The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA
The classical view of mRNA 3′-end processing involves the recruitment of the poly (A) polymerase by RNA-binding proteins. Here, a variant poly (A) polymerase, Star-PAP, contributes towards substrate specificity of some mRNAs by recruiting the CPSF complex involved in mRNA cleavage.
Keywords: 3′-end RNA processing; phosphoinositides; PI4,5P2; poly A polymerase; Star-PAP
Abstract
Star-PAP is a poly (A) polymerase (PAP) that is putatively required for 3′-end cleavage and polyadenylation of a select set of pre-messenger RNAs (mRNAs), including heme oxygenase (HO-1) mRNA. To investigate the underlying mechanism, the cleavage and polyadenylation of pre-mRNA was reconstituted with nuclear lysates. siRNA knockdown of Star-PAP abolished cleavage of HO-1, and this phenotype could be rescued by recombinant Star-PAP but not PAPα. Star-PAP directly associated with cleavage and polyadenylation specificity factor (CPSF) 160 and 73 subunits and also the targeted pre-mRNA. In vitro and in vivo Star-PAP was required for the stable association of CPSF complex to pre-mRNA and then CPSF 73 specifically cleaved the mRNA at the 3′-cleavage site. This mechanism is distinct from canonical PAPα, which is recruited to the cleavage complex by interacting with CPSF 160. The data support a model where Star-PAP binds to the RNA, recruits the CPSF complex to the 3′-end of pre-mRNA and then defines cleavage by CPSF 73 and subsequent polyadenylation of its target mRNAs.
Introduction
In eukaryotes, generation of messenger RNA (mRNA) is a multistep process. mRNA precursors (pre-mRNA) undergo capping at the 5′-end, splicing of introns, and processing at the 3′-end before they are exported as mature mRNA to the cytoplasm (Hirose and Manley, 2000; Maniatis and Reed, 2002; Auboeuf et al, 2005; Moore and Proudfoot, 2009). The 3′-end processing of pre-mRNA involves two tightly coupled reactions—endonucleolytic cleavage followed by the subsequent addition of a polyadenosine tail to the 3′-end of the cleaved RNA (Colgan and Manley, 1997; Wahle and Ruegsegger, 1999; Zhao et al, 1999; Edmonds, 2002; Proudfoot and O'Sullivan, 2002; Mandel et al, 2008).
Mammalian pre-mRNA contains three primary cis-acting elements that define the accuracy of cleavage and polyadenylation: the consensus hexamer (AAUAAA) polyadenylation signal, the cleavage site, and the GU-rich downstream element (Colgan and Manley, 1997; Zhao et al, 1999; Mandel et al, 2008). The 3′-end pre-mRNA processing reaction requires cleavage and polyadenylation stimulating factor (CPSF), which binds the hexamer AAUAAA (Keller et al, 1991), cleavage stimulating factor (CstF) that recognizes G/U-rich elements (Takagaki et al, 1990), cleavage factor Im (CF Im) (Brown and Gilmartin, 2003), cleavage factor IIm (CF IIm) (de Vries et al, 2000), poly A polymerase (PAP) (Takagaki et al, 1988; Ryner et al, 1989), and nuclear poly A-binding protein (PABP) (Wahle, 1991a). There is evidence that PAP, RNAPII, CPSF, CstF, CF Im and IIm are involved in the recognition and cleavage step whereas PAP, CPSF, PABP are involved in the polyadenylation step (Wahle and Ruegsegger, 1999; Zhao et al, 1999; Edmonds, 2002; Mandel et al, 2008).
Mammalian CPSF consists of five polypeptides—CPSF 160, CPSF 100, CPSF 73, CPSF 30 and hFip1 (Mandel et al, 2008). CPSF 160 recognizes the AAUAAA sequence on pre-mRNA (Murthy and Manley, 1995) and interacts with PAP and CstF (Murthy and Manley, 1995). CstF cooperates with CPSF for the formation of a stronger complex on pre-mRNA (Takagaki et al, 1990; Murthy and Manley, 1992, 1995). CPSF 73 acts as an endonuclease that cleaves the pre-mRNA at the cleavage site (Ryan et al, 2004; Mandel et al, 2006). The functions of CPSF 100 are not yet defined (Jenny et al, 1994), whereas the role of CPSF 30 may be to cooperate with CPSF 160 in RNA substrate recognition (Barabino et al, 1997). Another CPSF subunit, hFip1 also binds canonical PAP and is presumed to bring PAP close to the polyadenylation site (Kaufmann et al, 2004).
Recently, we reported the regulation of 3′-end processing of specific messages by phosphoinositide signalling mediated via a novel poly A polymerase named Star-PAP (for Speckle targeted PIPKIα-regulated poly A polymerase) (Mellman et al, 2008). Star-PAP has also been reported to have terminal uridylyl transferase activity toward U6 snRNA in vitro (Trippe et al, 2006). Star-PAP localized to nuclear speckles together with the type Iα phosphatidylinositol phosphate kinase (PIPKIα), and these two enzymes regulate expression of select messages including heme oxygenase-1 (HO-1) (Mellman et al, 2008). Star-PAP is distinct from other PAPs in that PI4,5P2, the product of PIPKIα, specifically stimulates PAP activity by >10-fold in vitro and is required in vivo after priming with oxidative stress signalling (Mellman et al, 2008). Star-PAP forms a complex with 3′-end processing components such as CPSF 73, CstF, RNAPII and Symplekin in vivo, which also associate with canonical polyadenylation complex. However, there was no detectable PAPα in the Star-PAP complex or vice versa demonstrating that these two PAPs assemble into distinct complexes. The knockdown of Star-PAP resulted in the accumulation of uncleaved HO-1 RNA signifying that Star-PAP was required for 3′-end cleavage (Mellman et al, 2008).
Canonical PAPα is recruited to the 3′-cleavage site by interactions with the CPSF complex that in turn binds to the pre-mRNA (Keller et al, 1991; Murthy and Manley, 1995). Here, the mechanism for Star-PAP in the cleavage of its target HO-1 pre-mRNA is defined in vivo and in vitro. The results demonstrate that Star-PAP binds to HO-1 3′-UTR RNA and is required for the stable assembly of CPSF complex on HO-1 RNA through its interactions with CPSF160 and 73, distinct from that of canonical PAPα. These data support a model where Star-PAP promotes the interaction of CPSF complex to pre-mRNA and is required for the assembly of the 3′-end processing complex and defines specificity of CPSF 73 cleavage.
Results
Star-PAP-dependent cleavage of its target mRNA in vivo
HO-1 expression is highly induced by oxidative stress and the agonist tert-butyl hydroquinone (tBHQ). To monitor changes in HO-1 mRNA, total RNA was isolated from HeLa cell transfected with control or siRNA Star-PAP or with tBHQ treatment, and subjected to 3′-RACE and primer extension analysis. 3′-RACE measured the amount of cleaved and polyadenylated (mature) mRNA and the primer extension analysis gives a measure of the downstream cleavage product (Figure 1A and C). RACE products (∼1000 bp) were observed with RNAs from both resting and tBHQ-treated cells (Figure 1B, lanes 1–2) and a loss of product with siRNA knockdown of Star-PAP (Figure 1B, lane 3). To confirm that this loss was not due to alternative splicing, the 3′-RACE analysis was repeated with a primer in the last exon of HO-1 (Supplementary Figure S1A). Consistent with enhanced HO-1 expression, an increase in intensity of HO-1 RACE product was detected in samples from tBHQ-treated cells (Figure 1B, lane 2; Supplementary Figure S1A and B), which was lost upon Star-PAP knockdown (Supplementary Figure S1B). GAPDH, a non-target RNA, was not affected by Star-PAP knockdown or by tBHQ treatment (Figure 1B, lanes 6–8).
Figure 1.
