Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing
The production and processing of ribosomal RNA is an essential and complex process. Here, a polynucleotide 5′-kinase, Nol9, is shown to have an important function in pre-rRNA processing and 60S ribosomal subunit biogenesis.
Keywords: nucleolus, polynucleotide kinase, ribosomal RNA, ribosomal RNA processing
Abstract
In a cell, an enormous amount of energy is channelled into the biogenesis of ribosomal RNAs (rRNAs). In a multistep process involving a large variety of ribosomal and non-ribosomal proteins, mature rRNAs are generated from a long polycistronic precursor. Here, we show that the non-ribosomal protein Nol9 is a polynucleotide 5′-kinase that sediments primarily with the pre-60S ribosomal particles in HeLa nuclear extracts. Depletion of Nol9 leads to a severe impairment of ribosome biogenesis. In particular, the polynucleotide kinase activity of Nol9 is required for efficient generation of the 5.8S and 28S rRNAs from the 32S precursor. Upon Nol9 knockdown, we also observe a specific maturation defect at the 5′ end of the predominant 5.8S short-form rRNA (5.8SS), possibly due to the Nol9 requirement for 5′>3′ exonucleolytic trimming. In contrast, the endonuclease-dependent generation of the 5′-extended, minor 5.8S long-form rRNA (5.8SL) is largely unaffected. This is the first report of a nucleolar polynucleotide kinase with a role in rRNA processing.
Introduction
Ribosomal RNA (rRNA) maturation represents a dramatic example of RNA processing, embracing a large number of enzymatic activities, small nucleolar RNAs (snoRNAs) and non-ribosomal proteins (Henras et al, 2008). rRNA processing is best understood in the yeast Saccharomyces cerevisiae (Venema and Tollervey, 1999). Analogous processing events and the high conservation of the proteins involved suggest substantial similarities among several organisms, yet individual steps were shown to be variable (Gerbi and Borovjagin, 2004).
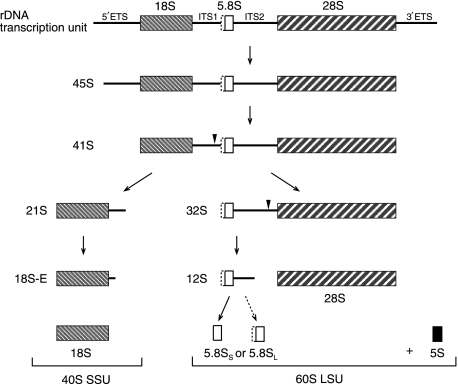
Three out of four human rRNAs, 18S, 5.8S and 28S, are transcribed from one polycistronic transcription unit (Gerbi and Borovjagin, 2004). In an ordered series of endo- and exonucleolytic events, external and internal transcribed spacers (ETS and ITS, respectively) are removed from the primary transcript and the mature rRNAs liberated (Figure 1) (Hadjiolova et al, 1993). Immediately after transcription, external spacer sequences are degraded, generating first 45S and then 41S intermediates. A subsequent endonucleolytic cleavage within ITS1 splits the 41S precursor into the 21S and 32S rRNAs. The 21S is further processed via the 18S-E intermediate into the mature 18S rRNA, the RNA component of the 40S small ribosomal subunit (SSU). Processing of 32S is more complex, involving an elusive endonuclease activity that cleaves within ITS2. Eventually, the mature 5.8S and 28S rRNAs are liberated and assemble, together with the independently transcribed and processed 5S rRNA, into the 60S large ribosomal subunit (LSU). Two forms of 5.8S have been described in yeast and mammals (Rubin, 1974; Bowman et al, 1983), a major short form (5.8SS) and a long, 5′-extended form (5.8SL).
Figure 1.
The 18S, 5.8S and 28S rRNAs are organized into a single polycistronic rDNA transcription unit, which also contains external transcribed spacers (ETS) on the 5′ and 3′ ends and two internal transcribed spacers (ITS). A series of endo- and exonucleolytic steps are required for proper maturation of rRNAs via various intermediates. The mature 18S rRNA is eventually assembled into the 40S small ribosomal subunit (SSU); 5.8S and 28S rRNAs together with the independently transcribed and processed 5S rRNA are core components of the 60S large ribosomal subunit (LSU). Two forms of 5.8S are reported to co-exist; the major 5.8SS(hort) and the 5′-extended 5.8SL(ong) form. Depicted are detectable intermediates of the major rRNA processing pathway in HeLa cells (derived from Hadjiolova et al, 1993; Rouquette et al, 2005). Boxes represent rRNAs, triangles mark relevant endonucleolytic cleavage sites.
Recent advances in large-scale mass spectrometry and high throughput screens have revealed a multitude of proteins to be involved in rRNA processing (Andersen et al, 2002; Scherl et al, 2002; Boisvert et al, 2010), yet detailed studies on their individual roles are missing and important enzymatic activities are still elusive.
Our laboratory previously identified Clp1, an RNA 5′-kinase phosphorylating tRNA exons and siRNAs in vitro (Weitzer and Martinez, 2007b). Clp1 was initially described as a component of the mRNA 3′ end formation and polyadenylation machinery (de Vries et al, 2000) and was later also implicated in the splicing of precursor tRNAs as a binding partner of the Sen endonuclease (de Vries et al, 2000; Paushkin et al, 2004). Bioinformatic analysis revealed a family of proteins closely related to Clp1, the ‘Grc3/Nol9 family' (Braglia et al, 2010), that contains Walker A and Walker B motifs, both implicated in ATP/GTP binding (Walker et al, 1982). Interestingly, human Nol9 was previously detected in proteomic analyses of the nucleolus (Andersen et al, 2002; Scherl et al, 2002). Temperature sensitive mutants of the yeast homologue of Nol9, Grc3, showed an rRNA processing defect in a global screen for non-coding RNA processing (Peng et al, 2003); yet, the role of Grc3 is not clarified.
Here, we identify Nol9 as a novel polynucleotide 5′-kinase that primarily co-sediments with nuclear pre-60S particles in HeLa cells. We show that the kinase activity of Nol9 is required for efficient processing of the 32S precursor into 5.8S and 28S rRNAs and present evidence for two different processing pathways generating the two forms of 5.8S, similar to the situation in yeast. This is the first implication of a polynucleotide kinase activity in the rRNA maturation pathway.
