Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum
Ero1 is thought to be the only oxidase that mediates the re-oxidation of protein disulphide isomerases (PDIs) during oxidative protein folding in the ER. This study reveals that peroxiredoxin IV can also directly oxidize PDI-family members and thus act as a second source of oxidizing equivalents for disulphide-bond formation in the ER.
Keywords: disulphide formation, hydrogen peroxide, peroxiredoxin IV, protein disulphide isomerase, protein folding
Abstract
Disulphide formation in the endoplasmic reticulum (ER) is catalysed by members of the protein disulphide isomerase (PDI) family. These enzymes can be oxidized by the flavoprotein ER oxidoreductin 1 (Ero1), which couples disulphide formation with reduction of oxygen to form hydrogen peroxide (H2O2). The H2O2 produced can be metabolized by ER-localized peroxiredoxin IV (PrxIV). Continuous catalytic activity of PrxIV depends on reduction of a disulphide within the active site to form a free thiol, which can then react with H2O2. Here, we demonstrate that several members of the PDI family are able to directly reduce this PrxIV disulphide and in the process become oxidized. Furthermore, we show that altering cellular expression of these proteins within the ER influences the efficiency with which PrxIV can be recycled. The oxidation of PDI family members by PrxIV is a highly efficient process and demonstrates how oxidation by H2O2 can be coupled to disulphide formation. Oxidation of PDI by PrxIV may therefore increase efficiency of disulphide formation by Ero1 and also allows disulphide formation via alternative sources of H2O2.
Introduction
Disulphide formation is an essential modification, which occurs within the endoplasmic reticulum (ER) of eukaryotic cells. Introduction of native disulphides can be a complex process, requiring not only oxidation of cysteine residues but also reduction and isomerization of non-native disulphides (Jansens et al, 2002). In keeping with this complexity, a number of oxidoreductases have been identified, which catalyse thiol-disulphide exchange reactions within the ER (Sevier and Kaiser, 2006). Protein disulphide isomerase (PDI) represents one of the most extensively studied ER oxidoreductases and is a member of a family of ER-resident thioredoxin-like proteins (Appenzeller-Herzog and Ellgaard, 2008). Around 20 PDI homologues have been described in mammalian cells, many of which exhibit distinct client-protein specificities (Jessop et al, 2009b). PDI-family proteins possess active sites containing cysteine residues, within a CXXC motif. Disulphides formed between these cysteine residues can be exchanged with client proteins leading to formation of disulphides within the client and reduction of the PDI-family member (Creighton et al, 1980). Moreover, disulphide exchange can occur in the opposite direction allowing the reduction of non-native disulphides within client proteins with the concomitant oxidation of the PDI-family member.
To allow continuous transfer of disulphides to client proteins, PDI must be re-oxidized. The oxidation of PDI can be catalysed by ER oxidoreductin 1 (Ero1) (Pollard et al, 1998; Frand and Kaiser, 1999). Ero1 can generate disulphides de novo by transfer of electrons to molecular oxygen, generating hydrogen peroxide (H2O2) in the process (Tu and Weissman, 2002; Gross et al, 2006). In higher eukaryotes, the H2O2 produced by Ero1 can be metabolized by an ER-resident enzyme peroxiredoxin IV (PrxIV, Prx4) (Tavender et al, 2008; Tavender and Bulleid, 2010b). The essential catalytic unit of PrxIV is a dimer with a peroxidatic cysteine residue (Cys124) in one chain being able to reduce H2O2 generating water and becoming oxidized to a cysteine sulphenic acid in the process. This sulphenylated cysteine subsequently reacts with a resolving cysteine residue (Cys245) of the partner chain within the dimeric subunit, leading to formation of an intermolecular disulphide. Hence, PrxIV converts the oxidizing potential of H2O2 into a disulphide bond.
To maintain activity towards H2O2, the interchain disulphide within PrxIV needs to be reduced. This reduction is achieved for other cellular peroxiredoxins by a member of the thioredoxin family of proteins (Kalinina et al, 2008). It is presently unknown which enzyme fulfils this role in the ER for PrxIV, although it is clear that a robust reducing system is present for recycling of the peroxidatic disulphide (Tavender and Bulleid, 2010b). There are several thioredoxin-like proteins within the ER that could fulfil the role of a reductase with the inevitable consequence that these enzymes would become oxidized. Hence, the recycling of PrxIV could provide an alternative pathway for the generation of oxidizing equivalents for disulphide formation in proteins entering the secretory pathway. Such an alternative pathway has been suggested to exist based upon the fact that, while Ero1 activity is essential in yeast, knockout of both Ero1 paralogues in mice (Zito et al, 2010) or Ero1 in Drosophila (Tien et al, 2008) does not cause a severe phenotype. In addition to PrxIV recycling, there are several other potential mechanisms whereby PDI proteins may become oxidized independent of Ero1. These include oxidation by the quiescin sulphydryl oxidases (Chakravarthi et al, 2007; Rancy and Thorpe, 2008) or direct oxidation by H2O2 (Karala et al, 2009), dehydroascorbate (Saaranen et al, 2010), oxidized vitamin K (Wajih et al, 2007; Schulman et al, 2010) or glutathione (GSH) disulphide (Appenzeller-Herzog et al, 2010). While Ero1 can provide the oxidizing equivalents for disulphide formation, the contribution of alternative pathways to oxidative protein folding is still to be determined.
To determine whether there is a potential role for PrxIV in disulphide formation, we tested the ability of several PDI-family members to reduce peroxidatic disulphides and return PrxIV to its peroxide reactive state. We demonstrate that PDI, P5 and ERp46 can directly reduce PrxIV in vitro and can also enhance recycling of peroxidatic disulphides within the ER of mammalian cells. Indeed, PDI itself was oxidized more efficiently by PrxIV than by Ero1α. In addition to establishing the pathways for maintaining the catalytic activity of PrxIV, these findings demonstrate an alternative mechanism for oxidation of PDI and its homologues. Moreover, the coupling by PrxIV of the reduction of H2O2 to the formation of a disulphide ensures that two disulphides are formed for every oxygen molecule reduced.
Results
PDI-family members reduce peroxidatic disulphides in PrxIV
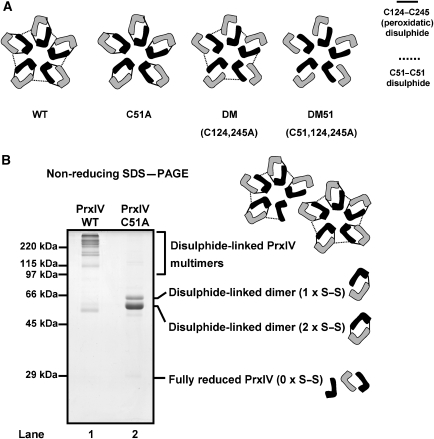
The overall oligomeric structure of PrxIV is a decamer consisting of five dimers linked by interchain disulphide bonds (Figure 1A, wild-type (WT)). PrxIV is a member of the 2-cys family of peroxiredoxins, and therefore, has a reaction cycle that involves conversion of a cysteine thiol to the sulphenylated form following reaction with H2O2 (Wood et al, 2003). The so-called peroxidatic cysteine (Cys124) then reacts with a second cysteine residue, called the resolving cysteine (Cys245), to form an interchain disulphide. As both polypeptides contain peroxidatic and resolving cysteines, two such interchain disulphides can form. To follow this reaction, we can separate the protein by SDS–PAGE under non-reducing conditions and follow the formation or loss of multimeric species (Figure 1B). However, the pattern of redox species is complicated by the presence of the interchain disulphide (Tavender et al, 2008), which forms between individual dimers (via Cys51) (Figure 1B, lane 1) (Tavender and Bulleid, 2010b). The exact role of the non-catalytic disulphides is unclear, although they are not necessary for peroxidase activity of PrxIV (Supplementary Figure S1). The presence of non-catalytic disulphides in purified PrxIV results in a complex pattern of multimeric species, which affects the interpretation of changes to the redox status of PrxIV using a gel-based assay (Supplementary Figure S2). To simplify changes to the redox status of PrxIV, we therefore use a PrxIV C51A mutant (Figure 1, C51A) that lacks the ability to form non-catalytic disulphides.