Star-PAP is required for HO-1 mRNA cleavage in vivo. (A) Schematic of 3′-RACE assay. (B) 3′-RACE assay of HO-1 (lanes 1–3) and GAPDH (lanes 6–8) with total RNA isolated from HeLa cells—resting with control RNAi (WT), treated with tBHQ (+tBHQ), and siRNA knockdown of Star-PAP (siStar-PAP). A RACE product (∼1000 bp) and DNA size markers (kilobases) is indicated. The western analysis of Star-PAP knockdown is shown in the right panel. (C) Schematic of primer extension analysis. (D) Primer extension analysis by reverse transcription reaction using a primer specific to HO-1 (lanes 1–4) or GAPDH (lanes 5–8) RNA that is 3′ of the cleavage site. The DNA size marker (lane 9), origin of the gel and the primer extension product are indicated.
In addition, extension of a 32P-radiolabelled primer specific to the HO-1 pre-mRNA downstream of the cleavage site (Figure 1C) also corroborates these results. Primer extension showed a signal (∼60 nucleotides) from resting cell RNA, an enhanced signal with the RNA isolated from tBHQ-stimulated cells (Figure 1D), and a loss upon Star-PAP knockdown (Figure 1D, lane 3), which was rescued by re-expression of wild-type (WT) Star-PAP containing silent mutations in the siRNA site (Figure 1D, lane 4). Consistently, extension with an additional downstream primer also showed a loss of cleavage by Star-PAP knockdown (Supplementary Figure S1C). There were no detectable differences in the 3′-end cleavage of GAPDH within the same conditions (Figure 1D, lanes 5–8, Supplementary Figure S1C). These results support a role for Star-PAP in 3′-cleavage of its target HO-1 pre-mRNA in vivo.
Star-PAP is essential for efficient cleavage of its target RNAs
To reconstitute Star-PAP-dependent cleavage, nuclear extracts were prepared (see Figure 2G for fractionation) from WT and siRNA Star-PAP knockdown (siStar-PAP) HeLa cells with or without tBHQ treatment (Dignam et al, 1983). An in vitro transcribed HO-1 UTR RNA encompassing the cis-acting elements required for 3′-end processing (Figure 2A; Supplementary Figure S2A for sequence) was assayed for 3′-end cleavage using the nuclear extract (Humphrey et al, 1987). The HeLa nuclear extracts displayed cleavage of HO-1 UTR RNA (Figure 2B) and this was increased (>3-fold) by treating the cells with tBHQ prior to the isolation of nuclear extracts (Figure 2B, lanes 4–5; Supplementary Figure S2C). However, PI4,5P2 addition did not impact the cleavage reaction (Figure 2B). To confirm the specificity of cleavage, the poly A signal on the HO-1 substrate was mutated from AAUAAA to AAGTAC (poly-A mutation, PAM) (Rigo and Martinson, 2008). The poly A site mutated HO-1 was not cleaved regardless of the conditions used (Figure 2C, lanes 1–4). The identity of the cleaved RNA was confirmed by comparison with an in vitro transcribed RNA from the 5′-end up to 3′-cleavage site of HO-1 UTR (Supplementary Figure S2B).
Figure 2.
Star-PAP specifically promotes cleavage of its target mRNA in a cell-free system. (A) Schematic of the HO-1 cleavage substrate. (B) Cleavage assay of radiolabelled HO-1 substrate reconstituted (+/− PI4,5P2) with nuclear extracts prepared from HeLa cells (+/− tBHQ treatment) (lanes 2–5). The control RNA (HO-1 RNA) (lane 1), pre-mRNA (P), cleaved RNA (C) are indicated. The approximate sizes of RNA are indicated by a DNA size marker. (C) Cleavage assay with poly A signal mutation (PAM) under the similar conditions as in B (lanes 1–4). Control—HO-1 RNA without treatment with nuclear extract. (D) Cleavage assay of HO-1 RNA with nuclear extracts from HeLa cells after the siRNA knockdown of Star-PAP (siStar-PAP) (lanes 6–9) and control HeLa cells (HeLa NE) (lanes 1–4). (E, F) Cleavage assay of Star-PAP non-target, GCLC (lanes 1–9) and GAPDH (lanes 1–8) UTRs with nuclear extracts prepared from control (HeLa NE) and siRNA Star-PAP knockdown (siStar-PAP) with or without tBHQ treatment (+/− tBHQ). Control—substrate RNA without treatment with nuclear extract. (G) Western blot analysis of nuclear fractionation, C, cytosolic fraction; N, nuclear fraction. (H) Western analysis of siRNA Star-PAP knockdown; cells transfected with control RNA, siRNA Star-PAP (siStar-PAP) and untransfected (extract WT) are indicated.
To investigate whether Star-PAP is required for efficient cleavage, Star-PAP was knocked down by siRNA (Figure 2H for western) and the nuclear extract prepared from these cells was assayed for cleavage of HO-1 substrate. Nuclear extracts from Star-PAP knockdown cells failed to cleave HO-1 UTR RNA both with and without tBHQ or PI4,5P2 stimulation (Figure 2D, lanes 6–9), demonstrating that Star-PAP is required for the cleavage of target pre-mRNAs. Non-target mRNA templates, GCLC (Figure 2E) and GAPDH (Figure 2F) were unaffected upon Star-PAP knockdown or tBHQ treatment, consistent with Star-PAP-dependent cleavage having specificity for its target messages.
In an approach to rescue the cleavage defect resulting from knockdown of Star-PAP, the nuclear extracts prepared from the knockdown cells were supplemented with functionally active recombinant His-tagged Star-PAP or His-tagged PAPα (Supplementary Figure S3C for PAPα activity). The cleavage defect was rescued by His-Star-PAP (Figure 3A, lanes 8–11) but not by His-PAPα (Figure 3A, lanes 12–15). Moreover, His-Star-PAP supplementation was not able to rescue the lack of cleavage from control poly A mutated HO-1 (Figure 3B, lanes 1–4). In a rescue experiment with increasing concentrations of Star-PAP, we observed a dose-dependent increase in cleavage with increasing Star-PAP (Figure 3C, lanes 4–10). These observations establish that Star-PAP specifically is required for the 3′-cleavage of its target mRNAs.
Figure 3.
Recombinant Star-PAP rescues the cleavage defect and Star-PAP interacts with subunits of CPSF. (A) Cleavage assay of HO-1 RNA with nuclear extracts prepared from siRNA Star-PAP knockdown HeLa cells (siStar-PAP) (lanes 4–7), supplemented with 20 nM recombinant His-tagged Star-PAP (lanes 8–11) or recombinant His-tagged PAPα (lanes 12–15) as indicated. (B) Cleavage assay of poly A signal mutant (PAM) with supplemented His-Star-PAP (lanes 1–4) under similar conditions as in (A). (C) Cleavage assay of HO-1 with siRNA Star-PAP nuclear extract supplemented with increasing amounts of His-Star-PAP (0.01–20 nM) (lanes 4–10) as indicated. The control RNA, pre-mRNA (P) and cleaved RNA (C) are indicated. (D) Western analysis of siRNA Star-PAP knockdown in Hela cells. (E) Western blot analysis of Flag Star-PAP purification for the indicated CPSF subunits, CstF and PAPα; Sup, supernatant; FT, flow through; Eluate, elution fraction; W, wash. (F) GST pull-down assay with GST-tagged Star-PAP, GST (as indicated on the top) with overexpressed CPSF subunits in E. coli (indicated on the right). Input shows 20% of the lysates used for binding.
Star-PAP associates with CPSF subunits
Previously, Star-PAP was shown to associate with components of 3′-end RNA processing machinery (Mellman et al, 2008). To further define the association of Star-PAP with CPSF subunits, Flag-tagged Star-PAP was isolated from stable expressing HEK 293 cells and the copurification of respective CPSF components were examined by immunoblotting. 5% each of supernatant and the elution fraction was used for the analysis by immunoblotting. The purification resulted in the copurification of a complex of proteins containing 3′-processing and transcriptional components (Mellman et al, 2008). All of the subunits of CPSF—160, 100, 73 and 30 copurify with Star-PAP (Figure 3E). As previously observed (Mellman et al, 2008), CstF 64 was detected in the complex but not PAPα (Figure 3E). The control purifications from parental HEK 293 cells (Supplementary Figure S3A) and HEK 293 cells expressing Flag E-70 (a cytoplasmic protein; Supplementary Figure S3B) did not show association with any of the CPSF subunits.