Results
Nol9 is a polynucleotide kinase
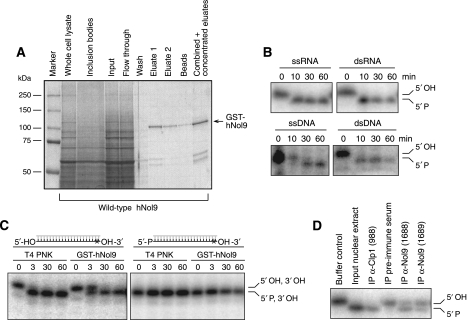
To determine whether Nol9 displays polynucleotide kinase activity, we expressed and purified GST-tagged human Nol9 from insect cells (Figure 2A). Recombinant Nol9 phosphorylated single-stranded and double-stranded (ds) RNA and DNA substrates with high efficiency (Figure 2B). In order to analyse which terminus becomes phosphorylated, we incubated GST-tagged Nol9 with dsRNA substrates displaying either a phosphate or a hydroxyl group at the 5′ or 3′ ends (Figure 2C). As a control, we used the well-studied T4 polynucleotide kinase (T4 PNK), which possesses kinase activity towards the 5′ end (Wang and Shuman, 2002). Nol9 was able to transfer a phosphate to 5′ ends of dsRNAs, but could not phosphorylate 3′ termini, thus displaying the same 5′-specificity as T4 PNK. We were also able to immunopurify Nol9 RNA 5′-kinase activity from nuclear extracts using specific antisera (Figure 2D). We conclude that Nol9 is a 5′ end-specific RNA/DNA kinase.
Figure 2.
Human Nol9 is a polynucleotide 5′-kinase. (A) Coomassie blue-stained SDS–PAGE to monitor Nol9 purification. GST-tagged wild-type Nol9 was expressed in sf9 insect cells for 4 days. Cells were lysed by sonication and whole cell lysates separated by centrifugation into non-soluble inclusion bodies and the soluble fraction, which was used as input in the purification procedure. After incubation with the soluble fractions, the glutathione beads were washed and subsequently the bound proteins eluted twice with glutathione. Only a minor fraction of the protein stayed associated with the beads. Before being used in kinase assays, the two eluates were combined and concentrated. (B) Nol9 phosphorylates single (ss)- or double-stranded (ds) RNA and DNA substrates. Purified GST-tagged hNol9 was incubated with the indicated substrates for the indicated time points. Phosphorylation was monitored by a mobility shift in electrophoresis. (C) Nol9 phosphorylates 5′ ends of dsRNA substrates. Purified GST-Nol9 was incubated with the indicated RNA substrates displaying either 5′ and 3′ hydroxyl or 5′ phosphate and 3′ hydroxyl groups. Kinase activity was monitored over time. T4 polynucleotide kinase (PNK) was used as a control. Asterisk indicates radioactive label within substrate RNA. (D) Endogenous Nol9 phosphorylates RNA substrates. 5′-phosphorylation of dsRNA substrates after immunoprecipitation (IP) from HeLa nuclear extracts using antisera against Nol9 or Clp1.
Nol9 co-sediments mainly with pre-60S rRNP particles
Nol9 was previously found in high-throughput proteomic analyses of the human nucleolus (Andersen et al, 2002; Scherl et al, 2002; Boisvert et al, 2010), the main site of rRNA maturation (Boisvert et al, 2007). Already during transcription, the nascent rRNA precursors are packaged into precursor ribosomal particles (pre-rRNP), containing a multitude of ribosomal and nucleolar proteins (Warner and Soeiro, 1967; Granneman and Baserga, 2004). These complexes allow for efficient and accurate rRNA maturation and also ensure quality control. Two major forms of pre-rRNP particles are reported for HeLa nuclear fractions; the pre-40S rRNP, containing the maturing 18S rRNA, and the pre-60S rRNP particles, which include the 5.8S and 28S rRNAs and their precursors (Vaughan et al, 1967; Warner and Soeiro, 1967).
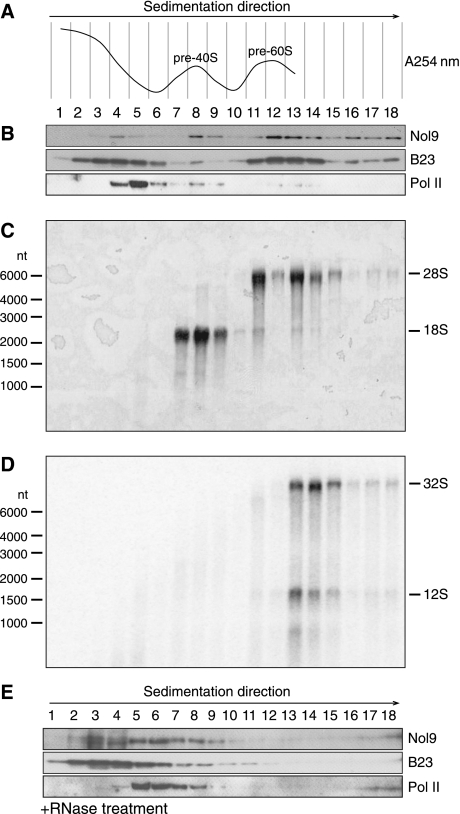
To investigate whether Nol9 associates with pre-rRNPs, we separated nuclear extracts from HeLa cells on a 10–30% sucrose density gradient and collected 18 fractions by downwards displacement. Each fraction was monitored by UV spectrometry at 254 nm to determine RNA content; yet, fractions 14–18 could not be measured due to the high sucrose concentration. Peaks comprising fractions 7–9 and 11–13 were observed, corresponding to the pre-40S and pre-60S rRNP particles, respectively (Figure 3A). Every fraction was tested for protein and RNA composition. Immunoblotting revealed that Nol9 accumulated mainly in fractions 12 and 13 (Figure 3B). Nucleophosmin/B23, which co-localizes with ribosomal proteins of the 60S ribosomal subunit in HeLa nucleoli (Maggi et al, 2008), was also over-represented in these fractions together with the mature 28S rRNA (Figure 3C) and 12S and 32S RNA, the precursors of 5.8S and 28S rRNAs, respectively (Figure 3D). RNA polymerase II was used as a control, as it is not part of rRNP particle, and sedimented at lower weight fractions (Figure 3B). A significant amount of Nol9 protein sedimented in fraction 8, in which mature 18S rRNA accumulated (Figure 3B). It appears that Nol9 is enriched to a certain extent within both the pre-40S and pre-60S particles; however, Nol9 seems to preferentially associate with the pre-rRNP containing 28S and its precursor.