Figure 1.
Determining PrxIV redox status using SDS–PAGE. (A) Depiction of interchain disulphides formed within the wild-type (WT) PrxIV decamer. For clarity, individual PrxIV molecules within each catalytic subunit are represented in different shades. Peroxidatic disulphides (solid lines) form between PrxIV molecules within dimeric subunits and C51–C51 disulphides (dotted lines) link adjacent dimers. Also depicted are disulphides formed within PrxIV cysteine to alanine mutants used in this study (C51A, DM and DM51). (B) Non-reducing SDS–PAGE analysis and Coomassie blue staining of purified PrxIV showing electrophoretic behaviour of disulphide-linked forms in wild-type and C51A mutant PrxIV.
Purified recombinant PrxIV C51A exhibits a distinct mobility pattern when separated by SDS–PAGE carried out under non-reducing conditions. The monomeric form migrates close to the 25 kDa marker, whereas two dimeric forms migrate close to the 58 kDa marker; the low-mobility dimer corresponds to a single disulphide-bonded form, whereas the high-mobility dimer corresponds to the double disulphide-bonded form (Figure 1B). We have previously shown (Tavender and Bulleid, 2010b) and show here that recombinant PrxIV C51A purified under aerobic conditions predominantly contains two interchain disulphides (Figure 2C, lane 1). Approximately 80% of the PrxIV contained two disulphides, with the rest containing a single disulphide. The purified PrxIV C51A therefore provides an ideal starting material with which to measure factors capable of recycling the protein to its reduced form.
Figure 2.
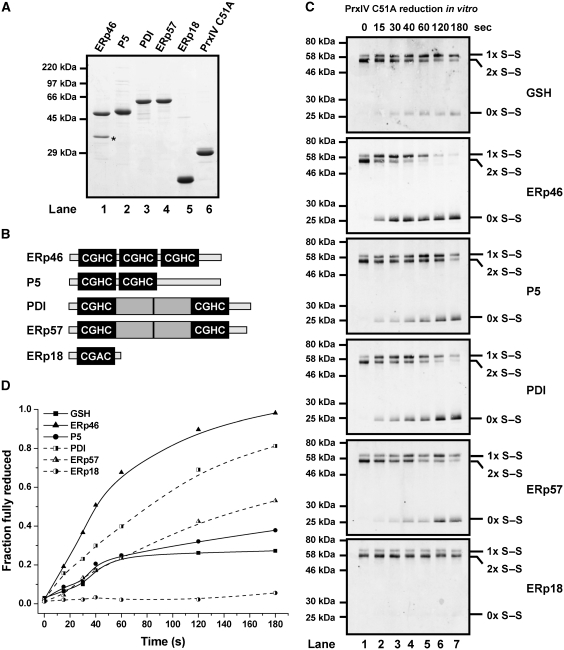
PDI-family members can reduce PrxIV peroxidatic disulphides in vitro. (A) SDS–PAGE analysis (reducing conditions) and Coomassie blue staining of purified proteins prepared for use in this study. An ERp46 degradation product is indicated (*). (B) Simple representation of domain architecture for PDI-family members used in this study. Catalytically active thioredoxin-like domains with CXXC motifs are depicted as black boxes, while grey boxes indicate inactive thioredoxin-like domains. (C) Fluorescent western blots showing disulphide-bonded status of purified PrxIV C51A (3 μM) during incubation with 10 mM GSH or 3 μM reduced PDI-family member. Reactions were quenched with TCA and precipitated proteins were re-dissolved in the presence of NEM before non-reducing SDS–PAGE and blotting with antibody to PrxIV. PrxIV dimers containing two disulphides (2 × S–S) or one disulphide (1 × S–S) as well as fully reduced PrxIV (0 × S–S) are indicated. (D) Western blots were used for quantification of fully reduced PrxIV C51A by densitometry. Intensity of fully reduced form is expressed as a proportion of the sum total intensity of 0 × S–S, 1 × S–S and 2 × S–S fractions. Data presented are representative experiments performed at least three times with similar results.
To test the ability of ER-resident thioredoxin-like proteins to reduce peroxidatic disulphides, we expressed and purified several human PDI-family members, namely ERp46, P5, PDI, ERp57 and ERp18 (Figure 2A). These PDI-family proteins display variation in terms of size, domain architecture and number of active sites (Figure 2B). PDI-family proteins were prepared in a reduced state (Supplementary Figure S3), and equimolar concentrations of each were incubated with PrxIV C51A before the addition of TCA to quench thiol-disulphide exchange. Equimolar concentrations were used because quantification of PrxIV levels in human HT1080 cells indicated PrxIV to be present at comparable concentrations to most of the oxidoreductases tested (Supplementary Figure S4). In addition, we tested the ability of GSH at physiological concentration to reduce purified PrxIV C51A (Figure 2C and D). Strikingly, both ERp46 and PDI reduced PrxIV more rapidly and more extensively than GSH during the brief incubation period. ERp46 almost completely removed the high-mobility dimer and accumulated around 70% of the total PrxIV in a fully reduced form within the first minute. P5 and ERp57 also served as effective reductases, initially reducing PrxIV C51A in a comparable manner to GSH but eventually accumulating a higher proportion of fully reduced material (Figure 2D), indicating that equimolar quantities of either protein can reduce peroxidatic disulphides at least as effectively as a 3300-fold excess of GSH. In contrast, ERp18 had a negligible effect generating barely perceptible quantities of fully reduced PrxIV (<10%). These results clearly show that the PDI family of oxidoreductases have specificity in their ability to reduce the peroxidatic disulphides in PrxIV.
Reduction of PrxIV results in oxidation of ER oxidoreductases
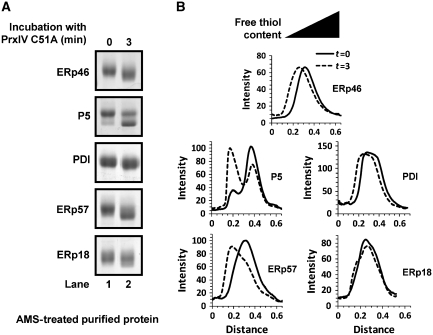
Reduction of PrxIV by a PDI-family member implies the occurrence of a thiol-disulphide exchange reaction between the two proteins. In this case, reduction of PrxIV should result in oxidation of the PDI-family member. To test this, we took samples of each ER oxidoreductase that had been incubated for 3 min with PrxIV C51A (samples are shown in Figure 2C, lane 7). Following TCA precipitation, proteins were re-suspended in a denaturing buffer containing the sulphydryl-modifying agent 4-acetamido-4′-maleimidylstilbene-2,2′-disulphonic acid (AMS). AMS reacts with thiol groups, increasing the molecular weight of the protein and retarding electrophoretic mobility. A protein with oxidized cysteine residues lacks free thiols. Hence, it is not modified by AMS and therefore migrates more rapidly than a protein with reduced cysteines (Supplementary Figure S3). Following incubation with PrxIV a clear decrease in AMS-modified protein was observed for ERp46, P5, PDI and ERp57 (Figure 3A and B), indicating substantial oxidation of cysteine residues in each protein. ERp18 remains predominantly reduced and therefore AMS modified. The small population of ERp18 that does become oxidized is attributable to the aforementioned reduction of up to 10% of PrxIV C51A. Thus, the recycling of the catalytic thiols of PrxIV by ERp46, PDI, P5 or ERp57 occurs concomitant with the oxidation of these proteins.