To define the direct interaction of Star-PAP with CPSF subunits, GST pull-down assays were performed using recombinant GST-tagged Star-PAP as bait and the CPSF subunits 160, 100 and 73 overexpressed in Escherichia coli as prey. From these studies, it was determined that GST-Star-PAP interacts directly with CPSF 160 and CPSF 73 but not with CPSF 100 (Figure 3F).
Star-PAP specifically binds the HO-1 UTR RNA
As Star-PAP is required for cleavage of its target pre-mRNA and also contains putative RNA-binding domains (zinc finger (ZF) and RNA recognition domain) (Mellman et al, 2008), this suggests that Star-PAP may directly associate with its target mRNA. To explore this possibility, an RNA immunoprecipitation (RIP) experiment was carried out by cross-linking the RNA protein complexes in the cell with formaldehyde as previously described (Gilbert et al, 2004). The cross-linked RNA was immunoprecipitated with Star-PAP-specific antibody and the associated RNA was identified using primers specific to HO-1 UTR equivalent of cleavage substrate (Figure 4A). Strikingly, an in vivo association of Star-PAP with HO-1 RNA similar to the control RNAPII (Mellman et al, 2008) was detected (Figure 4B). In contrast, PAPα did not associate with HO-1 RNA (Figure 4B). In case of non-target GCLC, no association of Star-PAP with the 3′-UTR RNA was detected (Figure 4B). These results demonstrate that Star-PAP specifically interacts with the target HO-1 RNA in vivo.
Figure 4.
Star-PAP associates and interacts directly with HO-1 RNA. (A) Schematic of HO-1 3′-UTR for RIP analysis. (B) RIP analysis of HO-1 and GCLC UTR by IP with antibodies specific to RNA Pol II (RNAPII), Star-PAP and PAPα followed by reverse transcription–PCR with primers specific to HO-1 and GCLC UTR regions. (C) RNA EMSA of HO-1 (lanes 1–6) or GAPDH (lanes 7–12) (substrate for cleavage assays) with increasing concentrations of recombinant His-Star-PAP (0–20 nM; from lanes 2–6, respectively, 1, 2, 4, 10 and 20 nM). The control RNA (Probe), unbound RNA (F) and Star-PAP–RNA complex (B) are indicated. (D) EMSA of HO-1 RNA with 10 nM Star-PAP in the presence of increasing Star-PAP antibody (antibody super shift) (lanes 2–7) or (E) β-tubulin antibody (lanes 2–5). Unbound RNA (F), Star-PAP-HO-1 RNA binary complex (B) and antibody, Star-PAP and RNA ternary complexes (T) are indicated. (F) Cold competitions of HO-1 RNA binding with Star-PAP. EMSA of HO-1 RNA with increasing Star-PAP concentrations (lanes 2–5, respectively, 10, 20, 30 and 40 nM) (lanes 1–5) and 20-fold molar excess of the respective competitor RNAs as indicated (lanes 6–9). Specific (S) and non-specific (NS) competitions are indicated. Lanes 6–7 and 8–9 correspond to specific and non-specific competitions equivalent of reactions in lanes 4–5, respectively. (G–J) EMSA of HO-1 RNA with increasing amounts of various His-Star-PAP deletions as indicated (ΔZF, zinc-finger deletion; ΔRRM, RNA recognition motif deletion; ΔZF-RRM, combine deletion of both ZF and RRM; FL, full-length Star-PAP), or (K) His-ZF-RRM peptide. The control (Probe), unbound (F) and binary complexes (B) are indicated.
To investigate the direct interaction, an in vitro RNA electrophoretic mobility shift assay (EMSA) of radiolabelled HO-1 UTR RNA was carried out with increasing amounts of recombinant His-Star-PAP (0–20 nM). A slower migrating band, indicating an interaction, was detected with increasing Star-PAP concentration (Figure 4C, lanes 1–6). However, the equivalent UTR region of the non-target GAPDH did not show any interaction with Star-PAP (Figure 4C, lanes 7–12). The interaction of Star-PAP with the HO-1 RNA was further supported by antibody super shift where the binary complex of Star-PAP (10 nM) and HO-1 was retarded by the addition of Star-PAP antibody (Figure 4D) but not by β-tubulin antibody (Figure 4E). Specific binding of Star-PAP to HO-1 was confirmed by cold RNA competition using 20-fold molar excess of the unlabelled RNAs (Figure 4F). While the HO-1 RNA-Star-PAP complex was out competed by cold HO-1 (lanes 6–7), a non-specific RNA of similar length could not compete with binding (lanes 8–9). PAPα, on the other hand, did not show distinct mobility shifts of the HO-1 RNA at the similar concentrations (Supplementary Figure S4A) consistent with previous reports that PAPα has low affinity for RNA (Wahle, 1991b). The results demonstrate that Star-PAP directly and specifically interacts with the 3′-UTR of HO-1 pre-mRNA.
To identify the domain required for Star-PAP binding to its target RNA, we deleted the ZF, RRM or both from His-Star-PAP (Supplementary Figure S6D for purified proteins) and tested for their ability to bind HO-1 RNA. Neither the deletion of ZF nor RRM alone could abolish interaction of Star-PAP although the affinity was lower than that of full-length (FL) Star-PAP (Figure 4G–J). Whereas FL showed mobility shift of HO-1 probe at a concentrations as low as 4–5 nM (Figure 4C and J), and a half-maximal binding of ∼10 nM (Figure 4C); ZF or RRM deletions exhibited weak mobility shifts from ∼10 nM (Figure 4G) or 20 nM (Figure 4H) with no saturation of binding reached even at 40 nM protein. The deletion of both ZF and RRM completely abolished Star-PAP binding to RNA (Figure 4I). These data demonstrate that both ZF and RRM domains are involved in the interaction of Star-PAP with HO-1 RNA.
We further assayed the binding of purified His-tagged ZF, RRM and ZF-RRM peptides of Star-PAP (Supplementary Figure S6E for purified peptides) with HO-1 RNA. The ZF or RRM domains did not exhibit significant interaction with the HO-1 RNA (Supplementary Figure S4B–D) and at higher concentration showed non-specific interactions (data not shown). The N-terminal region containing the ZF-RRM, interestingly, bound to the HO-1 RNA with almost a five-fold greater affinity than the FL Star-PAP with saturation at 4 nM (Figure 4K; Supplementary Figure S4D).
