Abstract
Purpose
Genetic tests for the most commonly mutated genes in dilated cardiomyopathy (DCM) can confirm a clinical diagnosis in the proband and inform family management. Presymptomatic family members can be identified, allowing for targeted clinical monitoring to minimize adverse outcomes. However, the marked locus and allelic heterogeneity associated with DCM have made clinical genetic testing challenging. Novel sequencing platforms have now opened up avenues for more comprehensive diagnostic testing while simultaneously decreasing test cost and turn around time.
Methods
By using a custom design based on triplicate resequencing and separate genotyping of known disease-causing variants, we developed the DCM CardioChip for efficient analysis of 19 genes previously implicated in causing DCM.
Results
The chip’s analytical sensitivity for known and novel substitution variants is 100% and 98%, respectively. In screening 73 previously tested DCM patients who did not carry clinically significant variants in 10 genes, 7 variants of likely clinical significance were identified in the remaining 9 genes included on the chip. Compared with traditional Sanger-based sequencing, test cost and turn around time were reduced by ~50%.
Conclusions
The DCM CardioChip is a highly efficient screening test with a projected clinical sensitivity of 26–29%.
Keywords: dilated cardiomyopathy, resequencing array, genetic test, locus heterogeneity
Dilated cardiomyopathy (DCM) is a disease of the cardiac muscle, characterized by left ventricular enlargement and systolic dysfunction. It is responsible for significant morbidity and mortality and is a leading indication for heart failure and transplant.1 The prevalence of idiopathic DCM is estimated to be ~1 in 2500. Approximately 30–50% of these patients have a family history, indicating an underlying genetic etiology in an important proportion (reviewed in Ref. 2). To date, mutations in >30 genes have been implicated in causing DCM, typically showing autosomal dominant inheritance. Unlike hypertrophic cardiomyopathy (HCM), which is considered to be mainly a disease of the cardiac sarcomere, the genetic etiology of DCM is more diverse. Affected structures include the sarcomere, the Z-disk, the nuclear lamina, the intermediate filaments, and the dystrophin-associated glycoprotein complex (reviewed in Ref. 3). Because of the high cost of dideoxy sequencing, most previous multigene studies have screened only a subset of genes or tested a small number of patients.4 – 12 Therefore, accurate mutation detection rates for any of these genes are difficult to define. Clinical genetic testing laboratories must attempt a balance between maximizing clinical sensitivity (i.e., the number of genes tested) and minimizing test price and turn around time (TAT). For disorders that are characterized by a high degree of locus and allelic heterogeneity, genes with low variant detection rates are, therefore, often not included in a clinical testing panel.
Because of the availability of novel sequencing platforms, it is now possible to screen a much larger number of genes simultaneously and, thereby, improve clinical sensitivity. One of these novel sequencing platforms is the Affymetrix Gene-Chip® Resequencing Array, a customizable high-density oligo-nucleotide microarray. This platform has been used successfully to sequence mitochondrial, bacterial, or small eukaryotic genomes and selected genes for specific diseases, such as retinitis pigmentosa, congenital myasthenic syndromes, and HCM.13–18 Although initial studies showed high false negative and positive rates, improvements including redundant tiling and custom bioinformatics algorithms have enabled adaptation into clinical services at a reasonable analytical sensitivity.19 Parallel processing of a large number of genes can result in a gain in labor, time, and cost efficiency as outlined for an HCM resequencing array.14 However, it is important to point out that this effect needs to be confirmed for every new test because individual parameters can vary greatly depending on the selected genes and/or the laboratory performing the test.
We used this platform to develop a novel resequencing test for DCM that screens the entire coding sequence and splice sites of 19 genes implicated in DCM. Ten of these genes were already part of existing clinical DCM tests in our laboratory and elsewhere, and included the sarcomere genes, MYBPC3,4,9,12MYH7,4–6,9,11,12 TNNT2,4–7,9,11,12 TNNI3,6 TPM1,4,6,12 andACTC,12 the Z-disk gene, LDB3,5,20,21 the nuclear lamina gene,LMNA,12,22–27 the cardiac Ca++ ATPase inhibitor gene, PLN,11 and the Barth syndrome gene, TAZ.28 Nine new genes were added based on published evidence, with a focus on mutation detection rate and additional supporting evidence, such as absence in healthy controls and/or familial segregation analyses (Tables 1 and 6 and references within). Emphasis was given to genes whose products are part of the continuum of structures already known to be affected in DCM patients (such as the Z-disk and the nuclear lamina). Specifically, we included emerin (EMD), a nuclear lamina protein known for its role in Emery-Dreifuss muscular dystrophy, which can present with DCM29–31 (reviewed in Ref. 32), and the intermediate filament gene desmin (DES), known for its role not only in myofibrillar myopathy but also in isolated DCM.33–36 Furthermore, we added several genes encoding the Z-disk proteins alpha actinin (ACTN28), cardiac muscle LIM protein (CSRP38,37–40), titin cap/telethonin (TCAP37,39,41), and metavinculin (VCL42–44). Dominant mutations in these genes have been found in DCM patients, and for TCAP, recessive inheritance has been described for patients with of limb girdle muscular dystrophy 2G.45 Another gene, delta sarcoglycan (SGCD) is part of the dystrophin-associated protein complex, and recessive mutations have been implicated in limb girdle muscular dystrophy 2F. More recently, dominantSGCD mutations have been described in isolated DCM.46,47 Finally, two genes whose products are not part of the above-mentioned structures were the cytokine cardiotrophin (CTF148) and the ATP sensitive potassium channel ABCC9.49,50 Mutations in several additional genes of interest have been described, but these genes were not included because of various reasons. For example, titin (TTN, reviewed in Ref. 51) could not be included because of the large size (~80 kb) of its coding sequence. Several other genes were not considered at this point because of promising but as of yet insufficient evidence that is awaiting further confirmation and characterization by the scientific community (e.g.,PSEN1,52 PSEN2,52 LAMA4,53 and DMN12).
Table 1.
Contents of the DCM CardioChip
| Gene | Name | Location/function | Locus | Reference sequence | Published studies | Exons sequenced |
Bases sequenced |
|---|---|---|---|---|---|---|---|
| MYBPC3 | Myosin binding protein 3 | Sarcomere | 11p11.2 | NM_000256 | 4, 9, 12 | 34 | 4,505 |
| MYH7 | Myosin heavy chain 7, beta | Sarcomere | 14q12 | NM_000257 | 4–6, 9, 11, 12 | 38 | 6,402 |
| TNNI3 | Troponin I | Sarcomere | 19q13.4 | NM_000363 | 6 | 8 | 793 |
| TNNT2 | Troponin T2 | Sarcomere | 1q32 | NM_001001430.1; Exon 2: NM_000364.2 | 4–7, 9, 11, 12 | 15 | 1,167 |
| TPM1 | Tropomyosin 1 | Sarcomere | 15q22.1 | NM_000366.5; Exon 6b: NM_001018005.1 | 4, 6, 12 | 11 | 1,151 |
| ACTC | Actin, alpha | Sarcomere | 15q14 | NM_005159 | 12 | 6 | 1,254 |
| LDB3 | LIM domain binding 3 | Z-disk | 10q22.2–q23.3 | NM_007078.2: Exons 5, 6 and 9: NM_001080116.1 | 5, 20, 21 | 16 | 2,828 |
| ACTN2 | Alpha actinin | Z-disk | 1q42–q43 | NM_005691 | 8 | 21 | 3,105 |
| CSRP3 | Cysteine- and glycine-rich protein 3 | Z-disk | 11p15.1 | NM_003476 | 8, 37–40 | 5 | 685 |
| TCAP | Titin-cap | Z-disk | 17q12 | NM_003673 | 37, 39, 41 | 2 | 544 |
| VCL | Vinculin | Z-disk | 10q22.1–q23 | NM_014000 | 42–44 | 22 | 3,625 |
| LMNA | Lamin A/C | Nuclear lamina | 1q21.2 | NM_170707.1 | 12, 22–27 | 12 | 2,235 |
| EMD | Emerin | Nuclear lamina | Xq28 | NM_000117 | 29–31 | 6 | 885 |
| PLN | Phospholamban | Inhibitor of cardiac Ca-ATPase | 6q22.1 | NM_002667.2 | 11 | 1 | 179 |
| TAZ | Tafazzin | Mitochondrial | Xq28 | NM_000116.2 | 28 | 11 | 1,099 |
| ABCC9 | ATP-binding cassette, subfamily C, member 9 | ATP-sensitive K channel | 12p12.1 | NM_005691 | 49, 50 | 39 | 5,568 |
| CTF1 | Cardiotrophin 1 | Cytokine | 16p11.2-p11.1 | NM_001330 | 48 | 3 | 666 |
| DES | Desmin | Intermediate filament | 2q35 | NM_001927 | 33–36 | 9 | 1,593 |
| SGCD | Sarcoglycan, delta | Dystrophin-glycoprotein complex | 5q33 | NM_172244; Exons 8b and 9: NM_000337 | 46, 47, 64 | 9 | 1,249 |
| Total | 19 | 268 | 39,533 | ||||
Table 6.