Figure 3.
PDI-family members are oxidized by PrxIV C51A in vitro. (A) AMS-dependent changes in electrophoretic mobility of purified reduced PDI-family members following incubation of each protein with equimolar oxidized PrxIV C51A. Reactions were quenched by TCA addition and precipitated proteins were re-dissolved in denaturing buffer containing AMS. Proteins were visualized by SDS–PAGE (reducing conditions) and Coomassie blue staining. (B) Densitometry profiles of stained bands for each gel-lane displayed in (A) illustrating relative electrophoretic mobilities. ‘Zero' distance value of graphs relates to bottom end of each lane. Relationship between gel position and relative free thiol content is indicated (black triangle).
GSH enhances PrxIV recycling by reducing PDI-family members
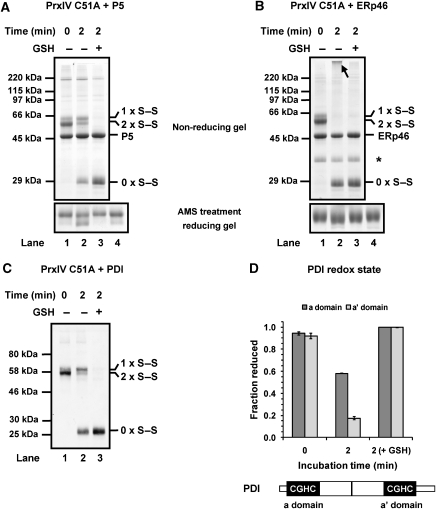
A role for GSH in maintaining ER oxidoreductases in a reduced state has been highlighted previously (Jessop and Bulleid, 2004). Addition of GSH to a reaction mixture containing PrxIV C51A and P5 resulted in complete reduction of peroxidatic disulphides within 2 min (Figure 4A). This reduction is in stark contrast with incubations lacking GSH where one third of total PrxIV is typically fully reduced. The complete reduction of peroxidatic disulphides observed with a combination of P5 and GSH suggests more than simply a cumulative effect of the reductants, as individually they only reduce PrxIV by ∼30 and 20%, respectively. When the redox state of P5 was assayed following incubation with PrxIV C51A and GSH, it was found to be completely reduced (Figure 4A, lane 3, lower panel). It therefore appears that GSH maintains P5 in a reduced state during incubation with PrxIV. In a similar manner, inclusion of GSH in reactions containing PrxIV C51A and ERp46 led to ERp46 remaining more reduced than in the absence of GSH (Figure 4B, bottom panel). Control samples of P5 or ERp46 incubated in buffer alone (Figure 4A and B, lane 4) remained reduced demonstrating that neither protein is oxidized in the absence of PrxIV during the incubation period.
Figure 4.
Glutathione reduces PDI-family members and enhances peroxidatic recycling. Purified PrxIV C51A (6 μM) was incubated with equimolar reduced P5 (A) or ERp46 (B) in the presence and absence of 10 mM GSH. Reactions were split in two and quenched by TCA addition. One sample for each treatment was precipitated, re-dissolved in SDS sample buffer containing NEM and analysed by non-reducing SDS–PAGE and Coomassie staining (top panel). The second sample for each was re-dissolved in denaturing buffer containing AMS and visualized by Coomassie blue staining following SDS–PAGE under reducing conditions (bottom panel). Lane 4 contains reduced P5 or ERp46 incubated alone for 2 min and modified with AMS as above. An arrow indicates high molecular weight material observed for ERp46 and an asterisk (*) demarcates the ERp46 degradation product. (C) PrxIV C51A (4μM) was incubated with equimolar reduced PDI plus or minus 10 mM GSH. Four replicates were prepared for each reaction and quenched by TCA addition. One replicate was precipitated, re-dissolved in SDS sample buffer containing NEM and analysed by non-reducing SDS–PAGE and western blotted with antibody to PrxIV. Remaining replicates were re-dissolved in denaturing buffer containing iodoacetic acid, separated using non-reducing SDS–PAGE and PDI excised and prepared for MALDI-TOF analysis of redox status as detailed in Chambers et al (2010). For a and a' domains of PDI, the proportion of active sites recorded as being reduced are expressed as a fraction of the whole population (D). Values are mean averages of triplicates with error bars representing s.d. A schematic depicting the positions of the a and a' domains of PDI is included.
Unlike the situation with P5, GSH addition did not enhance peroxidatic disulphide reduction by ERp46, simply because PrxIV C51A was fully reduced within the 2-min incubation even in the absence of GSH. However, an effect on the redox status of ERp46 was observed, which may indicate that GSH augments the efficacy of PrxIV recycling by ERp46. When analysed by Coomassie blue stain, a small amount of high molecular weight material was consistently visible during reduction of PrxIV C51A by ERp46 (Figure 4B, indicated with arrow). Whether this material consists of protein aggregates or intermediate complexes of PrxIV and ERp46 is unclear. Whatever the nature of this species, its appearance was curtailed by inclusion of GSH.
In a similar manner to that observed for P5, addition of GSH to a reaction mixture containing PrxIV C51A and PDI caused almost complete reduction of peroxidatic disulphides (Figure 4C). Within these reactions, the redox condition of each PDI active site could be quantitatively determined (Figure 4D) using a recently described differential alkylation and mass spectrometry approach (Chambers et al, 2010). Using this methodology, we demonstrated that about 65% of the active sites within PDI became oxidized within 2 min when incubated with PrxIV in the absence of GSH. This extent of PDI oxidation correlates with the extent of reduction of PrxIV (∼60%, Figure 2D). In the presence of GSH, both active sites of PDI remain in the reduced form. Intriguingly, it was also clear that in the absence of GSH, reduction of PrxIV results in the preferential oxidation of the PDI a' domain (Figure 2D). Such an effect has been demonstrated previously during the oxidation of PDI by Ero1α (Chambers et al, 2010). The reduction of peroxidatic disulphides could be mediated exclusively by the a' domain, with subsequent oxidation of the a domain via PDI intramolecular exchange as has been suggested for the reaction with Ero1α (Chambers et al, 2010).