Mapping of the Star-PAP-binding site on HO-1 UTR RNA
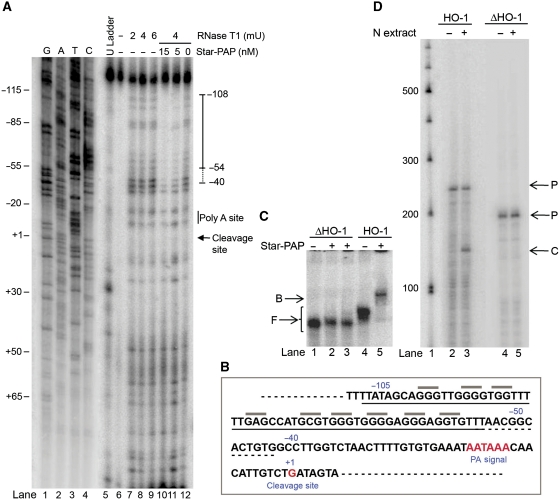
To map the binding site of Star-PAP on HO-1 UTR, we employed an RNA footprinting assay as described (Murakawa and Nierlich, 1989; Damgaard et al, 1998) using the HO-1 RNA substrate for cleavage (Figure 2A). The RNA was digested in the absence or presence of increasing His-Star-PAP with RNase T1 and the extent of cleavage was detected by extension of a primer complementary to the 3′-end of HO-1 RNA (Figure 5A). In the absence of Star-PAP, we obtained a ladder of digested fragments with three different concentrations of RNase T1 (Figure 5A, lanes 7–9 and 12). The site of protection by Star-PAP was distinguished after comparing the sample without Star-PAP (lanes 7–9 and 12) with that received 5 nM (lane 11) or 15 nM (lane 10) His-Star-PAP. Several positions within the HO-1 UTR became specifically protected against RNase T1 cleavage by Star-PAP. The primary site of protection extended from ∼108 to ∼55 nucleotides upstream of cleavage site (Figure 5A) with heightened efficiency of protection with increased Star-PAP (lanes 10–11). In addition, there were weaker (minor) protections, which extended further downstream to ∼45 nucleotides upstream of cleavage site (Figure 5A). The protection region and the putative reactive sites protected are shown in Figure 5B. A protection of similar region on HO-1 UTR RNA by Star-PAP was also observed from footprinting with RNase S (Supplementary Figure S5).
Figure 5.
Determination of Star-PAP-binding site on HO-1 RNA. (A) Footprint of Star-PAP on HO-1 UTR RNA by RNase T1 probing. The digestion pattern with different RNase T1 concentrations (mU) in the absence of Star-PAP (lanes 7–9, 12), or in the presence of 5 or 15 nM Star-PAP (lanes 10–11), and the untreated HO-1 RNA (lane 6) are indicated. Sequencing ladder generated using the primer for primer extension from the template of HO-1 RNA (lanes 1–4) and a dideoxy U ladder generated by primer extension reaction (lane 5) are indicated. Numbers refer to the position of nucleotides with respect to cleavage site taken as +1. (B) The primary region of Star-PAP protection (solid line) on HO-1 RNA, the extended minor protection (dashed line) and the putative reactive sites protected (grey lines) (C) RNA EMSA of HO-1 RNA deleted for the Star-PAP protection site from −110 to −50 (lanes 1–3) and control EMSA with HO-1 UTR RNA (lanes 4–5). (D) Cleavage assay of HO-1 UTR deletion as in (C) with HeLa nuclear extract (lanes 4–5) and control HO-1 UTR RNA (lanes 2–3).
Based on these observations, we deleted the region of protection from −110 to −50 with respect to the cleavage site (taken as +1) on HO-1 RNA and assayed for Star-PAP binding. Star-PAP was unable to bind the deleted HO-1 RNA (Figure 5C). Additionally, in an in vitro cleavage assay, HeLa nuclear extract did not cleave the HO-1 RNA with Star-PAP-binding site deletion (Figure 5D). These data suggest that Star-PAP-binding sequence in HO-1 lies within the region approximately from 108 to 50 nucleotides upstream of the cleavage site (this sequence is underlined Figure 5B).
Star-PAP recruits CPSF and stabilizes the processing complex
Star-PAP binds HO-1 pre-mRNA in vitro and in vivo and also interacts with CPSF subunits, it is possible that Star-PAP promotes the binding of CPSF on pre-mRNA. To test this hypothesis, the association of CPSF subunits onto the 3′-end of pre-mRNA was investigated by in vivo RIP analysis. Individual CPSF components were immunoprecipitated (IP'ed) from the cross-linked HeLa cell lysates with control RNAi or siRNA Star-PAP knockdown and the associated HO-1 pre-mRNA encompassing the poly A site (Figure 4A) was detected by reverse transcriptase (RT)–PCR. The results indicate an association of CPSF 160 and overall CPSF complex (which includes other subunits 100, 73, 30) with HO-1 UTR (Figure 6A, lanes 5–12). Remarkably, siRNA knockdown of Star-PAP eliminated the in vivo association of the CPSF subunits with the HO-1 UTR (Figure 6A). The control RIP using RNAPII antibodies did not change upon Star-PAP knockdown (Figure 6A, lanes 2–3). In addition, the association of CPSF subunits and RNAPII with the Star-PAP non-target GAPDH message was not influenced by Star-PAP knockdown (Figure 6A). A control using primers specific to the coding region of HO-1 mRNA where CPSF subunits were unlikely to get cross-linked also showed no association (Supplementary Figure S7A) with or without Star-PAP knockdown supporting the specificity at the 3′-end.
Figure 6.
Star-PAP promotes CPSF binding and stabilizes the cleavage complex on HO-1 RNA. (A) RIP analysis using antibodies specific to RNAPII (lanes 2–3) and CPSF subunits (lanes 5–12), followed by detection of associated RNA by RT–PCR using primers specific to the UTR region encompassing the poly A signal of HO-1 and GAPDH. (B) RNA EMSA of HO-1 UTR (Figure 4A) with the CPSF 160 complex. CPSF 160 was IP'ed from HeLa cell with control RNAi lysates (+) (lanes 2–3) or lysates of Star-PAP RNAi (Si) (lanes 4–5), and the IP eluate was incubated with radiolabelled HO-1 RNA (see Figure 4A). The unbound RNA (F) and complex of the 3′-RNA with CPSF 160 (S) are indicated. (C) EMSA of HO-1 RNA with nuclear extracts from HeLa cells with (lanes 6–10) or without (lanes 1–5) Star-PAP knockdown for various incubation times. The unbound fraction (F) and specific processing complex (S) are indicated. (D) RNA EMSA of poly A signal mutated HO-1 RNA (lanes 1–5) with HeLa nuclear extract. (E) EMSA of GAPDH UTR RNA with nuclear extracts control (lanes 1–5) or Star-PAP knockdown (lanes 6–10). F, unbound RNA fraction; S, specific processing complex.
To further study the requirement of Star-PAP for CPSF interaction with HO-1 RNA, CPSF 160 was IP'ed from nuclear lysates with or without Star-PAP knockdown. The CPSF 160 complex was eluted from the IP (Supplementary Figure S7C and E for western and SDS–PAGE) and was then assayed for binding to HO-1 RNA (Figure 2A). CPSF 160 complex showed the mobility shift of HO-1 RNA, indicating an interaction between the CPSF complex and HO-1 RNA that was absent in the control IgG eluates (Figure 6B). Knockdown of Star-PAP resulted in a loss of this complex formation (Figure 6B, lane 5). These results support the in vivo observation that the assembly of the CPSF complex on the HO-1 UTR requires Star-PAP.
Previous studies have shown that EMSA using L3 UTR RNA with nuclear extracts in the presence of either 3′-dATP for cleavage or ATP for polyadenylation specifically shows a processing complex (S) assembled on RNA (Humphrey et al, 1987). To investigate the role of Star-PAP in the assembly of the processing complex (S), EMSA of HO-1 RNA was carried out in the absence of both ATP and 3′-dATP with nuclear extracts prepared from HeLa cells with or without Star-PAP knockdown (Supplementary Figure S7D for SDS–PAGE). As reported for L3 UTR RNA (Humphrey et al, 1987), a slower migrating complex (S) was observed for both HO-1 (Figure 6C, lanes 1–5) and GAPDH (Figure 6E, lanes 1–5) that were stable during electrophoresis for at least 2 h. The nuclear extracts prepared from siRNA Star-PAP knockdown cells exhibited a loss of complex formation with HO-1 RNA, and when analysed by electrophoresis, this resulted in a diffuse banding pattern (Figure 6C, lanes 6–10), indicating the dissociation of the complex formed during gel electrophoresis. As expected, the poly A site mutation abolished the stable complex (S) formation (Figure 6D). For GAPDH, the assembly of the complex (S) was independent of Star-PAP knockdown (Figure 6E). These results illustrate that Star-PAP not only facilitates the stable association of CPSF on the HO-1 UTR RNA, but also promotes the assembly of a stable 3′-end processing complex. Together, these results suggested that Star-PAP's role in cleavage was to promote the stable association of the CPSF complex with RNA.
