Functional organization of the Sm core in the crystal structure of human U1 snRNP
The U1 small nuclear ribonucleoprotein initiates the assembly of the spliceosome. Here, the structure of the natively purified U1 small nuclear ribonucleoprotein particle reveals the core Sm protein ring and its interactions with the Sm site in the small nuclear RNA.
Keywords: pre-mRNA splicing, RNA–protein complex, Sm core RNP, U1 small nuclear ribonucleoprotein particle, X-ray crystallography
Abstract
U1 small nuclear ribonucleoprotein (snRNP) recognizes the 5′-splice site early during spliceosome assembly. It represents a prototype spliceosomal subunit containing a paradigmatic Sm core RNP. The crystal structure of human U1 snRNP obtained from natively purified material by in situ limited proteolysis at 4.4 Å resolution reveals how the seven Sm proteins, each recognize one nucleotide of the Sm site RNA using their Sm1 and Sm2 motifs. Proteins D1 and D2 guide the snRNA into and out of the Sm ring, and proteins F and E mediate a direct interaction between the Sm site termini. Terminal extensions of proteins D1, D2 and B/B′, and extended internal loops in D2 and B/B′ support a four-way RNA junction and a 3′-terminal stem-loop on opposite sides of the Sm core RNP, respectively. On a higher organizational level, the core RNP presents multiple attachment sites for the U1-specific 70K protein. The intricate, multi-layered interplay of proteins and RNA rationalizes the hierarchical assembly of U snRNPs in vitro and in vivo.
Introduction
Pre-mRNA splicing is an essential step in the expression of most eukaryotic genes. It is mediated by the spliceosome, a RNA–protein machinery comprised of five non-coding RNAs and more than 150 proteins (reviewed in Wahl et al, 2009). The spliceosome exhibits a particularly dynamic catalytic cycle, during which its constituents are recruited stepwise to the pre-mRNA substrate, are remodelled multiple times in order to properly locate reactive sites on the pre-mRNA and to generate functional active sites and are finally disassembled in an ordered manner (Staley and Guthrie, 1998; Wahl et al, 2009). To cope with the complexity of the assembly process, many spliceosomal factors are pre-organized as multi-factorial subunits. The main spliceosomal building blocks are the small nuclear ribonucleoprotein particles (snRNPs), named U1, U2, U4/U6 and U5 in the major spliceosome according to their unique uridine-rich snRNAs. Specific functions of the snRNPs include recognizing and pairing of the intron termini, providing RNP remodelling activities and building up the active sites. Thus, unravelling the functional architecture of the snRNPs is crucial for understanding of the splicing process at the molecular level.
The U1, U2, U4 and U5 snRNAs exhibit a conserved U-rich stretch within a single-stranded region, referred to as the Sm site (consensus sequence PuAU4−6GPu; Branlant et al, 1982). After synthesis by RNA polymerase II and m7G capping in the nucleus, the snRNAs are exported to the cytoplasm where their Sm sites are bound by a group of seven Sm proteins (D1, D2, F, E, G, D3 and B/B′; the two splice variants B/B′ differ by 11 residues at their C termini and are jointly referred to as B in the following; reviewed in Khusial et al, 2005). The U6 snRNA represents an exceptional case. It remains in the nucleus, does not contain a Sm site and associates with a group of seven Sm-like proteins (LSm 2–8; Seraphin, 1995; Achsel et al, 1999). Together, the Sm site RNA and the Sm proteins form the Sm core RNP (Branlant et al, 1982; Liautard et al, 1982), which in EM analyses appears doughnut-shaped (Kastner et al, 1990). Properly assembled Sm cores are a pre-requisite for hypermethylation of the snRNA caps and for the transport of the core particles to the nucleus (Kolb et al, 2007; Chari et al, 2009).
