FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages
The transcriptional regulator FoxO1 promotes inflammation by regulating TLR4-mediated signalling in macrophages. TLR4 activation as well as insulin signalling inactivates FoxO1, thereby limiting the inflammatory response.
Keywords: ChIP-Seq, FoxO1, macrophage, Tlr4, transactivation
Abstract
The macrophage-mediated inflammatory response is a key etiologic component of obesity-related tissue inflammation and insulin resistance. The transcriptional factor FoxO1 is a key regulator of cell metabolism, cell cycle and cell death. Its activity is tightly regulated by the phosphoinositide-3-kinase-AKT (PI3K-Akt) pathway, which leads to phosphorylation, cytoplasmic retention and inactivation of FoxO1. Here, we show that FoxO1 promotes inflammation by enhancing Tlr4-mediated signalling in mature macrophages. By means of chromatin immunoprecipitation (ChIP) combined with massively parallel sequencing (ChIP-Seq), we show that FoxO1 binds to multiple enhancer-like elements within the Tlr4 gene itself, as well as to sites in a number of Tlr4 signalling pathway genes. While FoxO1 potentiates Tlr4 signalling, activation of the latter induces AKT and subsequently inactivates FoxO1, establishing a self-limiting mechanism of inflammation. Given the central role of macrophage Tlr4 in transducing extrinsic proinflammatory signals, the novel functions for FoxO1 in macrophages as a transcriptional regulator of the Tlr4 gene and its inflammatory pathway, highlights FoxO1 as a key molecular adaptor integrating inflammatory responses in the context of obesity and insulin resistance.
Introduction
It is well established that chronic activation of proinflammatory pathways within insulin target cells can lead to insulin resistance. Recent studies, including those from our group, have further suggested that macrophages are the most prominent immune cells underlying the proinflammatory tissue response associated with obesity and type 2 diabetes mellitus (Patsouris et al, 2008; Schenk et al, 2008). In both humans and rodents, adipose tissue macrophages (ATMs) accumulate in adipose tissue (AT) with increasing body weight, and ATMs are a significant contributor to inflammation and insulin resistance in obesity (Kanda et al, 2006; Weisberg et al, 2006; Nomiyama et al, 2007; de Luca and Olefsky, 2008).
The search for proinflammatory signalling cascades activated in obesity and insulin resistance has recently focused on the Toll-like receptors (TLRs), a group of pattern recognition receptors that activate host defenses in response to microbial-derived ligands. These receptors are exemplified by Tlr4, which is activated by Gram-negative bacteria lipopolysaccharide (LPS) (Chow et al, 1999). Tlr4 may also function as a sensor for saturated fatty acids (SFAs), which are increased in obesity (Lee et al, 2001, 2003; Fessler et al, 2009). After binding ligands, TLRs use a downstream cascade of signalling molecules, including adaptor proteins, such as myeloid differentiation factor 88 (MyD88), and Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF), that ultimately regulate the activities of signal-dependent transcription factors, such as NFκB and interferon regulatory factor 3 (IRF3). These in turn induce the transcription of genes that encode cytokines, chemokines, and other effectors of the innate immune response. Dietary SFAs have been implicated in promoting the metabolic syndrome and atherosclerosis. While still under debate (Erridge and Samani, 2009), several independent laboratories have demonstrated that SFAs can activate Tlr4 (Fessler et al, 2009). SFAs-mediated activation of inflammation is prevented in macrophages and adipocytes that are deficient in Tlr2, Tlr4 or Tlr2/Tlr4 (Shi et al, 2006; Kim et al, 2007; Nguyen et al, 2007). Our recent study further indicated that Tlr4 signalling in haematopoietic-derived cells is essential for the development of hepatic and AT insulin resistance in obese mice (Saberi et al, 2009).
FoxO1, also known as FKHR, together with two other mammalian isoforms (FoxO3 and FoxO4), constitute the FoxO subfamily of the forkhead transcription factor family, a large array of transcription factors characterized by the presence of a conserved 110-amino acid winged helix DNA-binding domain (DBD) (Kops and Burgering, 1999). FoxO subfamily members have important functions in a wide range of cellular processes, such as DNA repair, cell cycle control, stress resistance, apoptosis and metabolism (Nakae et al, 2000; Barthel et al, 2005; Furukawa-Hibi et al, 2005). Phosphatidylinositol-3-kinase (PI3K)/Akt signalling, which is activated by insulin and certain cytokines and growth factors, phosphorylates each of the FoxO proteins at three different Ser/Thr residues (Nakae et al, 2000). The phosphorylated FoxO proteins are exported from the nucleus, and become sequestered in the cytoplasm, where they interact with 14-3-3 protein. FoxO1 is the most abundant FoxO isoform in insulin-responsive tissues including liver, fat and pancreas, and is negatively regulated by the insulin-induced PI3K-Akt signalling cascade. Impaired insulin signalling to FoxO1 is one mechanism for the metabolic abnormalities of type 2 diabetes.
FoxO1 has been shown to suppress adipogenesis at an early phase of the adipocyte differentiation program (Nakae et al, 2003). In addition, we and others have demonstrated that FoxO1 has important functions in mature adipose cells/tissue as well (Dowell et al, 2003; Fan et al, 2009; Kim et al, 2009). FoxO1 haploinsufficient mice are partially protected from high fat diet-induced insulin resistance and diabetes (Nakae et al, 2003; Kim et al, 2009). Moreover, we have proposed a potential mechanism to explain this effect by showing FoxO1-mediated transrepression of adipocyte PPARγ via direct protein–protein interactions (Fan et al, 2009; Kim et al, 2009).
Several recent studies have proposed a role for FoxO1 in hematopoietic and immune cells (Fabre et al, 2005; Arden, 2007; Tothova et al, 2007). For example, FoxO1 has been reported to stimulate expression of the proinflammatory cytokine IL-1β in macrophages (Su et al, 2009). FoxO1 has also been shown to regulate apoptosis in dendritic cells (Riol-Blanco et al, 2009).
In the present study, we applied the technique of chromatin immunoprecipitation (ChIP) combined with massively parallel sequencing (ChIP-Seq), and show that FoxO1 regulates the inflammatory response of mature macrophages by directly transactivating Tlr4 gene expression. While FoxO1 potentiates Tlr4 signalling, activation of the latter induces AKT phosphorylation and subsequently inactivates FoxO1, establishing a self-limiting mechanism of inflammation. These findings, coupled with our previous discovery of FoxO1 as an insulin-regulatable PPARγ transrepressor in mature adipocytes (Fan et al, 2009), suggest cell type specific functions of FoxO1 in macrophages and adipocytes that integrate the physiological effects of insulin and inflammatory signalling pathways within AT.
Results
FoxO1 enhances Tlr4 signalling in macrophages
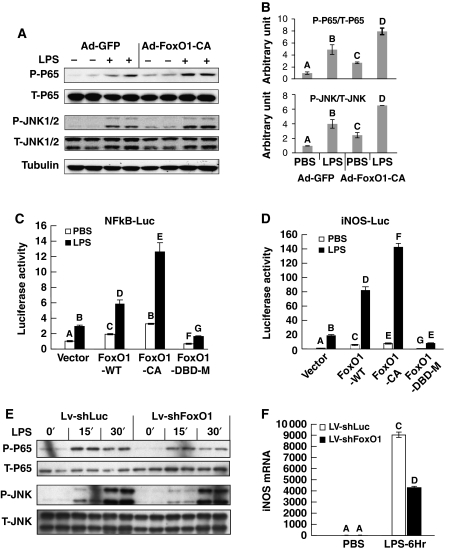
Chronic inflammation, characterized by increased macrophage infiltration and activation in AT, is an important mechanism underlying insulin resistance associated with obesity and diabetes (Schenk et al, 2008; de Luca and Olefsky, 2008; Olefsky and Glass, 2010). The monocyte/macrophage RAW264.7 cell line provides a convenient model to study macrophage function. We found that ectopic expression of constitutively active (CA) FoxO1 in RAW264.7 cells potentiates the effect of the Tlr4-specific ligand LPS to induce phosphorylation of NFκB (p65) and JNK1/2 (Figure 1A, quantitated in Figure 1B). This phenomenon was confirmed in elicited mouse peritoneal macrophages (Supplementary Figure S1A). We then introduced luciferase reporters, driven by either an NFκB response element (NFκB-Luc) or the native iNOS promoter (iNOS-luc) into RAW264.7 cells. Our results show that LPS-induced activation of these promoters was potentiated by wild-type (WT) FoxO1 and even more so by CA FoxO1, but not by the transactivationally incompetent DBD FoxO1 mutant (Figure 1C and D), demonstrating that the transactivational activity of FoxO1 is required for the proinflammatory effect of FoxO1. FFAs can also signal through Tlr4, and Figure 2G shows that FoxO1 potentiates FFA-induced iNOS promoter activity in RAW264.7 macrophages. Next, we depleted FoxO1 by transducing RAW264.7 cells with a lentivirus encoding shRNA against FoxO1 gene. This led to an ∼90% knockdown of endogenous FoxO1m (Supplementary Figure S2), and attenuated LPS-stimulated NFκB and JNK1/2 activation (Figure 1E), as well as iNOS expression (Figure 1F).