Star-PAP controls cleavage by promoting CPSF interaction with HO-1 RNA
Star-PAP interacts directly with both CPSF 160 and 73 and controls CPSF binding to RNA. Therefore, we examined whether Star-PAP regulates the in vitro cleavage of HO-1 RNA by CPSF subunits. The results demonstrate that in the presence of recombinant Star-PAP, CPSF 160 and 73 showed specific cleavage of the HO-1 RNA (Figure 7A, lanes 3 and 5). This required the intact poly A signal, when this sequence was mutated, the RNA was not cleaved under any of the assay conditions (lanes 6–8). The addition of PI4,5P2 did not affect the cleavage of the HO-1 RNA (lanes 2–5). The inactive CPSF 73 mutant (CPSF 73 (M)) where the Zn-binding site was mutated (Mandel et al, 2006), did not show any specific cleavage in the presence of Star-PAP and CPSF 160 (Figure 7B). Also the recombinant PAPα, CPSF 160 and 73, did not show cleavage of HO-1 RNA (Supplementary Figure S8A). The combination of Star-PAP, CPSF 160 and CPSF 73 were unable to cleave the control GAPDH RNA substrate (Supplementary Figure S8B). Consistent with the previous report (Mandel et al, 2006), CPSF 73 alone showed non-specific nuclease activity toward RNA whereas Star-PAP, CPSF 160 or CPSF 73 (M) did not exhibit any nuclease activity toward the HO-1 substrate (Figure 7C; Supplementary Figure S6A–C for SDS–PAGE).
Figure 7.
Star-PAP facilitates HO-1 cleavage by CPSF subunits, and PI4,5P2 stimulates coupled polyadenylation. (A) Cleavage reactions of HO-1 (lanes 1–5) and poly A mutation (lanes 6–8) with recombinant CPSF 160 (40 nM) and 73 (120 nM) with Star-PAP (20 nM) as indicated. The control HO-1 RNA, pre-mRNA (P) and cleaved RNA (C) are indicated. (B) Cleavage reaction as in (A) but in the presence of mutant CPSF 73 (lanes 3–4). (C) Cleavage assay of HO-1 RNA with individual proteins as indicated (D) Coupled cleavage and polyadenylation assay of HO-1 RNA (lanes 3–6) and poly A signal mutation (lanes 8–11) in the presence or absence of PI4,5P2 with HeLa nuclear extracts with or without tBHQ treatment as indicated. Polyadenylated (poly A) and pre-mRNA (P) are indicated. (E) Stable ternary complex formation on HO-1 RNA by EMSA (lanes 1–6) with CPSF 160–30 nM (lanes 2 and 4), 80 nM (lane 3) and Star-PAP (15 nM) (lanes 4 and 6). The control RNA (Probe), unbound RNA (F), binary complex of Star-PAP-HO-1 or CPSF 160-HO-1 (B) and ternary complex of CPSF, Star-PAP and HO-1 RNA (T) are indicated. (F) RNA EMSA experiment of HO-1 with increasing CPSF (0–120 nM) (lanes 1–6) (G) RNA EMSA of HO-1 with increasing CPSF (0–120 nM) in the presence of 15 nM Star-PAP (lanes 2–6). The approximate half-maximal binding was obtained from the mobility shifts. Symbols are as described in legend to (E). (H) EMSA of poly A signal mutant HO-1 RNA with increasing amount of His-CPSF 160 (left, lanes 1–3), or His-Star-PAP (right, lanes 4–6).
CPSF 160 binds the poly A site in combination with Star-PAP on HO-1 RNA. Therefore, the interaction of CPSF 160 and Star-PAP with HO-1 RNA was tested in vitro. Recombinant CPSF 160 exhibited a weak interaction with HO-1 UTR RNA (Figure 7E, lanes 2 and 3). Upon addition of His-tagged Star-PAP (15 nM), in the presence of 30 nM CPSF 160, a distinct super-shifted band appeared, representing a ternary complex of RNA, CPSF 160 and Star-PAP (Figure 7E, lane 4; Supplementary Figure S8C), which was absent in Star-PAP alone addition (Figure 7E, lane 6). In an EMSA of HO-1 RNA with increasing amount of CPSF 160 (from 0 to 120 nM), we observed, albeit weak, a dose-dependent binary complex formation with increased CPSF 160 with a half-maximal CPSF binding of ∼60 nM (Figure 7F, lanes 2–6). In the presence of Star-PAP (15 nM), the CPSF 160 concentration-dependent ternary complex of CPSF-Star-PAP-HO-1 formed with higher affinity, and a half-maximal binding of ∼20 nM was observed (Figure 7G, lanes 2–6). Increasing CPSF concentration in the presence of Star-PAP transitioned to a distinct ternary complex band (Figure 7G, lanes 4–6). These combined results illustrate that Star-PAP is required for stable interaction of CPSF 160 onto its target pre-mRNA, which in turn could recruit and position other cleavage factors.
Star-PAP control of cleavage is poly A signal dependent
For many pre-mRNAs, the poly A signal is critical for cleavage, and mutation of this sequence abolishes cleavage (Wickens and Stephenson, 1984; Sheets et al, 1990). Cleavage assays demonstrated that though Star-PAP promotes CPSF binding to pre-mRNA, the actual cleavage of HO-1 RNA is dependent on the presence of an intact poly A signal (Figures 2 and 7A). To further define the significance of this site in HO-1 UTR, the interaction of the CPSF 160 complex was investigated with poly A signal mutation. The mutation abolished the binding of CPSF 160 complex IP'ed from nuclear lysates (Supplementary Figure S7B). In addition, the mutation completely abolished recombinant CPSF 160 binding to HO-1 UTR RNA (Figure 7H, lanes 1–3). Conversely, the Star-PAP interaction with the HO-1 RNA was not affected by the mutation (Figure 7H, lanes 4–6). These results confirm that the poly A signal remains critical for cleavage and also for CPSF interactions with HO-1 RNA. These data also indicate that Star-PAP though promotes the CPSF 160 interaction with HO-1 RNA, the specificity for binding and cleavage is determined by the poly A signal.
HO-1 mRNA polyadenylation is stimulated by PI4,5P2
Star-PAP activity was previously reported to be activated by the addition of PI4,5P2. As HO-1 cleavage is not affected by PI4,5P2, the role of PI4,5P2 was investigated in a coupled cleavage and polyadenylation assay of the HO-1 UTR RNA using HeLa nuclear extracts. Although the role of Star-PAP in the cleavage reactions is independent of PI4,5P2, there was significant stimulation of the coupled polyadenylation reaction by PI4,5P2 in nuclear extracts from cells treated with tBHQ (Figure 7D, lanes 3–6). Most importantly, the coupled cleavage and polyadenylation was completely abolished with the poly A signal mutation (lanes 8–11). This supports a mechanism wherein HO-1 pre-mRNA cleavage is dependent upon Star-PAP which then is followed by polyadenylation that is stimulated by PI4,5P2.
Discussion
The results demonstrate that Star-PAP participates both in the cleavage and polyadenylation of its target pre-mRNA by a mechanism that is distinct from that of canonical PAPα. Previous studies demonstrated that CPSF binds the AAUAAA element on pre-mRNA and directly interacts with PAPα (Wilusz et al, 1990; Gilmartin and Nevins, 1991; Murthy and Manley, 1992, 1995). The CPSF interaction with PAPα results in recruitment of PAPα to the cleavage site (Murthy and Manley, 1992, 1995). In contrast, Star-PAP directly binds to both the pre-mRNA and the CPSF subunits 160 and 73. These associations promote the stable interaction of CPSF with the 3′-UTR of Star-PAP target HO-1 RNA and assemble a functional cleavage complex. The in vitro and in situ interactions between Star-PAP and CPSF 160 and 73 are confirmed by reconstitution of specific 3′-cleavage of the HO-1 RNA in vitro with recombinant Star-PAP and CPSF 160 and 73.