Clinical sensitivity
| Published detection rate | This study (n = 94) | ||||
|---|---|---|---|---|---|
| Gene | Average (%) | Range (%) | References | A (%) | B (%) |
| MYBPC3 | 5.0 | 1.0–10.5 | 4, 9, 10, 12 | ||
| MYH7 | 5.5 | 0–12.9 | 4–6, 9–12 | ||
| TNNI3 | 0.5 | 0–2.1 | 6, 9, 10, 65 | ||
| TNNT2 | 2.8 | 0–9.5 | 4–12 | ||
| TPM1 | 0.0 | 0 | 4, 6, 9, 10, 12, 42, 66 | ||
| ACTC | 0.0 | 0 | 8–10, 12, 42 | ||
| LMNA | 4.3 | 0–8.2 | 8–10, 22–26, 27, 67, 68 | ||
| PLN | 1.4 | 0–5.0 | 11, 69–72 | ||
| LDB3 | 2.3 | 0–6.0 | 5, 12 | ||
| Subtotal | 21.8 | ||||
| ABCC9 | 0.6 | 0.6 | 49 | 0.0 | 0.0 |
| ACTN2 | 0.9 | 0.9 | 8 | 3.2 | 3.2 |
| CSRP3 | 0.3 | 0–0.9 | 5, 8, 39 | 1.1 | 2.1 |
| CTF1 | 0.4 | 0.40 | 48 | 0.0 | 0.0 |
| DES | 0.5 | 0–2.3 | 35, 36 | 1.1 | 1.1 |
| EMD | a | a | 30, 31 | 0.0 | 0.0 |
| SGCD | 1.6 | 0–5.9 | 46, 47, 64 | 0.0 | 0.0 |
| TCAP | 0.5 | 0.3–0.7 | 5, 39, 41 | 0.0 | 0.0 |
| VCL | 0.1 | 0–0.6 | 42 | 2.1 | 4.3 |
| Subtotal | 4.9 | 7.5 | 10.7 | ||
| Total | 26.7 | 29.3 | 32.5 | ||
Case report.
A, likely clinically significant; B, likely significant + unknown significance.
Here, we describe the technical validation of the DCM CardioChip, its analytical performance parameters, and its significant impact on test cost and TAT. We also discuss the results obtained from testing 73 DCM probands and provide a projected clinical sensitivity.
MATERIALS AND METHODS
Samples
Patient samples
This study was approved by the Partners HealthCare and Brigham and Women’s Hospital Institutional Review Boards. Samples were used as follows: DCM CardioChip validation: (a) Positive controls: samples with known sequence alterations were anonymized and used to create a set of 7 “pooled positive controls” including 199 known sequence alterations. The DNA source for individual amplicons came from multiple patients with unique mutations allowing for each control to contain between 20 and 56 mutations per pool. (b) Additional technical validation: complete dideoxy sequencing data and chip-based sequencing data were generated for 25 anonymized samples that were previously tested and did not contain any disease causing variants in a subset of the genes present on the chip (MYH7, MYBPC3, TNNT2, TNNI3, TPM1, ACTC, LDB3, PLN, LMNA, and TAZ). Clinical sensitivity: 73 patient samples were tested to estimate the clinical sensitivity. Cohort 1 included the 25 samples used for technical validation, which represent patients with DCM from a broad referral population. Cohort 2 comprised 48 samples from well-characterized patients seen at the Brigham and Women’s Hospital in Boston. Patients were not included if there was significant coronary artery disease, uncontrolled hypertension, significant valvular disease, or other identifiable cause of DCM (e.g., peripartum and anthracycline exposure). Age at diagnosis was 40.5 ± 14.2 years, 76% were male gender;92% were provider or self-identified Caucasian, the remaining were African American (6%) or multiracial (2%). A definite or potential family history of DCM was present at 58% and 23% of probands, respectively. Left ventricular ejection fraction was reduced in all subjects (mean 29 ± 12%), and none had features suggestive of HCM, arrhythmogenic right ventricular cardiomyopathy, or left ventricular noncompaction. Eleven patients (23%) died or required cardiac transplantation or placement of a left ventricular assist device.
Healthy control samples
Novel variants were genotyped in ethnically matched control samples.
Caucasian (n = 200)
Subjects were disease free and older than the age of 18 years. “Disease” was defined as current or past diagnoses made by a medical care provider, which required medication or other forms of treatment or therapy. Surgery was considered an exclusionary criterion, as was the use of prescription medications or regularly used over the counter medications. The subjects completed a structured questionnaire that detailed current and past history of illness and treatment. All responses were confirmed by clinical interview by physicians. Physical examinations were completed at the study visit.
African American (n = 200)
These were anonymized samples collected through the Crimson sample collection mechanism at Brigham and Women’s Hospital (the Crimson collection is a large cohort of subjects, without a chronic disease diagnosis, who where seen at the Brigham and Women’s Hospital). All subjects were between the ages of 15 and 45 years and were originally chosen to be controls for an asthma genetics study.
Genotyping (healthy controls)
Sequenom I-Plex technology was used. Multiplex polymerase chain reaction (PCR) assays were designed using Sequenom Spectro DESIGNER software (version 3.0.0.3; Sequenom, SanDiego, CA) by inputting sequence containing the single nucleotide polymorphism (SNP) site and the 100-bp flanking sequences on either side of the variant. The variants were grouped into multiplexes, and reactions were performed using standard conditions.
PCR and Sanger sequencing
Exons were amplified by PCR using standard conditions (15 µL reactions containing 5 ng genomic DNA, 2 mM MgCl2, 0.75 µL dimethyl sulfoxide, 1 M betaine, 0.2 mM dNTPs, 20 pmol primers, and 0.2 µL AmpliTaq Gold® (Applied Biosystems) using the following cycling conditions: 95°C for 10 minutes; 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute for 30 cycles; and 72°C for 10 minutes. PCR products are purified with Ampure® Magnetic Beads (Agencourt) and then directly sequenced bidirectionally by dye-terminator sequencing using a universal primer. Sequencing products are purified using Cleanseq™ Magnetic Beads (Agencourt) and separated by capillary electrophoresis on an ABI 3730 DNA Analyzer (Applied Biosystems).
