Abstract
Regulation of gene expression during infection of the thermophilic bacterium Thermus thermophilus (T. th.) HB8 with the bacteriophage P23-45 was investigated. Macroarray analysis revealed host transcription shut-off and identified three temporal classes of phage genes: early, middle, and late. Primer extension experiments revealed that the 5′ ends of P23-45 early transcripts are preceded by a common sequence motif that likely defines early viral promoters. T. th. HB8 RNA polymerase (RNAP) recognizes middle and late phage promoters in vitro but does not recognize early promoters. In vivo experiments revealed the presence of rifampicin-resistant RNA polymerizing activity in infected cells responsible for early transcription. The product of the P23-45 early gene 64 shows a distant sequence similarity with the largest, catalytic subunits of multisubunit RNAPs and contains the conserved metal-binding motif that is diagnostic of these proteins. We hypothesize that ORF64 encodes rifampicin-resistant phage RNAP that recognizes early phage promoters. Affinity isolation of T. th. HB8 RNAP from P23-45-infected cells identified two phage-encoded proteins: gp39 and gp76, that bind the host RNAP and inhibit in vitro transcription from host promoters, but not from middle or late phage promoters, and may thus control the shift from host to viral gene expression during infection. To our knowledge, gp39 and gp76 are the first characterized bacterial RNAP-binding proteins encoded by a thermophilic phage.
Keywords: Thermus thermophilus, thermophage, phage promoters, RNA polymerase, RNAP-binding proteins
Introduction
Transcription is the first step and primary regulatory determinant of gene expression. Multisubunit DNA-dependent RNA polymerases (RNAPs) are complex, highly regulated molecular machines. RNAP, alone or in complex with regulatory factors, is central to the transcription process. Every stage of transcription is regulated via RNAP interactions with various transcription factors. Bacteriophages (phages) have evolved highly effective mechanisms to modify bacterial RNAP to serve the needs of the phage (reviewed in 1). Recent studies indicate that phages are the most abundant life form in the Biosphere 2; 3, and that the phage gene pool is the largest source of natural gene diversity. Consequently, while many phages use common strategies to subjugate their hosts, the number of phage-encoded regulatory mechanisms is virtually endless 4. At the time of writing, more than 580 complete phage genome sequences (NCBI, last modified July 2010) had been determined. Comparative genomic analysis provides important insights into the diversity and evolution of phages and their hosts. However, our understanding of gene expression regulation strategies utilized by phages is relatively limited. Classical studies of gene regulation mechanisms in a handful of model Escherichia coli (E. coli) phages (e.g. γ, T4, and T7) have led to key discoveries in molecular biology. More recently, transcription profiling and bioinformatic predictions have been successfully applied to aid our understanding of the molecular features of phage regulatory networks 5; 6; 7; 8. The ultimate understanding of transcription regulatory mechanisms is based on structure-function analysis of RNAP alone, and RNAP-regulator complexes. However, a high resolution structure of E. coli RNAP is absent, therefore, almost no structural information on the action of transcription factors encoded by model E. coli phages in complex with E. coli RNAP has been obtained. Currently, only the structures of bacterial RNAP from the thermophilic eubacteria Thermus aquaticus (T. aq.) 9; 10; 11 and T. th. 12; 13 have been determined. Thus, at present, the most reasonable way to structurally investigate phage-driven prokaryotic transcription regulation is to study phages that infect thermophilic eubacteria (i.e. thermophages). Despite the advances in our understanding of the structures of thermophilic eubacterial RNAPs, there is insufficient knowledge about thermophages, in particular their biology and the gene regulation mechanisms they employ during host bacterial infection. The genomes of several thermophages infecting Thermus species have been completely sequenced 14; 15; 16, and the gene expression strategy of one of these phage, φYS40, has been investigated in detail 8.
In this work we studied the temporal regulation of transcription of another T. th. HB8 phage, P23-45, whose genome has recently been sequenced 15. We identified three temporal classes of P23-45 genes and their corresponding promoters by a combination of gene macroarray, in vivo primer extension, and in vitro transcription experiments. P23-45 middle and late promoters have consensus elements that differ from those of T. th. housekeeping promoters and φYS40 thermophage promoters 8. Yet, P23-45 middle and late promoters are recognized by unmodified host T. th. RNAP in vitro or in vivo. In contrast, P23-45 early promoters are not recognized by host T. th. RNAP either in vitro or in vivo. The early promoters are defined by an unusual 11 bp conserved sequence motif, which is likely recognized by a unique phage-encoded RNAP. Affinity isolation of T. th. RNAP from P23-45-infected cells identified two phage-encoded T. th. RNAP-binding proteins: gp76 and gp39, the products of an early and a middle gene, respectively. These proteins bind the host RNAP in vitro and efficiently inhibit transcription from host bacterial promoters but not from middle or late phage promoters. Thus, these proteins may be responsible for the shut-off of host transcription and therefore the switch from host to viral gene expression. To our knowledge, this is the first description of thermophage-encoded thermophilic bacterial RNAP-binding proteins.
Results
Macroarray analysis of gene expression during P23-45 infection
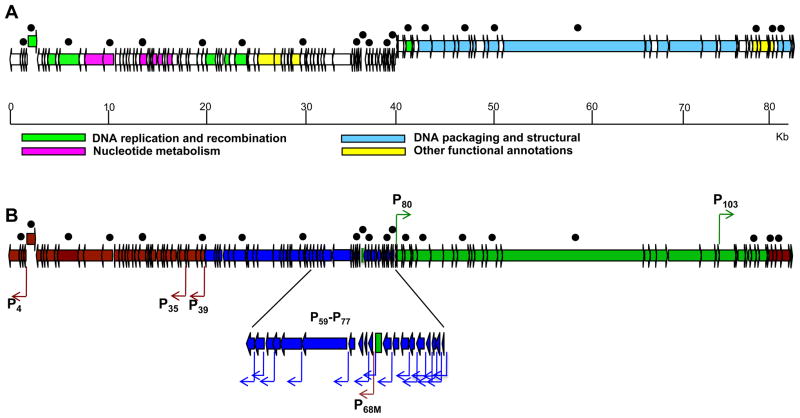
Almost half of the P23-45 genome (ORFs 1–78) is transcribed in the same direction (leftward in Fig. 1A). These genes form a single cluster in the left arm of the genome. The remaining P23-45 ORFs (ORFs 79–117) are transcribed in the rightward direction. These ORFs form a single cluster at the right arm of the genome. The only exception is ORF5, which is transcribed in the rightward direction, but is located in the left arm of the genome (Fig. 1A). To characterize the temporal profile of P23-45 gene expression, a macroarray of P23-45 phage genes was prepared. The array contained spots with equal amounts of PCR-amplified DNA fragments of 20 P23-45 genes and one non-coding region of the P23-45 genome (marked by black dots in Fig. 1A and B). The genes chosen for the array encode proteins of different functional classes. One group of spots represented the abundance of mRNA from a cluster of small genes with unknown functions from the left arm (genes 57, 68, 69, 76, 78 and the non-coding region of triplex-forming mirror repeats 15). The second group of spots represented left arm genes involved in DNA replication, recombination, and nucleotide metabolism (genes 4, 5, 11, 14, 24, 39, and 46). The third group of spots on the array represented right arm genes that encode the P23-45 virion structural proteins (genes 82, 89, 94, 96, and 114), and predicted DNA packaging (gene 85) and lysis proteins (genes 108 and 112). Since closely spaced or partially overlapping genes are usually transcribed from the same promoter, some spots on the array may likely report the abundance of transcripts of multiple P23-45 genes.
Figure 1. Genome and transcription map of the T. th. HB8 bacteriophage P23-45.
A. Functional annotation of the predicted P23-45 ORFs. The different colors of the arrows represent the different functions assigned for the proteins encoded by P23-45. Upper arrows denote a rightward orientation of the genes, while lower arrows denote a leftward orientation of the genes.
B. The different colors of the transcription map indicate different temporal classes of genes: blue, early; brown, middle; green, late. Promoters are colored corresponding to their temporal class and are depicted as bent arrows. A section of the early cluster is shown in more detail. In both A and B black dots indicate genes used in gene macroarray analysis of the temporal transcription program.
In order to determine whether P23-45 shuts off host transcription, PCR-amplified DNA fragments of seven housekeeping T. th. HB8 genes: rpoC (encoding the RNAP β′ subunit), sigA (encoding the primary sigma factor σA), dnaK (encoding a chaperone), TTHA0466 (encoding alcohol dehydrogenase), infB and infC (encoding translation initiation factors IF2 and IF3, respectively), and rpsA (encoding the ribosomal protein S1) were spotted on the membrane. The array also contained spots with total T. th. HB8 genomic DNA, P23-45 genomic DNA, and a PCR-amplified DNA fragment of the Drosophila melanogaster (D. me.) zfrp8 gene that was used as a normalizing and loading control.