Figure 3.
Nol9 co-fractionates mainly with pre-60S particles. (A) HeLa nuclear extracts were separated on a 10–30% sucrose density gradient and divided into 18 fractions by downwards displacement. UV absorption at 254 nm of fractions 1–13 was measured to follow RNA concentration and displayed in a graph. All fractions were analysed for RNA and protein content. (B) Fractions from (A) were loaded onto an SDS–PAGE to monitor Nol9 fractionation by immunoblotting using affinity-purified Nol9 antiserum. B23 and RNA polymerase II were used as controls. (C) RNA was extracted from fractions in (A) and separated by electrophoresis, transferred to a membrane and stained with methylene blue to visualize 18S and 28S rRNAs. RNA from fraction 12 was greatly lost during this procedure. (D) After methylene blue staining, the membrane from (C) was hybridized with probe NB39 (see Figure 4B) to detect 32S and 12S processing intermediates. (E) Nol9 sedimentation is RNA dependent. Experiment was carried out as in (B), but the nuclear extracts were treated with RNase A before ultracentrifugation.
The composition of pre-rRNP particles is dependent on the presence of RNA. Treating the nuclear extracts with RNase A before centrifugation resulted in a different UV absorption curve over the gradient, with no obvious peaks for the pre-rRNP particles (data not shown). RNase A treatment did not alter sedimentation of RNA polymerase II (Figure 3E). Interestingly, Nol9 and B23 were largely shifted from fractions around 12 to lighter fractions (Figure 3E). This result shows that Nol9 associates with RNAs in nuclear extracts and is possibly part of pre-rRNP particles.
Nol9 is required for rRNA processing
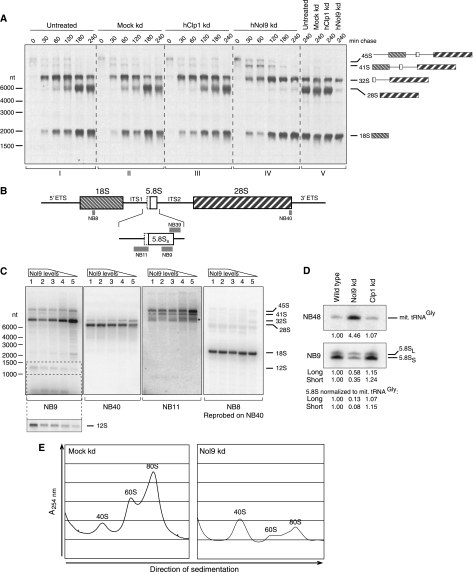
The above results together with the previous implication of the yeast homologue of Nol9, Grc3, in rRNA maturation (Peng et al, 2003) strongly suggested a role for Nol9 in the processing of human rRNA. Therefore, we metabolically labelled HeLa cells with 3H-methionine and chased rRNA maturation with non-radioactive methionine. As precursor rRNAs are heavily methylated early on, their processing can be tracked over time. While 45S and 41S precursors are rapidly processed in wild-type cells, the processing of 32S precursors is slow (Figure 4A, panels I and II). RNAi-mediated depletion (knockdown) of the RNA-kinase Clp1 did not interfere with the progress of rRNA maturation (panel III). Strikingly, depleting the cells of Nol9 for 6 days (91% knockdown efficiency, determined by qPCR, data not shown) led to a drastic decrease of mature 28S levels after 240 min of chase (panels IV and V), while the maturation of 18S was not affected. Additionally, processing of 45S and 41S precursors was slightly delayed. This suggests a role for Nol9 in the processing of rRNA, especially of the large subunit 28S rRNA.
Figure 4.
Depletion of Nol9 impairs ribosomal RNA processing. (A) Metabolic labelling of untreated (panel I) or mock-transfected HeLa cells (panel II), or HeLa cells transfected with siRNAs targeting Clp1 (panel III) or Nol9 (panel IV). Cells were starved in methionine-free medium followed by addition of 3H-methyl-methionine and chased in non-radioactive medium for the indicated time periods (in minutes). Panel V is shown to compare all four backgrounds after chasing for 240 min. (B) Illustration of the rRNA polycistronic transcript including the 5′ and 3′ external transcribed spacers (5′ETS and 3′ETS), the internal transcribed spacers 1 and 2 (ITS1 and ITS2) and the mature 18S, 5.8S and 28S rRNAs. Magnification of the 5.8S rRNA shows the short 5.8SS (white box) and the additional 5′ extension of the long 5.8SL form (dashed line). Probes for northern blot are indicated by grey boxes (NB8, NB9, NB11, NB39 and NB40). (C) Nol9 depletion affects maturation of 28S, but not 18S rRNA. A total of 0.5 μg total RNA containing wild-type levels of Nol9 mRNA (lanes 1 and 2), or obtained from different Nol9 knockdown experiments (efficiencies determined by qPCR, data not shown; 70% knockdown efficiency, lane 3; 83%, lane 4; 97%, lane 5) were separated by electrophoresis, transferred to a membrane and hybridized with the indicated probes. A longer exposure time was chosen for panel NB9 to monitor 12S levels. The asterisk in panel NB11 indicates 5′-elongated 32S intermediates. (D) Nol9 is required for maturation of the 5.8S rRNA. A total of 1 μg RNA from mock-transfected HeLa cells (lane 1) or cells transfected with siRNAs against Nol9 (lane 2) or Clp1 (lane 3) were hybridized with a probe against 5.8S (NB9); mitochondrial tRNAGly (NB48) was used as control. Signal intensities were quantified, normalized to lane 1 (wild type) and displayed beneath the blot. (E) Depletion of Nol9 results in a severe decrease of 60S and 80S ribosomes. Ribosome profiles were obtained from HeLa cells transfected with a mock control or siRNAs against Nol9. Cytoplasmic extracts were separated on a 10–45% sucrose density gradient. UV absorption along the gradient was measured.