Figure 1.
FoxO1 enhances Tlr4 signalling in macrophages. (A) RAW264.7 cells were infected with either Ad-GFP or Ad-FoxO1-CA at 100 MOI. After 48 h, cells were exposed to 100 ng/ml LPS for 30 min and then lysed for immunoblotting with the indicated antibodies. (B) Relative amount of phosphorylated versus total protein levels for P65 and JNK were quantitated by NIH-Image J; and the results are shown as average±s.d. (C, D) RAW264.7 cells were co-transfected with FoxO1-WT, FoxO1-CA or DBD mutant FoxO1 plasmid vectors together with NFκB-Luc (C) or iNOS-Luc (D), and then exposed to 100 ng/ml LPS or PBS control for 6 h before luciferase assay. (E) RAW264.7 cells were stably transduced with lentivirus encoding shRNA against luciferase (control) or FoxO1, then exposed to 100 ng/ml LPS for 15 and 30 min, and subsequently lysed for immunoblotting assays with indicated antibodies. (F) RAW264.7 cells were stably transduced with lentivirus encoding shRNA against luciferase or FoxO1 and then exposed to 100 ng/ml LPS for 6 h. iNOS mRNA expressions levels were assayed by real-time PCR. Data are presented as the average±s.d. Letters above the bars show statistical groups (ANOVA, P<0.05).
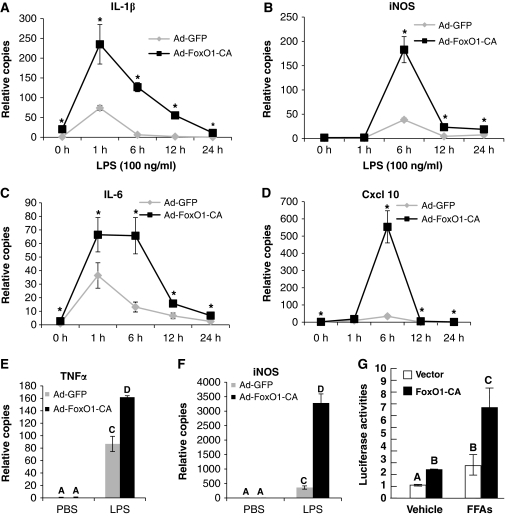
Figure 2.
FoxO1 enhances Tlr4 target genes expression in macrophages. (A–D) RAW264.7 cells were infected with Ad-GFP or Ad-FoxO1-CA at 100 MOI; 48 h posted infection, cells were exposed to 100 ng/ml LPS for 0–24 h as indicated. mRNA expressions of indicated inflammatory genes were quantitated by real-time quantitative PCR. Asterisks indicate statistically significant difference at each corresponding time point. (E, F) Elicited mouse peritoneal macrophages were infected as described above. Cells were then exposed to 100 ng/ml LPS for 6 h and mRNA expressions of TNFα (E) and iNOS (F) were quantitated by real-time PCR. (G) RAW264.7 cells were co-transfected with FoxO1-CA or control vector together with iNOS-Luc and TK-β-Gal, and were then exposed to 250 μM FFAs or vehicle in 0.1% BSA DMEM (LG) medium for 24 h before luciferase assay. Data are presented as the average±s.d. Letters above bars show statistical groups (ANOVA, P<0.05).
The LPS-induced increase in endogenous mRNA expression of inflammatory genes, such as iNOS, was also enhanced by WT and CA FoxO1 in RAW264.7 cells, and Figure 2A–D show the dynamic changes in several of these mRNAs in the presence or absence of CA-FoxO1 expression. Of note, expression of CA-FoxO1 exerted gene-specific effects on Tlr4 target genes, in some cases primarily affecting the magnitude of response (e.g. iNOS, Cxcl10), and in other cases affecting both the magnitude and duration of response (e.g. IL-1β and IL-6). FoxO1 therefore differentially modulates Tlr4 signalling. The FoxO1 enhancement of LPS-induced expression of various inflammatory genes in RAW 264.7 cells was also quantitated by ArrayPlate mRNA Assay (Supplementary Figure S1B). Figure 2E (iNOS) and F (TNFα) show that CA-FoxO1 produced comparable effects in elicited primary peritoneal macrophages.
LPS-induced inflammatory responses and glucose tolerance are reduced in FoxO1 haploinsufficient mice
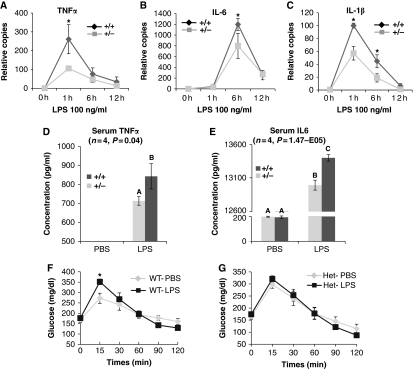
As an initial step to determine how our in vitro findings in Figures 1 and 2 translate to the in vivo setting, we studied primary bone marrow-derived macrophages (BMDMs) before and after stimulation with the Tlr4 ligand, LPS. As homozygous FoxO1 deletion is lethal, we prepared BMDMs from WT and FoxO1 haploinsufficient mice. As shown in Figure 3A–C, LPS-induced mRNA expression of TNFα (Figure 3A), IL-6 (Figure 3B) and IL-1β (Figure 3C) were all attenuated in primary macrophages from FoxO1+/− mice. LPS-induced phosphorylation of JNK and p65 were also decreased in the FoxO1+/− cells (Supplementary Figure S3).
Figure 3.
Effect of FoxO1 on Tlr4 signalling in vivo. (A–C) BMDMs derived from FoxO1+/− or WT mice were overnight starved in 0.1% BSA-containing medium, and treated with 100 ng/ml LPS. Cells were subsequently harvested at the indicated time points. mRNA expression of TNFα (A), IL-6 (B) and IL-1β (C) were quantitated. Asterisks indicate statistical significant difference at each corresponding time point. (D, E) Circulating plasma levels of TNFα (D) and IL-6 (E) 3 h post-LPS injection were measured. Data are presented as the average±s.d. Letters above the bars show statistical groups (ANOVA, P<0.05). (F, G) Following 6 h fast, WT (F) and FoxO1+/− (G) mice were injected with LPS (1 mg/kg) or vehicle 1 h before i.p. injection of glucose (1 mg/kg). Blood glucose levels were measured during a 2-h GTT. Asterisk indicates a significantly higher peak glucose level at 15 min in WT mice (n=4, P=0.039).
To further examine the role of FoxO1 in Tlr4-mediated inflammatory responses and insulin resistance in vivo, we administered a non-lethal dose of LPS (1 mg/kg, i.p.) to stimulate inflammatory pathways in FoxO1+/− and WT mice. Three hours after LPS injection, we measured circulating TNFα and IL-6 (Figure 3D and E), and both were lower in FoxO1+/− mice compared with WT mice. As LPS-induced acute inflammation leads to glucose intolerance (Arkan et al, 2005), we also performed glucose tolerance tests (GTTs), starting 1 h post-LPS injection. As shown in Figure 3F, LPS administration led to augmented hyperglycemia in WT mice, as indicated by a significantly elevated blood glucose level at 15′. In contrast, this LPS effect was not observed in FoxO1+/− mice, demonstrating a blunted response to the LPS challenge. Taken together, these data indicate that endogenous FoxO1 is required for full LPS-mediated Tlr4 proinflammatory signalling in vivo. As induction of inflammatory cytokines from myeloid cells is the mechanism for acute LPS-induced glucose intolerance (Arkan et al, 2005), and expression of cytokines such as TNFα, as well as the LPS receptor Tlr4, is mostly from macrophages in both AT (Weisberg et al, 2003; Oh et al, 2010) and liver (Supplementary Figure S4), we infer that protection from the LPS-induced glucose intolerance in FoxO1+/− mice is macrophage related.
FoxO1 and PPARγ transrepression
Recent evidence indicates that the anti-inflammatory effects of TZDs contribute to the insulin-sensitizing properties of this class of drugs (Ghisletti et al, 2007; Hevener et al, 2007). Ligand stimulation of the PPARγ nuclear receptor attenuates macrophage Tlr4 signalling by a transrepressional mechanism in which Tlr4-induced NFκB activity is blocked by PPARγ-mediated stabilization of NcoR on promoters of inflammatory pathway genes (Pascual et al, 2005; Ghisletti et al, 2007). Given that FoxO1 interferes with the transactivational function of PPARγ in adipocytes (Fan et al, 2009), it was important to determine whether the transrepressive properties of PPARγ are also inhibited by FoxO1, as this could be an alternative mechanism by which FoxO1 enhances macrophage Tlr4 signalling. In RAW264.7 cells co-transfected with an iNOS-Luc reporter and pcDNA-PPARγ, Rosiglitazone pretreatment inhibited LPS-induced promoter activity by 27% (Supplementary Figure S5A). As expected, LPS-induced iNOS promoter activity was enhanced by WT FoxO1, and repressed by Rosiglitazone. Importantly, FoxO1 did not inhibit the ability of Rosiglitazone to repress the iNOS promoter (Supplementary Figure S5A). Additionally, the FoxO1-DBD mutant (Supplementary Figure S5B), which is transactivationally inactive, but competent at binding to and transrepressing PPARγ (Fan et al, 2009), did not affect Rosiglitazone-induced suppression of the iNOS promoter. These data indicate that antagonism of PPARγ transrepression does not account for proinflammatory effect of FoxO1 in macrophages.