DCM CardioChip custom design
Gene chip specifications
The DCM CardioChip is a “49-format” array (accommodating up to 300 kb of DNA sequencing capacity) with 8-µm features and was designed using a shift mask design requiring17 masks.
Content
Coding exons and ±10 flanking intronic bases for 19 genes (39,533 bases) involved in DCM (Table 1).
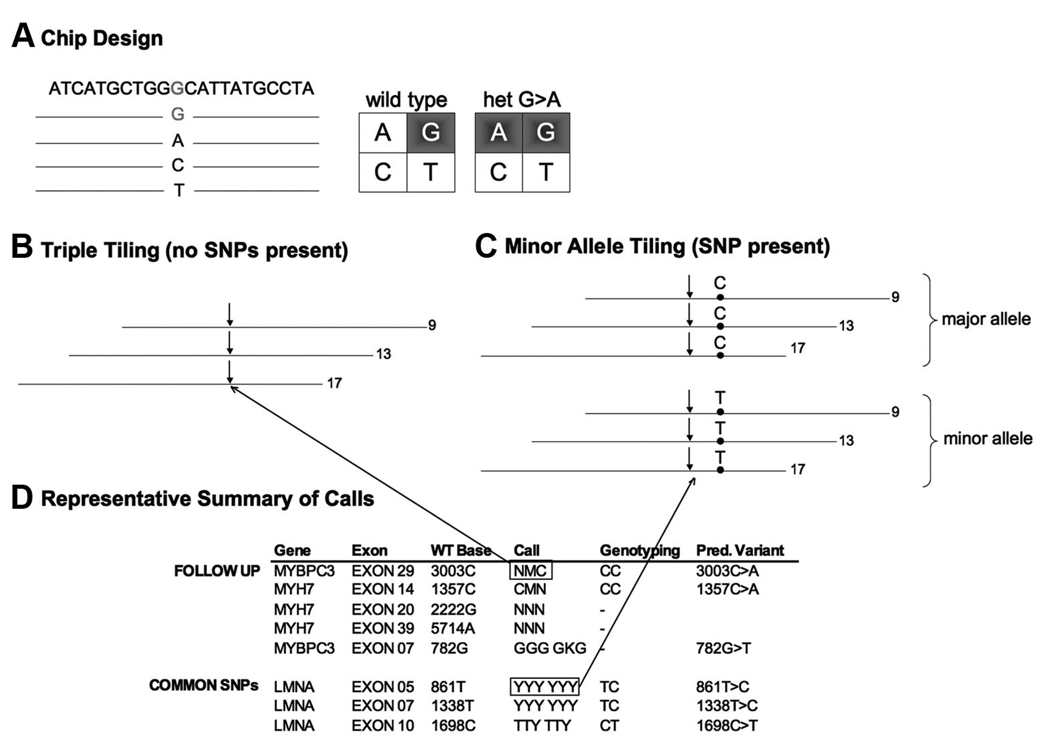
Redundant probe sets
To increase call rates, a triplicate set of probes were tiled for each base. This amounted to a minimum of 24 probes per base (4 probes per strand × 2 strands per base × triplicate tiling). In addition, to improve variant detection and call rates, each of the three probe sets contained a different position for interrogation (positions 9, 13, and 17 within the 25-nucleotide probe).
Minor allele tiling
To avoid allele dropout of mutations through lack of hybridization because of the presence of a nearby SNP, an additional triplicate set of sequencing probes surrounding common SNPs was tiled. These additional sequences contained the minor allele and allowed correct hybridization whenever the minor allele of a common variant was present in a patient (Fig. 1).
Fig. 1.
Design and data output. A, The basic design of a resequencing chip: each base is interrogated by four probes, containing the four possible bases, tiled in a quadrant (four additional probes query the reverse strand and are not shown). Typical hybridization patterns obtained from samples that are homozygous wild type or heterozygous for a variant are shown to the right. B, Triple tiling and minor allele tiling strategy: each probe is tiled in triplicate, with the interrogating position located at the 9th, 13th, and 17th positions on the 25-bp probe (arrows). C, When a known SNP (e.g., C>T) is located near the interrogating position, an additional set of triplicate probes were tiled containing the minor allele. D, Representative data output after the integrated GCOS, GSEQ, and BRLMM analysis. All positions requiring confirmatory analysis are listed in the “follow-up” section (top). For each position, the gene, exon, and wild type base is indicated. In the “call” column, triplicate calls are listed except when the presence of a nearby SNP triggered the use of the three additional minor allele tiling data sets.
Genotyping
The DCM CardioChip also contains genotyping probes for the variant allele of known disease-causing DCM variants and all common SNPs (see Tables, Supplemental Digital Content 1 and 2, http://links.lww.com/GIM/A98 and http://links.lww.com/GIM/A99). Together with the probe signals from wild type sequencing data, genotyping calls were made using the Bayesian Robust Linear Modeling using Mahalanobis distance (BRLMM) algorithm (http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf). Default parameters recommended by Affymetrix were used (quant-norm.sketch = 50,000, PM-only, brlmm.transform = ccs, K = 4, MS = 0.5).
Bioinformatics pipeline
The files generated by the Affymetrix Gene Chip Operating System (GCOS) software were processed with GSEQ version 4.1 batch analysis using default settings with the following exceptions: no signal threshold = 1.0, genome model = 0, quality score threshold = 1.0 (Affymetrix recommends 3), and base reliability threshold across samples = 0. BRLMM was used to derive genotypes for common SNPs and pathogenic variants and the use of the “minor allele” resequencing data sets was triggered whenever a heterozygous or homozygous SNP was detected. BRLMM was also used to derive genotyping calls for known disease-causing variants. The output was exported, triplicate tilings and genotyping calls were aligned, and a final list of variant calls and no-call bases was generated. All variant calls made by the chip and bases without call were confirmed by dideoxy sequencing. (See Figure, Supplemental Digital Content3, http://links.lww.com/GIM/A100 for a diagram of the bioinformatic pipeline.)
Chip assay
DNA was amplified using 101 multiplexes (2–3 primer pairs each) and 37 singleplex reactions (15 µL reaction volume containing 2.5 mM MgCl2, 1 M betaine monohydrate, 5.3% dimethyl sulfoxide, 133 µM dNTPs, and 1 U AmpliTaq Gold [ABI]). Primer sequences are available on request. Cycling conditions were as follows: 1 × (95°C for 10 minutes), 30 × (95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds), and 1 × (72°C for 10 minutes). Products were pooled using empirically determined amounts (generally 6–8 µL). PCR products were cleaned using the QIAquick PCR Purification Kit (Qiagen). Samples were eluted in 45 µL and then fragmented and labeled using the GeneChip Resequencing Assay Kit (Affymetrix) according to the manufacturer’s instructions. Hybridization was performed using a GeneChip Hybridization Oven 640 (Affymetrix). A GeneChip Fluidics Station 450 (Affymetrix) was used for the washing and staining protocols, and the GCOS version 1.4.0.036 software was used for fluidics and scanning.
RESULTS
Validation and test development approach
The DCM CardioChip was designed to resequence the coding region and splice junctions of 19 DCM genes (~40 kb) in triplicate. An additional set of triplicate sequences containing the minor alleles of common variants and a set of genotyping probes querying 281 known disease-causing variants were included to increase sensitivity. The chip’s technical performance was validated by comparing array-based sequencing data to the current “gold standard” (dideoxy sequencing), each generated for 25 samples and for a set of samples enriched for a large number of known sequence alterations. A bioinformatics pipeline integrating triplicate resequencing data with available genotyping calls was developed. The final clinical test consists of the array-based sequencing assay, followed by dideoxy sequencing-based confirmatory follow-up of all variant calls.