Previously, it had been reported that four factors (cleavage factors—CF Im and IIm, CPSF, PAP and CstF) were required for the reconstitution of cleavage in vitro (Takagaki et al, 1989). Although CPSF 160 recognizes the AAUAAA and cooperates with other factors to bind pre-mRNA (Murthy and Manley, 1995), CPSF 160 alone is not sufficient as the specificity and the binding requires intact CPSF (Bardwell et al, 1991; Keller et al, 1991; Murthy and Manley, 1995). CPSF interaction also requires cooperation with CstF and CF Im for the stable association with the RNA and vice versa (Gilmartin and Nevins, 1991; Weiss et al, 1991; MacDonald et al, 1994; Ruegsegger et al, 1996). In addition, recently it has been reported that both the CPSF 73 and 100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation (Kolev et al, 2008). CPSF 73 possesses the endonuclease activity and does not require CPSF 100 for its enzymatic activity for some substrates (Mandel et al, 2006). CPSF 100 does not show significant endonuclease activity (Jenny et al, 1994; Mandel et al, 2006). Thus, the mechanism of how CPSF 73 and 100 participate to assemble the active endonuclease for histone mRNA 3′-cleavage is unclear (Kolev et al, 2008). CPSF 100 might affect the assembly or positioning of CPSF 73 or, this might be unique for histone 3′-end maturation that contain distinct sequence elements and required influence from other factors that are absent in the canonical pre-mRNA 3′-end processing (Dominski and Marzluff, 2007). This raises the possibility that different combinations of CPSF subunits may participate with CPSF 73 in the 3′-processing of different pre-mRNAs.
In case of the Star-PAP target HO-1, Star-PAP binds to the HO-1 RNA upstream of the poly A signal. Star-PAP also interacts with CPSF 160 and enhances the interaction of CPSF 160 with HO-1 RNA. This results in a strong ternary complex formed on the RNA. In addition to CPSF 160 and RNA, Star-PAP also directly interacts with the endonuclease CPSF 73. The combined interactions between Star-PAP and CPSF 160 with CPSF 73 result in assembly of CPSF 73 on the RNA and define specifically for the cleavage site. CPSF 73 alone exhibited only non-specific cleavage of the HO-1 RNA, whereas the Star-PAP–CPSF 160–CPSF 73 complex exhibited specific 3′-end cleavage. The resulting 3′-cleavage is specific but weak, suggesting that optimum in vivo cleavage may require other processing factors as depicted in Figure 8. Therefore, the Star-PAP, CPSF 160 and CPSF 73 complex represent the minimal protein complex required for the specific cleavage of Star-PAP target mRNAs.
Figure 8.
Model of Star-PAP-mediated cleavage of target RNA. Direct contact of Star-PAP with HO-1 RNA recruits CPSF 160 to the poly A signal and association of other cleavage factors in the complex is also indicated.
Star-PAP differs from canonical PAPα in many aspects including stimulation by PI4,5P2 and functional specificity toward pre-mRNAs (Mellman et al, 2008). Mammalian PAPα is also required for the cleavage reactions in vitro and possibly in vivo (Zhao et al, 1999; Mandel et al, 2008), although the mechanism as to how PAPα is involved is not precisely defined. PAPα has very low affinity for RNA substrate (Wahle, 1991b; Martin and Keller, 1996) and lacks RNA-binding specificity (Zhao et al, 1999); and thus PAPα is thought to be recruited to pre-mRNA in part by the CPSF complex binding (Keller et al, 1991; Murthy and Manley, 1995). Star-PAP's specificity for pre-mRNAs on the other hand appears to be defined by its direct interaction with RNA. Star-PAP also directly interacts with both CPSF 160 and 73 and this promotes the CPSF 160 interaction with the poly A signal and recruitment of CPSF 73. The resulting complex has two contacts with the pre-mRNA and multiple protein–protein interactions. These interactions would enhance both affinity and specificity toward the RNA and may exclude PAPα from the cleavage complex. This model is outlined in Figure 8.
However, the exact mechanism as how PAPα is not able to access the Star-PAP target RNAs in the absence of Star-PAP is still unclear. One plausible explanation could be the loss of CPSF interaction with HO-1 RNA in the absence of Star-PAP rendering PAPα unable to be recruited onto the 3′-end of Star-PAP targets. There is a clear loss of in vivo 3′-end cleavage complex assembly upon Star-PAP knockdown on HO-1 RNA. This suggests that in the absence of Star-PAP, in spite of the other accessory factors such as CstF, CF Im and the intact poly A signal, CPSF subunits are unable to establish a stable cleavage complex. Analysis of the nucleotide sequence in this region of HO-1 UTR has not yet yielded unique or distinct sequence elements. However, the significance of trans-acting factors in the cooperative binding of CPSF on poly A signal of a pre-mRNA is well established (Lutz et al, 1996). In case of HO-1, despite of the intact poly A signal, CPSF requires Star-PAP to form a stable complex on the pre-mRNA. There are examples of similar suboptimal poly A sequences, where the existence of an intact AAUAAA is not sufficient for CPSF complex to stably bind the pre-mRNA (Gilmartin et al, 1995). This could also be analogous to cytoplasmic polyadenylation in oocytes, where phosphorylated CPEB directly interacts and recruits CPSF 160 to an AAUAAA on the pre-mRNA (Mendez et al, 2000). Previous studies indicate that the nucleotides required for CPSF interaction are confined to the AAUAAA hexamer (Bardwell et al, 1991; Keller et al, 1991) and could form specific complexes with RNAs as short as 10–18 nucleotides (Wigley et al, 1990; Keller et al, 1991). Interestingly, AAUAAA sequences occur commonly (∼16% of all pre-mRNAs in the cell) in the coding and intronic regions (Day, 1992). It is therefore possible that Star-PAP-directed recruitment enables CPSF to identify the authentic poly A site at the 3′-end by excluding those hexamers that reside elsewhere in target mRNAs.
Star-PAP binds HO-1 RNA and protects a region of ∼60 nucleotides approximately between 108 and 50 nucleotides upstream of cleavage site. Star-PAP contains two RNA-binding motifs, a ZF and an RNA recognition (RRM) motif (Mellman et al, 2008) both of which bind HO-1 RNA. Deletion of one motif results in lower affinity but the loss of binding was seen only after deletion of both the motifs. Therefore, the actual binding sequence of Star-PAP on the pre-mRNA could involve multiple RNA sequences that combinatorially interact with the ZF and/or RRM domains, indicating a bipartite or more complex binding sequence motif. This is consistent with Star-PAP's large footprint on the HO-1 RNA. In addition, EMSA of Star-PAP with HO-1 RNA at higher Star-PAP concentration showed more than one band. This could also suggest multiple recognition sequences of Star-PAP on the RNA or that Star-PAP might exist in different oligomeric state when it forms a complex with RNA. The multiple contact sites between Star-PAP and the pre-mRNA could enhance both the specificity and flexibility for targeting specific RNA sequences.
Star-PAP polymerase activity and the HO-1 expression is activated by oxidative stress signalling (Maines and Gibbs, 2005). The lipid messenger PI4,5P2, synthesized by PIPKIα, directly stimulates Star-PAP (Mellman et al, 2008). Star-PAP also associates with and is phosphorylated by protein kinases such CKIα (a PI4,5P2 effector) that are also required for expression of HO-1. However, some targets of Star-PAP are not regulated by PIPKIα and CKIα (Gonzales et al, 2008). The combined results suggest that Star-PAP is regulated by multiple signalling pathways that modulate Star-PAP activity (Barlow et al, 2009), its specificity toward mRNAs and/or its interaction with the processing factors.