Mechanism of FoxO1-mediated inflammation: global assessment of FoxO1 DNA-binding sites
To investigate the mechanisms of FoxO1 potentiation of Tlr4 signalling, we implemented and validated a biotin-tagging approach that enabled ChIP coupled to massively parallel sequencing (ChIP-Seq) (Supplementary Figures S6–S9). To provide sufficient enrichment required for ChIP-Seq, FoxO1 was fused at the N-terminus to the biotin ligase recognition peptide (BLRP) to facilitate biotinylation by the Escherichia coli biotin ligase BirA (Supplementary Figures S6 and S8). Tagging with BLRP did not alter FoxO1 transactivation or transrepression function (Supplementary Figure S7). RAW264.7 cell lines that expressed both BLRP-FoxO1 and BirA (‘double stable' clones) were established, and stable clones that expressed BLRP-FoxO1 at equivalent levels to the endogenous protein were selected (Supplementary Figure S9A). Cells were first subjected to overnight serum starvation and then to formaldehyde crosslinking. Chromatin was then fragmented and subjected to high affinity purification on Streptavidin magnetic beads. The bead complex was then subjected to TEV proteolysis to release the tagged FoxO1 protein and DNA adducts from the matrix, leaving behind any other biotinylated or non-specific proteins that were still tethered to the beads (Supplementary Figure S9B). Q–PCR was applied to confirm a high enrichment of known FoxO1 target gene sequences (p27 gene) over background (Supplementary Figure S9C). The DNA was then amplified and utilized to generate libraries for sequencing. Parallel sequencing was performed using the Illumina Genome Analyzer, and short reads (24 bp) were mapped to the mouse reference genome. Identical reads were combined, and peaks were defined using standard software packages robust statistical cutoffs (Valouev et al, 2008). Secondary analysis included computational motif discovery to identify enriched sequence elements in genomic-binding sites (Ogawa et al, 2005).
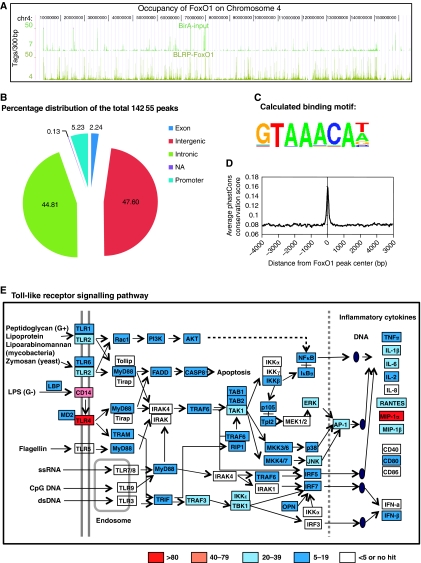
Figure 4A shows browser tracks from BirA cells (blank control) and BirA–BLRP-FoxO1 ChIP-Seq peaks on chromosome 4, illustrating high signal-to-noise ratios. FoxO1-binding location analysis revealed a large number of interaction sites. A total of 14 255 putative-binding sites (peaks) were identified, including 44.81% in intronic regions, 47.60% in intergenic regions, 5.23% in promoter regions and 2.24% in exon regions (Figure 4B). Figure 4C depicts the results of enriched motif analysis of the FoxO1 sites using TRANSFAC and JASPAR position weight matrices. The calculated DNA-binding motif (GTAAACAT/A) is very similar to published FoxO1 DNA-binding sequences ((A/C)(C/A)AAA(T/C)A).
Figure 4.
Genome-wide localization of FoxO1 in RAW264.7 macrophage cells and FoxO1 occupancy on genes involved in Tlr signalling. (A) Browser tracks of BirA cells (blank control) and BirA–BLRP-FoxO1 ChIP-Seq peaks in chromosome 4. Note the high signal-to-noise ratios. (B–D) Location analysis of FoxO1-binding sites (peaks). (B) FoxO1-binding regions were mapped relative to their nearest RefSeq genes. The promoter region was defined as <1 kb segment upstream from the transcription start site (TSS). (C) Web logo of consensus motif position weight matricies (PWMs) generated by a de novo motif search of FoxO-binding sites. (D) Sequences of FoxO1-binding sites are highly conserved among different vertebrate species. (E) Gene Ontology analysis (using Kyoto Encyclopedia of Genes and Genomes pathways (KEGG)) of the genes located near FoxO1-binding regions (peaks) has identified Toll-like receptor signalling as one of the most-highly enriched pathways (P=2.62E−08; Bonferroni cut point: 8.47E−04). Genes have been coloured according to the intensity (height) of the dominant FoxO1 peak on each corresponding gene.
Figure 4D shows a measure of evolutionary conservation of global FoxO1-binding sequences in 17 vertebrates, including mammalian, amphibian, bird and fish species, based on a phylogenetic hidden Markov model, phastCons (Siepel et al, 2005). Conservation scores rise sharply when they approach the centres of FoxO1-binding sites such that sequences in the centres of FoxO1-binding sites are ultraconserved among different vertebrate species.
Role of FoxO1 in Tlr4 signalling
The functional significance of DNA binding by FoxO1 was then assessed by examining the genes located near FoxO1-binding regions (peaks). For each FoxO1-binding peak, the nearest gene was determined, as well as the distance from the transcription start site (TSS) to the centre of the peak. Gene Ontology analysis (using Kyoto Encyclopedia of Genes and Genomes pathways (KEGG)) of the closest genes identified by this approach revealed a total of 31 significantly enriched pathways (Table I). Among these, TLR signalling was one of the most-highly enriched pathways (P=2.62E−08; Bonferroni cut point: 8.47E−04, Table I). Figure 4E depicts the occupancy of FoxO1 on genes in the KEGG TLR signalling pathway. Genes have been coloured according to the intensity (height) of the dominant FoxO1 peak on each corresponding gene. Clearly, a number of genes in this pathway are coordinately targeted by FoxO1.
Table 1. The functional significance of DNA binding by FoxO1 was assessed by examining the genes located near FoxO1-binding regions (peaks).