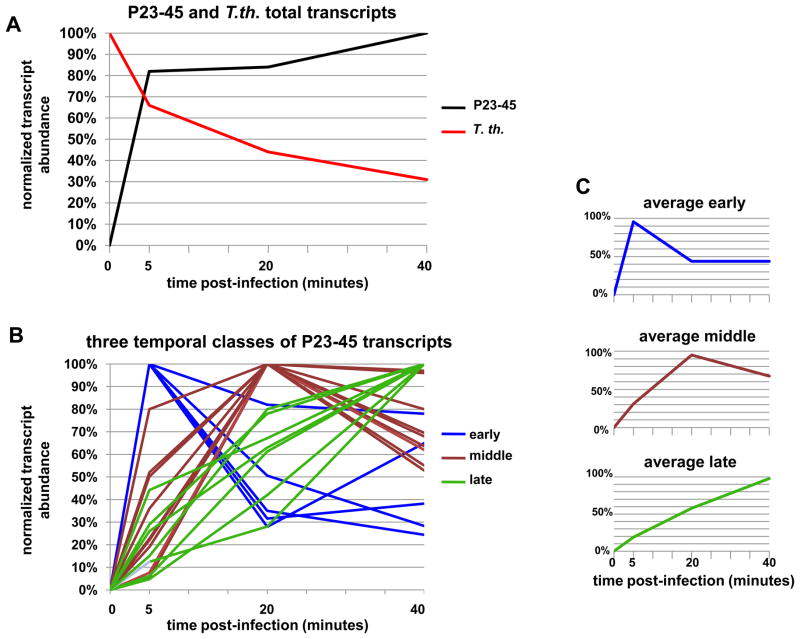
T. th. HB8 cells were infected with P23-45 at a multiplicity of infection of 10 and total RNA was extracted 5, 20, and 40 minutes post-infection. As a control, RNA was extracted from T. th. HB8 cells immediately prior to P23-45 infection. Equal amounts of total RNA were used to generate radioactively labeled cDNA by random priming/reverse transcription followed by hybridization to the macroarray membrane. The amount of radioactivity hybridized to different spots of the macroarray reflected the abundance of transcripts of each corresponding gene. For each time point, three independent macroarray experiments were performed. To quantitatively analyze the macroarray data, the radioactive signal from each spot was corrected for background and normalized according to the relative strength of the signal from the D. me. zfrp8 spot. The mean amount of radioactivity for each macroarray spot was plotted as a function of time post-infection (Fig. 2). As expected, the total amount of P23-45 transcripts increased with time post-infection relative to the control zfrp8 spots (Fig. 2A). In contrast, the total amount of T. th. HB8 transcripts decreased throughout the same period (Fig. 2A), indicating that P23-45 either executes host transcription shut-off or increases the rate of host RNA decay. The behavior of individual host transcripts during P23-45 infection was complex; while the abundance of most transcripts decreased throughout the infection cycle, the rates at which the transcripts decreased varied and in one case (sigA) the transcript abundance remained unchanged (data not shown). The reasons for the observed differences in the abundance of the host transcripts were not investigated; however, one explanation could be the interplay between the rates of host transcript synthesis and stability in the infected cell.
Figure 2. Macroarray data analysis.
A. Normalized abundances of total T. th. HB8-encoded transcripts and total P23-45-encoded transcripts are shown as red and black lines, respectively.
B. Normalized abundances for individual P23-45 transcripts are presented. Transcripts that belong to different temporal classes are colored: early, blue; middle, brown; late, green. The expression curves represent average results of three independent experiments.
C. Average normalized abundances of P23-45 transcripts of different temporal classes.
To compare the behavior of individual P23-45 transcripts, plots of normalized spot signal intensities as a function of time post-infection were scaled to equalize the mean abundance of each transcript. The accumulation of individual transcripts peaked at different times during the infection cycle, indicating the presence of different temporal classes of phage genes (Fig. 2B). Phage genes were clustered based on the time when their transcripts became most abundant 17. Analysis of transcript abundance patterns revealed three distinct temporal classes of genes: early, middle, and late (Fig. 2B). The early class gene transcripts peaked 5 minutes post-infection, while middle class gene transcripts peaked 20 minutes post-infection. Finally, the abundance of transcripts of the late class of genes increased dramatically by the end of infection (in our conditions the eclipse period of P23-45 was 35 minutes and lysis of T. th. HB8 by the phage was complete ~ 60 minutes post-infection). The average value of the scaled abundances calculated for each of the three temporal classes are shown as separate panels in Figure 2C.
P23-45 genes used in the macroarray analysis and the temporal classes they belong to are indicated by black dots and different colors, respectively, in Figure 1. Most centrally located genes with unknown functions belong to the early class, however, gene 78 and the non-coding triplex-forming region located in this part of the P23-45 genome belong to the middle and the late classes, respectively. Some genes encoding DNA replication and recombination components also belong to the early temporal class. Other genes from this functional group, as well as genes encoding nucleotide metabolism enzymes, comprise the middle class. As expected, genes of the late class were found exclusively in the right arm of the genome. They encode the P23-45 virion structural proteins, DNA packaging proteins, and lysis proteins.
Mapping P23-45 promoters
The previous automated annotation of P23-45 genes suggested that P23-45 does not encode its own RNAP 15 and must therefore rely on host RNAP to transcribe its early, middle, and late genes throughout the infection. Such temporal regulation can be achieved either by using specific sequences defining promoters of different temporal classes and/or by modification of RNAP promoter specificity by phage-encoded transcription factors. In the following section, we report our analysis of the P23-45 middle and late promoters followed by a discussion of the analysis of the P23-45 early promoters.
P23-45 middle and late promoters
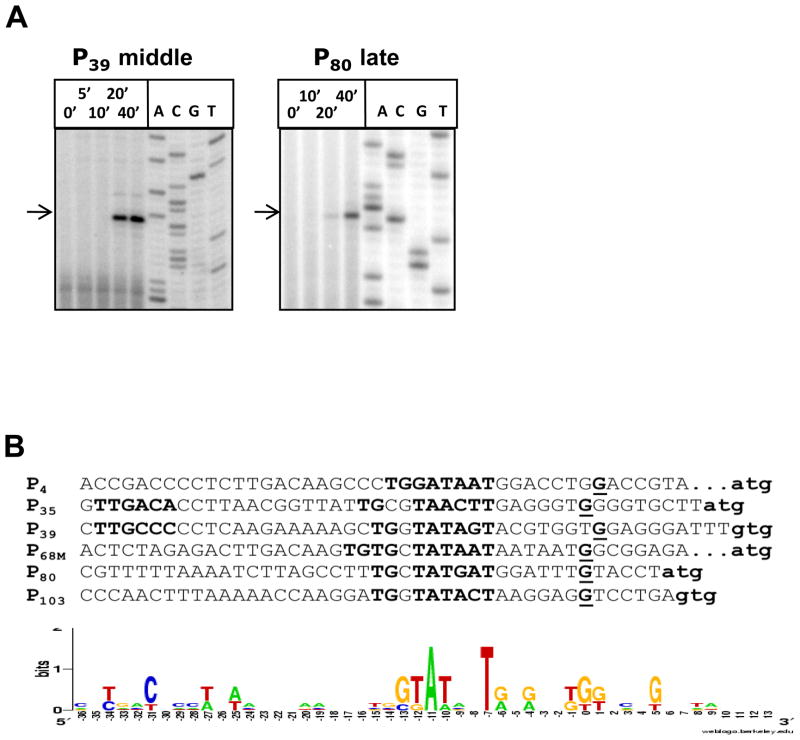
Non-coding P23-45 regions upstream of middle and late phage genes were examined by primer extension. Five primer extension products corresponding to the 5′ RNA ends of three middle (P4, P35, and P39) and two late (P80 and P103) putative promoters were detected. Additionally, one middle promoter, P68M, located in the early gene cluster was also identified by primer extension. The kinetics of primer extension product accumulation for these promoters during P23-45 infection matched the macroarray data for middle and late genes. Representative primer extension experiments with primers specific to middle and late genes are shown in Figure 3A.
Figure 3. Characterization of middle and late P23-45 promoters.
A. The kinetics of accumulation of in vivo primer extension products obtained with P23-45 transcripts from representative middle (P39) and late (P80) promoters during infection; the DNA products corresponding to 5′ RNA ends are labeled with arrows.
B. An alignment of P23-45 middle and late promoter sequences is shown. The putative −35 (where identified), −10 and TG/TGTG promoter elements are highlighted in bold uppercase text. Experimentally identified transcription start sites are highlighted in bold and underlined. The annotated translation initiation codons are shown in bold lowercase text. The corresponding sequence logos for P23-45 middle and late promoters are depicted below the alignment. The size of the letters indicates the degree of conservation; positions are defined with respect to identified transcription start sites.