The Sm proteins are characterized by two conserved sequence motifs, Sm1 and Sm2, separated by a variable spacer. None of the Sm proteins alone interact stably with RNA, but the proteins form specific hetero-oligomers involving their Sm motifs, which serve as building blocks during Sm core assembly (Raker et al, 1996). Crystal structures of D1-D2 and D3-B dimers (Kambach et al, 1999), together with biochemical (Raker et al, 1996; Urlaub et al, 2001) and yeast two-hybrid analyses (Fury et al, 1997), suggested that the seven Sm proteins associate in a ring-like manner around the Sm site RNA in the order D1, D2, F, E, G, D3 and B. A similar functional diversification is not found in prokaryotes, which typically exhibit one and exceptionally up to three different types of Sm/LSm proteins that form homo-hexameric or homo-heptameric ring structures (Scofield and Lynch, 2008). Thus, structures of the homo-oligomeric systems from prokaryotes (Törö et al, 2001) do not reveal all aspects of the organization of eukaryotic Sm core RNPs.
Assembly of a Sm core RNP follows a strict pathway in vitro (Raker et al, 1996) and in vivo (Kolb et al, 2007; Chari et al, 2009). Sm protein complexes D1-D2 and F-E-G initially form a stable sub-core with the snRNA that is subsequently joined by the D3-B dimer (Raker et al, 1996). To complete U snRNP biogenesis, varying numbers of snRNP-specific proteins are recruited to the assembled cores, and the Sm core RNP modulates the binding of some snRNP-specific proteins (Nelissen et al, 1994). The molecular mechanisms underlying the ordered assembly of the snRNPs are still largely unknown.
A recently reported crystal structure at 5.5 Å resolution of a reconstituted human U1 snRNP, assembled from recombinant, partially truncated proteins and an engineered RNA (Pomeranz Krummel et al, 2009), allowed the construction of an overall backbone model of the particle, verified the proposed arrangement of the Sm proteins and revealed unexpected modes of interaction of some U1-specific proteins with the Sm core RNP. However, details of the protein–RNA interactions in the Sm core RNP are still poorly understood. For example, while Sm proteins assemble on oligo-U RNA, a Sm site with flanking purines is required for the exceptional thermodynamic stability of the Sm core RNP, and neighbouring higher-order structural elements strongly increase the kinetics of assembly (Raker et al, 1999). Furthermore, despite their high conservation, Sm sites are surprisingly tolerant to mutations (Jones and Guthrie, 1990), yet a given Sm site is not necessarily functional in the context of all snRNPs (Jarmolowski and Mattaj, 1993). To gain further insight into the functional architecture of a Sm core RNP, we have solved the crystal structure of a chymotrypsin-trimmed human U1 snRNP, natively purified from HeLa nuclear extract at 4.4 Å resolution.
Results
Crystallization and structure solution of natively purified human U1 snRNP by in situ limited proteolysis
Initial crystallization screens with natively purified U1 snRNP produced poorly diffracting crystals after several months at 4°C. SDS–PAGE and mass spectrometric analysis of washed crystals revealed that several U1-associated proteins had been truncated by spurious amounts of contaminating proteases. To aid crystallization, we deliberately included traces of proteases in the crystallization screens, which yielded different crystal forms that exhibited varying diffraction power. Pre-treatment of U1 snRNP with proteases and purification of the trimmed particles also yielded crystals, which were, however, small and exhibited very weak diffraction. Thus, in situ limited proteolysis is a useful tool for the crystallization of complex RNPs.
Crystals that yielded a complete data set to 4.4 Å resolution (Table I) were obtained by inclusion of chymotrypsin and of a DNA nonamer, complementary to the 5′-splice site (SS)-binding region of U1 snRNA in the crystallization setup. Analysis of washed crystals revealed that a C-terminal RS-like region of U1-70K, a C-terminal RNA recognition motif (RRM) of U1-A, the U1-C protein as well as a C-terminal RG-repeat/Pro-rich region of Sm protein B were removed during the crystallization, while the RNA remained intact (Supplementary Figure S1). The structure of the trimmed U1 snRNP was solved by a combination of multiple isomorphous replacement and molecular replacement using a model of the seven-membered Sm protein ring (Kambach et al, 1999) and the crystal structure of the N-terminal RRM of the U1-A protein in complex with one stem-loop of U1 snRNA (Oubridge et al, 1994; Table I). A large part of the backbone phosphate groups of the RNA could be assigned to bulges in the electron density map. In addition, several transition regions between single and double-stranded regions provided clear landmarks. The RNA register was locally consolidated by the position of iridium hexammine ions specifically bound at the G79•U60 and G56•U83 wobble base pairs (Keel et al, 2007; Supplementary Figure S2). Model building was further guided by known protein–RNA neighbourhoods, indicated by UV-induced crosslinking between residues Tyr112 and Leu175 of U1-70K and U1 snRNA nucleotides G28 and U30, respectively (Urlaub et al, 2000), and between Sm proteins G and B and the first and third nucleotides of the Sm site, respectively (Urlaub et al, 2001). During multiple cycles of model building, phase combination and refinement, the published backbone model of the recombinant U1 snRNP (Pomeranz Krummel et al, 2009) served as a useful guide.