| No. of pathway | GO | Term | P-value | Bonferroni cut point | No. of genes in term | No. of target genes in term | No. of target genes | No. of total genes |
|---|---|---|---|---|---|---|---|---|
| 1 | mmu:04010 | MAPK signalling pathway | 2.29E−14 | 3.55E−04 | 258 | 141 | 1634 | 5061 |
| 2 | mmu:05220 | Chronic myeloid leukaemia | 1.87E−10 | 9.80E−04 | 75 | 51 | 1634 | 5061 |
| 3 | mmu:04620 | Toll-like receptor signalling pathway | 2.62E−08 | 8.47E−04 | 100 | 59 | 1634 | 5061 |
| 4 | mmu:05214 | Glioma | 8.03E−08 | 1.22E−03 | 63 | 41 | 1634 | 5061 |
| 5 | mmu:04510 | Focal adhesion | 1.34E−07 | 5.26E−04 | 189 | 95 | 1634 | 5061 |
| 6 | mmu:04210 | Apoptosis | 2.09E−07 | 1.00E−03 | 84 | 50 | 1634 | 5061 |
| 7 | mmu:04662 | B-cell receptor signalling pathway | 5.77E−07 | 1.25E−03 | 64 | 40 | 1634 | 5061 |
| 8 | mmu:05211 | Renal cell carcinoma | 4.66E−06 | 1.22E−03 | 70 | 41 | 1634 | 5061 |
| 9 | mmu:04660 | T-cell receptor signalling pathway | 4.90E−06 | 9.80E−04 | 93 | 51 | 1634 | 5061 |
| 10 | mmu:04370 | VEGF signalling pathway | 7.62E−06 | 1.22E−03 | 71 | 41 | 1634 | 5061 |
| 11 | mmu:05222 | Small cell lung cancer | 8.54E−06 | 1.06E−03 | 85 | 47 | 1634 | 5061 |
| 12 | mmu:04012 | ErbB signalling pathway | 8.54E−06 | 1.06E−03 | 85 | 47 | 1634 | 5061 |
| 13 | mmu:05212 | Pancreatic cancer | 1.22E−05 | 1.22E−03 | 72 | 41 | 1634 | 5061 |
| 14 | mmu:04670 | Leukocyte transendothelial migration | 1.41E−05 | 8.77E−04 | 110 | 57 | 1634 | 5061 |
| 15 | mmu:04060 | Cytokine–cytokine receptor interaction | 1.99E−05 | 4.55E−04 | 246 | 110 | 1634 | 5061 |
| 16 | mmu:05210 | Colorectal cancer | 2.26E−05 | 1.09E−03 | 85 | 46 | 1634 | 5061 |
| 17 | mmu:04664 | Fc epsilon RI signalling pathway | 2.61E−05 | 1.19E−03 | 76 | 42 | 1634 | 5061 |
| 18 | mmu:04630 | Jak-STAT signalling pathway | 3.97E−05 | 6.85E−04 | 153 | 73 | 1634 | 5061 |
| 19 | mmu:05223 | Non-small cell lung cancer | 7.04E−05 | 1.61E−03 | 53 | 31 | 1634 | 5061 |
| 20 | mmu:04115 | p53 signalling pathway | 0.00014 | 1.39E−03 | 66 | 36 | 1634 | 5061 |
| 21 | mmu:05215 | Prostate cancer | 0.000145 | 1.09E−03 | 90 | 46 | 1634 | 5061 |
| 22 | mmu:04912 | GnRH signalling pathway | 0.00015 | 1.04E−03 | 95 | 48 | 1634 | 5061 |
| 23 | mmu:05221 | Acute myeloid leukaemia | 0.000159 | 1.56E−03 | 57 | 32 | 1634 | 5061 |
| 24 | mmu:04810 | Regulation of actin cytoskeleton | 0.000252 | 5.56E−04 | 205 | 90 | 1634 | 5061 |
| 25 | mmu:04910 | Insulin signalling pathway | 0.000291 | 7.94E−04 | 135 | 63 | 1634 | 5061 |
| 26 | mmu:05218 | Melanoma | 0.000388 | 1.35E−03 | 71 | 37 | 1634 | 5061 |
| 27 | mmu:04520 | Adherens junction | 0.000476 | 1.32E−03 | 74 | 38 | 1634 | 5061 |
| 28 | mmu:00530 | Aminosugars metabolism | 0.000817 | 2.63E−03 | 31 | 19 | 1634 | 5061 |
| 29 | mmu:04930 | Type II diabetes mellitus | 0.001015 | 2.00E−03 | 45 | 25 | 1634 | 5061 |
| 30 | mmu:00640 | Propanoate metabolism | 0.001589 | 2.78E−03 | 30 | 18 | 1634 | 5061 |
| 31 | mmu:04150 | mTOR signalling pathway | 0.001715 | 1.85E−03 | 51 | 27 | 1634 | 5061 |
| For each FoxO1-binding peak, the nearest gene was determined, as well as the distance from the transcription start site to the centre of the peak. Gene Ontology analysis (using Kyoto Encyclopedia of Genes and Genomes pathways (KEGG)) of the closest genes revealed a total of 31 significantly enriched pathways (Bonferroni cut point less than the P-value). Shown are Gene Ontology (GO) ID, term name, P-value, Bonferroni cut point, number of target genes in term, number of total target genes, number of genes in term and number of total genes. | ||||||||
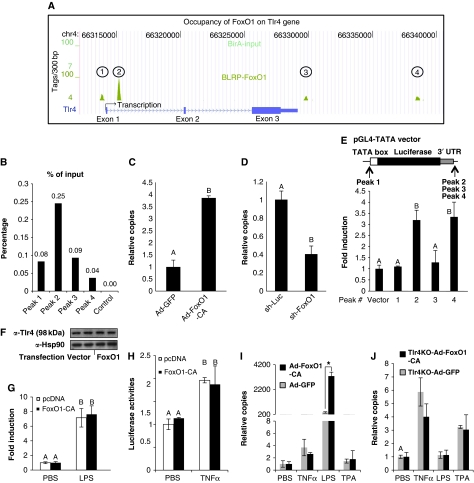
Tlr4 showed the most abundant FoxO1 signature and as Tlr4 is obviously the most proximal component of this signalling pathway, we focused our experiments on this gene. Four FoxO1 peaks were identified either within or close to the Tlr4 gene (Figure 5A). One was located in the promoter region, another within the first intron, and two in the intergenic region distal to the 3′ UTR. To validate these ChIP-Seq peaks, FoxO1 enrichment was assayed by ChIP-qPCR (quantitative PCR), and all four peaks were shown to be positive (data not shown). As additional validation, ChIP-qPCR was performed to quantify copy number of each peak relative to input genomic DNA. As shown in Figure 5B, DNA fragment abundance of the four binding sites was well correlated with the height of each peak as revealed by ChIP-Seq, whereas the control fragment (a non-related DNA region) showed no enrichment.
Figure 5.
FoxO1 regulates Tlr4 gene transcription. (A) Localization of FoxO1-binding sites on the mouse Tlr4 gene. The three exons, two introns, 3′ UTR and TSS of the mouse Tlr4 gene on chromosome 4 are shown. The four FoxO1-occupaying sites (peaks) are indicated. The height of each peak represents the number of tags per 300 bp of genomic DNA. The blank control (BirA cells track) is also shown. (B) Relative copy numbers of DNA segments of each of the four peaks and a non-related control chromatin region to input genomic DNA were quantitated by real-time PCR. (C, D) Tlr4 mRNA expression in RAW264.7 cells transduced with either Ad-FoxO1-CA (C, 100 MOI × 48 h) or Lv-shRNA (D) against FoxO1 was assayed by real-time PCR. (E) DNA fragments of each FoxO1 peak were cloned and put into the pGL4-TATA vector at the indicated positions. FoxO1-CA was co-transfected with each of the four resultant constructs into RAW264.7 cells to assay luciferase induction as a measure of transcriptional responsiveness. (F, G) An artificial Tlr4 signalling cascade was set up in HEK293 cells by co-expression of cDNAs for Tlr4, CD14 and MD2. The Tlr4 cDNA is driven by the CMV promoter and is therefore unregulatable by FoxO1. Responses of NFκB-luc to LPS stimulation were assayed in the presence and absence of FoxO1 co-expression. (H) RAW264.7 cells were co-transfected with FoxO1-CA or vector together with NFκB-Luc, and then exposed to 20 ng/ml TNFα for 6 h before luciferase assay. (I, J) WT (I) or Tlr4KO (J) primary peritoneal macrophages were infected with either Ad-GFP or Ad-FoxO1-CA at 100 MOI. After 48 h, cells were exposed to LPS (100 ng/ml), TNFα (20 ng/ml), TPA (100 ng/ml) or PBS for 6 h. iNOS mRNA was quantitated. Data are presented as the average±s.d. Letters above the bars show statistical groups (C–H) and asterisk (I) indicates statistical significant difference (ANOVA, P<0.05).
We demonstrated further that the occupancy of FoxO1 on the Tlr4 gene is functional by showing that Tlr4 mRNA expression was upregulated by ectopic expression of CA-FoxO1 in RAW264.7 cells (Figure 5C). Likewise, siRNA-mediated knockdown of FoxO1 led to decreased Tlr4 expression, indicating that endogenous levels of FoxO1 are necessary to maintain normal Tlr4 levels (Figure 5D). To further determine the functional relevance of each of the four individual FoxO1-binding sites, DNA fragments (around 500 bp) encompassing the centre of each FoxO1 peak were generated by PCR amplification of genomic DNA, and were cloned into pGL4-TATA (pGL4.10 containing a minimal TATA box in the 5′ region of the luciferase gene). Based on their location in the Tlr4 gene, peak 1 was placed in the 5′ promoter region of the TATA-luciferase cassette, and the other three in the 3′ region distal to the 3′ UTR of the luciferase gene (Figure 5E). The resultant four constructs, along with an empty pGL4-TATA vector were transiently transfected into RAW264.7 cells, and the effects of CA-FoxO1 were then examined. The ability of CA-FoxO1 to enhance luciferase expression for each construct is shown in Figure 5E. As seen, binding sites 2 and 4 confer transcriptional responsiveness to the TATA promoters, suggestive of functional relevance. The results were replicated when the DNA fragments for peak 2 and 4 were placed in the reverse orientation (data not shown). In addition, the two binding site constructs did not have response to transactivationally incompetent DBD FoxO1 (data not shown). These results suggest that these two sites are functional intronic/intergenic transcriptional enhancers of FoxO1 gene.