The DCM CardioChip added 9 new genes to the 10 for which clinical resequencing was already offered. To project the chip’s overall clinical sensitivity and to increase the likelihood of identifying novel variants in the new genes, we selected 73 samples from patients in whom no clinically significant variants were identified in the 10 previously tested genes (MYH7, MYBPC3, TNNT2, TNNI3, TPM1, ACTC1, LMNA, LDB3, PLN, and TAZ).
Analytical performance parameters
Call rate
The raw data were processed using the Affymetrix GCOS software, followed by the GSEQ version 4.1 batch analysis. The average call rate was 98.47% (n = 42, 95.47–99.00%), which is comparable with other published resequencing arrays.13–18
Confirmatory follow-up
Each final base call was derived from a set of three independent calls (generated by the three redundant probe sets, including three additional sets of minor allele probe sets when common heterozygous or homozygous SNPs were present) and the genotyping calls for known DCM mutations whenever available. All ambiguous calls, variant calls, and bases with no call were sequenced by dideoxy sequencing leading to a final assay call rate of 100%.
Analytical sensitivity
A series of 7 “pooled positive controls” containing 199 unique sequence alterations (166 substitutions and 33 insertions, deletions, or insertion/deletions) across the 268-coding exons covered by the DCM CardioChip were generated to determine the analytical sensitivity of the test. Because of the presence of several variants in more than 1 sample, the total number of variants analyzed was 1109 (989 substitutions and 120 insertions and deletions).
Substitutions
Of the unique substitutions, 5 of 166 (3.0%) were missed by the chip using only the resequencing portion (Table 2). Consistent with previously published results, the GC content in all three probe sets was high at these positions (56–84%; Table 3).13,54 When all substitutions present were included, the false negative rate was 18 of 989 (1.82%). This corresponds to the false negative rate for novel substitutions variants (for which no additional genotyping probes are present on the chip). When the genotyping portion of the chip was added to the analysis and only variants were analyzed for which genotyping probes were present (n = 124), the false negative substitution rate dropped to 0% (Table 3). This constitutes the false negative rate for known substitution variants. In summary, the analytical sensitivity of the DCM CardioChip is 100% for known substitutions and 98.0% for novel substitutions.
Table 2.
Analytical sensitivity
| Percent false negatives | |||
|---|---|---|---|
| N | Genotyping calls not considered |
Genotyping calls considered |
|
| Substitutions (unique) | 166 | 3.01 | 0 |
| Substitutions (all) | 989 | 1.82 | 0 |
| Insertion/deletion—worst case | 33 | 84.85 | 18.18 |
| Insertion/deletion—best case | 33 | 30.30 | 9.09 |
| Insertion/deletion—all tested | 58.33 (70/120) | 6.33 (74/79) | |
| Total | 199 | ||
Table 3.
False negative substitutions
| Gene | Exon | DNA | Protein | Resequencing call | Genotyping call | Percent GC contenta |
|---|---|---|---|---|---|---|
| MYBPC3 | 05 | c.537C>T | p.Ala179Ala | CNC | CT | 80/84/80 |
| TNNT2 | 10 | c.400C>G | p.Arg134Gly | CCC | CG | 80/80/76 |
| MYH7 | 37 | c.5380C>G | p.Gln1794Glu | CNC | CG | 68/60/60 |
| MYH7 | 15 | c.1549C>A | p.Leu517Met | CCC | NDb | 60/60/60 |
| MYBPC3 | 30 | c.3330+2T>G | TTT | NNc | 68/60/56 |
For probe sets 9/13/17.
Genotyping probes were not present for this variant.
NN call at this position targets amplicon for confirmatory follow-up.
ND, not determined.
Insertions and deletions (insertion/deletions)
It is well established that novel insertions and deletions are difficult to detect on hybridization-based resequencing arrays.13,19 However, because they often lead to aberrant calls in neighboring bases, they are sometimes detected, especially, in a clinical setting, where every variant call or no-call is followed up on by an independent test.19 Our validation set consisted of 33 unique (120 total) insertions, deletions, or insertion/deletions (23 deletions, 8 insertions, and 2 insertion/deletions), ranging from 1 to 20 bases, with most being <5 bases (n = 25; Table 4). When all 120 insertion/deletions across all experiments were analyzed, the false negative rate was 58.33% (Table 2). However, we noticed that there was a high degree of variation with regard to how often variants that were present in more than one sample were missed. Hence, we performed a “best case/worst case” analysis of the detection rates. Under the worst case scenario, a variant was scored as “not detected” when it was missed once (but detected several other times, e.g., “No-Yes-Yes-Yes”). Conversely, under the best case scenario, a variant was scored as “detected” if it was detected at least once, regardless of how many times it was missed (e.g., “Yes, No, No, No”). Using this approach, the false negative rate for insertions and deletions ranged from 30.30% to 84.85% (Tables 2 and 4), which is consistent with previously described detection rates.55,56 Inclusion of genotyping data for known insertion/deletions reduced the false negative rate to 6.33–18.18%. Of note, the detectability did not correlate with the size of the insertion or deletion (Table 4). Whether a given insertion or deletion will be detected by the array is highly dependent on the sequence content of the variant and the surrounding sequence. The false negative rates described here can, therefore, only serve as an estimate, although the true false negative rate for insertions and deletions is expected to lie in between the two extremes.
Table 4.
Detection of insertions and deletions
| Resequencing only | Genotyping included | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Exon/ intron |
DNA | Protein | Size (bp) |
No. times tested |
No. times detected |
Best casea |
Worst caseb |
No. times detected |
Best casea |
Worst caseb |
| Deletions | |||||||||||
| ACTN2 | 20 | c.2520del | p.Asp841fs | 1 | 1 | 0 | N | N | NT | N | N |
| MYBPC3 | 18 | c.1628del | p.Lys453fs | 1 | 2 | 0 | N | N | 2 | Y | Y |
| MYBPC3 | 25 | c.2558del | p.Gly853fs | 1 | 2 | 1 | Y | N | 2 | Y | Y |
| MYBPC3 | 33 | c.3735del | p.Phe1246fs | 1 | 2 | 1 | Y | N | 2 | Y | Y |
| MYH7 | 5 | c.457del | p.His153fs | 1 | 4 | 1 | Y | N | 4 | Y | Y |
| TNNI3 | 2 | c.12–7del | 1 | 2 | 1 | Y | N | 2 | Y | Y | |
| MYBPC3 | 15 | c.1357_1358del | p.Pro453fs | 2 | 3 | 3 | Y | Y | 3 | Y | Y |
| MYBPC3 | 27 | c.2780_2781del | p.Thr927fs | 2 | 3 | 1 | Y | N | 2 | Y | N |
| MYBPC3 | 27 | c.2833_2834del | p.Arg945fs | 2 | 3 | 1 | Y | N | 3 | Y | Y |
| MYBPC3 | 31 | c.3476_3477del | p.Phe1159fs | 2 | 3 | 0 | N | N | 3 | Y | Y |
| MYH7 | 15 | c.1477_1478del | p.Met493fs | 2 | 3 | 3 | Y | Y | NT | Y | Y |
| MYBPC3 | 17 | c.1513_1515del | p.Lys505del | 3 | 3 | 1 | Y | N | 3 | Y | Y |
| MYBPC3 | 19 | c.1814_1816del | p.Asp405del | 3 | 3 | 3 | Y | Y | 3 | Y | Y |
| MYH7 | 33 | c.4522_4524del | p.Glu1508del | 3 | 2 | 0 | N | N | 2 | Y | Y |
| MYH7 | 36 | c.5186_5188del | p.Lys1729del | 3 | 3 | 0 | N | N | 2 | Y | N |
| TNNT2 | 9 | c.285_287del | p.Glu96del | 3 | 3 | 1 | Y | N | 3 | Y | Y |
| TNNT2 | 11 | c.487_489del | p.Glu163del | 3 | 3 | 0 | N | N | 3 | Y | Y |