Nol9 functions in the processing of large subunit rRNAs
To further investigate processing defects, we conducted a series of northern blots, probing for different processing intermediates and products of the rRNA maturation pathway (Figure 4B). Three distinct strategies were used to knockdown Nol9, leading to different knockdown efficiencies of 70, 83 and 97% (Figure 4C, lanes 3, 4 and 5, respectively). The effect on rRNA maturation correlated with the Nol9 knockdown efficacy. Probes against the 5.8S (NB9) and 28S (NB40) showed elevated levels of the 32S processing intermediate and lower levels of all detectable downstream processing products, including the 12S intermediate and the mature 28S and 5.8S rRNAs (Figure 4C, panels NB9 and NB40; Figure 4D, panel NB9). As observed in the pulse chase experiment (Figure 4A), the early intermediates 45S and 41S precursor rRNA accumulated in correlation with the knockdown efficiency of Nol9 (Figure 4C, panel NB11). Maturation of the 18S rRNA was not influenced by Nol9 knockdown (Figure 4C, panel NB8). When we used probes against the U6 snRNA (data not shown) or the mitochondrial Glycine-tRNA (Figure 4D, NB48) as loading controls for the northern blots, we observed unequal signals, although total RNA amounts loaded were always the same. We suggest that this is due to an alteration of the overall composition of extracted RNA as a consequence of Nol9 depletion, resulting from the vast absence of the 28S and 5.8S rRNAs. As rRNAs make up the majority of cellular RNAs, decreasing rRNA levels might result in a relative enrichment of other RNA classes in total RNA extracts. Acknowledging this theory, we normalized the 5.8S levels observed in Figure 4D (NB9) to those of the mitochondrial Glycine-tRNA (NB48). Nol9 knockdown decreased the absolute levels of 5.8SS to 0.08 (8%) and of 5.8SL to 0.13 (13%) of mock-transfected cells.
The 5.8S and 28S rRNAs are core components of the large 60S ribosomal subunit, whereas the 18S is assembled into the small 40S ribosomal subunit (Venema and Tollervey, 1999; Fatica and Tollervey, 2002; Fromont-Racine et al, 2003). To investigate whether the levels of 40S SSU, 60S LSU and 80S ribosomes were imbalanced upon depletion of Nol9, we carried out HeLa cytoplasmic extract sedimentation studies. We observed a clear defect in 80S ribosome biogenesis in HeLa cells depleted of Nol9 as a direct effect of the vast absence of 60S ribosomal subunit (Figure 4E). This clearly indicates a requirement of Nol9 for the maturation of the LSU and hence the ribosome.
The polynucleotide kinase activity of Nol9 is required for rRNA processing
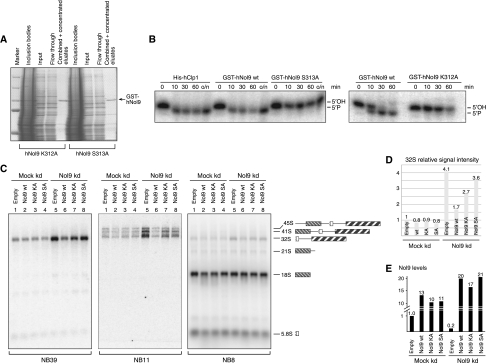
To differentiate whether the kinase activity of Nol9 itself is essential for rRNA maturation or Nol9 only serves as a scaffold protein within a larger complex, we expressed and purified kinase-inactive Nol9 mutants (Figure 5A). Nol9 contains Walker A and a Walker B motifs which are both implicated in ATP/GTP binding (Walker et al, 1982). Mutations within these domains often impair enzymatic activity (Wang and Shuman, 2001). The Walker A motif consists of the peptide stretch GxxxxGKS/T, which we altered by exchanging the lysine or serine (positioned at amino-acid 312 and 313 in human Nol9, respectively) with an alanine (referred to as K312A or S313A, respectively). These mutations efficiently abolished kinase activity in recombinant Nol9 confirming that the Walker A motif is essential for Nol9 activity (Figure 5B). To investigate the role of the kinase activity of Nol9 in rRNA processing, we first depleted Nol9 from HeLa cells and then expressed either wild-type or Walker A mutant human Nol9. The 32S precursor levels were monitored by northern blot and subsequently quantified by signal intensity measurement. Upon Nol9 knockdown, the levels of 32S were increased by a factor of 4.1 (Figure 5C, panel NB39, and D). Whereas concurrent over-expression of wild-type Nol9 diminished the elevated 32S levels to only 1.7, both mutated forms of hNol9, K312A and S313A, could mitigate the effect of the Nol9 knockdown on 32S levels only slightly (Figure 5C, panel NB39, and D). The S313A mutation seemed to be more severe than the K312A mutation which showed a mild rescue effect on the levels of 32S precursor rRNA. Equally, 41S and 45S precursor levels could be efficiently decreased by over-expressing wild type but not kinase-inactive Nol9 (Figure 5C, panel NB11). Probing for 18S levels was used to monitor equal loading, knowing that Nol9 does not alter its maturation. Wild-type and mutant forms of Nol9 were expressed with equal efficiency as determined by qPCR (Figure 5E), excluding that the above results were due to different expression levels. Therefore, our data strongly suggest that the kinase activity of Nol9 is required for efficient rRNA processing.
Figure 5.
The kinase activity of Nol9 is required for efficient rRNA processing. (A) Coomassie blue-stained SDS–PAGE to monitor purification of the Nol9 mutants, as Figure 2A. (B) Nol9 Walker A mutations K312A and S313A abolish kinase activity. Equal amounts of purified GST-Nol9 (wild type or mutant) were incubated with double-stranded RNA substrates for indicated time points in minutes; o/n, over night; His-tagged Clp1 was used as control. (C) HeLa cells were first transfected with an empty vector (mock kd) or a short hairpin construct targeting the 3′UTR of Nol9 (Nol9 kd). In the second round of transfection, either an empty vector or a vector encoding wild-type Nol9, mutant version K312A or S313A of Nol9 was co-transfected. RNA was extracted, separated by electrophoresis, transferred to a membrane and hybridized using indicated probes. (D) Signal intensities of 32S precursor from the above northern blots in (C), panel NB39, were quantified and displayed in a graph. (E) qPCR analysis of Nol9 mRNA levels in the RNA extracts used in (C). Values were normalized to the mock knockdown/empty vector control.