Table 1. Crystallographic data.
| Data set | Native | Ir hexammine | Os hexammine | [Ir3N(SO4),(H2O)3] | KAuCN | PIPa | K5CoW12O40 |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Wavelength (Å) | 0.999 | 1.0 | 1.1364 | 1.10475 | 0.976 | 1.0703 | 1.2125 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Space group | C2 | C2 | C2 | C2 | C2 | C2 | C2 |
| Unit cell parameters | |||||||
| a, b, c (Å) | 358.4, 88.2, 150.9 | 357.5, 86.8, 150.6 | 360.9, 86.6, 151.0 | 358.8, 88.1, 150.9 | 358.0, 88.9, 151.9 | 358.9, 88.1, 150.9 | 357.4, 87.6, 150.8 |
| β (deg) | 111.9 | 112.6 | 113.2 | 112.0 | 111.8 | 111.9 | 112.6 |
| Resolution (Å)b | 90–4.4 (4.6–4.4) | 50–5.5 (5.7–5.5) | 100–7.9 (8.1–7.9) | 100–5.5 (5.7–5.5) | 50–6.3 (6.5–6.3) | 50–7.2 (7.5–7.2) | 100–7.2 (7.4–7.2) |
| Reflections | |||||||
| Unique | 28 079 | 14 396 | 4984 | 14 590 | 9838 | 6610 | 6550 |
| Completeness (%) | 99.3 (99.3) | 99.7 (100) | 99.4 (100) | 99.6 (97.3) | 99.1 (100) | 99.7 (99.2) | 99.5 (95.2) |
| Redundancy | 3.3 (3.5) | 3.6 (3.7) | 6.1 (6.3) | 3.7 (3.7) | 3.7 (3.8) | 7.3 (7.2) | 6.8 (4.8) |
| I/σ(I) | 12.0 (2.0) | 17.7 (1.4) | 13.6 (2.8) | 18.3 (1.5) | 13.1 (1.6) | 23.5 (2.7) | 12.9 (0.7) |
| Rsym(I)c | 0.054 (0.73) | 0.041 (0.78) | 0.066 (0.79) | 0.082 (0.79) | 0.080 (0.72) | 0.033 (0.73) | 0.051 (0.92) |
| Phasing | |||||||
| Resolution (Å) | 90–4.4 | 35–5.5 | 50–7.9 | 50–5.5 | 35–6.3 | 47–7.2 | 35.2–7.2 |
| Sites | 6 | 2 | 2 | 4 | 1 | 1 | |
| Phasing power (acentric)d | |||||||
| Isomorphous | 0.65 | 0.47 | 0.28 | 0.45 | 0.57 | 0.61 | |
| Anomalouse | 0.20 | 0.39 | 0.10 | 0.17 | 0.20 | 0.21 | |
| RCullis (acentric)f | |||||||
| Isomorphous | 0.63 | 0.78 | 0.60 | 0.74 | 0.72 | 0.71 | |
| Anomalous | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | |
| FOMg | 0.56 | ||||||
| Refinement | |||||||
| Resolution (Å) | 90–4.4 | ||||||
| Reflections | |||||||
| Number | 28 077 | ||||||
| Completeness (%) | 99.3 | ||||||
| Test set (%) | 5.0 | ||||||
| Rhwork | 0.299 | ||||||
| Rhfree | 0.348 | ||||||
| Contents of A.U. | |||||||
| Protein residues | 1716 | ||||||
| Nucleic acid residues | 346 | ||||||
| Ramachandran ploti | |||||||
| Favoured | 85.8 | ||||||
| Allowed | 11.6 | ||||||
| Outliers | 2.6 | ||||||
| r.m.s.d. Geometry | |||||||
| Bond lengths (Å) | 0.004 | ||||||
| Bond angles (deg) | 1.201 | ||||||
| Abbreviations: A.U., asymmetric unit; r.m.s.d., root-mean-square deviation. | |||||||
| aPIP—[Pt2I2(H2NCH2CH2NH2)2]. | |||||||
| bData for the highest resolution shell in parentheses. | |||||||