In the cytosol, PI4,5P2 is a membrane lipid that modulates enzyme activities and also induces the assembly of protein complexes (Heck et al, 2007). In the nucleus, PI4,5P2 is at the inner nuclear envelop and also in a nuclear compartment separate from known membrane (Boronenkov et al, 1998; Barlow et al, 2009). The Star-PAP-mediated 3′-end processing of HO-1 requires PIPKIα that generates PI4,5P2. Spatially, the nuclear organization of the PI4,5P2 that regulates Star-PAP is not known. One possibility is that the PI4,5P2-regulated processing occurs at the nuclear envelop close to the nuclear pore complex. This concept would be consistent with a model proposed for transcriptional memory in yeast (Tan-Wong et al, 2009) that involves 3′-processing complexes (Ansari and Hampsey, 2005). In such a model, PI4,5P2 would facilitate the assembly of the processing complex containing Star-PAP or potentially other unique PI4,5P2-regulated proteins. Such a model would also be consistent with a putative role for PI4,5P2 in the regulation of mRNA export (Okada et al, 2008) such that the nuclear PI4,5P2 could modulate both 3′-end processing and export of certain pre-mRNAs and their coupling.
Materials and methods
Cell culture, transfections and cell stimulation
HEK 293 and HeLa cells were obtained from ATCC and maintained in DMEM with 10% FBS at 37°C in 5% CO2. HeLa cells were transfected using Oligofectamine for siRNA knockdown (Invitrogen). In HEK 293 cells, transfections were accomplished by calcium phosphate method with 5 μg of plasmid DNA. siRNA oligos were used at a final concentration of 120 nM oligo/ml. The growth media was exchanged 4–6 h after transfection and the transfection was repeated 24 h later. Cells were harvested for analysis 48 h after second transfection. The Star-PAP siRNA oligo used was GUGUGUUUGUCAGUGGCUU. When required, cells were treated with 100 μM of tBHQ in DMSO for 4 h to stimulate the antioxidant response. Control cells were treated with DMSO.
Protein purifications
Affinity purified human Flag-Star-PAP was obtained from HEK 293 cells with stably expressed Flag-Star-PAP as described previously (Mellman et al, 2008). Recombinant His-tagged Star-PAP and PAPα purifications were performed as described previously (Mellman et al, 2008). The individual as well as the combine deletions of ZF and RRM of Star-PAP were made on the pET 28 construct of Star-PAP (Mellman et al, 2008). These deletions or His-tagged CPSF 160 and 73 (pET 28 construct) were obtained by overexpression in E. coli BL21(DE3) after induction with 0.5 mM IPTG at 37°C for 4 h or 18°C overnight. The proteins were purified using Ni-NTA affinity chromatography as described previously (Laishram and Gowrishankar, 2007). The proteins were concentrated with PEG, snap frozen and stored in −80°C.
Immunoprecipitations and immunoblottings
Immunoprecipitation and immunoblotting experiments were performed as described previously (Mellman et al, 2008).
Preparation of nuclear fraction
The nuclear extracts from Hela cells were prepared as in Dignam et al (1983) except that the crude nuclei were prepared using NE-PER Nuclear and Cytoplamic Extraction Kit (PIERCE Biotechnology). The nuclei were then extracted with 20 mM HEPES (pH 7.9), 25% glycerol (v/v), 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA and 0.5 mM DTT supplemented with protease inhibitor cocktail (Roche) as described in Dignam et al (1983). A 95% confluent 10 cm dish of cells were extracted into 40 μl of nuclear fraction, and 10 μl of this fraction was used for each cleavage reaction.
GST pull down
GST-Star-PAP fusions (Mellman et al, 2008) and GST were immobilized on glutathione-sepharose beads by incubating the respective overexpressed E. coli lysates with glutathione-sepharose beads pre-equilibriated in TEN 100 buffer (20 mM Tris, pH 7.5, 0.1 mM EDTA and 100 mM NaCl). The beads were again washed three times with TEN 100 and separately incubated with the E. coli extracts of overexpressed CPSF subunits at 4°C for 2 h. After incubation, the beads were washed three times with NETN buffer (20 mM Tris, pH 7.4, 300 mM NaCl, 0.1 mM EDTA and 0.5% NP40). Buffers were supplemented with protease inhibitor cocktail (Roche), DNaseI and RNase A. The bound proteins were visualized by western blotting. The input shows 20% of the lysate used for pull down.
RNA isolation
Total RNA from the cell was isolated using the Trizol (Invitrogen) method. A measure of 1 ml of Trizol reagent was used for a 10-cm dish of 95% confluent cells.
3′-RACE assay
The cDNAs for 3′-RACE assays were synthesized using the SMART RACE KIT (Clontech) with 1 μg of total RNA isolated from HeLa cells. The gene-specific forward primers of HO-1 and GAPDH used for RACE PCR were 5′-GACCTGCCCAGCTCTGGCGAG-3′ and 5′-GTATCGTGGAAGGGGACTCATGG-3′, respectively. PCR reactions were performed as described by the manufacturer. The RACE products were confirmed by sequencing.
Primer extension analysis
For primer extension analysis, oligos were end labelled with 5 units of poly nucleotide kinase (New England Biolabs) in 1 × buffer in the presence of 50 μCi of γ32P-ATP at 37°C for 1 h. Reactions were then cleaned with the Nucleotide Removal Kit (Qiagen). Approximately 1 × 106 c.p.m. equivalent of radiolabelled primer was reverse transcribed from 2 μg of total RNA at 50°C with AMV RT in 1 × RT buffer for 2 h for primer extension. The reaction was stopped by addition of EDTA (5 mM) and extracted with phenol chloroform. The extended products were analysed on a 6% sequencing gel. The primers used were 5′-ACCAGGGAGGAAAAGGTCAGTTC-3′ for HO-1 and 5′-AAGAATGTCTCACCTTGACAC-3′ for GAPDH.
RNA immunoprecipitation
RIP experiments were performed as described previously (Gilbert et al, 2004; Mellman et al, 2008). The primers specific to HO-1 used to study Star-PAP and CPSF associations were 5′-TTCTGTTGTTTTTATAGCAG-3′ as forward and 5′-TCAAACAGACCAGCTCCTG-3′ as reverse. For GCLC, the primers used were, respectively, 5′-GATGATTAAAGAATGCCTGGTT-3′ (forward) and 5′-TATGCTTCTTTCTAGAAACATC-3′ (reverse).
Construction and in vitro synthesis of substrate RNAs
HO-1 RNA substrate was obtained from the plasmid pTZ-HO-1, harboring HO-1 3′-UTR under the T7 promoter. The plasmid contained a region of HO-1 UTR encompassing the poly A signal, extending from 120 bp upstream to 122 bp downstream of cleavage site cloned in PstI and BamHI sites of pTZ19R (Fermentas). Similarly, pTZ-GCLC and pTZ-GAPDH has the corresponding UTR regions in pTZ19R.
Uniformly radiolabelled RNA substrates were prepared by in vitro transcription of the templates linearized with BamHI using the T7 transcription Kit (Fermentas). A typical 20 μl reaction contained 1 × reaction buffer, 20 units of RNase inhibitor, 1 μg of DNA, 500 μM each of ATP and CTP, 50 μM UTP and 100 μM GTP in the presence of 50 μCi α-32P-UTP and 400 μM m7G cap analogue and 40 units of T7 RNA polymerase (Nielsen and Shapiro, 1986). The RNA from the in vitro transcription reaction was cleaned up using the Qiagen Mini RNeasy Kit. Poly A signal mutation was introduced by Quik Change site-directed PCR mutagenesis.