Nol9 knockdown affects the ratio between short and long 5.8S rRNA
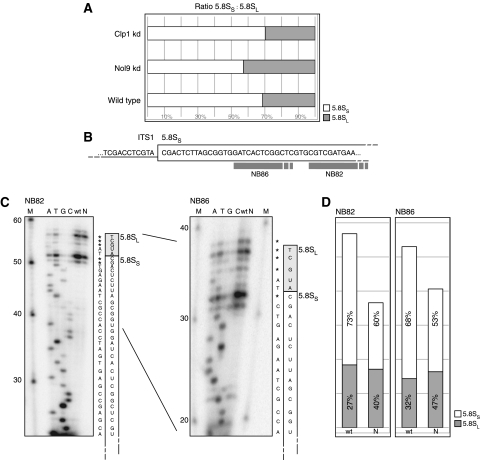
In yeast, two alternate processing pathways are described to generate the 5′end of 5.8S rRNAs producing either the ‘short' (‘5.8SS') or the extended form of 5.8S rRNA bearing six additional nucleotides on the 5′ end (‘long' or ‘5.8SL') (Schmitt and Clayton, 1993; Henry et al, 1994; Faber et al, 2006). In mammals, 5′ heterogeneity of 5.8S rRNA was also described (Khan and Maden, 1977; Bowman et al, 1983; Smith et al, 1984). Aside the decrease of total 5.8S rRNA upon knockdown of Nol9 (Figure 4D, panel NB9), we also observed an alteration of the ratio between the short and the long 5.8S rRNAs (Figure 6A). Whereas in wild-type HeLa cells 5.8SS comprised 70% and 5.8SL only 30% of all mature 5.8S rRNAs, Nol9 knockdown reproducibly shifted this ratio towards the long 5.8SL form, resulting in ∼55% 5.8SS versus ∼45% 5.8SL. Primer extension analysis using two individual primers near the 5′ end of 5.8S (Figure 6B) confirmed this observation; the majority of transcripts terminated at the predicted 5′ end of 5.8S in wild-type RNA preparations, while a smaller fraction of 5′-extended transcript was detected (Figure 6C and D). Although we could not predict the exact sequence of the 5′ extension from the primer extension analysis, it still allowed us to conclude that 5.8SL was 5 nucleotides longer than 5.8SS in HeLa cells and that Nol9 knockdown decreased the short form, but did not alter the relative levels of the long form of 5.8S (Figure 6D). Altogether, we conclude that, similar to the situation in yeast, in HeLa cells, two alternate pathways for the generation of the 5′ end of 5.8S co-exist, resulting in either the short or long 5.8S rRNA. Affecting the generation of the major short form is not completely rescued by an increase of the levels of the long form, suggesting that the pathway for 5.8SL cannot complement the loss of the 5.8SS maturation machinery. Therefore, the polynucleotide kinase Nol9 is primarily involved in the generation of the major 5.8SS form.
Figure 6.
Nol9 knockdown affects the ratio between 5.8SS and 5.8SL. (A) Signal intensities of 5.8SS and 5.8SL in Figure 4D were measured, calculated ratios are depicted in a bar graph. (B) Genomic sequence of the ITS1-5.8S junction in the rDNA locus derived from NCBI (HSU13369). Grey boxes depict location of primers used in primer extension. (C) A total of 1 μg RNA from wild-type cells (‘wt') or Nol9 knockdown cells (‘N'; identical to the RNA used in Figure 4C, lanes 1 and 5, respectively) were used in primer extension with the indicated primer (NB82 or NB86). In parallel, sequencing reactions were performed using ddNTPs (A, ddATP; T, ddTTP; G, ddGTP; C, ddCTP) M, decade marker with size indications on the left side. The sequencing results are depicted on the right side of both gels, asterisks indicate unreadable results. According 5.8SS/L sequences as derived from the NCBI database (HSU13369) are illustrated inside the rectangles. (D) Signal intensities of 5.8SS and 5.8SL from (C) were quantified and plotted against the y axis. Calculated ratios between the short and long forms are stated within the columns.
Discussion
We report the involvement of Nol9, a novel polynucleotide 5′-kinase, in the maturation of the 60S ribosomal subunit. Nol9 is a potent RNA/DNA 5′-kinase (Figure 2) that mainly co-sediments with pre-60S rRNPs in nuclear extracts (Figure 3). Absence of Nol9 decreases 5.8S and 28S rRNA levels to a great extent and increases the levels of the 32S, 41S and 45S precursors (Figure 4). Interestingly, the mere presence of Nol9 in the cell is not enough to ensure efficient rRNA processing; only the wild-type, kinase-active form of Nol9 is capable to overcome rRNA processing defects upon prior depletion of Nol9 (Figure 5). Nol9 specifically impairs maturation of the major 5.8SS form, but does not seem to be involved in generating the minor 5.8SL form (Figure 6). As we could not identify the direct substrate for Nol9 in rRNA processing, we can only speculate on the role of the kinase activity. The obvious block of 32S processing implies that cleavage within ITS2, separating 5.8S and 28S, cannot occur. This can be due to (1) a direct effect of Nol9 on the endonuclease activity itself (Figure 7A, box 1) or (2) an upstream quality control check that detects improperly completed 32S processing in ITS1 (Good et al, 1997) (Figure 7A, box 2). Either option would lead to a transient accumulation of 32S as well as the upstream precursors 41S and 45S followed by premature degradation, as we observed upon Nol9 knockdown (Figure 4A).
Figure 7.
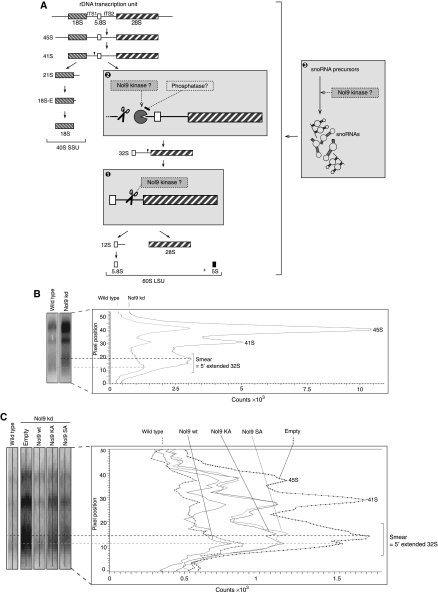
A model for the function of Nol9 in rRNA processing. (A) Possible roles of the polynucleotide kinase Nol9 in rRNA processing. (1) Nol9 might act directly on the endonuclease cleaving within ITS2. (2) Nol9 together with an elusive phosphatase activity might regulate 5′ end formation of 32S RNA by an exonuclease. (3) Nol9 might be required for snoRNA processing. Scissors indicate endonucleolytic cleavage sites, the ‘pacman' represents the exonuclease. RNAs are depicted in 5′>3′ orientation. (B) Nol9 might function in the 5′ end formation of 32S through 5′>3′ degradation. Lanes 1 and 5 of the northern blot from Figure 4C, panel NB11, were analysed using the ImageQuant analysis software by GE Healthcare. The signal intensities of the area depicted on the left side were gradually measured and illustrated in a graph. Dashed lines mark the pixel position of the intensity peaks in the smear. (C) Over-expression of wild-type Nol9 can rescue the impaired 5′ end formation of 32S rRNA upon prior Nol9 depletion. Lanes 5 to 8 of the northern blot from Figure 5C, panel NB11, were analysed as in (B).