| MYBPC3 | 22 | c.2148+6_2148+9del | 4 | 3 | 1 | Y | N | 3 | Y | Y | |
| MYBPC3 | 28 | c.2943_2947del | p.Gln981fs | 5 | 2 | 1 | Y | N | 2 | Y | Y |
| TNNT2 | 4 | c.53–11_53–7del | 5 | 28 | 16 | Y | N | NT | Y | N | |
| MYBPC3 | 32 | c.3600_3609del | p.Cys1201fs | 10 | 5 | 3 | Y | N | 5 | Y | Y |
| MYBPC3 | 2 | c.177_187del | p.Glu60fs | 11 | 10 | 1 | Y | N | 10 | Y | Y |
| MYBPC3 | 16 | c.1457+6_1457+25del | 20 | 2 | 1 | Y | N | 2 | Y | Y | |
| Insertions | |||||||||||
| MYBPC3 | 24 | c.2373_2374insG | p.Trp792fs | 1 | 3 | 0 | N | N | 3 | Y | Y |
| MYBPC3 | 29 | c.3166_3167insG | p.Ala1056fs | 1 | 3 | 2 | Y | N | NT | Y | N |
| MYBPC3 | 30 | c.3226_3227insT | p.Asp1076fs | 1 | 3 | 0 | N | N | 3 | Y | Y |
| MYBPC3 | 3 | c.333_334insT | p.Glu112X | 1 | 1 | 1 | Y | Y | 0 | N | N |
| MYBPC3 | 4 | c.506–12_506–11insC | 1 | 1 | 0 | N | N | 0 | N | N | |
| MYH7 | 26 | c.3337–3_3337–2insC | 1 | 3 | 1 | Y | N | 3 | Y | Y | |
| MYBPC3 | 31 | c.3332_3335dup | p.Trp1112fs | 4 | 2 | 0 | N | N | 2 | Y | Y |
| MYBPC3 | 33 | c.3742_3759dup | p.Gly1248_Cys1253dup | 18 | 3 | 2 | Y | N | 2 | Y | N |
| Insertion/deletions | |||||||||||
| MYBPC3 | 21 | c.1999_2000delinsG | p.Leu667fs | 1 | 3 | 1 | Y | N | NT | Y | N |
| MYBPC3 | 2 | c.239delCinsGAGG | p.Ala80delinsGlyGly | 3 | 3 | 3 | Y | Y | NT | Y | Y |
Best case = variant was detected at least once.
Worst case = variant was missed at least once.
Y, fits criteria; N, does not fit criteria; NT, not tested.
Specificity
The resequencing portion of the chip had an average false positive rate of 0.09%, (36 of 39,533 bases queried per experiment) and, therefore, an analytical specificity of 99.91%. Because in a clinical setting, each positive call is followed up by an independent assay (dideoxy sequencing in our laboratory), the final false positive rate of the test is close to 0% (equal to the false positive rate of dideoxy sequencing).
Cost and TAT analysis
Because of the complexity of many genetic tests, their cost and TAT can be extremely high compared with other clinical tests. For genetically heterogeneous diseases such as DCM, diagnostic laboratories must strive to increase clinical sensitivity by including more genes. However, in the past, the limiting factor has been the high labor and reagent cost of Sanger sequencing. Fokstuen et al.14 reported substantial cost savings comparing a similar array covering 12 HCM genes with conventional dideoxy-based sequencing. Because laboratory practices and workflows, array designs, and analytical performance parameters, all of which impact test cost, can differ greatly, we conducted a similar analysis and compared the test cost and TAT of the DCM CardioChip with two Sanger-based tests: the previously offered DCM panels (10 genes, 2 tiers of tests) and a hypothetical test covering all of the 19 genes present on the DCM chip. Our laboratory considers the three following factors the main drivers of test cost: labor, reagents/laboratory ware, and test development expenses. Compared with Sanger sequencing of all 19 genes, the greatest savings using the DCM Cardiochip occurred with labor (~55% reduction) and reagents/laboratory ware (~51% reduction). Besides the savings that occur through parallel processing of a large number of genes, the main factor leading to a reduction in labor is the reduced amount of manual data review, which is analyzed automatically by an informatics pipeline. In our laboratory, the only manual review left is that associated with the confirmatory analysis of calls made by the chip, which is still done by Sanger sequencing. Overall, the DCM CardioChip is still almost 50% cheaper compared with a hypothetical Sanger sequencing test covering the same genes and almost 30% cheaper than the previously offered two DCM panels, which covered only 10 of the 19 genes present on the chip. The TAT of the DCM CardioChip since test launch has averaged at about 7 weeks. This is a 50% reduction compared with the hypothetical Sanger sequencing test and a 30% reduction compared with the previously offered DCM panels.
Detected variants and clinical sensitivity
Sixteen missense, splice, and frame shift variants (4 of them previously reported) were identified in 17 patients (Table 5). All other patients remained negative for clinically significant variants. All novel variants were genotyped in a minimum of 300 ethnically matched control chromosomes (black and/or white, see Materials and Methods). Variants identified in individuals of Hispanic origin or of unknown race were genotyped in both cohorts, although, here, absence from controls has to be interpreted with more caution as it remains possible that the individuals were of neither racial origin. Silent variants were assumed to be benign and were not genotyped in controls (Table 5).
Table 5.
Variants identified
| Gene | Exon/ intron |
DNA | Protein | No. samples (n = 73) |
Racea | No. control chromosome (C) |
No. control chromosome (B) |
dbSNP | MAF | Cons.e | Splicingb | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants of likely clinical significance | ||||||||||||
| DES | 04 | c.897+4_897+5del | — | 1 | C | 0/364 | ND | — | — | Reduced | ||
| CSRP3 | 03 | c.148G>A | p.Ala50Thr | 1 | C | 0/344 | 0/376 | — | High | |||
| ACTN2 | 03 | c.343G>A | p.Val115Met | 1 | C | 0/380 | 0/380 | — | High | |||
| ACTN2 | 20 | c.2520delT | p.Asp841fs | 1c | 0/368 | ND | — | — | ||||
| ACTN2 | 19 | c.2323C>T | p.His775Tyr | 1 | C | 0/326 | 0/358 | — | High | |||
| VCL | 03 | c.314G>A | p.Arg105Gln | 1 | C | 0/326 | 0/376 | — | High | |||
| VCL | 22 | c.3373C>T | p.Arg1125Cys | 1 | C | 0/356 | 0/364 | — | High | |||
| Variants of unknown clinical significance | ||||||||||||
| ACTN2 | 18 | c.2194G>A | p.Ala732Thr | 1c | C | 0/372 | ND | — | High | |||
| VCL | 14 | c.1907A>G | p.His636Arg | 2 | H | 0/324 | 0/318 | rs71579374 | — | High | ||
| CSRP3 | 06 | c.535A>G | p.Thr179Ala | 1 | C | 0/328 | 0/376 | — | Low | |||
| Variants of unlikely clinical significance | ||||||||||||
| ABCC9 | 15 | c.2093–7T>C | — | 2 | W, U | 1/356 | 0/358 | — | NA | No effect | ||
| ABCC9 | 17 | c.2200G>A | p.Val734Ile | 1 | B | 3/346 | 0/374 | rs61688134 | > 0.005 | High | 50 | |
| ABCC9 | 27 | c.3409G>A | p.Val1137Ile | 1 | B | 0/366 | 4/366 | — | High | |||
| CSRP3 | 02 | c.10T>C | p.Trp4Arg | 1 | W | 4/334 | 2/376 | rs45550635 | 0.003 | Med | 8, 37–40 | |
| DES | 02 | c.638C>T | p.Ala213Val | 1 | U | 6/570d | ND | rs41272699 | — | High | 34, 36, 60 | |
| DES | 09 | c.1375G>A | p.Val459Ile | 1 | B | 0/300d | 2/200d | rs73991549 | — | High | 36 | |
| Novel silent variants | ||||||||||||
| ABCC9 | 18 | c.2262T>C | p.Tyr754Tyr | 1 | — | |||||||
| ABCC9 | 21 | c.2523C>T | p.Ala841Ala | 1 | — | |||||||
| ABCC9 | 21 | c.2631G>A | p.Thr877Thr | 1 | — | |||||||
| ABCC9 | 30 | c.3768T>C | p.Leu1256Leu | 2 | — | |||||||
| ACTN2 | 08 | c.762C>T | p.His254His | 1 | — | |||||||
| DES | 01 | c.9G>A | p.Gln3Gln | 2 | — | |||||||
| DES | 03 | c.669T>C | p.Ile223Ile | 1 | — | |||||||
| EMD | 02 | c.144C>T | p.Leu48Leu | 2 | — | |||||||
| SGCD | 03 | c.123C>G | p.Leu41Leu | 1 | — | |||||||
| VCL | 06 | c.768T>C | p.Asp256Asp | 4 | — | |||||||
| VCL | 15 | c.2025G>A | p.Val675Val | 1 | — | |||||||
Race of individual carrying listed variant.