We then determined whether upregulation of Tlr4 gene expression accounts for the FoxO1 enhancement of LPS-induced inflammatory signalling. To do this, we took advantage of the fact that HEK293 cells do not express endogenous Tlr4 and generated an artificial Tlr4 signalling cascade in these cells by transfecting them with cDNAs for Tlr4 and two co-factors, CD14 and MD2. The resulting HEK293TCM cells demonstrated robust responses to LPS with a significant induction of inflammatory gene expression, whereas HEK293 cells were non-responsive to LPS (Supplementary Figure S10). Importantly, Tlr4 expression in HEK293TCM cells was not enhanced by exogenous FoxO1 expression (Figure 5F), as the Tlr4 cDNA was driven by the CMV promoter, which does not contain a FoxO1 response element. Finally, in this system, LPS stimulation of the NFκB-responsive promoter (NFκB-luc) was not enhanced by CA-FoxO1 (Figure 5G), showing that FoxO1 is unable to enhance the LPS-induced NFκB response pathway when Tlr4 gene expression is no longer regulated by FoxO1. It is possible that upregulation of Tlr4 is not the exclusive mechanism for FoxO1 modulation of Tlr4 action, as the KEGG analysis revealed that FoxO1-binding sites were located on the DNA of 58 genes downstream of Tlr signalling. Nevertheless, our data demonstrate that direct upregulation of the Tlr4 receptor is at least one major mechanism for FoxO1 potentiation of LPS-mediated inflammation in macrophages.
Selective effect of FoxO1 on Tlr4 versus TNFα signalling
We next tested whether FoxO1 can potentiate TNFα-induced proinflammatory signalling in macrophages. In either RAW264.7 (Figure 5H) or HEK293TCM (data not shown) cells, TNFα stimulation of the NFκB-responsive promoter (NFκB-luc) was not enhanced by FoxO1. It should be clarified that the use of NFκB-luc only assesses the impact of FoxO1 on signalling, and not on the regulation of genes that may contain FoxO1-binding sites in regulatory elements.
CA-FoxO1 also substantially enhanced LPS-induced expression of iNOS (Figure 5I), IL-6 (Supplementary Figure S11A) and MCP1 (Supplementary Figure S11C) in WT primary peritoneal macrophages, but CA-FoxO1 was without effect on TNFα- or TPA-induced inflammatory gene expression. In contrast, in Tlr4 knockout (Tlr4−/−) peritoneal macrophages, LPS was without effect (as expected) and the CA-FoxO1 had no effect to enhance LPS-, TNFα- or TPA-induced gene expressions (Figure 5J; Supplementary Figure S11B and D).
Finally, gene ontology analysis of the ChIP-Seq data indicated no enrichment of genes involved in TNFα signalling. As Tlr4 and TNFα cascades converge and share a significant number of common downstream effecter molecules, the lack of effect of FoxO1 on TNFα stimulation indicates that the locus of FoxO1 interaction is at an early step in Tlr4 signalling, consistent with FoxO1 potentiation of the Tlr4 inflammatory pathway through direct transactivation of the Tlr4 gene.
Feedback regulation of Tlr4 signalling on FoxO1
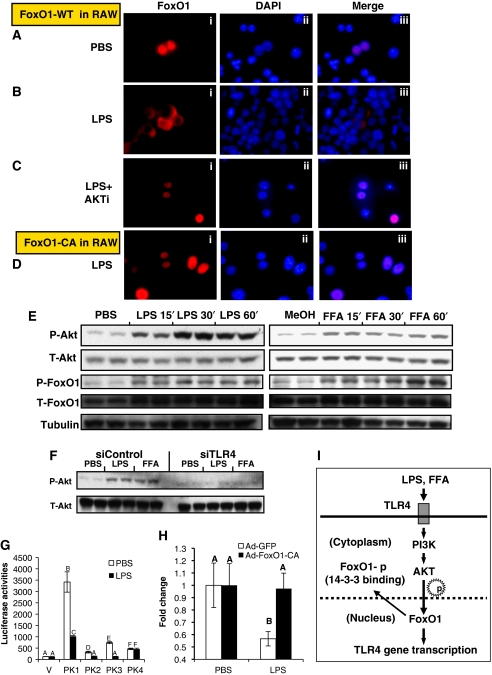
AKT-mediated phosphorylation of FoxO1 leads to nuclear exclusion, the key regulatory event controlling FoxO1 activity. We observed that FoxO1 was localized to the nucleus in overnight-starved RAW264.7 cells (Figure 6A), whereas LPS (Figure 6B) or FFA (Supplementary Figure S12A) treatment induced nuclear export. This effect was AKT dependent, as LPS or FFA treatments-induced AKT phosphorylation (Figure 6E), whereas, co-treatment with a specific AKT inhibitor (Logie et al, 2007) abolished the LPS (Figure 6C) or FFA (Supplementary Figure S12A) effect. The actions of LPS and FFA on AKT phosphorylation were coupled to FoxO1 phosphorylation (Figure 6E), and the effects of individual FFAs are shown in Supplementary Figure S12B and C. Furthermore, the AKT-resistant CA-FoxO1 remained in the nucleus, even in the presence of LPS treatment (Figure 6D). An identical pattern of FoxO1 subcellular localization was observed in peritoneal macrophages (Supplementary Figure S13). LPS-induced nuclear export of endogenous FoxO1 was also verified by immunostaining studies in RAW264.7 cells (Supplementary Figure S14). These results also show that Tlr4 is essential for both LPS- and FFA-induced AKT activation in RAW264.7 cells, as siRNA knockdown of endogenous Tlr4 largely eliminated the effects on AKT phosphorylation (Figure 6F).
Figure 6.
Feedback regulation of Tlr4 on FoxO1 in macrophages. (A–C) RAW264.7 cells were infected with Ad-FoxO1-WT. After 48 h, cells were starved overnight and subsequently exposed to PBS (A), 100 ng/ml LPS (B) or LPS+5 μM AKTi (C) for 30 min. Cells were then fixed for immunostaining with HA probe (153) antibody and DAPI DNA staining. (D) Ad-FoxO1-CA-infected RAW264.7 cells were treated with LPS and stained with HA probe antibody. (E) RAW264.7 cells were starved overnight in 0.1% BSA DMEM (LG) medium and subsequently exposed to 100 ng/ml LPS or 300 μM FFAs for 15′, 30′ and 60′. Immunoblottings were performed with the indicated antibodies. (F) After overnight starvation, RAW264.7 cells electroporated with control (siControl) or Tlr4 (siTlr4) siRNA were stimulated with 100 ng/ml LPS or 300 μM FFAs for 30′. AKT activation was assayed by immunoblotting. (G) pGL4-TATA empty vector and those vectors containing DNA fragment of each FoxO1 peak were transfected into RAW264.7 cells and luciferase activities were assayed 6 h post-treatment of 100 ng/ml LPS. (H) RAW264.7 cells were infected with either Ad-GFP or Ad-FoxO1-CA at 100 MOI. After 48 h, cells were exposed to LPS (100 ng/ml) for 6 h. Tlr4 mRNA was then assayed by real-time qPCR. Tlr4 mRNA levels at time 0 in each group were set to 1. (I) Proposed model for the FoxO1-mediated regulation of Tlr4 signalling in macrophages. FoxO1 enhances LPS or FFA-triggered Tlr4-mediated inflammatory signalling by direct activation of Tlr4 gene transcription activation. FoxO1 has four DNA-binding sites on the Tlr4 gene, and their occupancy is associated with increased Tlr4 gene expression. LPS or FFAs, intriguingly, inactivate macrophage FoxO1 by Tlr4-dependent induction of AKT, which triggers phosphorylation and nuclear exclusion of FoxO1. The Tlr4-AKT-FoxO1 axis provides a self-limiting mechanism by which macrophages avoid inappropriate long-term overactivation of inflammation after initiation of the inflammatory response, limiting this inflammatory burst may induce a state of sustained, low-grade inflammation.
It is possible that activation of AKT leading to inactivation of ‘proinflammatory' FoxO1 could represent a self-limiting feedback mechanism to prevent inappropriate long-term overactivation of inflammatory responses. Consistent with this idea, LPS treatment inhibited the enhancer activity of three of the four FoxO1 elements identified in the Tlr4 gene (Figure 6G). LPS treatment also reduced Tlr4 mRNA expression in RAW264.7 cells, and this reduction was prevented by CA-FoxO1, which is non-phosphorylatable by LPS stimulation of AKT (Figure 6H).
Anti-inflammatory effect of insulin
AKT1 is the exclusive isoform in macrophages (Supplementary Figure S15A), as opposed to AKT2, which is the dominant isoform in insulin target tissues, such as liver, AT and skeletal muscle (Garofalo et al, 2003). LPS-induced AKT phosphorylation was enhanced by CA-FoxO1 in macrophages (Supplementary Figure S15B), consistent with the finding that FoxO1 enhances Tlr4 signalling. As the Tlr4-AKT1-FoxO1 cascade represents a self-limiting mechanism for Tlr4 inflammatory signalling in macrophages, non-Tlr4-mediated activation of AKT should also attenuate Tlr4 inflammatory responses. Insulin is an important AKT activator in many tissues, and we therefore, determined whether a functional insulin signalling system exists in macrophages. As shown in Supplementary Figure S16A, phosphorylation of IRS1, IR and AKT were induced by insulin in serum-starved RAW264.7 macrophages. These insulin effects were associated with attenuated LPS responses, as shown by blunted LPS-induced inflammatory cytokine expression in cells pretreated with insulin (Supplementary Figure S16B–E). To assess whether FoxO1 is involved in the anti-inflammatory effect of insulin, BMDMs were prepared from FoxO1+/− and WT mice. Supplementary Figure S17A shows that pretreatment of BMDMs with insulin led to decreased IKK, and JNK activation by LPS in WT macrophages. Whereas, this effect was blurred in FoxO1+/− cells. These effects on Tlr4 signalling translated to inflammatory gene expression patterns. Thus, insulin pretreatment inhibited LPS-mediated expression of TNFα, IL-6, MCP1 and iNOS, whereas FoxO1 haploinsufficiency attenuated the effect of insulin (Supplementary Figure S17B–E).