Increased expression of PDI-family proteins enhances PrxIV recycling in the ER
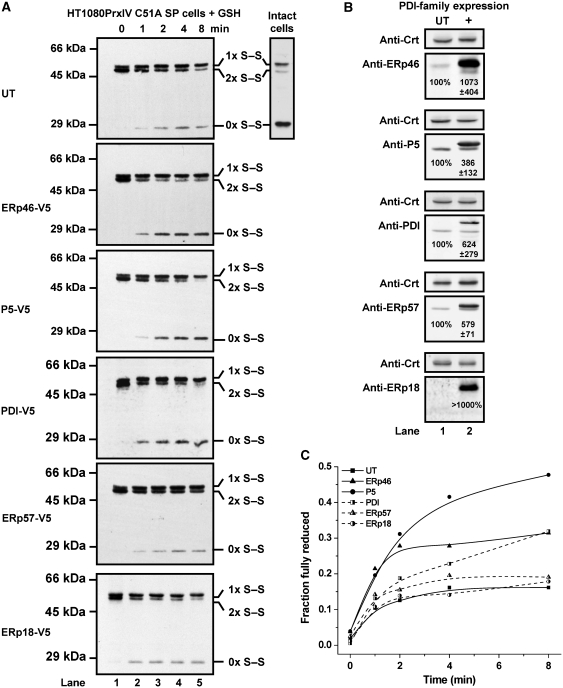
Having demonstrated that ERp46, PDI, P5 and ERp57 can directly reduce peroxidatic disulphides of PrxIV in vitro, we set out to determine if they could exert an influence over this process within the ER. For these experiments, we used a HT1080 cell line stably overexpressing PrxIV C51A. The intracellular form of this protein exists as a mixture of oxidized and reduced species (Figure 5A, side panel) (Tavender and Bulleid, 2010b). When the cytosol from this cell line was removed by digitonin permeabilization of the plasma membrane, PrxIV C51A becomes nearly completely oxidized (Figure 5A, lane 1). Hence, during the preparation of semipermeabilized (SP) cells, factors required for maintaining reducing pathways within the ER are removed. We have shown previously that a reducing pathway can be re-established by addition of GSH to SP cells resulting in re-equilibration of the PrxIV redox status to reflect that seen for intact cells (Tavender and Bulleid, 2010b).
Figure 5.
Increased PDI-family expression enhances peroxidatic recycling in SP cells. (A) Non-reducing SDS–PAGE and anti-PrxIV western blot analysis. SP cells were prepared from HT1080 PrxIV C51A cells transiently transfected to overexpress ER oxidoreductases (as indicated) or without DNA (UT). Following incubation with 10 mM GSH, SP cells were treated with NEM, harvested and lysed. The side panel for sample ‘UT' shows a similar anti-PrxIV western blot for intact HT1080 PrxIV C51A cells treated with NEM prior to lysis. (B) SP cells prepared in (A) were used to determine oxidoreductase content by fluorescent western blotting. Samples transfected with PDI-family constructs (+) were probed alongside control samples (UT) first with the relevant anti-PDI-family antibody, then stripped and probed with anticalreticulin (Crt). Samples presented within each individual panel are from the same gel. Anti-Crt panels presented are from the same gel as the corresponding anti-PDI-family blot. Intensities for oxidoreductase samples were calculated by densitometry and adjusted to reflect Crt values. Intensities for control samples were taken as 100% and levels of overexpression for transfected cells given relative to this. Values are mean averages of triplicate quantifications±s.d. Endogenous ERp18 was not detected although intensity values for transfectants indicated at least 1000% increased expression. Ectopically expressed proteins exhibit some altered mobility due to epitope tags. (C) Appearance of fully reduced PrxIV for blots as in (A) was quantified as in Figure 2D.
To demonstrate the kinetics of GSH-dependent reduction of PrxIV C51A in SP cells, GSH was added to a final concentration of 10 mM, and at various time points, the redox status of PrxIV was frozen by the addition of the membrane-permeable thiol-alkylating agent N-ethylmaleimide (NEM) (Figure 5A, top panel). During the time course, a shift from high-mobility to low-mobility dimer was evident along with the appearance of some fully reduced PrxIV. It is unclear whether reduction of peroxidatic disulphides in PrxIV involves direct reduction by GSH or is caused by GSH first reducing endogenous ER oxidoreductases necessary for recycling PrxIV. If the reduction of peroxidatic disulphides in the ER relies principally upon an enzymatic process, we hypothesized that increasing the concentration of relevant oxidoreductases in the ER would enhance the rate of GSH-initiated reduction of PrxIV. Consequently, SP cells were prepared using HT1080 PrxIV C51A cells that had been transfected to transiently overexpress ERp46, P5, PDI, ERp57 or ERp18 (Figure 5B). Ectopic expression was typically between 4- and 10-fold higher than endogenous levels although this could not be accurately determined for ERp18 on account of our inability to detect the endogenous protein.
The increase in concentration of ERp46, P5 and PDI each caused an enhanced recovery of reduced PrxIV C51A following the addition of GSH to SP cells (Figure 5A and C). The differences between these treatments and untransfected cells are best visualized by comparing the relative proportions of each PrxIV species at 2 and 4 min following GSH addition (Figure 5A). Despite exhibiting reductase activity towards PrxIV disulphides in vitro, elevated concentrations of ERp57 displayed no discernible effect upon reduction of PrxIV C51A in SP cells. This lack of effect is perhaps not surprising given that ERp57 interacts with calnexin and calreticulin (Oliver et al, 1997) and, therefore, is unlikely to act on non-glycoprotein clients such as PrxIV. The fact that ERp57 does not enhance PrxIV reduction in the ER therefore suggests that ERp57 may be adequately sequestered by calnexin or calreticulin even when ERp57 is overexpressed. Overexpression of ERp18 also failed to increase the appearance of fully reduced PrxIV C51A, which is again not surprising considering the negligible activity of ERp18 as a reductase of PrxIV disulphides in vitro (Figure 2A and B). Hence, it would appear that in the mammalian ER, ERp46, P5 and PDI can catalyse the reduction of the peroxidatic disulphides within PrxIV.
Mixed disulphides indicate direct reduction of peroxidatic disulphides by PDI-family members in the ER
To establish that ERp46, P5 and PDI act directly on PrxIV, we investigated whether we could trap mixed disulphides that should form during the disulphide exchange reaction. The reduction of disulphides by PDI-family members requires the nucleophilic attack on a disulphide by the first cysteine residue of the conserved CXXC motif. A transient mixed disulphide is formed between the oxidoreductase and the target molecule, which is rapidly resolved by the second cysteine residue of the CXXC motif. Trapping of mixed disulphides is facilitated by cysteine to alanine mutation to modify the CXXC motif to CXXA. Without the second cysteine, resolution of the mixed disulphide is dependent on an alternative thiol source, substantially increasing the lifespan of this normally short-lived intermediate (Dick and Cresswell, 2002).