Using SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer.
Identified in same individual.
Numbers obtained from listed references.
Evolutionary conservation.
ND, not determined; C, Caucasian; H, Hispanic; B, black; U, unspecified; MAF, minor allele frequency in Caucasians; NA, not applicable.
Variants of likely clinical significance
A variant was considered likely to be clinically significant if it was absent from healthy controls and showed a high degree of evolutionary conservation or was predicted to impact splicing or truncate/degrade the protein through a frameshift or nonsense mutation mechanism. Seven variants (c.897 + 4_897 + 5del in DES, p.Ala50Thr in CSRP3, p.Val115Met, p.Asp841fs, and p.His775Tyr in ACTN2, and p.Arg105Gln and p.Arg1125Cys in VCL) met these criteria (Table 5 and Table, Supplemental Digital Content 2, http://links.lww.com/GIM/A99). It is important to point out that current genetic tools may be insufficient to adequately identify rare benign variants. On the one hand, recent genome-wide analyses suggest that the majority of missense variants have a deleterious effect of some magnitude (although it may be insufficient to cause overt disease) and that a variant’s population frequency correlates with its potential clinical significance.57 However, another recent study estimated that the number of control chromosomes used by us and the majority of similar studies (200–400) will likely only identify 80% of variants with a frequency of 0.1% or higher and that at least 3000 control individuals are necessary to rule out that a variant is not a rare benign change.58 Therefore, additional genetic evidence such as functional studies or segregation with disease would be needed to confirm the significance of variants for which pathogenicity is implicated based on evolutionary conservation and absence from a small control cohort alone.
Variants of unknown clinical significance
An additional three variants were also absent from controls but were considered to be of unknown clinical significance (Table 5 and Table, Supplemental Digital Content 2, http://links.lww.com/GIM/A99), because the affected amino acid was less conserved (p.Thr179Ala in CSRP3; Figure, Supplemental Digital Content 4, http://links.lww.com/GIM/A101), the variant (p.Ala732Thr in ACTN2) was found in the presence of another variant of likely clinical significance (p.Asp841fs in ACTN2), or the variant was present in dbSNP (p.His636Arg in VCL), although no clinical information or population frequency was available.
Variants of unlikely clinical significance
Finally, five missense variants and one potential splice variant were present in controls at varying frequencies and are most likely benign when present in isolation. Four of them (p.Val734Ile in ABCC9, p.Ala213Val, and p.Val459Ile in DES and p.Trp4Arg in CSRP3) have been previously published as potentially disease causing and warrant a more detailed discussion here. The p.Val734Ile variant (ABCC9) has been detected in 1.7–3.2% of Caucasians (rs61688134). Although it was associated with an increased risk of myocardial infarction,50 its high population frequency excludes it from being a highly penetrant mutation for DCM (the population prevalence of DCM is much lower than the population frequency of this variant). The p.Ala213Val variant (DES) has been reported by several studies and was initially thought to be disease causing.33,34,59 However, two subsequent studies identified this variant in a total of 6 of 570 control chromosomes (~1% population frequency).36,60 In addition, it did not segregate with disease in one family.36 This variant is also present in dbSNP (rs41272699) although no frequency information is available. The p.Val459Ile variant (DES) is one of two previously reported desmin variants located in the tail domain of the protein (the majority of all disease-causing desmin variants are located in the central rod domain). It has been reported in dbSNP (rs73991549, no frequency information available). In addition, it has been reported in 2 of 69 black patients with DCM (2 of138 chromosomes) and in 2 of 200 black Coriell control chromosomes (but was absent from 300 Caucasian control chromosomes). The authors concluded that it is a low penetrance variant restricted to the black population based on a subtle effect on desmin filament assembly in vitro.36 We identified this variant in one black DCM proband as part of our validation study. In our opinion, its presence in 1% of black control chromosomes, presence in black patients with DCM with a similar frequency (1.4%), and location outside the domain harboring the majority of reported desmin variants suggest that it is insufficient to cause disease when present in isolation although a modifying effect cannot be ruled out. The p.Trp4Arg variant (CSRP3) has been reported, but the available data surrounding it is conflicting. One study detected this variant in10 of 536 DCM probands, not in 640 control chromosomes, and provided functional data supporting a pathogenic role.39 However, additional studies detected the p.Trp4Arg variant in healthy control chromosomes (0.02% Caucasian,8,37,38,40 4 of 334 Caucasian and 2 of 376 black chromosomes; our own data). In addition, the variant was detected in patients with HCM and DCM,8,37,38,40 which further argues against a primary disease-causing role, because the molecular mechanisms underlying the two cardiomyopathies are fundamentally different.
Clinical sensitivity
Because no single published study analyzed all 19 genes present on the DCM CardioChip, it is difficult to derive an accurate clinical sensitivity for this test. However, a meta-analysis of all published studies (each having tested overlapping subsets of these genes) combined with the data presented here allows a projection.
The DCM CardioChip added 9 new genes to 10 previously available clinical tests, for which combined published data suggest a detection rate of ~22% (Table 6). Therefore, our cohort of 73 patients, who did not carry clinically significant variants in these 10 previously tested genes, corresponds to 94 untested patients. Ten of these patients carried variants of likely (n = 7) or unknown clinical significance (n = 3, one present with another variant). Taking both variants of likely and unknown significance into account, the projected maximum detection rate is 10.7% (10 of 94). However, it is likely that some of these variants are rare benign variants, particularly in light of the fact that the sizes of our control cohorts were moderate. Therefore, a more conservative detection rate for the nine newgenes on the DCM CardioChip, including only the variants of likely significance, is 7.5% (7 of 94). It is important to point out that, because of the small size of our cohort and most previously published cohorts, this number remains an estimate and that the true clinical sensitivity for these nine genes will emerge overtime. In summary, these data combined with all previously published data project the overall clinical sensitivity of the DCM CardioChip to be 26–29%.