To confirm that the in vivo identified 5′ RNA end points are transcription start points, we tested the ability of P23-45 genomic DNA fragments containing putative middle and late P23-45 promoters to serve as templates for in vitro abortive transcription initiation with purified T. th. HB8 σA-associated holoenzyme. For each promoter tested, combinations of nucleotide substrates that should have permitted transcription initiation (based on the results of in vivo primer extension analysis) were used. In all cases, robust transcription was detected (for example, see Fig. 5B, lanes 14 and 15). Therefore, we conclude that in vivo primer extension products correspond to transcription start points and that unmodified host RNAP-σA holoenzyme can recognize P23-45 middle and late promoters in vitro.
Figure 5. Characterization of putative early P23-45 promoters.
A. The kinetics of accumulation of representative in vivo primer extension products obtained with P23-45 transcripts from putative early promoters P64 and P68E during infection; the DNA products corresponding to 5′ RNA ends are labeled with arrows.
B. The results of abortive transcription initiation by T. th. HB8 RNAP-σA holoenzyme at representative P23-45 early promoters (lanes 1–13) are shown. Dinucleotide RNA substrates corresponding to the −2/−1 and/or −1/+1 positions with respect to the putative transcription start sites (see Fig. 4) were used as the primers. Radioactively labeled NTP corresponding to the putative +1 or +2 positions was used to form the trinucleotide RNA product. As a positive control, abortive transcription at representative P23-45 middle and late promoters is shown (lanes 14 and 15).
Comparisons of sequences upstream of the transcription start points of P23-45 middle and late transcripts revealed a motif that was common to promoters of both classes (Fig. 3B). The middle and late P23-45 promoters are characterized by a −10-like element (consensus sequence 5′-GTATanT-3′) with the highest conservation at positions −11 (A) and −7 (T) relative to the experimentally determined transcription start point (Fig. 3B). In addition, an extended −10 “TG/TGTG” motif is present 0–2 bp upstream of the −10 element. In two cases, appropriately positioned motifs similar to the consensus T. th. −35 promoter element were identified in two middle phage promoters, P35 and P39 (Fig. 3B, for the consensus T. th. −35 promoter element see Fig. 4). Our failure to differentiate between the middle and late P23-45 promoter sequences may be due to the small number of promoter sequences examined. Alternatively, the distinct temporal patterns of activity from these promoters may be caused not by differences in basal promoter elements or by binding of transcription factors to specific regulatory sites, but by the phage-dependent modification of host T. th. HB8 RNAP and/or differences in intrinsic promoter strengths.
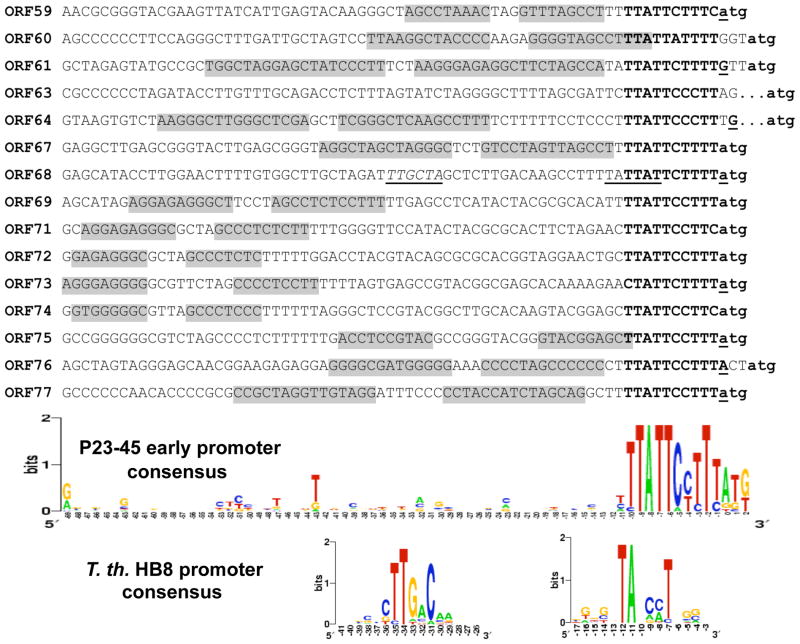
Figure 4. Alignment of DNA sequences upstream of the annotated translation initiation start codons of P23-45 early genes.
The novel promoter elements are highlighted in bold uppercase text. Experimentally identified 5′ RNA ends are highlighted in bold text and underlined. The annotated translation initiation start codons are highlighted in bold lowercase text. The grey boxes indicate stem-forming regions of putative rho-independent terminators. The sequence logos for the early P23-45 promoters and the independently aligned −35 and −10 regions of T. th. HB8 −10/−35 promoters are depicted below the alignment. The size of the letters indicates the degree of conservation; positions are defined with respect to putative or identified transcription start sites.
P23-45 early promoters
Based on previous studies of other phages, one would expect that transcription of early P23-45 genes should be driven by strong promoters recognized by the housekeeping form of the host RNAP holoenzyme: T. th. HB8 σA-associated holoenzyme. Early phage promoters need to be strong to efficiently compete with host promoters for host RNAP. Thus, early phage promoters would be expected to have a good match to the host σA-associated holoenzyme consensus promoter sequence. Indeed, such an expectation was fulfilled by the phage φYS40, a T. th. HB8 phage previously studied in our laboratory 8.
We utilized a T. th. RNAP-σA holoenzyme promoter bioinformatic profile described in 8 to search P23-45 DNA upstream of early P23-45 genes. To our surprise, with the exception of a likely σA-dependent promoter upstream of gene 68 located in the middle of the early cluster (P68E), no high-matching candidate sequences were found. Visual inspection of non-coding regions separating P23-45 early genes revealed the presence of a common 11 bp sequence motif (5′-TTATTCcTTTA-3′) located immediately upstream of annotated start codons (Fig. 4). Copies of this motif were identified upstream of early genes 59–61, 63, 64, 67–69, and 71–77. No additional copies of the 11 bp motif are present in any other region of the P23-45 genome. The logo of the motif is shown below the alignment in Figure 4 and compared to the T. th. promoter consensus logo. As can be seen, the two logos are clearly distinct from each other.
We hypothesized that this 11 bp motif may define early P23-45 promoters. To test this hypothesis, RNA samples used in macroarray experiments were subjected to primer extension analysis using primers specific to eight genes from the early gene cluster (see the alignment in Fig. 4, the experimentally identified 5′ ends are underlined). In all cases, an identical result was obtained: a primer extension product whose end corresponds to the last nucleotide of the 11 bp motif. Moreover, the kinetics of primer extension product accumulation for early P23-45 genes was in agreement with the macroarray data. Representative primer extension experiments with primers specific to genes 64 and 68 are shown in Figure 5A.
The results presented above suggest that the 11 bp motif may define the early P23-45 promoters that are obviously distinct from known host or phage promoters. Alternatively, the 11 bp motif could be the site of post-transcriptional processing of an early P23-45 polycistronic transcript(s) that initiates elsewhere upstream. The following experiments were performed to distinguish between these possibilities. First, putative P23-45 early promoters whose transcription start sites had been identified in vivo were tested in an in vitro abortive transcription initiation assay using purified T. th. HB8 RNAP-σA holoenzyme. For each putative promoter tested, combinations of nucleotide substrates that should have permitted transcription initiation (based on the results of in vivo primer extension analysis) were used. Unexpectedly, T. th. RNAP-σA holoenzyme was either unable to transcribe or yielded only small amounts of product for almost every transcription template tested (Fig. 5B; lanes 1–3 and 6–13); the only exception was P68E (lanes 4–5). However, robust transcription from this promoter could be explained by the fact that the 11 bp motif is embedded in a recognizable σA-dependent promoter (as revealed by bioinformatic analysis; Fig. 4 alignment, the −35-like and −10-like elements are underlined). For comparison, T. th. HB8 RNAP-σA holoenzyme actively transcribed from P23-45 middle and late promoters (P68M and P103, see Fig. 5B, lanes 14 and 15). Therefore, we conclude that unmodified host T. th RNAP-σA holoenzyme is unable to initiate transcription from DNA fragments containing the 11 bp motif in vitro.