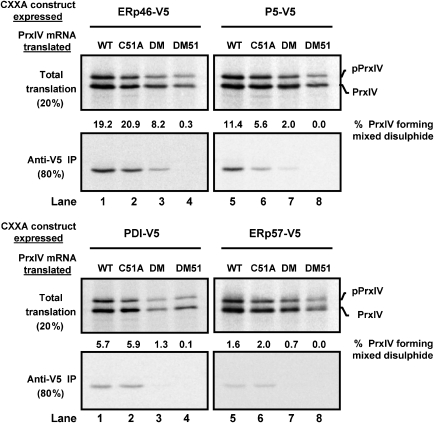
We have previously created a range of HT1080 cell lines stably expressing PDI-family CXXA mutants and used these to identify PrxIV as one of the predominant endogenous proteins forming mixed disulphides with ERp46 in HT1080 cells (Jessop et al, 2009b). Likewise, when translated in vitro in the presence of SP cells produced from these cell lines, radioactively labelled PrxIV formed prevalent mixed disulphides with ERp46 and P5. In each case, the native PrxIV sequence was translated, potentially containing both peroxidatic disulphides and non-catalytic disulphides. Consequently, it was not possible to conclude whether the mixed disulphides were formed during the nucleophilic attack by the PDI-family members on peroxidatic disulphides or on non-catalytic disulphides within PrxIV. To address this issue, a similar experiment was undertaken using PrxIV cysteine to alanine mutants, which could only form one of the two species of disulphide. SP cells were prepared from HT1080 cell lines stably expressing V5-tagged ERp46, P5, PDI or ERp57 in which all active site CXXC motifs were mutated to CXXA. The SP cells were then added to in vitro translation reactions with mRNA encoding one of four PrxIV variants (depicted in Figure 1A). C51A cannot form the non-catalytic disulphide, DM cannot form the peroxidatic disulphide and DM51 cannot form any disulphides. Once the incubation was complete, thiol-disulphide exchange was inhibited with NEM, SP cells were harvested and V5-tagged PDI-family members were immunoisolated along with any covalently linked mixed disulphide partners (Figure 6). The CXXA mutants of ERp46, P5 and PDI accumulated mixed disulphides more readily with PrxIV C51A (i.e. via nucleophilic attack of the peroxidatic disulphide) than with PrxIV DM (Figure 6, compare lanes 2 and 3 for all samples). In keeping with the apparent lack of ERp57 activity towards PrxIV C51A in the ER, ERp57 failed to form significant mixed disulphides with PrxIV (Figure 6, bottom right). These results demonstrate that ERp46, P5 and PDI but not ERp57 can directly reduce the peroxidatic disulphides in PrxIV within the ER.
Figure 6.
Substrate-trapping PDI-family mutants target peroxidatic disulphides in PrxIV. SP cells prepared from HT1080 cell lines expressing substrate-trapping PDI-family CXXA mutants were added to an in vitro translation system. Wild-type (WT) or mutant (C51A, DM, DM51) PrxIV was translated and radioactively labelled, treated with NEM and SP cells harvested. One fifth of cells were lysed directly with SDS sample buffer for analysis of total translation products (top panels for each set). Remaining cells were used for anti-V5 immunoisolation of PDI-family mutants along with any trapped substrate. PrxIV was visualized by SDS–PAGE (reducing conditions) and phosphorimaging. PrxIV isolated with each PDI-family mutant was measured by densitometry and is expressed as a percentage of the total translated product forming a mixed disulphide. ‘pPrxIV' indicates full-length preprotein, whereas ‘PrxIV' indicates protein that has been translocated to the ER and had the signal peptide removed. Samples are representative of repeat experiments.
Lowering the intracellular concentration of ER oxidoreductases compromises reduction of PrxIV
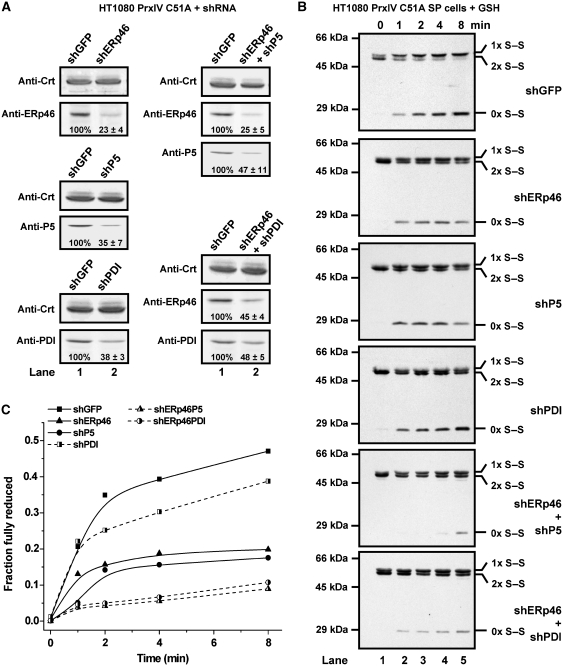
From the above results, it is clear that at least three members of the PDI family of ER oxidoreductases are capable of directly reducing the peroxidatic disulphides in PrxIV. It is not apparent whether these enzymes have overlapping functions or whether one enzyme preferentially reduces PrxIV in the ER. To address this question, we used shRNA treatment of the PrxIV C51A overexpressing cell line to lower the levels of ERp46, P5 or PDI either individually or in combination. Quantification of the levels of these proteins following treatment of cells with shRNA revealed knockdowns to between 20 and 50% of normal levels (Figure 7A). The consequence of this knockdown on the reduction of ER PrxIV C51A in the presence of GSH was then determined using the SP-cell assay (Figure 7B and C). In comparison to control cells that had been treated with a non-specific shRNA, the rate of reduction of PrxIV C51A was compromised with each individual shRNA. A modest slowing of the rate was seen with the PDI shRNA-treated cells with a more dramatic effect seen with the shRNA constructs targeting ERp46 and P5. In addition, the rate of recovery of reduced PrxIV C51A with cells treated with ERp46 shRNA in combination with P5 or PDI shRNA was severely perturbed (Figure 7B and C). Such a dramatic effect following the lowering of the levels of these proteins demonstrates that they contribute to the overall rate of recycling of PrxIV. These results further support the hypothesis that ERp46, P5 and PDI are required to maintain PrxIV in an enzymatically active form.
Figure 7.
Decreased PDI-family expression compromises peroxidatic recycling in SP cells. (A) HT1080 PrxIV C51A cells were transfected with shRNA to PDI-family member as indicated or control shRNA (shGFP). After selection for the shRNA-expressing population, SP cells were prepared and analysed by SDS–PAGE (reducing conditions) and fluorescent western blotting. Blots were probed with relevant antibodies against ER oxidoreductases then stripped and probed with anti-Crt. Intensities of oxidoreductase bands were calculated by densitometry and adjusted to reflect loading controls. Values for shGFP samples were taken as 100% and knockdown of ER oxidoreductase expression calculated relative to this. Values are mean averages of triplicate quantifications±s.d. (B) SP cells generated above were incubated with 10 mM GSH as indicated then treated with 40 mM NEM. Cells were harvested, lysed and subjected to non-reducing SDS–PAGE analysis prior to western blotting with antibody to PrxIV. (C) Appearance of fully reduced PrxIV was quantified as in Figure 2D.
Discussion
The concept that Ero1 is the sole source of oxidizing equivalents for disulphide formation in proteins entering the secretory pathway has recently been questioned due to a lack of a dramatic phenotype when both Ero1 paralogues are deleted in mice (Zito et al, 2010). Several additional pathways have been suggested to compensate for a lack of Ero1 (Margittai and Banhegyi, 2010), though their relevant contribution to disulphide formation remains to be established. Here, we have demonstrated that the oxidation of members of the ER oxidoreductase family by PrxIV is a robust and efficient mechanism to provide oxidizing equivalents for these enzymes. Importantly, the product of the reaction of Ero1 with oxygen, H2O2, is used as the electron acceptor during disulphide formation in PrxIV meaning that the combination of reduction of oxygen by Ero1 and PrxIV results in the oxidation of two molecules of ER oxidoreductase. Coupling these two enzymes together will result in a much more efficient oxidative pathway and may go some way to explain why the oxidation of PDI by Ero1 in vitro is a relatively inefficient process (Sevier et al, 2007; Baker et al, 2008; Chambers et al, 2010). Such a pathway for disulphide formation via PrxIV is not exclusively dependent upon Ero1 as there are other enzymes, which can generate H2O2 in the ER. These include the NADPH oxidase Nox4 (Van Buul et al, 2005) and the quiescin sulphydryl oxidase (Hoober et al, 1999). In addition, H2O2 produced by the mitochondria could also potentially enter the ER due to the close association of these two organelles (Simmen et al, 2010). Hence, in the absence of Ero1, PrxIV could fuel disulphide formation by providing the necessary oxidizing equivalents. It is of interest to note that yeast do not contain an equivalent ER-localized peroxiredoxin and that, unlike in mammals, Ero1p in yeast is an essential gene (Frand and Kaiser, 1998; Pollard et al, 1998).