DISCUSSION
The combination of marked locus and allelic heterogeneity of DCM poses a substantial challenge to sequencing-based clinical genetic testing to identify disease-causing mutations. Although traditional dideoxy-based sequencing technology is still considered the gold standard for molecular diagnostics, it is too costly to allow comprehensive screening of the myriad of DCM genes. To reduce test cost and TAT, many laboratories have offered reflexive testing strategies, where genes are grouped into panels allowing a sequential analysis starting with the panel offering the highest detection rate and proceeding with additional panels only if this initial test is negative. Although cost effective for those patients carrying a disease-causing variant in the commonly mutated genes, this strategy is extremely costly and time consuming for all other patients. In addition, rare cases of compound or double heterozygotes may be missed by traditional analysis if the first panel identifies a clinically significant variant and the subsequent/second panel is not performed. Our analyses show that the DCM CardioChip is significantly cheaper (almost 50%) and quicker (7 weeks TAT vs. an estimated 14 weeks) compared with a Sanger-based sequencing test covering the same 19 genes. Parallel processing of all genes also eliminates the danger of missing compound and double heterozygotes.
Our array-based resequencing test contains 19 DCM genes,10 of which were previously offered in 2 separate test panels. Based on available published data, variants in the nine newly added genes (ACTN2, VCL, CSRP3, TCAP, ABCC9, CTF1, SGCD, DES, and EMD) are generally rare and less well studied, which previously prevented inclusion into a clinical test. Our results suggest that the overall added sensitivity for these genes may be at least 7.5%, which is in the same range as what has been reported across the few available studies so far (Table 6 and references within). However, because our cohort and most other cohorts analyzed to date are small, the true clinical sensitivity for this set of genes is going to emerge over time.
The DCM CardioChip has a high analytical sensitivity for substitution variants but a reduced ability to detect novel insertions and deletions, which is a well-established limitation of array-based resequencing tests. Although it is a general limitation of the technology, it may not be significantly limiting for clinical testing of DCM because insertion/deletion mutations are generally rare among the main, well-characterized DCM genes (MYBPC3, MYH7, TNNT2, TNNI3, TPM1, ACTC, LMNA, LDB3, and PLN). Only MYBPC3 is known to have a significant prevalence of insertions and deletions, but this seems to be true for only HCM (internal unpublished observations). The variant spectrum of all other genes (ACTN2, VCL, CSRP3, TCAP, ABCC9, CTF1, SGCD, DES, and EMD) has not been well characterized, and mutations in these genes are generally rare. As such, the DCM CardioChip is a worthwhile compromise considering that cost-effective screening of these genes could otherwise not be offered at this point. The genetic basis for DCM is only partially understood today, and new genes are being discovered at a rapid pace. For example, promising candidates that have been discovered since the DCM CardioChip was designed include the cardiac ankyrin repeat protein, ANKRD1,61 the RNA-binding protein RBM20,62 and the Z-disk protein nexilin, NXN.63 Together with other genes that were not included into this design of the DCM CardioChip for various reasons outlined earlier (see Introduction), there is already a large and growing number of genes under consideration for an expanded DCM test. However, given the large number of additional genes, the size limitations of array-based sequencing chips, and the relative difficulty of adding new content, it is likely that further genes will be added when new sequencing platforms such as Next Generation technologies are established in clinical testing laboratories.
Supplementary Material
ACKNOWLEDGMENTS
C.Y.H. was supported in part by the J. Ira and Nicki Harris Family Research Award in the Division of Cardiovascular Medicine, Brigham and Women’s Hospital, and J.G.S. was supported by NIH Grants 5R01HL080494 and 1R01HL084553. The authors thank Raju Kucherlapati for his continued support in developing clinical array based sequencing tests and Alison Brown at the Partners HealthCare Center for Personalized Genetic Medicine for performing the healthy control studies.
Footnotes
Disclosure: The authors declare no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.geneticsinmedicine.org).
REFERENCES
- 1.Taylor DO, Edwards LB, Boucek MM, et al. Registry of the International Society for Heart and Lung Transplantation: twenty-fourth official adult heart transplant report–2007. J Heart Lung Transplant. 2007;26:769–781. doi: 10.1016/j.healun.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–981. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Cowan J, Morales A, Siegfried JD. Progress with genetic cardiomyopathies: screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Heart Fail. 2009;2:253–261. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daehmlow S, Erdmann J, Knueppel T, et al. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;298:116–120. doi: 10.1016/s0006-291x(02)02374-4. [DOI] [PubMed] [Google Scholar]
- 5.Hershberger RE, Parks SB, Kushner JD, et al. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1:21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamisago M, Sharma SD, DePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 7.Mogensen J, Murphy RT, Shaw T, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Mohapatra B, Jimenez S, Lin JH, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 9.Moller DV, Andersen PS, Hedley P, et al. The role of sarcomere gene mutations in patients with idiopathic dilated cardiomyopathy. Eur J Hum Genet. 2009;17:1241–1249. doi: 10.1038/ejhg.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimizu M, Ino H, Yasuda T, et al. Gene mutations in adult Japanese patients with dilated cardiomyopathy. Circ J. 2005;69:150–153. doi: 10.1253/circj.69.150. [DOI] [PubMed] [Google Scholar]
- 11.Villard E, Duboscq-Bidot L, Charron P, et al. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 12.Zeller R, Ivandic BT, Ehlermann P. Large-scale mutation screening in patients with dilated or hypertrophic cardiomyopathy: a pilot study using DGGE. J Mol Med. 2006;84:682–691. doi: 10.1007/s00109-006-0056-2. [DOI] [PubMed] [Google Scholar]
- 13.Denning L, Anderson JA, Davis R, Gregg JP, Kuzdenyi J, Maselli RA. High throughput genetic analysis of congenital myasthenic syndromes using resequencing microarrays. PLoS One. 2007;2:e918. doi: 10.1371/journal.pone.0000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fokstuen S, Lyle R, Munoz A, et al. A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Hum Mutat. 2008;29:879–885. doi: 10.1002/humu.20749. [DOI] [PubMed] [Google Scholar]
- 15.Mandal MN, Heckenlively JR, Burch T, et al. Sequencing arrays for screening multiple genes associated with early-onset human retinal degenerations on a high-throughput platform. Invest Ophthalmol Vis Sci. 2005;46:3355–3362. doi: 10.1167/iovs.05-0007. [DOI] [PubMed] [Google Scholar]
- 16.Waldmuller S, Muller M, Rackebrandt K, et al. Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clin Chem. 2008;54:682–687. doi: 10.1373/clinchem.2007.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S, Kassauei K, Cutler DJ, et al. An oligonucleotide microarray for high-throughput sequencing of the mitochondrial genome. J Mol Diagn. 2006;8:476–482. doi: 10.2353/jmoldx.2006.060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwick ME, McAfee F, Cutler DJ, et al. Microarray-based resequencing of multiple Bacillus anthracis isolates. Genome Biol. 2005;6:R10. doi: 10.1186/gb-2004-6-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothiyal P, Cox S, Ebert J, Aronow BJ, Greinwald JH, Rehm HL. An overview of custom array sequencing. Curr Protoc Hum Genet. 2009;Chapter 7(Unit 7):17. doi: 10.1002/0471142905.hg0717s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arimura T, Hayashi T, Terada H, et al. A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J Biol Chem. 2004;279:6746–6752. doi: 10.1074/jbc.M311849200. [DOI] [PubMed] [Google Scholar]