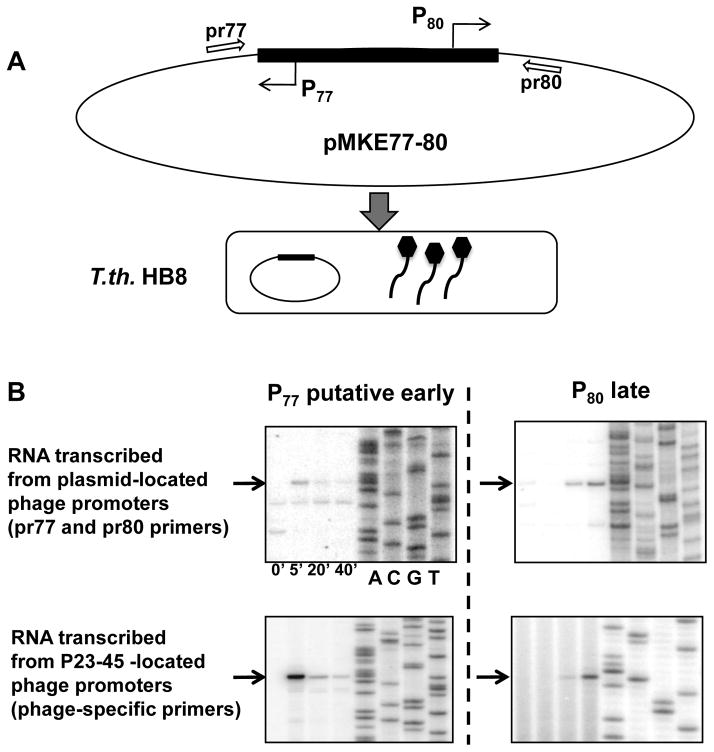
If processing of an early polycistronic precursor transcript were responsible for the appearance of primer extension products whose 5′ ends are located just downstream of the 11 bp motif, then transcription initiation of this precursor transcript most likely originate in the non-coding region of the phage genome that separates the divergently transcribed early and late gene clusters (Fig. 1). This region, between genes 77 (an early gene) and 80 (a late gene) contains an 11 bp motif upstream of gene 77. In an effort to identify a hypothetical early P23-45 promoter located upstream of gene 77 and responsible for early viral transcription, the following experiment was performed. The intergenic region between P23-45 genes 77 and 80 was cloned into the T. th.-E. coli shuttle plasmid pMKE1 18; to generate pMKE77-80, a plasmid containing the validated late promoter, P80, and the putative divergent early promoter, P77 (Fig. 6A). T. th. HB8 was transformed with pMKE77-80 and the resulting strain was infected with P23-45 (Fig. 6A). Total RNA was extracted from infected cells at various times post-infection and in vivo primer extension analysis with primers complementary to plasmid sequences upstream and downstream of the P23-45 insert was performed. As a control, primer extension reactions with P23-45 phage genome-specific primers that reported the transcriptional activity of P77 and P80 located on the P23-45 genome were also performed (Fig. 6B). Control reactions revealed the expected late accumulation of the P80 transcript and the early accumulation of a transcript whose 5′ end coincided with the last nucleotide of the 11 bp motif; i.e. P77 (Fig. 6B, two lower panels). A primer extension product corresponding to plasmid-located P77 was absent from uninfected cells, peaked 5 minutes post-infection and steadily decreased afterwards (Fig. 6B, upper left panel). A primer extension product corresponding to plasmid-located late P80 was detectable in the absence of infection and continuously increased throughout the infection (Fig. 6B, right upper panel). The appearance of the P80 transcript in the absence of the phage shows that this promoter is recognized by host RNAP in uninfected cells in vivo and is consistent with our findings that unmodified host T. th. RNAP initiates transcription from P80 (and other P23-45 middle and late promoters) in vitro. The absence of this transcript immediately post-infection may be due to phage-dependent modification(s) of the host RNAP that regulates the coordinated temporal transcription program of P23-45 and prohibits premature recognition of late promoters via unknown mechanism. No primer extension product upstream of P77 was detected in uninfected cells. Our attempts to identify a hypothetical promoter located upstream of P77 with several additional primers, both P23-45 phage genome-specific and pMKE77-80 plasmid-specific, were similarly unsuccessful (data not shown). These data make us conclude that it is very unlikely that there is a strong viral promoter from which early P23-45 genes are transcribed by host RNAP to produce a single precursor transcript.
Figure 6. Putative P23-45 early promoters are functional in vivo.
A. Schematic diagram illustrating an experiment to probe the functionality of P23-45 promoters in vivo. T. th. HB8 harboring the plasmid pMKE77-80 was infected with P23-45 and total RNA was extracted throughout the infection at the time points indicated.
B. The results of in vivo primer extension analysis of RNA transcribed from the divergent plasmid-located P23-45 early (P77) and late (P80) promoters (upper panels) and from the genome-located P77 and P80 promoters (lower panels). The DNA products corresponding to 5′ RNA ends are labeled with arrows.
An “internal” putative early promoter P59 was also cloned into pMKE1 and the activity of this promoter in P23-45 infected and uninfected cells was monitored. An identical result to that described for P77 (no activity in uninfected cells and an “early” pattern of activity during infection) was observed (data not shown). Taken together, these data suggest that despite the lack of in vitro activity, P59 and P77 (and, likely, other genomic sites defined by the presence of the 11 bp motifs) may function as early phage promoters that are recognized immediately after infection by either a phage-encoded, but as yet unidentified RNAP, or a phage-modified host RNAP.
Transcription of P23-45 early genes is Rifampicin-resistant
Rifampicin (Rif), a strong inhibitor of bacterial RNAPs, including the T. th. RNAP, binds to a pocket formed by the RNAP β subunit and efficiently blocks the synthesis of transcripts longer than 2–3 nt 19. Conversely, all phage-encoded RNAPs studied thus far are resistant to Rif 6; 20. Therefore, if early P23-45 genes were transcribed by host RNAP, then the appearance of these transcripts shall be suppressed by the addition of Rif. On the other hand, if a phage RNAP were responsible for early P23-45 transcription, the appearance of these transcripts shall not be affected by Rif.
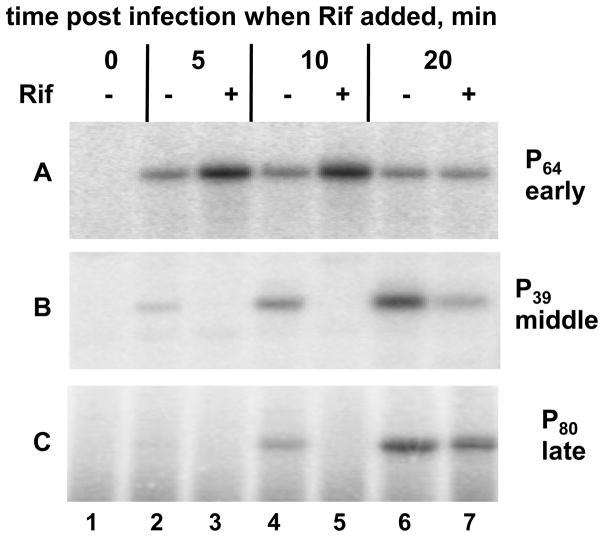
We performed P23-45 infection followed by the addition of Rif at different time points of infection followed by additional 10 minutes incubation and primer extension analysis of selected phage transcripts. Since middle and late P23-45 mRNAs are transcribed by host RNAP, we expected that Rif will inhibit accumulation of these transcripts. This expectation was fulfilled (Fig. 7, panels B and C): the addition of Rif 5, 10, and 20 minutes post-infection inhibited accumulation of a middle (P39) and late (P80) promoter transcripts compared to a control infection that was not treated with Rif. The effect was less pronounced when Rif was added 20 minutes post-infection, since by this time P39 and P80–originated transcripts started to accumulate prior to Rif addition. For 5 and 10 minutes time points, Rif addition led to complete disappearance of primer extension bands corresponding to either P39 or P80. In contrast, the addition of Rif had a completely different effect on the abundance of primer extension product corresponding to the early P64 phage transcript (Fig. 7, panel A). The addition of Rif 5 or 10 minutes post-infection led to increase of this transcript abundance compared to untreated control cells. Addition of Rif 20 minutes post-infection, when early transcription ceases based on the kinetics of early transcript accumulation, had no effect on the P64 transcript abundance. The difference in the abundances of the P64-originated transcript in total RNA extracted from Rif-treated and Rif-untreated cells may be explained by increased proportion of the P64 transcript in Rif-treated cells, where host RNAP-dependent transcripts are not accumulating. We conclude that the synthesis of early P23-45 transcripts is Rif-independent and may therefore be due to activity of Rif-resistant RNAP that is distinct from Rif-sensitive host RNAP.
Figure 7. Transcription from P23-45 early promoters is Rif-resistant and does not depend on host RNAP.
The results of in vivo primer extension analysis of RNA extracted from phage-infected cells in the absence of Rif (-) or in the presence of Rif (+) added at different time points post-infection. The panels A, B, and C demonstrate the kinetics of accumulation of transcripts from representative early, middle, and late P23-45 promoters, respectively.
Identification of P23-45-encoded T. th. RNAP-binding proteins
Based on our macroarray data, P23-45 executes shut-off of T. th. HB8 transcription, expresses its genes in a coordinated manner, and must rely on the host transcription machinery for the expression of middle and late genes. Therefore, we hypothesized that P23-45 may encode transcription factors to alter T. th. HB8 RNAP promoter specificity and activity. To identify such proteins, we affinity isolated T. th. HB8 RNAP genomically-tagged with a protein A (4PrA) tag from T. th. HB8 cells infected with P23-45 and identified RNAP co-isolating proteins in the sample using the MudPIT technique 21 (Fig. 8). As a control, analysis of proteins affinity isolated from P23-45-infected wild-type T. th. HB8 cells (untagged RNAP) was performed. Proteins present in both the T. th. HB8/P23-45-infected (untagged) and the T. th. HB8 RNAP-4PrA/P23-45-infected (tagged) samples were filtered out of the data set obtained for the RNAP-4PrA tagged strain.