Comparing the relative efficiencies of Ero1 and PrxIV in oxidizing ER oxidoreductases could provide some indication as to the relative contribution of each pathway to disulphide formation. When the physiological concentrations of the components are taken into account, then the oxidation of PDI by PrxIV seems much more efficient than that by Ero1α. PDI and PrxIV are present in the ER of HT1080 cells at roughly equivalent concentrations, whereas Ero1α is a relatively low abundance protein (Gess et al, 2003). About 20% of the PDI active sites become oxidized by Ero1α after 30 min incubation when combined in a ratio of 1:25 (Chambers et al, 2010). In comparison, 60% of the PDI active sites were oxidized by PrxIV within 2 min when incubated at a ratio of 1:1. Ideally, we would have liked to couple the activity of the two enzymes in vitro to oxidize PDI, but the inability to maintain reduced PrxIV for subsequent reaction in aerobic buffers precluded this approach. Indeed, it is the exquisite sensitivity of PrxIV to oxidation by low concentrations of H2O2 that would provide an extremely efficient mechanism to convert any H2O2 produced in the ER into disulphides.
During the course of this study, we consistently found that ERp46 and P5 were more efficiently oxidized by PrxIV than PDI. In addition, all three ER oxidoreductases were oxidized to a greater extent than GSH by PrxIV. However, when GSH was included in the incubations, the oxidoreductases were completely reduced. In addition, the presence of GSH increased the rate of reduction of PrxIV by P5 and PDI and prevented the formation of high molecular weight aggregates. Together, these results suggest that the ER oxidoreductases first reduce the peroxidatic disulphide in PrxIV and are then reduced by GSH. Given that the ER oxidoreductases are predominantly in a reduced form (Jessop and Bulleid, 2004), these results suggest that any disulphide formed within these enzymes is rapidly exchanged with either substrate proteins, or with GSH to form GSSG. Indeed, recent work looking at the Ero1α-dependent recovery of cells from treatment with the reducing agent DTT shows an initial increase in the GSSG concentration, suggesting that the ultimate target for disulphide exchange under these conditions is GSH (Appenzeller-Herzog et al, 2010). The concentration of GSSG then recovers to lower levels, indicating that GSSG is either used for disulphide formation in newly synthesized proteins or is reduced by an as yet unidentified reduction pathway. The fact that GSH efficiently reduces the ER oxidoreductases suggests that GSSG may well be a major source of disulphides for newly synthesized proteins.
ERp46 outperformed P5 and PDI in the ability to reduce PrxIV in vitro and in the formation of mixed disulphides with PrxIV species in SP cells. Moreover, both elevation and knockdown of ERp46 substantially influenced reduction of PrxIV in the ER. ERp46 may therefore be a preferred substrate for thiol-disulphide exchange with PrxIV. The preferential oxidation of ERp46 by PrxIV could be due to the presence of three rather than two active thioredoxin domains in this protein. ERp46 has been previously described as endoPDI due to its upregulation in endothelial cells, where it can act as a stress survival factor (Sullivan et al, 2003). In addition, it is upregulated along with a number of other ER oxidoreductases as well as PrxIV in differentiating B cells (van Anken et al, 2003). These previous studies are consistent with a role for ERp46 in regulating ER redox balance.
Such a role for ER oxidoreductases would be distinct from, or in addition to, their role in disulphide formation in proteins entering the secretory pathway. The study of the roles for the individual ER oxidoreductases in the past has focused on the idea that each may act as an oxidase or reductase on specific substrates. The association of individual oxidoreductases with ER chaperones, such as ERp57 with the calnexin/calreticulin cycle (Oliver et al, 1997; Jessop et al, 2009a) and P5 with BiP (Meunier et al, 2002; Jessop et al, 2009b), indicates specific roles for these enzymes on subsets of proteins and to some extent may explain substrate specificity. Further dissection of the mechanisms of specificity for P5 and ERp46 are essential for full understanding of their individual roles in recycling PrxIV. The fact that we show only limited specificity of ER oxidoreductases towards PrxIV would suggest that some of these proteins may also act as a conduit for oxidizing equivalents to produce GSSG rather than acting directly with client proteins. As each of these proteins is present at significant concentrations in the ER, their ability to transfer disulphides between H2O2 and GSH would establish an efficient redox buffer to balance the ER redox status.
It is now well established that the activity of Ero1α is tightly regulated by the presence of non-catalytic disulphides that need to be reduced to activate the enzyme (Tavender and Bulleid, 2010a). As PrxIV is able to very efficiently react with any H2O2 produced in the ER, is there a requirement to regulate the activity of PrxIV? Other cellular peroxiredoxins can be inhibited in the presence of high concentrations of H2O2 by the over oxidation of the peroxidatic cysteine residue to form sulphinic or sulphonic derivatives (Woo et al, 2003). The expression of a deregulated mutant of Ero1α results in a marked increase in the concentration of GSSG in the ER (Appenzeller-Herzog et al, 2008), and results in the formation of over oxidized PrxIV (Tavender and Bulleid, 2010b). In addition, treating cells with DTT causes a rapid over oxidation of PrxIV presumably due to an acute increase in H2O2 caused by a rapid reduction of the regulatory disulphides in Ero1α and in substrate proteins. Hence, it may well be the case that, under certain stress conditions, PrxIV becomes over oxidized thereby attenuating peroxidase activity and allowing H2O2 to act as a signal for ER oxidative stress.
There are potentially several mechanisms for maintaining a redox balance and preventing oxidant build-up in the ER (Gorlach et al, 2006), so it is possibly not surprising that the mouse knockout of PrxIV, like the Ero1α knockouts, is still viable with the only notable phenotype being an effect on testicular morphology (Iuchi et al, 2009). The lack of broad effects of PrxIV knockout upon mouse physiology indicates that a robust antioxidant defence remains even in the absence of PrxIV. This may be consistent with PrxIV—like Ero1—forming one of several integrated systems to facilitate disulphide formation and to regulate ER redox homeostasis.
Materials and methods
Reagents and antibodies
All reagents were acquired from Sigma-Aldrich (Dorset, UK) and enzymes were acquired from Promega (Southampton, UK) unless otherwise stated. Rabbit polyclonal anti-PrxIV was purchased from Ab Frontier (Seoul, Korea), rabbit polyclonal anti-calreticulin was purchased from Stressgen (Exeter, UK) and rabbit polyclonal anti-ERp46 was purchased from AbCam (Cambridge, UK). Rabbit polyclonal antibodies raised against PDI, P5 and ERp57 have been described previously (Jessop and Bulleid, 2004), while purified protein prepared for this study was used to raise a rabbit polyclonal antibody to ERp18. Mouse monoclonal anti-V5 was obtained from Invitrogen (Paisley, UK).