As we show that the kinase activity of Nol9 itself is required for 32S processing (Figure 5C and D), Nol9 does not only serve as a scaffolding protein, but actually contributes to the rRNA maturation pathway with its kinase activity. Interestingly, the RNA-kinase Clp1, a homologue of Nol9, was found associated to the tRNA endonuclease complex (Paushkin et al, 2004). In yeast, the 3′ end of 18S is generated by the endonuclease Nob1 (Pertschy et al, 2009), which binds to the putative Walker A-type kinase Fap7 (Granneman et al, 2005). Although to date no definitive role of a polynucleotide kinase within an endonucleolytic event is identified, the recurring observations of complexes containing both kinase and endonuclease activities are intriguing (Figure 7A, box 1).
rRNA processing follows a series of events, and only the correct order assures proper maturation of the ribosomes. For yeast it was shown that the accurate removal of spacer sequences introduces conformational changes that allow the pathway to proceed to the next step (Cote et al, 2002). Most processing mutants only showed mild accumulations of precursors due to rapid and complete degradation by the exosome (Allmang et al, 2000), very similar to our observations for Nol9 depletion in HeLa cells. In yeast, the 5′ end of the 32S equivalent is mainly produced by the combined action of an endonucleolytic cleavage event upstream and subsequent exonucleolytic digestion by Rat1. A second, less frequent pathway involves a single endonucleolytic cleavage generating a 32S form containing a 5′ extension of a few nucleotides (Schmitt and Clayton, 1993; Chu et al, 1994; Henry et al, 1994). In metazoans, the mechanism is assumed to be similar (Tycowski et al, 1994). For HeLa cells, the endonucleolytic cleavage site of the major pathway was mapped between nucleotides 942 and 949 within ITS1, ∼150 nucleotides upstream of the 32S precursor (Idol et al, 2007). Figure 4C, panel NB11, shows a blot hybridized to a probe against the very 3′ end of the spacer ITS1, visualizing all 5′-extended precursors of 32S RNA. A prominent smear can be observed (indicated by an asterisk), which resembles the 5′-elongated 32S RNA after the endonucleolytic cleavage of the main pathway in ITS1, but before complete exonucleolytic degradation. Comparing lanes 1 (wild-type cells) and 5 (the most efficient depletion of Nol9) by linear density analysis, the pattern shows a very low signal intensity for wild-type cells with peaks in the lower parts of the smear and the 41S and 45S RNAs (Figure 7B). RNAi depletion of Nol9 not only increased the signal intensities of 41S and 45S RNAs, but also shifted the intensity peak of the smear upwards, indicating that the majority of the precursor is not efficiently 5′ trimmed. Over-expression of wild-type Nol9 after Nol9 depletion eliminated this upwards shift and decreased the signal intensities of 5′-extended 32S precursors to almost wild-type levels (Figure 7C). Kinase-inactive mutants could not restore wild-type patterns, arguing for the importance of the kinase activity of Nol9 in the formation of the 5′ end of 32S. In the course of rRNA processing, cleavage within ITS1 might generate an entry point for the RNA-kinase Nol9, providing the 5′ phosphate and thus a favourable substrate for the subsequent 5′>3′ exonucleolytic activity (Stevens, 1980). Knocking down Nol9 would impair this phosphorylation event, thereby blocking efficient 5′ end trimming to produce the actual 32S precursor and consequently resulting in an rRNA maturation defect specifically of the large subunit rRNAs (Figure 7A, box 2).
Whereas in humans the endonuclease of the major pathway cleaving within ITS1 is elusive, this process is well characterized in yeast, in which the RNase MRP cleaves the rRNA precursor at the corresponding A3 site, providing an entry point for Rat1 (Schmitt and Clayton, 1993; Chu et al, 1994; Henry et al, 1994). Efforts to show this function for metazoan MRP homologues have been unsuccessful so far. It is worth noting that MRP belongs to the RNase P family and generates 5′ phosphorylated cleavage products (Reilly and Schmitt, 1995). In case metazoan and yeast endonucleases are similar, Nol9-kinase activity would not be required. As we do not observe a distinct cleavage intermediate upon Nol9 knockdown, but a smear of exonucleolytic degradation intermediates, our data could argue against a single entry point for the kinase Nol9, but rather suggest constant phosphorylation events after removal of each individual nucleotide (Figure 7A, box 2). The following possibilities could support our hypothesis; either the metazoan exonuclease generates 5′ hydroxyl ends or a phosphatase activity continuously de-phosphorylates the 5′ ends and thus slows down 5′>3′ degradation. The existence of a counteracting phosphatase activity in human cells has been a matter of speculation (Weitzer and Martinez, 2007a; Braglia et al, 2010), yet to date, no such activity could be attributed to a known protein in vivo or reconstituted in an in vitro system. However, it is interesting to note that Grc3, the yeast homologue of Nol9, was found in purifications of Rai1, an important partner and enhancer of the 5′>3′ exonuclease Rat1 (Sydorskyy et al, 2003). Depletion of Rai1 or Grc3 impairs Rat1-dependent degradation of the 3′ trailing transcript in RNA polymerase I termination (El Hage et al, 2008; Kawauchi et al, 2008; Braglia et al, 2010). In addition, we show that Nol9 depletion mainly impairs maturation of the major 5.8SS form, but not of 5.8SL (Figure 6). In yeast, 5.8SL is generated by a distinct endonucleolytic event, but does not involve the action of an exonuclease (Faber et al, 2006). Our data present evidence that two independent processing pathways also co-exist in human cells generating either the short or long form of 5.8S. Nol9 seems to be involved in the exonuclease-dependent processing of 5.8SS rRNA.