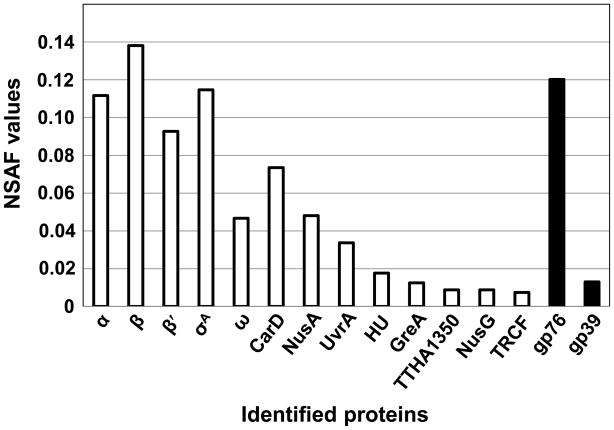
Figure 8. MudPIT analysis of proteins that co-isolate with T. th. HB8 RNAP affinity isolated from P23-45 infected cells.
Normalized Spectral Abundance Factor (NSAF) values for RNAP subunits and co-isolated T. th. HB8- and P23-45-encoded proteins are shown. T. th. HB8-encoded proteins are shown as white colored bars and P23-45-encoded proteins are shown as black colored bars.
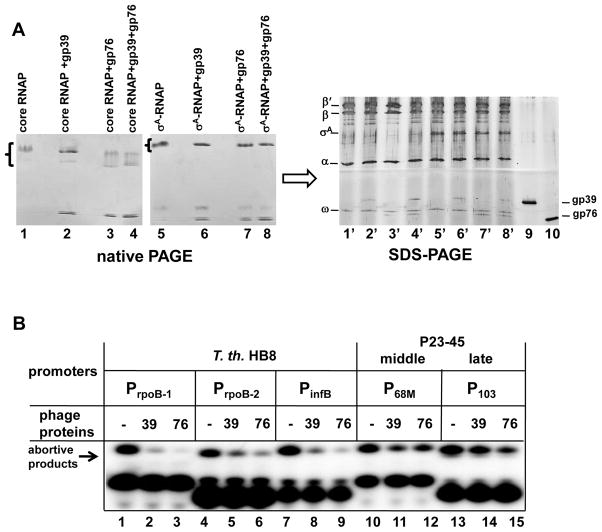
Analysis of the material affinity isolated from the T. th. HB8 RNAP-4PrA/P23-45-infected cells revealed the presence of the core RNAP subunits (αββ′ω) and the primary σ factor, σA. The RNAP subunits and σA were present at stoichiometric levels, as estimated by normalized spectral counts (Fig. 8), and another protein present at stoichiometric levels was CarD (TTHA0168). In a recent study, it was demonstrated that T. th. HB8 CarD could interact with the N-terminus of the RNAP β subunit in a bacterial two-hybrid assay 22, 23. Several known transcription elongation and anti-termination factors (NusA, NusG, and GreA), the transcription-repair coupling factor (TRCF), the nucleoid associated protein HU and the exonuclease ABC subunit A (UvrA) were also detected, although these proteins were present at lower levels. An uncharacterized protein, TTHA1350, was also identified; although TTHA1350 was detected at low levels, this result indicates that TTHA1350 may play a role in the T. th. HB8 transcription cycle. In addition to the host bacterial proteins, two P23-45-encoded proteins: gp39 (16.2 kDa; detected at a level substoichiometric to the core RNAP subunits) and gp76 (5.8 kDa; detected at a level stoichiometric to the core RNAP subunits) were identified (Fig. 8). To corroborate the affinity isolation results, the DNA encoding these two proteins was cloned into an E. co. pET28-derived expression vector 24 and recombinant gp39 and gp76 proteins were purified to homogeneity and assayed for their ability to bind to host T. th HB8 RNAP core and σA-associated holoenzymes (Fig. 9A). The phage proteins were incubated with T. th. RNAP core or σA-holoenzymes and the mixtures were resolved by native gradient PAGE (Fig. 9A, left panel). The results of subsequent denaturing SDS-PAGE indicated that gp39 and gp76 can interact with both the core and the σA-holoenzymes (Fig. 9A, for gp39 lanes 2 and 6, and lanes 2′ and 6′; for gp76 lanes 3 and 7, and lanes 3′ and 7′). Thus, we conclude that gp39 and gp76 are able to bind T. th. HB8 RNAP in vivo and in vitro. As can be seen from Figure 9, they can interact with RNAP simultaneously (Fig. 9A, lanes 4 and 8, and lanes 4′ and 8′) but do not interact with each other (ZB and LM, unpublished results), suggesting that they bind to distinct sites on RNAP. To our knowledge, this is the first documentation of thermophage-encoded thermophilic host bacterial RNAP-binding proteins. Gp39 (a middle phage protein) and gp76 (an early phage protein) have no recognizable conserved motifs or similarities with other proteins in the public databases 15. Thus, they are novel bacterial RNAP-binding proteins.
Figure 9. The P23-45 phage proteins gp39 and gp76 bind to T. th. HB8 RNAP and inhibit transcription initiation from host promoters but not from middle and late phage promoters.
A. Gp39 and gp76 bind to both core RNAP and RNAP-σA holoenzyme. The proteins alone or together were incubated with RNAP core (lanes 2–4) or with RNAP-σA holoenzyme (lanes 6–8) and analyzed by native gradient PAGE. Next, the native gels were stained with Coomassie stain, and the bands due to proteins (indicated by brackets in Fig. 9A) were excised from the gels and their composition were determined by denaturing SDS-PAGE (lanes 1′-8′). Gp39 and gp76 were loaded as markers (lanes 9 and 10, respectively).
B. The results of abortive transcription initiation by T. th. HB8 RNAP-σA holoenzyme at several representative host T. th. HB8 (lanes 1–9) and middle/late phage (lanes 10–15) promoters in the presence or absence of either gp39 or gp76 are shown.
In order to elucidate the possible role(s) of gp39 and gp76 in P23-45 infection, we tested their ability to influence transcription by the host RNAP. In vitro abortive transcription initiation reactions using DNA fragments containing three different T. th. HB8 σA-dependent promoters (PrpoB-1, PrpoB-2, and PinfB) and two P23-45 promoters, a middle promoter (P68M) and a late promoter (P103), were performed in the presence or in the absence of either gp39 or gp76 (Fig. 9B). As can be seen, both proteins efficiently inhibited transcription from the host bacterial promoters that belong to the −10/−35 promoter class (Fig. 9B, lanes 1–9). When added together, gp39 and gp76 demonstrated a strong additive effect in transcription inhibition at these promoters (data not shown). In contrast, both gp39 and gp76 were much less efficient at inhibiting transcription from the phage middle and late promoters that belong to the extended −10 class of promoters that lack the −35 promoter element (Fig. 9B, lanes 10–15, see also P23-45 middle and late promoter alignment in Fig. 3B). Thus, the binding of gp39 and gp76 to T. th. HB8 RNAP leads to promoter-specific transcription inhibition. Both proteins were purified as polyhistidine-tagged versions. To check if the tags may introduce non-physiological activities, we compared his-tagged and untagged (with his tag removed by thrombin) proteins and found that the tag does not interfere with their RNAP binding and transcription inhibition activities in vitro. Our data suggests that gp39 and gp76 may be responsible for host transcription shut-off during P23-45 infection and may likely act by interfering with −35 promoter element-RNAP interactions. However, the molecular mechanisms underlying host transcription inhibition remain to be fully elucidated.
Discussion
In this work, we investigated the transcription strategy of P23-45, a lytic thermophilic siphovirus infecting the thermophilic eubacterium T. th. HB8. To study the P23-45 gene expression pattern, we used a combination of bioinformatic and biochemical methods, an approach that was successfully used by us to study host and viral gene expression during infection of T. th. HB8 by an unrelated thermophage, a large myovirus φYS40 8. In the case of P23-45, macroarray analysis and in vivo primer extension revealed that i) host bacterial transcription is shut-off during the P23-45 infection cycle and ii) three temporal classes of viral genes: early, middle, and late, exist. Most of the known phages do not encode their own RNAP for expression of early genes and must therefore rely on host RNAP to transcribe their genes. Early promoters of such phages tend to be very strong, with a good match to the host promoter consensus, to successfully compete with host promoters for host RNAP at the onset of infection. Initially, automated bioinformatic analysis of the P23-45 genome did not reveal a recognizable RNAP gene 15; leading us to hypothesize that P23-45 early promoters are similar to host promoters and are recognized by the host T. th. RNAP-σA holoenzyme. Contrary to this expectation, no early P23-45 promoters with homology to host σA-dependent promoters were also identified. Instead, we determined that many early phage genes are preceded by a common sequence motif. This highly conserved 11 bp motif has a consensus sequence 5′-TTATTCcTTTA-3′, with the highest conservation at positions −9 (T), −8 (A), −7 (T), −6 (T), and −2 (T) relative to the experimentally determined 5′ end of the transcript. In 13 of the 15 early P23-45 promoters, the 11 bp motif is located immediately upstream of the annotated translation start codon. Thus, many early phage transcripts appear to be leaderless. A similar situation was observed during the analysis of late and middle transcripts of the φYS40 thermophage 8.