Preparation of purified proteins
All proteins (minus secretory signal peptides) were expressed and purified using Escherichia coli BL21-DE3. His-tagged PrxIV purification has been described previously (Tavender and Bulleid, 2010b), and ERp18 was cloned into pRSFDuet-1 (Novagen, Nottingham, UK) for His-tagged protein purification in an identical manner. His-tagged PDI was similarly expressed and purified by Ni2+ column chromatography as described previously (Baker et al, 2008) as was His-tagged ERp57, following expression from a plasmid kindly donated by Stephen High (University of Manchester, UK), and His-tagged P5 following cloning into pET21a(+) (Novagen). A construct expressing a GST-ERp46 fusion protein was a kind gift from David Thomas (McGill University, Montreal, Canada). GST-ERp46 was expressed for 4 h using 0.5 mM IPTG induction, bound to GSH-sepharose (GE Healthcare, Buckinghamshire, UK) by standard batch purification and ERp46 eluted by on-column cleavage with precision protease (GE Healthcare) in accordance with the manufacturer's instructions. Proteins were reduced by incubation with 5 mM DTT for 15 min at room temperature then DTT removed by buffer exchange into reaction buffer (50 mM NaH2PO4, 150 mM NaCl, 1 mM EDTA, pH 7.4) using a PD10 column or Superdex 200 gel filtration column (both GE Healthcare). Each protein was quantified using the relevant 280 nm absorption extinction coefficient.
Reduction of PrxIV in vitro
PrxIV and ER oxidoreductases were incubated in reaction buffer at 37°C at the concentrations and time periods indicated in the text. Where required, GSH was added from a 10 × stock (dissolved in reaction buffer and adjusted to pH 7 with NaOH). Reactions were quenched by addition of four volumes of 12.5% w/v TCA in H2O, incubated on ice for 15 min and precipitated proteins pelleted by centrifugation for 15 min at 15 000 g, 4°C. Pellets were vortexed to re-suspend in ice-cold acetone and pelleted again for 5 min. Following a repeat acetone wash pellets were air dried for 15 min and re-suspended in either SDS sample buffer (31.25 mM Tris–HCl pH 6.8, 2% w/v SDS, 5% v/v glycerol, 0.01% w/v bromophenol blue) containing 40 mM NEM or alkylation buffer (200 mM Tris–HCl pH 8, 0.5% SDS, 6 M urea) containing 10 mM AMS or 50 mM IAA as appropriate. In the case of SDS sample buffer plus NEM, samples were vortexed and immediately heated at 100°C for 5 min. For alkylation buffer plus AMS/IAA, samples were incubated for 30 min in the dark at room temperature then 0.25 volumes of 5 × SDS sample buffer added and boiled as above.
Mass spectrometry determination of PDI redox condition
This analysis was performed exactly as recently described (Chambers et al, 2010).
GSH-dependent reduction of PrxIV in SP cells
Where required, cells were initially transfected with recombinant DNA to modulate ER oxidoreductase levels. For overexpression, cells were seeded to 60% confluence in 15 cm cell culture dishes and transfected with 12 μg plasmid DNA encoding the appropriate V5-tagged PDI-family member (construct creation described elsewhere (Jessop et al, 2009b)). Transfection was performed using polyethylenimine exactly as established previously (Gleghorn et al, 2010), with volumes increased three-fold to reflect increased quantity of DNA. Cells were harvested for SP-cell preparation at 48 h post-transfection. For knockdown of PDI-family expression, cells were transfected as above using DNA constructs encoding shRNA against the relevant PDI-family members (Origene, Rockville, MD, USA). At 16 h post-transfection, puromycin selection (1 μg/ml) was introduced to remove cells not expressing shRNA and maintained for a further 4–5 days before preparation of SP cells. SP cells were prepared as detailed previously (Wilson et al, 1995) and suspended in KHM buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM magnesium acetate, pH 7.2 with KOH) at a final concentration of 107 SP cells/ml. For protein expression controls, samples containing 3 × 105 SP cells were immediately transferred to ice. GSH was added to remaining suspension from a 10 × stock (dissolved in KHM buffer and adjusted to pH 7 with NaOH) and incubated at 37°C. At required times, samples containing 3 × 105 SP cells were removed to ice and NEM added to 40 mM final concentration. After a minimum of 5 min, nine volumes of KHM were added and cells were pelleted by centrifugation for 2 min at 4000 g. Supernatants were removed and cells were lysed by addition of 30 μl SDS sample buffer and heated at 100°C for 5 min.
Trapping mixed disulphides with PrxIV translated in vitro
PrxIV translation and translocation into SP cells was performed essentially as described previously (Tavender et al, 2008). RNA transcripts were prepared by T7 transcription from XhoI linearized pcDNA3.1/hygro(+) constructs encoding PrxIV or mutants thereof. Following translation for 1 h at 30°C, cells were treated with 40 mM NEM for 5 min on ice then diluted with nine volumes of ice-cold KHM buffer. One fifth of suspension was centrifuged for 1 min at 10 000 g, supernatant was removed and SP cells were lysed in SDS sample buffer for analysis of total translation products. Remaining suspension was centrifuged as above, supernatant was removed and cells were lysed by vortexing with 50 μl IP buffer (50 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 0.5 mM PMSF, 1% v/v Triton X-100). Lysates were clarified by centrifugation for 2 min at 16 000 g and supernatant was mixed with 10% w/v SDS for final concentration 1% w/v SDS. After 3 min at 100°C, samples were diluted to 1.2 ml with IP buffer and incubated with 50 μl 10% w/v Protein A sepharose (PAS) for 30 min at 4°C with rotation. PAS was removed by centrifugation for 1 min at 16 000 g and supernatants were mixed with 50 μl fresh PAS and 1 μl anti-V5 antibody before incubation for 16 h at 4°C with rotation. PAS was harvested by centrifugation as above, and supernatants were discarded. PAS was washed three times with IP buffer and bound proteins were released by addition of SDS sample buffer and heating at 100°C for 5 min.
Electrophoresis, western blotting and densitometry
For Coomassie blue staining of purified proteins, 2–4 μg of protein was typically loaded. For western blot analysis, 400 ng was used. For western blots of SP-cell lysates, an equivalent of 105 cells were used per sample under reducing conditions, 1.5 × 105 cells under non-reducing conditions. For SDS–PAGE under reducing conditions, DTT was added to samples at 50 mM concentration before boiling, for non-reducing conditions DTT was omitted. Gels were stained overnight using colloidal Coomassie blue (10% w/v phosphoric acid, 10% w/v ammonium sulphate, 0.12% Coomassie brilliant blue G250, 20% v/v methanol) followed by destain in distilled water or were transferred to nitrocellulose for western blotting. Due to problems with high background observed using alternative western blotting protocols, non-reducing anti-PrxIV blots of SP cell derived material were performed exactly as described previously (Tavender et al, 2008). For all other samples, a system was used incorporating fluorescent secondary antibodies. For fluorescent western blots, proteins were transferred to Odyssey nitrocellulose (Li-Cor Biosciences, Cambridge, UK), for traditional western blots, standard Protran membrane (Whatman, UK) was used. Blocking and primary antibody incubations for fluorescent blots were as previously detailed (Tavender et al, 2008). Secondary antibody differed only in dilution factor (1 in 2500) and in being performed in a light-shielded box. Following final wash steps, blots were scanned using an Odyssey SA imaging system (Li-Cor Biosciences). For quantification of fluorescent western blots, scans were performed at minimum intensities required to visualize all relevant species. Densitometry was then performed upon unmodified output images using ImageJ (NIH, USA). For traditional blots, multiple exposures were performed and blots were selected in which samples exhibited a linear response to the chemiluminescent substrate. Band intensities were again quantified using ImageJ. In all cases, identical sized boxes were used for all protein samples within a given blot and a local background also subtracted for each individual sample. Due to the absence of cytosolic proteins, Crt was used as a loading control for quantification of SP-cell protein content. During initial experiments, samples were first probed with anti-Crt, then western blot membranes stained using amido-black to visualize total protein. Crt levels were reliably unaffected by modulation of PDI-family expression and therefore suitable as a reference standard. Where required, bound antibodies were stripped from nitrocellulose membranes by 2 × 5 min washes in 0.2 M NaOH followed by 5 min in distilled water. Primary antibodies were then re-applied without the need for further blocking.