| cRsym(I)=ΣhklΣI∣Ii(hkl)−〈I(hkl)〉∣/ΣhklΣi∣Ii(hkl)∣; for n independent reflections and i observations of a given reflection, where 〈I(hkl)〉=average intensity of the i observations. | |||||||
| dPhasing power=∣FH,calc∣/Σhkl∣∣FPH±FP∣−∣FPH,calc∣∣. | |||||||
| eAlthough the anomalous signals were weak, they were included in phasing and visibly improved the electron density. | |||||||
| fRCullis=Σhkl∣∣FPH±FP∣−∣FPH,calc∣∣/Σhkl∣FPH−FP∣. | |||||||
| gFOM=mean figure of merit=〈cos(αbest−αcalc)〉, where α=phase angle. | |||||||
| hR=Σhkl∣∣Fobs∣−∣Fcalc∣∣/Σhkl∣Fobs∣, where Rwork=hkl∉T; Rfree=hkl∈T; Rall=all reflections; T=test set. | |||||||
| iCalculated with MolProbity (http://molprobity.biochem.duke.edu/). | |||||||
Overall structure
The present model of the chymotrypsin-trimmed U1 snRNP (hereafter referred to as U1 snRNP) includes residues 1–164 of native U1 snRNA, a region covering the RRM and an N-terminal extension of the U1-70K protein (residues 34–183 out of 437 amino acids total), the N-terminal RRM of the U1-A protein (residues 1–114 out of 282 amino acids total) and the Sm folds with varying terminal appendices of all seven Sm proteins but lacks U1-C (see also Supplementary Figure S3 and Supplementary Table SI for protein residues included in the model). The very C-terminal RG-repeat regions of D1 and D3 could not be located in the electron density, although these proteins apparently remained intact during limited proteolysis (Supplementary Figure S1). The regions that are protease sensitive or lack electron density are either intrinsically unstructured (for example, the RS-like domain of U1-70K, RG-repeat/Pro-rich regions of Sm proteins) or flexibly attached to the folded core (for example, the C-terminal RRM of U1-A) in the framework of intact U1 snRNP. An asymmetric unit of the present crystal form contains two U1 snRNPs, whose overall structures are very similar (r.m.s.d. of 1.54 Å between all phosphorus and Cα atoms).
As concluded from solution studies (Krol et al, 1990; Duckett et al, 1995) and from the crystallographic analysis of recombinant U1 snRNP (Pomeranz Krummel et al, 2009), U1 snRNA exhibits four stem-loops (SL1, SL2, SL3 and SL4) and a short base-paired region between nucleotides 12–16 and 118–122 (helix H; Figure 1A). SL1–3 and helix H are assembled as a four-way junction, in which SL1 stacks coaxially on SL2 and SL3 on helix H. The coaxial stacks cross each other at almost right angles. A single-stranded region (residues 123–136) encompassing the Sm site (residues 126–132) connects the base of helix H and SL4 (residues 137–164). SL4 is capped by a canonical UUCG tetraloop.
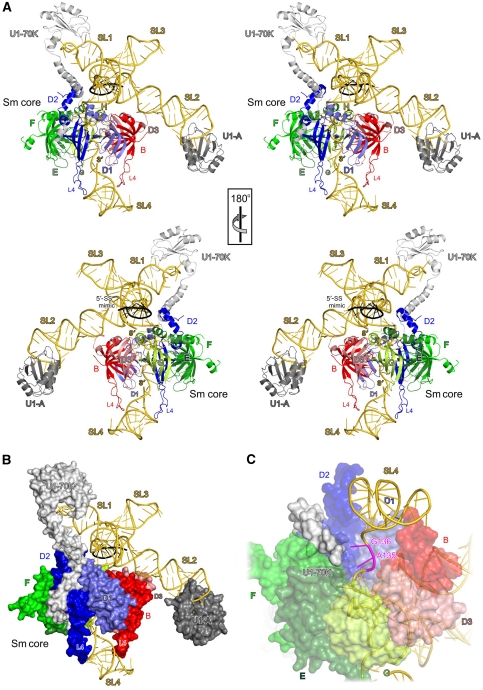
Figure 1.