Other possible substrates for Nol9 are the snoRNAs. This class of RNAs guides rRNA modifications (Kiss-Laszlo et al, 1996; Ganot et al, 1997; Ni et al, 1997) and can be required for rRNA cleavage itself (Kass et al, 1990; Chu et al, 1994). More than 90% of human snoRNAs are encoded in the introns of coding and non-protein-coding genes (Dieci et al, 2009). As for rRNAs, maturation of these snoRNAs involves the concerted action of endo- and exonucleases (Filipowicz and Pogacic, 2002). In yeast, Rat1 and Xrn1 are reported to provide the 5′>3′ exonuclease activity generating the 5′ end of intronic snoRNAs (Petfalski et al, 1998). Nol9 could be involved in generating the 5′ ends of snoRNAs in a similar manner as described above for the 5′ end formation of 32S RNA (Figure 7A, box 3). As Nol9 depletion specifically blocks generation of only the 5.8S and 28S rRNA, but does not impair 18S maturation or RNA methylation as observed with metabolic labelling (Figure 4A), it is unlikely that Nol9 is involved in the general processing of snoRNAs.
In the light of the emerging evidence of new ‘un-traditional' roles of the nucleolus (Olson et al, 2002; Gerbi et al, 2003), further studies could reveal additional functions for Nol9. A growing number of RNPs and their components are reported to localize to the nucleolus in some stages of their life cycle, including not only SRP (Jacobson and Pederson, 1998) and RNase P (Jacobson et al, 1997), but also RNA editing (Desterro et al, 2003), genome integrity (Kobayashi, 2008) and RNA viruses (Hiscox, 2007) were previously linked to the nucleolus. Given that Nol9 can efficiently phosphorylate RNA and DNA substrates, a future line of research will be to find endogenous substrates of Nol9. It will, therefore, be interesting to reveal direct interacting partners of Nol9, which could lead to new roles for the polynucleotide 5′-kinase Nol9 in RNA metabolism.
Materials and methods
Expression and purification of wild type and mutant Nol9
The open reading frame of human Nol9 (NM_024654) was cloned into pDEST20 using the Gateway technique (Invitrogen) and expressed in sf9 insect cells. After harvesting, the cells were lysed in 100 mM NaCl, 50 mM Tris–Cl pH 8, 5 mM MgCl2, 0.1 mM AEBSF, 1 mM DTT and sonicated. Purification was performed using Glutathione Sepharose 4B (GE Healthcare). Protein was eluted from the beads twice with 20 mM Glutathione pH 8, concentrated on Vivaspin 500 columns (Sartorius) and dialysed against 30 mM Hepes pH 7.4, 5 mM MgCl2, 100 mM KCl, 10% Glycerol, 0.1 mM AEBSF before use in kinase assays. Site-specific mutations were introduced using the QuikChange Site-directed mutagenesis kit (Agilent Technologies) and primers hNol9_K312A_rev (GGTATCTATTAAATGTTGACGCTCCAACATCCTGGG) and hNol9_K312A_fw (CCCAGGATGTTGGAGCGTCAACATTTAATAGATACC) or hNol9_S313A_rev (GGTATCTATTAAATGTTGCCTTTCCAACATCCTGGG) and hNol9_S313A_fw (CCCAGGATGTTGGAAAGGCAACATTTAATAGATACC).
In vitro kinase assay
Kinase assays were carried out as described in Weitzer and Martinez (2007b) in the presence of 5 mM ATP.
Immunoprecipitation of endogenous Nol9
Antisera were coupled to Protein A Sepharose 4FF (GE Healthcare) and incubated with HeLa cell extracts for 2 h at 4°C. Kinase assays were carried out directly on the beads. Antisera were produced by Gramsch Laboratories, Germany and Clp1 antiserum 988 as described in Weitzer and Martinez (2007b). Nol9 antiserum 1687/1688 and 1689 were targeted against peptides EEAHKEKPYRRPKFC and CITPRNRESHNKILR, respectively. Pre-immune serum was taken from the rabbit before production of antiserum 1688.
Sucrose density studies
Ribosomal profiles were obtained as described in Strezoska et al (2000) on a 10–45% sucrose gradient.
Pre-ribosomes were analysed as described in Pestov et al (2008) on a 10–30% sucrose gradient. Gradients were fractionated using downwards replacement and UV measurement was carried out using NanoDrop spectrophotometer. Fractions were analysed for RNA and protein content by northern blot and western blot, respectively. Primary antibodies used were Nol9, affinity-purified antiserum 1687 (Gramsch Laboratories); Nucleophosmin/B23, B 0456 (Sigma) and Polymerase II antibody H14 (Covance). RNAse A treatment was carried out incubating the nuclear extracts for 10 min at 30°C with 100 μg/ml RNAse A (Promega).
Cell culture, knockdown and transfection
HeLa cells were cultured as described in Weitzer and Martinez (2007b). siRNA-directed knockdown was performed using Lipofectamine 2000 reagent (Invitrogen) as recommended by the manufacturer and siRNAs against Clp1 (Dharmacon, L-019895-00) or Nol9 (Dharmacon, L-019418-02 for 97% knockdown efficiency; where indicated we used single siRNA targeting the 3′ UTR of Nol9, J-019418-18 for 70% knockdown efficiency). Optionally, we used the pSUPER vector system (oligoengine) to produce a short hairpin RNA targeting the 3′ UTR of Nol9 using oligonucleotides GATCCCCCCGATGCCCAAACCACTTTTTCAAGAGAAAAGTGGTTTGGGCATCGGTTTTTA and AGCTTAAAAACCGATGCCCAAACCACTTTTCTCTTGAAAAAGTGGTTTGGGCATCGGGG, as determined by RNAxs (Tafer et al, 2008), resulting in 83% knockdown efficiency. Knockdown was generally performed two consecutive times to assure effectivity. Cells were transfected with the siRNA for 3 days, split and re-transfected for another 3 days. Knockdown efficiency was determined by qPCR using standard conditions and QIAGEN QuantiTect Primer Assays (Nol9, QT00011305; GAPDH for normalization, QT01192646). In the rescue experiments, the pSUPER vector system was used for knockdown. In the second round of transfection with the pSUPER vector, we co-transfected wild type or mutant Nol9 cloned into the pcDNA-DEST53 Gateway vector (Invitrogen).
Metabolic labelling
Cells were labelled with L-[methyl-3H]-methionine as described in Strezoska et al (2000).