Comparison of putative P23-45 early promoters with the T. th. −10/−35 consensus promoter elements used in bioinformatic searches indicated that they are clearly different (Fig. 4, compare logos). Thus, it was not at all surprising that T. th. RNAP-σA holoenzyme did not recognize these sequences as promoters in vitro, or in the absence of P23-45 infection in vivo. Nevertheless, DNA fragments containing the 11 bp motifs, when positioned on a T. th. plasmid, led to the appearance of “correctly” initiated RNAs that behaved as early transcripts. We take these results as a strong indication that the 11 bp conserved sequences define early phage promoters. Experiments conducted in the presence of host RNAP inhibitor Rif clearly show that early P23-45 transcription is resistant to Rif, while middle and late transcription, which is carried out by T. th. RNAP, is Rif-sensitive. The data suggest that P23-45 encodes a Rif-resistant RNAP that transcribes its early genes from promoters defined by the 11 bp consensus motif.
Given that biochemical data strongly suggest the existence of P23-45-encoded RNAP, a more thorough bioinformatic analysis of the P23-45 genome was undertaken. All known RNAPs are divided into two unrelated families based on sequence, structure, and subunit composition. One family includes large multisubunit RNAPs of bacteria, archaea, and eukaryotes 25; 26; 27. The other family consists of small single-subunit RNAPs related to phage T7 RNAP and found in some bacteriophages, in mitochondria, and in chloroplasts 28; 29. The principal enzymatic activities are performed in both families of RNAPs via the same two-metal catalytic mechanism. All multisubunit RNAPs share a universal metal-binding signature motif: NADFDGD, in their largest subunits that together with additional conserved domains forms the active site. All T7-related RNAPs have other catalytic motifs and conserved domains/amino acids involved in the active center formation. In principle, it would seem more likely that P23-45 would encode a single-subunit RNAP that is present in many other bacteriophages. However, despite careful searches, we did not observe identify any sequence similarity to single-subunit RNAPs in the P23-45 genome. In contrast, a BLASTP search 30 with the ORF64 sequence used as the query retrieved as the best hit (after the closely related ortholog from bacteriophage P74-26) the A subunit of RNAP I from the stramenopile Thalassiosira pseudonana (T. ps.)
The detected region of similarity included a 65 amino acid segment which aligned with 32% amino acid identity and 47% similarity; the similarity was not statistically significant (expect value of 1.4). However, it was notable that the alignment encompassed a portion of the catalytic double-psi beta-barrel (DPBB) domain of RNAP, the most conserved portion of both the β′ and β subunits of all multisubunit RNAPs that is directly involved in nucleotide polymerization 26; 27; 31. Moreover, the RNAP amino acid signature that includes the three invariant and essential aspartates required for the coordination of the two Mg2+ ions that participate in catalysis was fully conserved in ORF64 ([NA]AD[FY]DGD, the magnesium-chelated aspartates are underlined). When the ORF64 sequence was combined with the T. ps. RNAP sequence to generate a position-specific scoring matrix (PSSM), the second iteration of the PSI-BLAST search 30 readily retrieved numerous RNAP sequences. Using the PHI-BLAST program 32, we found that, when the ORF64 sequence was compared to the non-redundant protein sequence database at the NCBI under the additional requirement that the signature Mg2+-binding motif was matched, the only retrieved sequences were those of RNAP subunits. Using the HHPred program 33, we found that the ORF64 sequence produced the best hit with the DPBB domain of the T. th. RNAP β′ subunit, with the E-value 0.091 when the Interpro collection of HMMs was searched. When the HHPred search was initiated with the isolated sequence of the predicted DPBB domain of ORF64, the same best hit was obtained, with a statistically significant E-value of 0.0031.
A multiple alignment of the putative DPBB domain of ORF64 and a representative set of eukaryotic, archaeal and bacterial RNAPs is shown in Figure 10. The alignment suggests that ORF64 contains a short version of the DPBB domain similar to the highly diverged β′ homologs of RNAPs from baculoviruses and fungal mitochondrial plasmids 31. The DPBB domain consists of six β-strands 27; 31. In addition to the (NA)D(FY)DGD signature motif, ORF64 retains several other invariant and essential amino acid residues of the DPBB domain such as the arginine at the end of S3 (corresponding to β′ R704 in T. aq. RNAP), and the proline and leucine in the loop downstream of S3 (β′ P706 and L708, respectively, in T. aq. RNAP) (Fig. 10).
Figure 10.
Alignment of the double-psi beta-barrel (DPBB) domains of ORF64 from P23-45 and ORF62 from closely related P74-26 and β′ orthologs from selected multisubunit RNAPs. The alignment was generated using the MUSCLE program 41, and the N-terminal part was manually modified on the basis of secondary structure predictions. The range of amino acid residues of the aligned segments in the respective protein precedes each sequence. Inserts in RNAP subunit sequences are shown as the numbers of amino acids. The residues conserved in all aligned sequences are denoted by asterisks; similar residues are denoted by colons. The most conserved segments of the alignment including the NADFDGD motif are shown as grey boxes. The predicted secondary structure of ORF64 (consensus of the PredictProtein and JPred predictions) is shown above the alignment. The secondary structure from two crystal structures, those of the RNAPs of the archaeon Sulfolobus shibatae (PDB code 2WAQ 44) and the bacterium T. aq. (2GHO 45) are shown underneath the alignment along with the consensus structure of the DPBB domain 31. In the secondary structure lines, e stands for extended conformation (β-strand) and h stands for α-helix; the S1–6 numbering is from 31. The organism name abbreviations: Thaps, Thalassiosira pseudonana; Phyin, Phytophthora infestans; Strpu, Strongylocentrotus purpuratus; Homsa, Homo sapiens; Micpu, Micromonas pusilla; Oryco, Oryza coarctata; Sulsh, Sulfolobus shibatae; Metsp, Methanocaldococcus sp.; Bacsu, Bacillus subtilis; Theaq, Thermus aquaticus.
We propose that ORF64 is a distant member of the multisubunit family of RNAPs. However, the putative P23-45 RNAP contains no counterparts to several other essential residues of the β′ RNAP subunit, suggesting that this enzyme could be mechanistically distinct from the known RNAPs. In our previous work highly sensitive mass spectrometric analysis of pure P23-45 virions has not detected ORF64 15. More recently, Western blot analysis revealed no ORF64 traces among P23-45 virion proteins using anti ORF64 polyclonal antibodies (Minakhin et al., unpublished data). It remains to be determined whether ORF64 functions on its own or requires additional host or phage-encoded factors for activity.
Macroarray and in vivo primer extension analyses also revealed P23-45 middle and late genes and the corresponding P23-45 promoters upstream of these genes; at present we cannot distinguish between the middle and the late promoters based on their consensus element sequences. Similarly to φYS40, the P23-45 middle/late promoters are characterized by a −10 consensus element supplemented with a “TG/TGTG” motif (Fig. 3B). Middle and late promoters of P23-45 are recognized by unmodified T. th. HB8 RNAP in vitro; as these promoters are inactive early in infection, it follows that their activity is somehow repressed until later stages of the infection cycle.
A combined view that emerges from our work thus indicates that the P23-45 transcription strategy is a simplified version of the transcription strategy employed E. coli phage N4. In the case of N4 infection, early genes are transcribed by the Rif-resistant phage-encoded RNAP that is encapsulated in the virion and injected into the infected cell with the N4 genome 20; 29. The middle genes are transcribed by another phage-encoded RNAP, a product of early N4 genes 34. Late N4 genes are transcribed by phage-modified host RNAP. In the case of P23-45, early genes are likely transcribed by the Rif-resistant RNAP encoded by ORF64, while the middle and late genes are transcribed by host RNAP. Previous analysis did not reveal the presence of ORF64 in P23-45 virions 15. This could have been caused by a low copy number of the putative phage RNAP in the virion. Alternatively, ORF64 may be initially transcribed by the host RNAP from P68, an upstream early promoter that contains an 11 bp motif embedded into a host RNAP-σA promoter (see alignment in Fig. 4, the −10/−35 elements are underlined). These possibilities are currently being investigated in our laboratory.
Using one-step affinity isolation of host T. th. HB8 RNAP and subsequent mass spectrometric analysis (MudPIT) of the affinity isolated sample, we identified two P23-45-encoded T. th. HB8 RNAP-binding proteins: gp76 and gp39. To the best of our knowledge, this is the first documentation of thermophage-encoded thermophilic bacterial RNAP-binding proteins. Gp76 and gp39 are encoded by early and middle P23-45 genes, respectively, and both proteins efficiently inhibit in vitro transcription by host RNAP from host promoters, but are less effective at inhibiting transcription from P23-45 middle and late promoters (Fig. 9B). Taken together, these data suggest that gp76 and gp39 may be involved in the shut-off of host transcription during the P23-45 infection program. Further biochemical and structural analysis of these proteins, in complex with T. th. RNAP, should make it possible to obtain a structure-based model of the action of a phage-encoded transcription regulator.