We also observed that in ATMs from lean mice, endogenous FoxO1 is predominantly located in nuclei, whereas in ATMs from HFD/obese mice, endogenous FoxO1 is predominantly cytoplasmic (Supplementary Figure S18A and B). Interestingly, we have shown in previous studies that compared with lean mice, FoxO1 accumulates in nuclei of obese adipocytes due to impaired insulin action in these cells. Consistent with the current results showing that nuclear FoxO1 upregulates Tlr4, we now show that Tlr4 mRNA levels are higher in adipocytes from HFD compared with lean mice (Supplementary Figure S18C).
Discussion
Chronic low-grade tissue inflammation is recognized as a necessary contributor to insulin resistance associated with obesity and diabetes. In obese states, macrophages are recruited to AT where they secrete proinflammatory cytokines, which can induce insulin resistance in neighbouring adipocytes via paracrine effects (Schenk et al, 2008). Although FoxO1 proteins have been shown to have pivotal functions in the transcriptional cascades that control metabolism in liver, muscle, pancreas, brain and ATs (Nakae et al, 2002; Kamei et al, 2004; Fan et al, 2009; Kim et al, 2009), FoxO1 has not yet been shown to regulate macrophage function. In this paper, we demonstrate that FoxO1 promotes inflammatory signalling in macrophages and have used ChIP-Seq and other approaches to demonstrate that FoxO1 directly transactivates the Tlr4 gene.
The FoxO1 forkhead transcription factor serves as a critical negative regulator of insulin action. Insulin resistance in insulin-responsive tissues, such as liver, adipose and muscle, results in impaired phosphorylation and increased nuclear accumulation of FoxO1 (Nakae et al, 2008; Fan et al, 2009). In the present study, we used both gain- and loss-of-function approaches in RAW264.7 macrophages, thioglycolate-elicited peritoneal primary macrophages and BMDMs, to show that nuclear FoxO1 promotes inflammation in these cell types. In cultured macrophages, FoxO1 potentiates Tlr4 signalling, with increased LPS-induced phosphorylation of the Tlr4 downstream signalling proteins (JNK and NFκB), as well as increased gene expression of major inflammatory effector molecules, such as iNOS and TNFα. A functional DBD of FoxO1 is critical to this enhanced inflammatory response, demonstrating that the transactivational activity of FoxO1 is responsible for enhanced Tlr4 signalling. Furthermore, studies in primary macrophages from FoxO1+/− mice, as well as in vivo measurements of inflammation and glucose tolerance in these animals, indicate that the role of FoxO1 in macrophages to regulate Tlr4 signalling translates to the in vivo situation. Our recent report that FoxO1+/− mice are partially protected from HFD-induced insulin resistance are also consistent with this concept (Kim et al, 2009).
The requirement of a functional DBD indicates that the transactivational function of FoxO1 accounts for its Tlr4-enhancing effect. We therefore used ChIP-Seq analysis to identify genome-wide FoxO1-binding sites in macrophages. Consistent with ChIP-Seq studies of other transcriptional factors (Robertson et al, 2007; Visel et al, 2009), the majority of FoxO1-binding sites are located in either intronic or intergenic regions (44.8 and 47.60%, respectively) on the mouse genome, in contrast to only 5.23% of peaks that are located near promoter regions. This finding implies that FoxO1 might have distinct roles in both transcription initiation and as an enhancer.
Ontology analysis of genes closest to each of the FoxO1-binding sites showed that FoxO1 occupancy is significantly enriched on genomic DNA of genes involved in Tlr signalling, which ranked third by P-value in a total of 31 KEGG pathways that were identified as being significantly enriched. In particular, ChIP-Seq revealed four functional FoxO1-binding sites in the Tlr4 gene region. When cloned and assayed in vitro, DNA segments of two of the four binding sites conferred transcriptional responsiveness to an otherwise non-responsive TATA promoter to FoxO1. Subsequent functional analysis further confirmed Tlr4 as a direct target gene that is upregulated by ectopic expression of FoxO1, and downregulated by siRNA-mediated knockdown of FoxO1. We further demonstrated that FoxO1-induced upregulation of Tlr4 expression is necessary for this enhanced LPS-induced NFκB-responsive promoter activity, as this effect was absent in cells where Tlr4 expression is no longer regulated by FoxO1. Finally, FoxO1 does not augment TNFα signalling, indicating a relatively specific effect of FoxO1 on an early event in Tlr4 signalling. Together, these data support a novel mechanism in which the proinflammatory effects of FoxO1 in macrophages largely result from direct transactivation of Tlr4.
Our study shows that FoxO1 can enhance Tlr4 stimulation by SFAs. Interestingly, Tlr4 can function as a sensor for SFAs (Lee et al, 2001, 2003; Nguyen et al, 2007; Fessler et al, 2009), and the recent finding that CD36 can mediates Tlr4–6 heterodimerization provides an interesting new mechanism by which FFAs can activate Tlr4 signalling (Stewart et al, 2010).
Upregulation of Tlr4 is unlikely to be the exclusive mechanism for FoxO1 modulation of Tlr4 signalling. In addition to the Tlr4 gene, our ChIP-Seq results suggest other modalities whereby FoxO1 can regulate inflammatory gene expression. Thus, we have identified several other FoxO1 target genes within the Tlr4 signalling cascade, including IL-1β, Cxcl10, MIP1α, etc. For these genes, FoxO1 not only augments their responses to LPS by enhancing Tlr4 signalling, but can also be a direct transcriptional enhancer.
We also performed a motif analysis of the FoxO1 cistrome, which showed that FoxO1 DNA-binding sites were enriched for PPAR (P=1.167E−22) and Androgen receptor (P=3.497E−31) response elements, consistent with our previous findings that FoxO1 can form protein complexes with PPARγ (Fan et al, 2009) and AR (Fan et al, 2007). We also found that response motifs for other transcriptional factors, such as LXR, IRF and NFκB, were significantly enriched across the FoxO1 cistrome (see Supplementary Table I), suggesting potential interactions between FoxO1 and these proteins.
While this manuscript was being prepared, an involvement of FoxO protein in innate immune regulation has been reported in Drosophila, where FoxO was shown to directly induce anti-microbial peptides (AMPs), an important class of immune effector molecules (Becker et al, 2010). This is fully consistent with our present finding that FoxO1 enhances innate immunity in mammalian macrophages by regulating Tlr4 signalling.
Several post-translational modifications have been shown to alter FoxO1 activity (Yamagata et al, 2008). However, AKT-mediated phosphorylation of FoxO1 with subsequent nuclear export and degradation represents its most important functional regulatory mechanism. In RAW 264.7 macrophage cells, Akt is phosphorylated on serine 473 after stimulation of Tlr4 by LPS (Ojaniemi et al, 2003; Laird et al, 2009). Consistent with these findings, we observed that LPS-induced Tlr4 activation led to rapid phosphorylation and nuclear export of FoxO1 in macrophages. These effects were apparently dependent on AKT induction, as they were not seen with AKT-resistant mutant FoxO1, or in cells treated with an AKT inhibitor. Hence, while FoxO1 upregulates Tlr4-mediated inflammatory signalling, the Tlr4-PI3K-AKT pathway may in turn inactivate FoxO1 transactivation and limit the inflammatory response. Further, a negative feedback role for the PI3K-AKT pathway on Tlr4-mediated inflammation is strongly supported by two recent genetic studies (Luyendyk et al, 2008; Tsukamoto et al, 2008). Tlr4-mediated AKT activation and subsequent inactivation of FoxO1 may ultimately serve to avoid inappropriate overactivation of the innate immune response (action model depicted in Figure 6I). This is consistent with the downregulation of Tlr4 by LPS and with the time course studies (Figure 2) in which gene induction is extinguished at later time points, as well as the finding that this tapering effect is attenuated for selected genes (Figure 2A and C) in the presence of non-phosphorylatable CA FoxO1. An additional anti-inflammatory mechanism may involve mOTR (Schmitz et al, 2008; Weichhart et al, 2008). As mTOR is activated by a Tlr4-PI3K-AKT cascade, and activated mTOR can attenuate LPS responsiveness. These concepts led us to explore the potential anti-inflammatory role of insulin signalling to FoxO1 in macrophages. Interestingly, we find that the canonical insulin signalling pathway is readily measured in macrophages. This causes Akt activation, whereupon FoxO1 becomes phosphorylated and excluded from the nucleus where it is inactive. These events are coupled to insulin-mediated anti-inflammatory effects and our findings reveal that FoxO1 has a key mechanistic function in these anti-inflammatory actions of insulin.