Northern blot analysis
RNA was extracted from HeLa cells using TRIzol reagent (Invitrogen), transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare) in 20 × SSC by capillary forces, UV-cross-linked and incubated with Ambion Ultrahyb oligo buffer. Hybridization to 5′ (32P)-phosphorylated oligonucleotides was carried out over night at 62°C, followed by consecutive washing steps in 5% SDS/5 × SSC, 1% SDS/1 × SSC and 0.1% SDS/1 × SSC. Radioactive signals were analysed by phosphorimaging. For analysis of the 5.8S rRNA, total RNA was separated on an 8% urea polyacrylamide gel (PAGE) (SequaGel system, National Diagnostics) and transferred to a nylon membrane in 0.5 × TBE with 300 mA for 2 h. Probes used: NB8 CCGGCCGTGCGTACTTAGACATGCATGGC,
NB9 CCGGGGCCGCAAGTGCGTTCGAAGTGTCG,
NB11 GGTCGATTTGGCGAGGGCGCTCCCGACG,
NB39 GGCGATTGATCGGCAAGCGACGCTCAGAC,
NB40 CTCCTTGGTCCGTGTTTCAAGACGGGTCGGG,
NB48 TACTCTTTTTTGAATGTTGTCAAAACTAGTTAATTGGAAGTTAACGGTACTATTTATACTAAAAGAGT,
For quantification of signal intensities in northern blot analyses the ImageQuant analysis software by GE Healthcare was used.
Primer extension analyses
Primer extension analysis was adapted from Beltrame and Tollervey (1992). A total of 1 μg RNA from wild-type or Nol9 knockdown cells were shortly incubated with 0.5 pmoles primer (NB82 CTAGCTGCGTTCTTCATCGACG or NB86 CGCACGAGCCGAGTGATC) at 85°C for denaturation. Consequently, primer extension was performed with the Invitrogen Superscript III system for 50 min at 50°C in the presence of 1 mM dNTPs. For the sequencing reaction, the indicated ddNTPs (A, ddATP; T, ddTTP; G, ddGTP; C, ddCTP) were added to the primer extension reaction at a final concentration of 0.25 mM. Reactions were loaded on an 8% urea PAGE and visualized by phosphorimaging.
Supplementary Material
Acknowledgments
We thank A Schleiffer for the bioinformatic analysis of Clp1 and Nol9; T Noto and K Mochizuki for sharing the northern blot protocol and reagents; O Vesper and I Moll for help with the ribosome profile; S Weitzer, J Popow, S Westermann, JM Peters, NJ Proudfoot, S Granneman, D Tollervey, NJ Watkins and C Schneider for discussion and/or comments on the paper. KH was funded by FWF-P20502-B11 and the Gen-Au program from the Austrian Government. JM is a Junior Group Leader supported by IMBA.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allmang C, Mitchell P, Petfalski E, Tollervey D (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28: 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J 11: 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 9: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman LH, Goldman WE, Goldberg GI, Hebert MB, Schlessinger D (1983) Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol 3: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P, Heindl K, Schleiffer A, Martinez J, Proudfoot NJ (2010) Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep 11: 758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L (1994) The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA 91: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote CA, Greer CL, Peculis BA (2002) Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8: 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O′Connell M, Carmo-Fonseca M (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci 116 (Part 9): 1805–1818 [DOI] [PubMed] [Google Scholar]
- Dieci G, Preti M, Montanini B (2009) Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 94: 83–88 [DOI] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D (2008) Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AW, Vos HR, Vos JC, Raue HA (2006) 5′-end formation of yeast 5.8SL rRNA is an endonucleolytic event. Biochem Biophys Res Commun 345: 796–802 [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14: 319–327 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV, Lange TS (2003) The nucleolus: a site of ribonucleoprotein maturation. Curr Opin Cell Biol 15: 318–325 [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV (2004) Pre-ribosomal RNA processing in multicellular organisms. In The Nucleolus, Olson MOJ (eds) New York, NY, USA: Kluwer Academic/Plenum Publishers [Google Scholar]
- Good L, Intine RV, Nazar RN (1997) Interdependence in the processing of ribosomal RNAs in Schizosaccharomyces pombe. J Mol Biol 273: 782–788 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Granneman S, Nandineni MR, Baserga SJ (2005) The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol 25: 10352–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP (1993) Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem 212: 211–215 [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA (2007) RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ (2007) Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis 39: 35–43 [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T (1997) Nuclear domains of the RNA subunit of RNase P. J Cell Sci 110 (Part 7): 829–837 [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T (1998) Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA 95: 7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B (1990) The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60: 897–908 [DOI] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ (2008) Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev 22: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Maden BE (1977) Nucleotide sequence relationships between vertebrate 5.8 S ribosomal RNAs. Nucleic Acids Res 4: 2495–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kobayashi T (2008) A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays 30: 267–272 [DOI] [PubMed] [Google Scholar]
- Maggi LB Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD (2008) Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol 28: 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Olson MO, Hingorani K, Szebeni A (2002) Conventional and nonconventional roles of the nucleolus. Int Rev Cytol 219: 199–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR (2004) Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321 [DOI] [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, Mitsakakis N, Ryan OW, Datta N, Jojic V, Pal C, Canadien V, Richards D, Beattie B, Wu LF, Altschuler SJ et al. (2003) A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933 [DOI] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E (2009) RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol Chapter 22: Unit 22 11 [DOI] [PubMed] [Google Scholar]
- Petfalski E, Dandekar T, Henry Y, Tollervey D (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol 18: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly TH, Schmitt ME (1995) The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Mol Biol Rep 22: 87–93 [DOI] [PubMed] [Google Scholar]
- Rouquette J, Choesmel V, Gleizes PE (2005) Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 24: 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM (1974) Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem 41: 197–202 [DOI] [PubMed] [Google Scholar]
- Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA (1993) Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Banerjee N, Sitz TO (1984) Gene heterogeneity: a basis for alternative 5.8S rRNA processing. Biochemistry 23: 3648–3652 [DOI] [PubMed] [Google Scholar]
- Stevens A (1980) Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem 255: 3080–3085 [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF (2000) Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol 20: 5516–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD (2003) Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol Cell Biol 23: 2042–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL (2008) The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol 26: 578–583 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA (1994) Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266: 1558–1561 [DOI] [PubMed] [Google Scholar]
- Vaughan MH, Warner JR, Darnell JE (1967) Ribosomal precursor particles in the HeLa cell nucleus. J Mol Biol 25: 235–251 [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Shuman S (2001) Domain structure and mutational analysis of T4 polynucleotide kinase. J Biol Chem 276: 26868–26874 [DOI] [PubMed] [Google Scholar]
- Wang LK, Shuman S (2002) Mutational analysis defines the 5′-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res 30: 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Soeiro R (1967) Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA 58: 1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J (2007a) hClp1: a novel kinase revitalizes RNA metabolism. Cell Cycle 6: 2133–2137 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Martinez J (2007b) The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447: 222–226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.