Cleavage assay
Cleavage assays were performed as described previously (Humphrey et al, 1987). Reactions contained 1 × 105 c.p.m. of 32P-labelled pre-mRNA substrate (∼0.5 nM) in a 25-μl reaction volume of cleavage buffer (20 mM creatinin phosphate, 0.5 mM MgCl2, 10% glycerol, 10 mM HEPES, pH 7.9, 50 mM KCl, 0.05 mM EDTA, 1% PVA) with 0.8 mM 3′-dATP and 10 μl of the nuclear extract. When required, PI4,5P2 was added at a concentration of 100 μM. The reactions were incubated at 30°C for 2 h and stopped by the addition of proteinase K mixture (2% sarcosyl, 100 mM Tris–Cl, pH 7.5, 20 mM EDTA and 400 μg/ml proteinase K) followed by incubation at 30°C for 10 min. RNA was extracted with phenol chloroform, precipitated with absolute alcohol in the presence of 3 M ammonium acetate and 1 μg carrier tRNA, and analysed on a 6% urea denaturing gel.
To assess cleavage with recombinant proteins, the cleavage assay was reconstituted in a similar buffer and conditions with His-CPSF 160 (40 nM), 73 (120 nM) and Star-PAP (20 nM).
For the coupled in vitro polyadenylation reaction, similar buffers and conditions were used as in the cleavage assay except that 3′-dATP was replaced by ATP.
RNA EMSA experiment
EMSA experiments were carried out each in 20 μl EMSA-binding buffer (10 mM Tris–Cl, pH 7.5; 1 mM EDTA, 50 mM NaCl, 0.5 mM MgCl2; 1 mM DTT, 10% glycerol) containing 0.5 nM of uniformly labelled RNA, 1 μg/ml bovine serum albumin and Star-PAP or other proteins at the indicated concentrations. After incubation at RT for 30 min, it was analysed on a non-denaturing 4% polyacrylamide gel.
For EMSA experiments with nuclear extracts, the substrate RNA was incubated in cleavage buffer with 1.0 μg protein of nuclear extract from either control or Star-PAP knockdown HeLa cells at 30°C for various time points as indicated, and the complex assembled on the RNA was analysed on a non-denaturing PAGE as described above. For EMSA with multiple proteins, the indicated concentrations of CPSF 160 and Star-PAP were added together before incubation of the reaction.
For EMSA with the IP CPSF 160 complex, CPSF 160 was IP'ed with 6 μg of CPSF 160 antibody or control IgG from the nuclear extracts as described (Mellman et al, 2008). After the IP, the CPSF 160 complex bound to the antibody was eluted from the antibody bound beads with 40 μl of Tris, pH 7.5 containing 200 mM NaCl. The EMSA reaction was carried out with 8 μl eluate from each IP or control IgG for the respective binding reactions in EMSA buffer as described above.
RNA footprinting
The RNA footprinting was carried out by probing with either RNase T1 or RNase S and detecting the cleaved fragments by primer extension as described (Murakawa and Nierlich, 1989; Damgaard et al, 1998). Around 0.5 nM of RNA was incubated in each of 20 μl reaction volume with or without indicated amounts of His-Star-PAP in RT for 20 min in EMSA buffer. Each sample was then digested with the indicated amounts of either RNase T1 or RNase S at RT for 15 min. The reaction was terminated with stop solution (Ambion) and then recovered by phenol extraction followed by precipitation.
For primer extension reaction, the air-dried RNA pellets were re-dissolved in 6 μl of annealing buffer (10 mM Tris, pH 7.0; 40 mM KCl; 0.5 mM EDTA) and mixed with 0.2 pmol (106 c.p.m. equivalent) of 5′-end labelled Primer. After incubation at 95°C for 1 min, the samples were transferred to 50°C for 10 min for annealing and stored on ice. To each sample, 4 μl of extension mix containing 1 μl of 10 × RT buffer (Fermentas), 2 mM dNTPs and 2 U of AMV RT (Fermentas) was added. The reaction mixture was incubated at 50°C for 45 min for reverse extension of the primer followed by incubation at 85°C for 10 min to inactivate the RT. The cDNAs were resolved on a 6% urea denaturing polyacrylamide gel. A marker sequence was generated by dideoxy sequencing of the template DNA of HO-1 UTR RNA using the same primer used for primer extension. A dideoxy U ladder was also generated using the similar primer extension reaction in the presence of dideoxy TTP with untreated RNA. The primer used for extension and sequencing was 5′-GGATCCCCCTATCACCACTTC-3′.
Supplementary Material
Acknowledgments
We thank Walter Keller for CPSF subunit constructs. We thank Christy Barlow, Mark Schramp, Nicholas Schill and Wiemin Li for discussions and critically reading the manuscript. This work is supported by NIH grant GM057549 to RAA and an American Heart Association (AHA) fellowship (award # 0920072G) to RSL.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ansari A, Hampsey M (2005) A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19: 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O'Malley BW (2005) A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol Cell Biol 25: 5307–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716 [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Wickens M, Bienroth S, Keller W, Sproat BS, Lamond AI (1991) Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell 65: 125–133 [DOI] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA (2009) Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol 20: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronenkov IV, Loijens JC, Umeda M, Anderson RA (1998) Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell 9: 3547–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM (2003) A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Dyhr-Mikkelsen H, Kjems J (1998) Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res 26: 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day IN (1992) Analysis of the 5′-AAUAAA motif and its flanking sequence in human RNA: relevance to cDNA library sorting. Gene 110: 245–249 [DOI] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF (2007) Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M (2002) A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ (2004) Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell 14: 457–464 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR (1995) CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev 9: 72–83 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Nevins JR (1991) Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol 11: 2432–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, Mellman DL, Anderson RA (2008) CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem 283: 12665–12673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA (2007) A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol 42: 15–39 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Humphrey T, Christofori G, Lucijanic V, Keller W (1987) Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3′ processing complexes. EMBO J 6: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hauri HP, Keller W (1994) Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol 14: 8183–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G (1991) Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA (2008) Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, Gowrishankar J (2007) Environmental regulation operating at the promoter clearance step of bacterial transcription. Genes Dev 21: 1258–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC (1996) Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14: 6647–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines MD, Gibbs PE (2005) 30 some years of heme oxygenase: from a ‘molecular wrecking ball' to a ‘mesmerizing' trigger of cellular events. Biochem Biophys Res Commun 338: 568–577 [DOI] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L (2008) Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J 15: 2593–2603 [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA (2008) A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD (2000) Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell 6: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Murakawa GJ, Nierlich DP (1989) Mapping the lacZ ribosome binding site by RNA footprinting. Biochemistry 28: 8067–8072 [DOI] [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1992) Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem 267: 14804–14811 [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Shapiro DJ (1986) Preparation of cap RNA transcript using t7 RNA polymerase. Nucleic Acid Res 14: 5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Jang SW, Ye K (2008) Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc Natl Acad Sci USA 105: 8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N, O'Sullivan J (2002) Polyadenylation: a tail of two complexes. Curr Biol 12: R855–R857 [DOI] [PubMed] [Google Scholar]
- Rigo F, Martinson HG (2008) Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 28: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger U, Beyer K, Keller W (1996) Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113 [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL (2004) Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Takagaki Y, Manley JL (1989) Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol Cell Biol 9: 4229–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP (1990) Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18: 5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T (1990) A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL (1988) Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell 52: 731–742 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL (1989) Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev 3: 1711–1724 [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ (2009) Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 23: 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ (2006) Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E (1991a) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66: 759–768 [DOI] [PubMed] [Google Scholar]
- Wahle E (1991b) Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 266: 3131–3139 [PubMed] [Google Scholar]
- Wahle E, Ruegsegger U (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev 23: 277–295 [DOI] [PubMed] [Google Scholar]
- Weiss EA, Gilmartin GM, Nevins JR (1991) Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J 10: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Stephenson P (1984) Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science 226: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Wigley PL, Sheets MD, Zarkower DA, Whitmer ME, Wickens M (1990) Polyadenylation of mRNA: minimal substrates and a requirement for the 2′ hydroxyl of the U in AAUAAA. Mol Cell Biol 10: 1705–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J, Shenk T, Takagaki Y, Manley JL (1990) A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol Cell Biol 10: 1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.