- 21.Vatta M, Mohapatra B, Jimenez S, et al. Mutations in Cypher/ZASP inpatients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Hermida-Prieto M, Monserrat L, Castro-Beiras A, et al. Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am J Cardiol. 2004;94:50–54. doi: 10.1016/j.amjcard.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Parks SB, Kushner JD, Nauman D, et al. Lamin A/C mutation analysis in acohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrot A, Hussein S, Ruppert V, et al. Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy. Basic Res Cardiol. 2009;104:90–99. doi: 10.1007/s00395-008-0748-6. [DOI] [PubMed] [Google Scholar]
- 25.Sebillon P, Bouchier C, Bidot LD, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–567. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sylvius N, Bilinska ZT, Veinot JP, et al. In vivo and in vitro examination of the functional significances of novel lamin gene mutations in heart failure patients. J Med Genet. 2005;42:639–647. doi: 10.1136/jmg.2004.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Tintelen JP, Hofstra RM, Katerberg H, et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154:1130–1139. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 28.D’Adamo P, Fassone L, Gedeon A, et al. The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet. 1997;61:862–867. doi: 10.1086/514886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manilal S, Recan D, Sewry CA, et al. Mutations in Emery-Dreifuss muscular dystrophy and their effects on emerin protein expression. Hum Mol Genet. 1998;7:855–864. doi: 10.1093/hmg/7.5.855. [DOI] [PubMed] [Google Scholar]
- 30.Sakata K, Shimizu M, Ino H, et al. High incidence of sudden cardiac death with conduction disturbances and atrial cardiomyopathy caused by a nonsense mutation in the STA gene. Circulation. 2005;111:3352–3358. doi: 10.1161/CIRCULATIONAHA.104.527184. [DOI] [PubMed] [Google Scholar]
- 31.Talkop UA, Talvik I, Sonajalg M, et al. Early onset of cardiomyopathy in two brothers with X-linked Emery-Dreifuss muscular dystrophy. Neuromuscul Disord. 2002;12:878–881. doi: 10.1016/s0960-8966(02)00134-7. [DOI] [PubMed] [Google Scholar]
- 32.Emery AE. Emery-Dreifuss muscular dystrophy—a 40 year retrospective. Neuromuscul Disord. 2000;10:228–232. doi: 10.1016/s0960-8966(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 33.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Desmin myopathy. Brain. 2004;127(Pt 4):723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 34.Kostareva A, Gudkova A, Sjoberg G, et al. Desmin mutations in a St. Petersburg cohort of cardiomyopathies. Acta Myol. 2006;25:109–115. [PubMed] [Google Scholar]
- 35.Li D, Tapscoft T, Gonzalez O, et al. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation. 1999;100:461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 36.Taylor MR, Slavov D, Ku L, et al. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115:1244–1251. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 37.Bos JM, Poley RN, Ny M, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Genet Metab. 2006;88:78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geier C, Gehmlich K, Ehler E, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17:2753–2765. doi: 10.1093/hmg/ddn160. [DOI] [PubMed] [Google Scholar]
- 39.Knoll R, Hoshijima M, Hoffman HM, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 40.Newman B, Cescon D, Woo A, et al. W4R variant in CSRP3 encoding muscle LIM protein in a patient with hypertrophic cardiomyopathy. Mol Genet Metab. 2005;84:374–375. doi: 10.1016/j.ymgme.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi T, Arimura T, Itoh-Satoh M, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 42.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 43.Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87:169–174. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Zemljic-Harpf AE, Miller JC, Henderson SA, et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol. 2007;27:7522–7537. doi: 10.1128/MCB.00728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira ES, Wiltshire TJ, Faulkner G, et al. Limb-girdle muscular dystrophytype 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat Genet. 2000;24:163–166. doi: 10.1038/72822. [DOI] [PubMed] [Google Scholar]
- 46.Karkkainen S, Miettinen R, Tuomainen P, et al. A novel mutation, Arg71Thr, in the delta-sarcoglycan gene is associated with dilated cardiomyopathy. J Mol Med. 2003;81:795–800. doi: 10.1007/s00109-003-0480-5. [DOI] [PubMed] [Google Scholar]
- 47.Tsubata S, Bowles KR, Vatta M, et al. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest. 2000;106:655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erdmann J, Hassfeld S, Kallisch H, Fleck E, Regitz-Zagrose V. Genetic variants in the promoter (g983G>T) and coding region (A92T) of the human cardiotrophin-1 gene (CTF1) in patients with dilated cardiomyopathy. Hum Mutat. 2000;16:448. doi: 10.1002/1098-1004(200011)16:5<448::AID-HUMU19>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minoretti P, Falcone C, Aldeghi A, et al. A novel Val734Ile variant in the ABCC9 gene associated with myocardial infarction. Clin Chim Acta. 2006;370:124–128. doi: 10.1016/j.cca.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–666. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 52.Li D, Parks SB, Kushner JD, et al. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet. 2006;79:1030–1039. doi: 10.1086/509900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knoll R, Postel R, Wang J, et al. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 54.Cutler DJ, Zwick ME, Carrasquillo MM, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebet T, Chiles R, Hsu AP, Mansfield ES, Warrington JA, Puck JM. Mutations causing severe combined immunodeficiency: detection with a custom resequencing microarray. Genet Med. 2008;10:575–585. doi: 10.1097/gim.0b013e31818063bc. [DOI] [PubMed] [Google Scholar]
- 56.Hartmann A, Thieme M, Nanduri LK, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30:115–122. doi: 10.1002/humu.20816. [DOI] [PubMed] [Google Scholar]
- 57.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ionita-Laza I, Lange C, Laird NM. Estimating the number of unseen variants in the human genome. Proc Natl Acad Sci U S A. 2009;106:5008–5013. doi: 10.1073/pnas.0807815106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bar H, Strelkov SV, Sjoberg G, Aebi U, Herrmann H. The biology of desmin filaments: how do mutations affect their structure, assembly, and organisation? J Struct Biol. 2004;148:137–152. doi: 10.1016/j.jsb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Goudeau B, Rodrigues-Lima F, Fischer D, et al. Variable pathgenic potentials of mutations located in the desmin alpha-helical domain. Hum Mutat. 2006;27:906–913. doi: 10.1002/humu.20351. [DOI] [PubMed] [Google Scholar]
- 61.Moulik M, Vatta M, Witt SH, et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54:325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brauch KM, Karst ML, Herron KJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hassel D, Dahme T, Erdmann J, et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 64.Sylvius N, Duboscq-Bidot L, Bouchier C, et al. Mutational analysis of the beta- and delta-sarcoglycan genes in a large number of patients with familial and sporadic dilated cardiomyopathy. Am J Med Genet A. 2003;120A:8–12. doi: 10.1002/ajmg.a.20003. [DOI] [PubMed] [Google Scholar]
- 65.Carballo S, Robinson P, Otway R, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105:375–382. doi: 10.1161/CIRCRESAHA.109.196055. [DOI] [PubMed] [Google Scholar]
- 66.Tesson F, Sylvius N, Pilotto A, et al. Epidemiology of desmin and cardiac actin gene mutations in a european population of dilated cardiomyopathy. Eur Heart J. 2000;21:1872–1876. doi: 10.1053/euhj.2000.2245. [DOI] [PubMed] [Google Scholar]
- 67.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathydue to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 68.Arbustini E, Pilotto A, Grasso M, et al. Novel human pathological mutations. Gene symbol: LMNA. Disease: cardiomyopathy, dilated with conduction defects. Hum Genet. 2009;125:350. [PubMed] [Google Scholar]
- 69.DeWitt MM, MacLeod HM, Soliven B, McNally EM. Phospholamban R14 deletion results in late-onset, mild, hereditary dilated cardiomyopathy. J Am Coll Cardiol. 2006;48:1396–1398. doi: 10.1016/j.jacc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 70.Haghighi K, Kolokathis F, Gramolini AO, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A 31. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haghighi K, Kolokathis F, Pater L, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 111:869–876. doi: 10.1172/JCI17892. 2003l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmitt JP, Kamisago M, Asahi M, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.