Overall structure of U1 snRNP. (A) Orthogonal stereo ribbon plots of native U1 snRNP. snRNA, gold; DNA nonamer mimicking a 5′-SS, black; U1-70K, light grey; U1-A, dark grey; D1, steel blue; D2, blue; F, green; E, dark green; G, lime; D3, light red; B, red. SLs and termini of the RNA and the long L4 loops of D2 and B are labelled. (B) Native U1 snRNP with the proteins shown in surface representation. The Sm proteins provide a platform for the four-way junction and 5′-SS-binding region. U1 proteins package the central region of the snRNA and the tips of SL1 and SL2, but leave the core of the cruciform, the 5′-SS-binding region, SL3 and the tip of SL4 open. These RNA regions may provide docking sites for other spliceosomal factors. The view is the same as in panel A. (C) Contacts between the N-terminal extension of the U1-70K protein (light grey surface) at the underside of the Sm ring (semitransparent surfaces) and U1 snRNA (gold) in the region of residues A135 and G136 (magenta). Rotated 135° about the x axis compared with panel A.
The global structure of U1 snRNP resembles a figurine (Figure 1A), with SL3 defining the neck and head, SL1 and SL2 corresponding to the two arms, the single-stranded region comprising the waist and SL4 making up the leg and foot. The loops of SL1 and SL2 are bound by the central RRM of U1-70K and the N-terminal RRM of U1-A, respectively (Figure 1A). In our structure, the N-terminal RRM (residues 10–89) of U1-A is followed by a positively charged C-terminal helix spanning residues 102–112 that binds the RNA backbone opposite the face contacted by the core RRM and thereby clamps the tip of SL2 (Supplementary Figure S4). This region was not included in the isolated crystal structure of the U1-A N-terminal RRM-SL2 complex (Oubridge et al, 1994) and was disordered in the absence of RNA in an NMR-structure of a U1-A fragment spanning residues 1–117 (Avis et al, 1996), demonstrating that its fold is induced on RNA binding.
Below helix H, the waist is encircled by a ring of the seven Sm proteins (Figure 1A). Using an elaborate selenomethionine-scanning approach, Pomeranz Krummel et al (2009) showed that the extended N terminus of U1-70K wraps around the Sm core RNP and contacts the U1-C protein at the opposite side of the Sm ring. We observed similar features in the present structure. N-terminal of its RRM, U1-70K folds into a long α-helix that runs along SL1 towards the Sm ring. Positively charged residues (Arg63, Arg66, Lys70, Lys74, Arg78) line the side of the helix facing stem 1. While we cannot locate their side chains, they most likely contact the sugar–phosphate backbone along the RNA. A region of irregular structure of U1-70K lies in a furrow between Sm proteins D2 and F and continues on the underside of the Sm ring (Figure 1B). In our structure, the electron density for the U1-70K N-terminal extension fades out beyond the centre of the Sm ring, indicating that the very N terminus of U1-70K is degraded or disordered. This finding underscores the importance of the U1-C protein, which is lacking in our structure, as a site of attachment for the very N terminus of U1-70K. On the underside of the Sm core RNP, we see the region around His34 of U1-70K directly interacting with the single-stranded U1 snRNA exiting the Sm core RNP in the vicinity of residues A135 and G136 (Figure 1C). Thus, U1-70K provides a U1-specific element that helps to guide the snRNA through the Sm core RNP.