Macrophages are a key cell type mediating chronic tissue inflammation, hepatic steatosis and insulin resistance. In this context, Tlr4 has emerged as a key transducing molecule, sensing extrinsic stimuli and converting them to cellular inflammatory responses. For example, depletion of Tlr4 from macrophages by adaptive transfer of Tlr4 KO bone marrow into WT mice, preventing obesity-induced tissue inflammation leading to amelioration of hepatic steatosis and insulin resistance (Saberi et al, 2009). Our current results highlight FoxO1 as a new transcriptional regulator of Tlr4 and its proinflammatory pathway.
Materials and methods
Materials
Mouse monocyte/macrophage RAW264.7 cells were cultured in Dulbecco's modified Eagle's medium (1 g/l glucose) supplemented with 10% low endotoxin FBS (Hyclone, Logan, UT). HEK293 cells and Rat-1 fibroblasts overexpressing WT human insulin receptors (HIRc-B cells) were maintained as previously reported (Fan et al, 2009). Peritoneal macrophage cells were harvested from C57Bl/6 male mice 72 h after intraperitoneal administration of 3% thioglycollate broth (Difco). Cells were then seeded onto tissue culture plates. After 2 h, non-adherent cells were removed by washing with PBS. Adherent peritoneal macrophages were then cultured in RPMI 1640 medium containing 10% low endotoxin FBS, supplemented with 2 mM L-glutamine (Invitrogen) and antibiotics (penicillin+streptomycin). Cells were cultured for 4 days before exposure to any treatment.
Rosiglitazone was provided by Pfizer, Inc. (La Jolla, CA). LPS, AKTi and free fatty acids (a cocktail FFA mixture and individual FFA) were obtained from Sigma-Aldrich. The following items were obtained commercially: insulin (Sigma-Aldrich); anti-FKHR (H128) antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-AKT (sc-1618, gives two bands, Santa Cruz Biotechnology), anti-Tlr4 (sc-10741, Santa Cruz Biotechnology); anti-FoxO1 (C29H4) rabbit mAb (Cell Signaling Technology); HA probe (153) (Santa Cruz Biotechnology) monoclonal; anti-NFκB p65 and anti-phospho-NFκB p65 (Ser536) antibodies, anti-SAPK/JNK and anti-phospho-SAPK/JNK (Thr183/Tyr185) antibodies, anti-AKT (cat. No. 9272, gives one band), anti-phospho-AMP kinase (Thr172) (cat. no. 2531), anti-AMP kinase (AMPK) (cat. no. 2532) antibodies, anti-AKT1, anti-AKT2, anti-phospho-AKT (Ser473) antibodies and anti-cleaved PARP antibody were from Cell Signaling Technology.
The firefly luciferase reporters, iNOS promoter reporter (iNOS-Luc), NFκB response element reporter (NFκB-Luc), as well as expression vectors for PPARγ and expression vectors for FLAG-FoxO1-WT, FLAG-FoxO1-CA (T24A, S253A and S316A) and FoxO1-DBD-mutant, were described previously (Fan et al, 2005, 2007, 2009). Adenoviruses encoding WT or CA mutant were donated by Dr Domenico Accili. Viral transductions were performed by incubation of target cells (RAW264.7 and peritoneal macrophages) at a multiplicity of infection (MOI) of 100 PFU/cell for 16 h.
Animals, GTT and primary macrophages
FoxO1 haploinsufficient mice on a C57BL/6J background, Tlr4KO and WT C57BL/6J mice were maintained under specific pathogen-free conditions. All animal experiments were performed humanely under protocols approved by the University of California, San Diego Animal Subjects Committee. A GTT was performed on mice that were fasted for 7 h. At 6 h of fasting, the mice were injected intraperitoneally with 1 mg/kg body weight of LPS. One hour later, the GTT was initiated with i.p. injection of 1 g/kg body weight of glucose. BMDMs were generated from FoxO1+/− and WT mice following established protocols (Nguyen et al, 2007).
Relative luciferase reporter assays and transfection
Relative luciferase reporter assays were performed as previously described (Fan et al, 2007). Transfections were carried out using either Lipofectamine 2000 (for HEK293 cells) or Lipofectamine LTX (for RAW264.7 cells) (Invitrogen). Each transfection experiment was independently repeated at least three times.
ChIP-sequencing assays
ChIP-Seq uses ChIP and massively parallel sequencing to identify genome-wide protein–DNA associations. However, the availability of adequate antibodies is a major limitation for providing the enrichment required for identification of genomic-binding sites using ChIP-Seq. We introduced a biotin-tagging system for rapid, high affinity purification of FoxO1 in the ChIP-Seq assay. This methodology (de Boer et al, 2003) is based on the ability of the E. coli biotin ligase BirA to efficiently biotinylate a short 23 amino acid BLRP fused to a protein of interest in eukaryotic cells. BLRP is a highly effective substrate for BirA and is not represented in the mouse genome. An expression vector was designed to place the BLRP peptide at the amino terminus of FoxO1 and to include a polyglycine spacer and TEV cleavage site (Supplementary Figure S6). We confirmed that placing the BLRP tag at the N-terminus of FoxO1 did not affect transcriptional function (both transactivation and transrepression), as assayed by transient reporter gene assays (Supplementary Figure S7A–C). We also confirmed that the BLRP-FoxO1 fusion protein was sufficiently biotinylated upon co-expression of BirA, and that biotinylation was not affected by intracellular AKT activity (Supplementary Figure S8). The efficient biotinylation enabled subsequent purification with streptavidin matrices. The BLRP-FoxO1 vector was made puromycin resistant, and then expressed in a RAW264.7 macrophage cell line that stably expressed BirA (neomycin resistant). Stable cell lines expressing both BirA and BLRP-tagged FoxO1 were selected in the presence of puromycin and neomycin, and clones that expressed the tagged protein at equivalent levels to the endogenous protein were selected (Supplementary Figure S9A).
Cells grown under the described experimental conditions were subjected to formaldehyde crosslinking. Chromatin was prepared, fragmented into 200–300 bp fragments by sonication, and subjected to high affinity purification on a streptavidin affinity matrix (NanoLink Streptavidin Magnetic Beads, Solulink, San Diego, CA). The matrix was subjected to TEV (AcTEV Protease, Invitrogen, San Diego, CA) proteolysis to release the tagged protein and DNA adducts, leaving any other biotinylated or non-specific proteins bound to the matrix (Supplementary Figure S9B). Protein–DNA crosslinks were reversed by heating, and the resulting DNA was purified. Q-PCR was applied to confirm high enrichment of a known FoxO1 target gene sequence (p27 gene) over background (an 11-fold increase, Supplementary Figure S9C). The DNA was then amplified to generate libraries for sequencing. Parallel sequencing was performed using the Illumina Genome Analyzer and short reads (24 bp) were mapped to the mouse reference genome. Identical reads were subsequently combined, and peaks were defined using standard software packages with robust statistical cutoffs (Valouev et al, 2008). Secondary analyses included computational motif discovery to identify enriched sequence elements in genomic-binding sites (Ogawa et al, 2005).
RNA interference
The siRNA target sequence against mouse FoxO1 is indicated in Supplementary Figure S2. An absence of homology to any other gene was confirmed by a BLAST search (National Center for Biotechnology Information, National Institutes of Health). The following siRNA sequence was used for against mouse Tlr4: rUrUrGrGrArCrCrUrGrArGrGrArGrArArCrArArArATT. The siRNA duplexes and a negative control (siRNA against luciferase gene) were purchased from Dharmacon Research Inc. (Lafayette, CO). RAW264.7 cells were electroporated with siRNA using the Gene Pulser XCell (Bio-Rad Laboratories, Hercules, CA). Electroporated cells were incubated for 48 h at 37°C before assays.
Real-time PCR
Total RNA was isolated and purified from cells using RNeasy columns and RNase-free DNase according to the manufacturer's instructions (Qiagen, Valencia, CA). First-strand cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The samples were run in 20 μl reactions using an, MJ Research PTC-200 96-well thermocycler coupled with the Chromo 4 Four-Color Real-Time System (GMI, Inc., Ramsey, MN). Gene expression levels were calculated after normalization to the standard housekeeping gene RPS3 using the ΔΔCT method as described previously (Fan et al, 2009), and expressed as relative mRNA compared with control. Primer information is available upon request.
Immunofluorescence
RAW264.7 or elicited peritoneal macrophage cells were cultured on 13 mm diameter glass coverslips and treated with PBS, LPS or LPS+AKTi. Cells were then fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton-X100, and stained with HA-probe antibodies against FoxO1. Secondary antibodies consisted of anti-mouse AlexaFluor 594, anti-rabbit AlexaFluor 594 and anti-rabbit AlexaFluor 488 (Invitrogen). Nuclei were visualized using DAPI staining (Invitrogen). Images were captured using a microscope (Nikon ECLIPSE TE2000-U, Nikon Microscope, San Diego, CA) running RS Image Version 1.9.1. software. Image processing was performed with Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Statistical analysis
Data were expressed as means±s.d. and evaluated by Student's two-tailed t-test or ANOVA, followed by post hoc comparisons with Fisher's protected least significant difference test. A P-value of 0.05 was used to determine significance.