Supplementary Material
Acknowledgments
We acknowledge the generosity of Stephen High (University of Manchester, UK) and Professor David Thomas (McGill University, Montreal, Canada) for their contribution of reagents and Gordon Lindsay (University of Glasgow) and other members of the Bulleid group for critical reading of the manuscript. This work was funded by the Wellcome Trust grant number, 88053.
Footnotes
The authors declare that they have no conflict of interest.
References
- Appenzeller-Herzog C, Ellgaard L (2008) The human PDI family: versatility packed into a single fold. Biochim Biophys Acta 1783: 535–548 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Riemer J, Christensen B, Sorensen ES, Ellgaard L (2008) A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J 27: 2977–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Riemer J, Zito E, Chin KT, Ron D, Spiess M, Ellgaard L (2010) Disulphide production by Ero1alpha-PDI relay is rapid and effectively regulated. EMBO J 29: 3318–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KM, Chakravarthi S, Langton KP, Sheppard AM, Lu H, Bulleid NJ (2008) Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J 27: 2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ (2007) Intracellular catalysis of disulfide bond formation by the human sulfhydryl oxidase, QSOX1. Biochem J 404: 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Tavender TJ, Oka OB, Warwood S, Knight D, Bulleid NJ (2010) The reduction potential of the active site disulfides of human protein disulfide isomerase. J Biol Chem 285: 29200–29207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE, Hillson DA, Freedman RB (1980) Catalysis by protein-disulphide isomerase of the unfolding and refolding of proteins with disulphide bonds. J Mol Biol 142: 43–62 [DOI] [PubMed] [Google Scholar]
- Dick TP, Cresswell P (2002) Thiol oxidation and reduction in major histocompatibility complex class I-restricted antigen processing and presentation. Methods Enzymol 348: 49–54 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA (1998) The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell 1: 161–170 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA (1999) Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell 4: 469–477 [DOI] [PubMed] [Google Scholar]
- Gess B, Hofbauer KH, Wenger RH, Lohaus C, Meyer HE, Kurtz A (2003) The cellular oxygen tension regulates expression of the endoplasmic oxidoreductase ERO1-Lalpha. Eur J Biochem 270: 2228–2235 [DOI] [PubMed] [Google Scholar]
- Gleghorn LJ, Trump D, Bulleid NJ (2010) Wild-type and missense mutants of retinoschisin co-assemble resulting in either intracellular retention or incorrect assembly of the functionally active octamer. Biochem J 425: 275–283 [DOI] [PubMed] [Google Scholar]
- Gorlach A, Klappa P, Kietzmann T (2006) The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal 8: 1391–1418 [DOI] [PubMed] [Google Scholar]
- Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D (2006) Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA 103: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober KL, Sheasley SL, Gilbert HF, Thorpe C (1999) Sulfhydryl oxidase from egg white. A facile catalyst for disulfide bond formation in proteins and peptides. J Biol Chem 274: 22147–22150 [DOI] [PubMed] [Google Scholar]
- Iuchi Y, Okada F, Tsunoda S, Kibe N, Shirasawa N, Ikawa M, Okabe M, Ikeda Y, Fujii J (2009) Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem J 419: 149–158 [DOI] [PubMed] [Google Scholar]
- Jansens A, van Duijn E, Braakman I (2002) Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science 298: 2401–2403 [DOI] [PubMed] [Google Scholar]
- Jessop CE, Bulleid NJ (2004) Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem 279: 55341–55347 [DOI] [PubMed] [Google Scholar]
- Jessop CE, Tavender TJ, Watkins RH, Chambers JE, Bulleid NJ (2009a) Substrate specificity of the oxidoreductase ERp57 is determined primarily by its interaction with calnexin and calreticulin. J Biol Chem 284: 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ (2009b) Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci 122: 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina EV, Chernov NN, Saprin AN (2008) Involvement of thio-, peroxi-, and glutaredoxins in cellular redox-dependent processes. Biochemistry (Mosc) 73: 1493–1510 [DOI] [PubMed] [Google Scholar]
- Karala AR, Lappi AK, Saaranen MJ, Ruddock LW (2009) Efficient peroxide-mediated oxidative refolding of a protein at physiological pH and implications for oxidative folding in the endoplasmic reticulum. Antioxid Redox Signal 11: 963–970 [DOI] [PubMed] [Google Scholar]
- Margittai E, Banhegyi G (2010) Oxidative folding in the endoplasmic reticulum: towards a multiple oxidant hypothesis? FEBS Lett 584: 2995–2998 [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, van der Wal FJ, Bulleid NJ, High S (1997) Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science 275: 86–88 [DOI] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1: 171–182 [DOI] [PubMed] [Google Scholar]
- Rancy PC, Thorpe C (2008) Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry 47: 12047–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaranen MJ, Karala AR, Lappi AK, Ruddock LW (2010) The role of dehydroascorbate in disulfide bond formation. Antioxid Redox Signal 12: 15–25 [DOI] [PubMed] [Google Scholar]
- Schulman S, Wang B, Li W, Rapoport TA (2010) Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci USA 107: 15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2006) Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal 8: 797–811 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA (2007) Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129: 333–344 [DOI] [PubMed] [Google Scholar]
- Simmen T, Lynes EM, Gesson K, Thomas G (2010) Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim Biophys Acta 1798: 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DC, Huminiecki L, Moore JW, Boyle JJ, Poulsom R, Creamer D, Barker J, Bicknell R (2003) EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem 278: 47079–47088 [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Bulleid NJ (2010a) Molecular mechanisms regulating oxidative activity of the ero1 family in the endoplasmic reticulum. Antioxid Redox Signal 13: 1177–1187 [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Bulleid NJ (2010b) Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J Cell Sci 123: 2672–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender TJ, Sheppard AM, Bulleid NJ (2008) Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem J 411: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien AC, Rajan A, Schulze KL, Ryoo HD, Acar M, Steller H, Bellen HJ (2008) Ero1 L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J Cell Biol 182: 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Weissman JS (2002) The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell 10: 983–994 [DOI] [PubMed] [Google Scholar]
- van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJ (2003) Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity 18: 243–253 [DOI] [PubMed] [Google Scholar]
- Van Buul JD, Fernandez-Borja M, Anthony EC, Hordijk PL (2005) Expression and localization of NOX2 and NOX4 in primary human endothelial cells. Antioxid Redox Signal 7: 308–317 [DOI] [PubMed] [Google Scholar]
- Wajih N, Hutson SM, Wallin R (2007) Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J Biol Chem 282: 2626–2635 [DOI] [PubMed] [Google Scholar]
- Wilson R, Allen AJ, Oliver J, Brookman JL, High S, Bulleid NJ (1995) The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J 307 (Part 3): 679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG (2003) Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem 278: 47361–47364 [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schroder E, Robin Harris J, Poole LB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40 [DOI] [PubMed] [Google Scholar]
- Zito E, Chin KT, Blais J, Harding HP, Ron D (2010) ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol 188: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.