer S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc Natl Acad Sci U S A. 2004;101:12610–5. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK Signaling Confers Tolerance to Oxidative Stress and Extends Lifespan in Drosophila. Dev Cell. 2003;5:811–6. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–25. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Waskar M, Landis GN, Shen J, Curtis C, Tozer K, Abdueva D, Skvortsov D, Tavare S, Tower J. Drosophila melanogaster p53 has developmental stage-specific and sex-specific effects on adult life span indicative of sexual antagonistic pleiotropy. Aging (Albany NY) 2009;1:903–36. doi: 10.18632/aging.100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Tetrault JM, Dellavalle RP, Lindquist SL. A new method for manipulating transgenes: engineering heat tolerance in a complex, multicellular organism. Curr Biol. 1993;3:842–53. doi: 10.1016/0960-9822(93)90218-d. [DOI] [PubMed] [Google Scholar]
- Wheeler JC, Bieschke ET, Tower J. Muscle-specific expression of Drosophila hsp70 in response to aging and oxidative stress. Proc Natl Acad Sci U S A. 1995;92:10408–12. doi: 10.1073/pnas.92.22.10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, King V, Tower J. Sequence requirements for upregulated expression of Drosophila hsp70 transgenes during aging. Neurobiol Aging. 1999;20:545–53. doi: 10.1016/s0197-4580(99)00088-3. [DOI] [PubMed] [Google Scholar]
- Wu D, Rea SL, Yashin AI, Johnson TE. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp Gerontol. 2006;41:261–70. doi: 10.1016/j.exger.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Yang J, Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. J Gerontol A Biol Sci Med Sci. 2009;64:828–38. doi: 10.1093/gerona/glp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeyati PL, van Heyningen V. Incapacitating the evolutionary capacitor: Hsp90 modulation of disease. Curr Opin Genet Dev. 2008;18:264–72. doi: 10.1016/j.gde.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sun H, Lu J, Li X, Chen X, Tao D, Huang W, Huang B. Lifespan extension and elevated hsp gene expression in Drosophila caused by histone deacetylase inhibitors. J Exp Biol. 2005;208:697–705. doi: 10.1242/jeb.01439. [DOI] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13726–31. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]