Supplementary Material
Acknowledgments
We thank Dr Domenico Accili for providing the various adenoviral FoxO1 constructs, and Dr Kun-Liang Guan for the FoxO1-DBD mutant plasmid. We thank Ms Elizabeth J Hansen for editorial assistance. This study was funded in part by National Institutes of Health Grants DK 033651 and DK 074868 (to JMO), DK 075479 (to JJK), and by University of California Discovery Program Project bio03-10383 (BioStar) with matching funds from Pfizer Incorporated. W Fan is supported by a Mentor-Based Postdoctoral Fellowship from the American Diabetes Association awarded to JM Olefsky.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arden KC (2007) FoxOs in tumor suppression and stem cell maintenance. Cell 128: 235–237 [DOI] [PubMed] [Google Scholar]
- Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M (2005) IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med 11: 191–198 [DOI] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG (2005) FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 16: 183–189 [DOI] [PubMed] [Google Scholar]
- Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, Hoch M (2010) FOXO-dependent regulation of innate immune homeostasis. Nature 463: 369–373 [DOI] [PubMed] [Google Scholar]
- Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F (1999) Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem 274: 10689–10692 [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 100: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luca C, Olefsky JM (2008) Inflammation and insulin resistance. FEBS Lett 582: 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell P, Otto TC, Adi S, Lane MD (2003) Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem 278: 45485–45491 [DOI] [PubMed] [Google Scholar]
- Erridge C, Samani NJ (2009) Saturated fatty acids do not directly stimulate Toll-like receptor signaling. Arterioscler Thromb Vasc Biol 29: 1944–1949 [DOI] [PubMed] [Google Scholar]
- Fabre S, Lang V, Harriague J, Jobart A, Unterman TG, Trautmann A, Bismuth G (2005) Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J Immunol 174: 4161–4171 [DOI] [PubMed] [Google Scholar]
- Fan W, Imamura T, Sonoda N, Sears DD, Patsouris D, Kim JJ, Olefsky JM (2009) FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J Biol Chem 284: 12188–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Yanase T, Morinaga H, Mu YM, Nomura M, Okabe T, Goto K, Harada N, Nawata H (2005) Activation of peroxisome proliferator-activated receptor-gamma and retinoid X receptor inhibits aromatase transcription via nuclear factor-kappaB. Endocrinology 146: 85–92 [DOI] [PubMed] [Google Scholar]
- Fan W, Yanase T, Morinaga H, Okabe T, Nomura M, Daitoku H, Fukamizu A, Kato S, Takayanagi R, Nawata H (2007) Insulin-like growth factor 1/insulin signaling activates androgen signaling through direct interactions of Foxo1 with androgen receptor. J Biol Chem 282: 7329–7338 [DOI] [PubMed] [Google Scholar]
- Fessler MB, Rudel LL, Brown JM (2009) Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol 20: 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N (2005) FOXO transcription factors in cell-cycle regulation and the response to oxidative stress. Antioxid Redox Signal 7: 752–760 [DOI] [PubMed] [Google Scholar]
- Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, McNeish JD, Coleman KG (2003) Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clin Invest 112: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK (2007) Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell 25: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, Watt MJ, Benner C, Febbraio MA, Nguyen AK, Folian B, Subramaniam S, Gonzalez FJ, Glass CK, Ricote M (2007) Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest 117: 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O (2004) Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 279: 41114–41123 [DOI] [PubMed] [Google Scholar]
- Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, Kitazawa S, Miyachi H, Maeda S, Egashira K, Kasuga M (2006) MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest 116: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim F, Pham M, Luttrell I, Bannerman DD, Tupper J, Thaler J, Hawn TR, Raines EW, Schwartz MW (2007) Toll-like receptor-4 mediates vascular inflammation and insulin resistance in diet-induced obesity. Circ Res 100: 1589–1596 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Li P, Huntley J, Chang JP, Arden KC, Olefsky JM (2009) FoxO1 haploinsufficiency protects against high-fat diet-induced insulin resistance with enhanced peroxisome proliferator-activated receptor gamma activation in adipose tissue. Diabetes 58: 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Burgering BM (1999) Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J Mol Med 77: 656–665 [DOI] [PubMed] [Google Scholar]
- Laird MH, Rhee SH, Perkins DJ, Medvedev AE, Piao W, Fenton MJ, Vogel SN (2009) TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J Leukoc Biol 85: 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Sohn KH, Rhee SH, Hwang D (2001) Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J Biol Chem 276: 16683–16689 [DOI] [PubMed] [Google Scholar]
- Lee JY, Ye J, Gao Z, Youn HS, Lee WH, Zhao L, Sizemore N, Hwang DH (2003) Reciprocal modulation of Toll-like receptor-4 signaling pathways involving MyD88 and phosphatidylinositol 3-kinase/AKT by saturated and polyunsaturated fatty acids. J Biol Chem 278: 37041–37051 [DOI] [PubMed] [Google Scholar]
- Logie L, Ruiz-Alcaraz AJ, Keane M, Woods YL, Bain J, Marquez R, Alessi DR, Sutherland C (2007) Characterization of a protein kinase B inhibitor in vitro and in insulin-treated liver cells. Diabetes 56: 2218–2227 [DOI] [PubMed] [Google Scholar]
- Luyendyk JP, Schabbauer GA, Tencati M, Holscher T, Pawlinski R, Mackman N (2008) Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol 180: 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Barr V, Accili D (2000) Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J 19: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Biggs WH III, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D (2002) Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet 32: 245–253 [DOI] [PubMed] [Google Scholar]
- Nakae J, Cao Y, Oki M, Orba Y, Sawa H, Kiyonari H, Iskandar K, Suga K, Lombes M, Hayashi Y (2008) Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 57: 563–576 [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH III, Arden KC, Accili D (2003) The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell 4: 119–129 [DOI] [PubMed] [Google Scholar]
- Nguyen MT, Favelyukis S, Nguyen AK, Reichart D, Scott PA, Jenn A, Liu-Bryan R, Glass CK, Neels JG, Olefsky JM (2007) A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem 282: 35279–35292 [DOI] [PubMed] [Google Scholar]
- Nomiyama T, Perez-Tilve D, Ogawa D, Gizard F, Zhao Y, Heywood EB, Jones KL, Kawamori R, Cassis LA, Tschop MH, Bruemmer D (2007) Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J Clin Invest 117: 2877–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK (2005) Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122: 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh da Y, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM (2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142: 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaniemi M, Glumoff V, Harju K, Liljeroos M, Vuori K, Hallman M (2003) Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur J Immunol 33: 597–605 [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK (2010) Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 72: 219–246 [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK (2005) A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsouris D, Li PP, Thapar D, Chapman J, Olefsky JM, Neels JG (2008) Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab 8: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riol-Blanco L, Delgado-Martin C, Sanchez-Sanchez N, Alonso CL, Gutierrez-Lopez MD, Del Hoyo GM, Navarro J, Sanchez-Madrid F, Cabanas C, Sanchez-Mateos P, Rodriguez-Fernandez JL (2009) Immunological synapse formation inhibits, via NF-kappaB and FOXO1, the apoptosis of dendritic cells. Nat Immunol 10: 753–760 [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Saberi M, Woods NB, de Luca C, Schenk S, Lu JC, Bandyopadhyay G, Verma IM, Olefsky JM (2009) Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab 10: 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S, Saberi M, Olefsky JM (2008) Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 118: 2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F, Heit A, Dreher S, Eisenacher K, Mages J, Haas T, Krug A, Janssen KP, Kirschning CJ, Wagner H (2008) Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur J Immunol 38: 2981–2992 [DOI] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS (2006) TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest 116: 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, Lacy-Hulbert A, El Khoury J, Golenbock DT, Moore KJ (2010) CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol 11: 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Coudriet GM, Kim DH, Lu Y, Perdomo G, Qu S, Slusher S, Tse HM, Piganelli J, Giannoukakis N, Zhang J, Dong HH (2009) FoxO1 links insulin resistance to proinflammatory cytokine IL-1beta production in macrophages. Diabetes 58: 2624–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Hazeki K, Hoshi M, Nigorikawa K, Inoue N, Sasaki T, Hazeki O (2008) Critical roles of the p110 beta subtype of phosphoinositide 3-kinase in lipopolysaccharide-induced Akt activation and negative regulation of nitrite production in RAW 264.7 cells. J Immunol 180: 2054–2061 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A (2008) Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, Muller M, Saemann MD (2008) The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29: 565–577 [DOI] [PubMed] [Google Scholar]
- Weisberg SP, Hunter D, Huber R, Lemieux J, Slaymaker S, Vaddi K, Charo I, Leibel RL, Ferrante AW Jr (2006) CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest 116: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112: 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 32: 221–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.