Experimental Procedures
Bacterial growth and phage infection
Bacteriophage P23-45 was generously provided by Dr. Michael Slater, Promega Corporation (Madison, WI). To isolate individual P23-45 plaques, 150 μL of a freshly grown T. th. HB8 culture (OD600~0.4), grown in the TB medium (0.8% [w/v] tryptone, 0.4% [w/v] yeast extract, 0.3% [w/v] NaCl, 1 mM MgCl2, and 0.5 mM CaCl2), was combined with a 100 μL dilution of phage stock, incubated for 10 minutes at 65°C, plated in soft TB agar (0.75 % [w/v]), and incubated overnight at 65°C. To prepare a phage stock suspension, an individual plaque was picked and subjected to two more rounds of plaque purification. The phage lysate was prepared using a previously described procedure 15.
E. coli strains XL-1Blue (New England Biolabs) and BL21(DE3) (Novagen) were used for molecular cloning and recombinant protein expression, respectively.
Total DNA purification and molecular cloning
P23-45 genomic DNA was extracted with a Lambda Midi kit (Qiagen) using the procedure recommended by the manufacturer. T. th. HB8 genomic DNA was purified by extraction with phenol-chloroform and subsequent precipitation with ethanol.
Plasmids encoding either polyhistidine-tagged gp39 or gp76 for recombinant protein production and purification were constructed as follows: the DNA encoding gp39 or gp76 was PCR-amplified using primers that appended NdeI and EcoRI sites at the 5′ and 3′ ends of each gene, respectively. The resultant PCR products were cleaved with NdeI and EcoRI and cloned between the NdeI and EcoRI sites of a pET28a-based plasmid, creating pSKB2-39HIS and pSKB2-76HIS. The plasmid pMKE77-80 was constructed as follows: a 504 bp DNA fragment of the P23-45 genome comprising the divergent P77 and P80 promoters together with the proximal parts of ORF77 and ORF80 coding regions was PCR-amplified using primers that appended NdeI and HindIII sites at the 5′ and 3′ end, respectively. The resultant PCR fragment was digested with NdeI and HindIII and cloned between the NdeI and HindIII sites of the E. coli – T. th.-E. coli shuttle plasmid pMKE1 18. The resultant plasmid, pMKE77-80, was used in in vivo primer extension experiments.
Strain construction and affinity isolation of Protein A-tagged T. th. HB8 RNAP
A similar strategy to that used to construct a T. th. HB8 strain encoding a polyhistidine tag ([His]10) appended to the 3′ end of the rpoC gene (encoding the RNAP β′ subunit) was used to construct a T. th. HB8 strain encoding a genomic β′-Protein A (4PrA) fusion protein 8. The resultant strain, T. th. HB8rpoC::4PrA, demonstrated a slightly slower growth rate compared to wild-type cells, but was infected with P23-45 as efficiently as wild-type.
To prepare P23-45-infected biomass, wild-type T. th. HB8 or T. th. HB8rpoC::4PrA cells were grown at 65 °C in 4 L of TB medium until OD600 ~ 0.35 and were infected with P23-45 at a multiplicity of infection of 10. Infection was ceased 20 minutes post-infection by rapidly cooling the samples in an ice water bath. Cells were harvested by centrifugation and washed once with ice-cold 10% (v/v) glycerol. Next, 1 mL of lysis buffer (20 mM Hepes [pH 7.5], 0.2 mg/mL phenylmethylsulfonyl fluoride (PMSF), 4 mg/mL pepstatin) was added to every 10 g of cell pellet, and the cells were frozen in liquid nitrogen. Both tagged and untagged T. th. HB8 cells were cryogenically lysed using the MM 301 Mixer Mill (Retsch; 24) and stored at −80°C until use. Affinity isolation of RNAP-4PrA and co-isolating proteins from P23-45 infected T. th. HB8 cells was performed as described for E. coli RNAP and co-isolating proteins 24; 35.
MudPIT analysis
Elution of T.th. HB8 RNAP-4PrA and co-isolating proteins from the IgG conjugated Dynabeads (Invitrogen) was achieved with 0.5 M ammonium hydroxide and 0.5 mM ethylenediaminetetraacetic acid (EDTA). The eluted proteins were frozen in liquid nitrogen and evaporated to dryness in a SpeedVac (Thermo Savant). The dried protein pellets were denatured, reduced, alkylated, and digested with endoproteinase LysC (Roche Applied Science) followed by digestion with trypsin (Promega). The peptide mixtures were pressure-loaded onto triphasic microcapillary columns, installed in-line with a Quaternary Agilent 1100 series HPLC pump coupled to a Deca-XP ion trap tandem mass spectrometer (ThermoElectron) and analyzed via ten-step chromatography 36. The MS/MS data sets were searched using SEQUEST 37 against a database of 117 P23-45 predicted gene products, combined with 2238 protein sequences from T. th. HB8 as described previously 15. Lists of detected proteins were established and compared using DTASelect/CONTRAST 38 as described previously 15. Protein levels across different samples were compared using Normalized Spectral Abundance Factor (NSAF) values 39; 40.
Proteins
Polyhistidine-tagged T. th. HB8 core RNAP and σA were used in native gel electrophoresis protein-protein interaction and in vitro transcription assays. The proteins were purified essentially as described in 8. Either recombinant polyhistidine-tagged gp39 or gp76 were produced as follows; the expression plasmids either pSKB2-39HIS or pSKB-76HIS were transformed into E. coli BL21 (DE3) cells, and transformants were selected in the presence of 50 μg/mL kanamycin. Cultures (4 L) were grown at 37 °C to an OD600 ~ 0.8 and recombinant protein overexpression was induced with 1 mM IPTG for 4 hours at 37 °C. Cells containing overexpressed recombinant proteins were harvested by centrifugation, and disrupted by sonication in buffer A (10 mM Tris-HCl [pH 8.0], 500 mM NaCl, 5 mM imidazole [pH 8.0], 5% glycerol, 0.2 mg/mL PMSF). Inclusion bodies were dissolved in buffer B (10 mM Tris-HCl [pH 8.0], 500 mM NaCl, 2 mM imidazole [pH 8.0], 7 M urea); loaded onto a 5 mL nickel-chelated Hi-Trap sepharose column (GE Healthcare) equilibrated in buffer B, and the column was washed with buffer B supplemented with 25 mM imidazole. The bound proteins, either gp39 or gp76, were eluted from the column with buffer B supplemented with 200 mM imidazole, and dialyzed against buffer C (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.5 mM EDTA). The proteins were loaded onto a MonoQ column (GE Healthcare) equilibrated in TGE buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 5% [v/v] glycerol), eluted with a linear gradient of NaCl from 150 mM to 450 mM, dialyzed against buffer D (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 50% [v/v] glycerol) and stored at −80°C.
Macroarray membrane preparation and data analysis
DNA fragments corresponding to each of the selected P23-45 ORFs, T. th. HB8 housekeeping genes and the D. me. zfrp8 gene (control) were PCR-amplified from the corresponding genomic DNA using gene-specific primer pairs (the sequences of the primers are available from the authors upon request). Membrane preparation, cDNA synthesis, and macroarray hybridization were performed as described 7. After hybridization, the amount of radioactivity from each spot was quantified using the ImageQuant software (Molecular Dynamics) and the background signal was subtracted from signals corresponding to every ORF spot. To allow comparison between the signals on different membranes, the background-corrected signals were normalized relative to the average of the two D. me. zfrp8 spot signals; the normalized spot signals were used for data analysis.
Primer extension
Primer extension reactions were performed essentially as described in our previous work 8. Exponential phase T. th. HB8 cells were infected with P23-45 and harvested at the same time points post-infection as for the macroarray experiments. In experiments utilizing Rif, Rif (Sigma-Aldrich) was added to T. th. HB8 cells infected with P23-45 at the designated time points to yield a final concentration of 2 mg/mL followed by 10 minutes incubation prior to RNA extraction. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s procedure. For each primer extension reaction, 10 μg of total RNA was reverse-transcribed with 100 units of SuperScript III enzyme from the First-Strand Synthesis kit for RT-PCR (Invitrogen) in the presence of 10 pmol of γ-32P end-labeled primer. The reactions were treated with RNase H, precipitated with ethanol and dissolved in formamide loading buffer. To identify the 5′ ends of the primer extension products, DNA sequencing reactions, accomplished using the fmol DNA Cycle Sequencing kit (Promega), containing both the correspoding PCR-amplified P23-45 genome fragments and end-labeled primers used for the primer extension reaction were performed. The reaction products were resolved on 6–8 % (w/v) polyacrylamide sequencing gels and visualized using a PhosphorImager (Molecular Dynamics).
Protein complex analysis
Either T. th. HB8 core RNAP or σA-holoenzyme (reconstituted with 1 μM core RNAP and 1 μM σA) was incubated with either gp39 (~ 5 μM) or gp76 (~ 5 μM) in 10 μL of transcription buffer (30 mM Tris-HCl [pH 7.9], 40 mM KCl, 10 mM MgCl2, 2 mM β–mercaptoethanol) for 10 minutes at 65°C. Subsequently, 4 μL of the reaction mixture was resolved on a native 4–15% (w/v) Phast gradient polyacrylamide gel (GE Healthcare); bands due to proteins were visualized by Coomassie blue staining. To interrogate the protein composition of the bands resolved by native gel electrophoresis, the bands were excised from the native gel and placed into the wells of a gradient 12–16% (w/v) gradient polyacylamide denaturing SDS gel, followed by electrophoresis and staining with silver.
In vitro transcription
A typical abortive transcription reaction was perfomed in a final volume of 10 μL and contained 200 nM of T. th. HB8 σA-holoenzyme and between 20–40 nM of a PCR-amplified DNA fragment containing either a T. th. HB8 or a P23-45 promoter in standard transcription buffer (30 mM Tris-HCl [pH 7.9], 40 mM KCl, 10 mM MgCl2, 2 mM β–mercaptoethanol). Reactions (where indicated) were supplemented with either gp39 or gp76 (15 μM), incubated for 10 minutes at 65 °C, followed by the addition of various RNA dinucleotides (100–500 μM), [α-32P] NTPs (3000 Ci/mmol) and the corresponding cold NTPs (100 μM). The reactions were incubated for a further 10 minutes at 65 °C prior to being terminated by the addition of an equal volume of urea-formamide loading buffer. The reaction products were resolved on a 20% (w/v) polyacrylamide denaturing gel and visualized using a PhosphorImager.
Sequence analysis
The sequences of the predicted proteins encoded in the P23-45 genome were searched against the non-redundant protein sequence database at the NCBI using the iterative PSI-BLAST 30 and the pattern-hit-initiated BLAST (PHI-BLAST) 32 programs searches with P23-45 deduced ORFs. Additional searches were performed using the HHPred program that implements pairwise comparison of hidden Markov models 33. The multiple alignment of the putative DPBB domains of ORF64 from P23-45, ORF62 from P74-26, and a representative set of multisubunit RNAPs was constructed using the MUSCLE program 41 Results of secondary structure prediction made using the PredictProtein 42 and JPred 43 programs were taken into consideration to manually refine the alignment.
Acknowledgments
Bacteriophage P23-45 was generously provided by Dr. Michael Slater from Promega Corporation. We thank Dr. E. Peter Geiduschek for critical reading of the manuscript and helpful advice. This work was supported by NIH grant R21 AI074769 (to L.M.), by NIH grant R01 GM61898 (to S.A.D.), by NIH grants RR00862 and RR022220 (to B.T.C.), by the Stowers Institute for Medical Research (L.F. and M.P.W.) by NIH grant R01 GM59295 (to K.S.), by Molecular and Cell Biology grant from the Presidium of Russian Academy of Sciences, Russian Foundation for Basis Research grant 07-04-00366-a, and by a National Center for Biotechnology of the Republic of Kazakhstan grant (to K.S.); E.V.K. is supported by the US Department of Health and Human Resources (National Library of Medicine, National Institutes of Health).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–22. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 2.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–7. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–82. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 5.Ventura M, Foley S, Bruttin A, Chennoufi SC, Canchaya C, Brussow H. Transcription mapping as a tool in phage genomics: the case of the temperate Streptococcus thermophilus phage Sfi21. Virology. 2002;296:62–76. doi: 10.1006/viro.2001.1331. [DOI] [PubMed] [Google Scholar]
- 6.Semenova E, Djordjevic M, Shraiman B, Severinov K. The tale of two RNA polymerases: transcription profiling and gene expression strategy of bacteriophage Xp10. Mol Microbiol. 2005;55:764–77. doi: 10.1111/j.1365-2958.2004.04442.x. [DOI] [PubMed] [Google Scholar]
- 7.Minakhin L, Semenova E, Liu J, Vasilov A, Severinova E, Gabisonia T, Inman R, Mushegian A, Severinov K. Genome sequence and gene expression of Bacillus anthracis bacteriophage Fah. J Mol Biol. 2005;354:1–15. doi: 10.1016/j.jmb.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 8.Sevostyanova A, Djordjevic M, Kuznedelov K, Naryshkina T, Gelfand MS, Severinov K, Minakhin L. Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage phiYS40. J Mol Biol. 2007;366:420–35. doi: 10.1016/j.jmb.2006.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–24. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 10.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–4. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 11.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–90. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 12.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–9. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 13.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 14.Naryshkina T, Liu J, Florens L, Swanson SK, Pavlov AR, Pavlova NV, Inman R, Minakhin L, Kozyavkin SA, Washburn M, Mushegian A, Severinov K. Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J Mol Biol. 2006;364:667–77. doi: 10.1016/j.jmb.2006.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minakhin L, Goel M, Berdygulova Z, Ramanculov E, Florens L, Glazko G, Karamychev VN, Slesarev AI, Kozyavkin SA, Khromov I, Ackermann HW, Washburn M, Mushegian A, Severinov K. Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23–45 and P74–26: siphoviruses with triplex-forming sequences and the longest known tails. J Mol Biol. 2008;378:468–80. doi: 10.1016/j.jmb.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jalasvuori M, Jaatinen ST, Laurinavicius S, Ahola-Iivarinen E, Kalkkinen N, Bamford DH, Bamford JK. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J Virol. 2009;83:9388–97. doi: 10.1128/JVI.00869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulose-Murphy M, Ha NK, Xiang C, Chen Y, Gillim L, Yarchoan R, Meltzer P, Bittner M, Trent J, Zeichner S. Transcription program of human herpesvirus 8 (kaposi’s sarcoma-associated herpesvirus) J Virol. 2001;75:4843–53. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno R, Zafra O, Cava F, Berenguer J. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid. 2003;49:2–8. doi: 10.1016/s0147-619x(02)00146-4. [DOI] [PubMed] [Google Scholar]
- 19.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–12. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 20.Falco SC, Zehring W, Rothman-Denes LB. DNA-dependent RNA polymerase from bacteriophage N4 virions. Purification and characterization. J Biol Chem. 1980;255:4339–47. [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–59. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, Darst SA. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq692. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savalia D, Westblade LF, Goel M, Florens L, Kemp P, Akulenko N, Pavlova O, Padovan JC, Chait BT, Washburn MP, Ackermann HW, Mushegian A, Gabisonia T, Molineux I, Severinov K. Genomic and proteomic analysis of phiEco32, a novel Escherichia coli bacteriophage. J Mol Biol. 2008;377:774–89. doi: 10.1016/j.jmb.2007.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severinov K. RNA polymerase structure-function: insights into points of transcriptional regulation. Curr Opin Microbiol. 2000;3:118–25. doi: 10.1016/s1369-5274(00)00062-x. [DOI] [PubMed] [Google Scholar]
- 26.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 2009;395:671–85. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. J Mol Biol. 2009;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, Cedergren R. On the evolution of the single-subunit RNA polymerases. J Mol Evol. 1997;45:671–81. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierczak KM, Davydova EK, Mustaev AA, Rothman-Denes LB. The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. EMBO J. 2002;21:5815–23. doi: 10.1093/emboj/cdf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer LM, KEV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and Eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Structural Biology. 2003;3:1–23. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Schaffer AA, Miller W, Madden TL, Lipman DJ, Koonin EV, Altschul SF. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–90. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willis SH, Kazmierczak KM, Carter RH, Rothman-Denes LB. N4 RNA polymerase II, a heterodimeric RNA polymerase with homology to the single-subunit family of RNA polymerases. J Bacteriol. 2002;184:4952–61. doi: 10.1128/JB.184.18.4952-4961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westblade LF, Minakhin L, Kuznedelov K, Tackett AJ, Chang EJ, Mooney RA, Vvedenskaya I, Wang QJ, Fenyo D, Rout MP, Landick R, Chait BT, Severinov K, Darst SA. Rapid isolation and identification of bacteriophage T4-encoded modifications of Escherichia coli RNA polymerase: a generic method to study bacteriophage/host interactions. J Proteome Res. 2008;7:1244–50. doi: 10.1021/pr070451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–75. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 37.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 38.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 40.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–33. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–3. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 44.Korkhin Y, Unligil UM, Littlefield O, Nelson PJ, Stuart DI, Sigler PB, Bell SD, Abrescia NG. Evolution of Complex RNA Polymerases: The Complete Archaeal RNA Polymerase Structure. PLoS Biol. 2009;7:e102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuznedelov K, Lamour V, Patikoglou G, Chlenov M, Darst SA, Severinov K. Recombinant Thermus aquaticus RNA polymerase for structural studies. J Mol Biol. 2006;359:110–21. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]