Scaffolding of the U1 snRNP structure by the snRNA
Structural elements of a U1 snRNA model predicted based on solution studies (Krol et al, 1990; Duckett et al, 1995) could be well fitted as rigid bodies to the electron density (Figure 2A; Supplementary Figure S2). The tips of SL3 and SL4 are less well defined in the electron density compared with the remainder of the RNA, suggesting that they are flexible. A major structuring element of the RNA is the four-way junction 5′ of the Sm site, which in other U snRNAs is replaced by simpler stem-loop structures. Global organization of the four-way junction in the framework of the U1 snRNP is very similar to its structure in isolated U1 snRNA deduced from structure probing in solution (Krol et al, 1990; Duckett et al, 1995). The interaction with common and specific proteins seems to fine tune and presumably stabilize the relative orientation of the double helical stacks. In the structure of recombinant U1 snRNP, the apical loop of SL2 was replaced by a kissing loop in order to promote crystal packing (Pomeranz Krummel et al, 2009). As a consequence, the direction of SL2 in that structure deviates from the path of SL2 in the present U1 snRNP structure (Figure 2B).
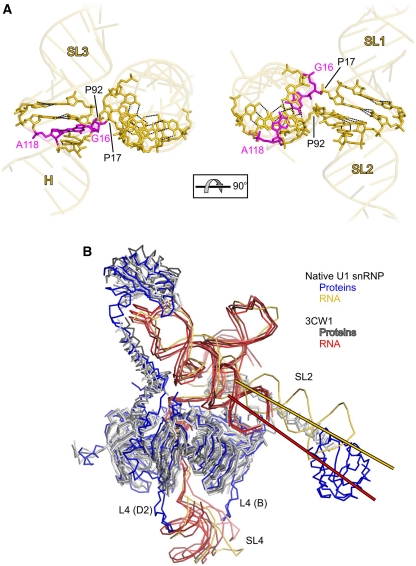
Figure 2.
RNA elements and structural comparison. (A) Orthogonal views of the U1 four-way junction. The view in the left panel is from the upper left corner of Figure 1A. The RNA is in gold, with nucleotides at the crossover shown as sticks. Dashed lines, hydrogen bonds. Phosphates 17 and 92, at which the chain changes direction, and the non-canonical G16-A118 base pair (magenta) are pointed out. (B) Superposition of the backbone models of the present native U1 snRNP structure and of the engineered, recombinant U1 snRNP structure (PDB ID 3CW1; Pomeranz Krummel et al, 2009). Cα-atoms of the proteins and phosphorus atoms of the RNAs of all four complexes of the recombinant U1 snRNP were superimposed on the native U1 snRNP structure excluding the U1-A N-terminal RRM, SL2 (RNA residues 47–91) and SL4 (RNA residues 137–164). Proteins and RNA of the native U1 snRNP, blue and gold, respectively; proteins and RNA of the recombinant U1 snRNP, shades of grey and red colours, respectively. Lines indicate the helical axes of SL2 (present structure, gold; 3CW1, red). Landmark elements are labelled. The view is the same as in Figure 1A.
At the phosphate groups of residues G17 and C92, the RNA chain changes direction and connects the branches of the cruciform (Figure 2A). Phosphate 17 links the ascending strand of helix H to the ascending strand of SL1. Phosphate 92 connects the descending strand of SL2 to the ascending strand of SL3. These turning points are in perfect agreement with modelling based on structure probing of naked U1 snRNA (Krol et al, 1990; nucleotide numbering in Krol et al, 1990 is off by plus one residue compared with the present numbering scheme). As also suggested based on the solution model, there are no unpaired bases around the four-way junction. All bases at the crossovers, except G16, are involved in Watson–Crick-type pairing. G16 forms an elongated purine–purine pair, with A118 involving the Watson–Crick faces of the bases (Figure 2A), facilitating the step from helix H to SL1 without requiring intervening unpaired nucleotides.
Modular RNA recognition by the Sm proteins
The RNA element connecting helix H at the base of the four-way junction to the 3′-terminal SL4 passes through a ring formed by the seven Sm proteins (Figure 1A) and can be divided into several segments (Figure 3A). A three-residue linker (A123-A125; brown in Figure 3A, which we refer to as the ‘RNA entry') connects helix H to the Sm site residues (A126-G132; gold). Below the Sm site, the RNA leaves the Sm ring via an extended region formed by four nucleotides (G133-G136; red-brown in Figure 3A, the ‘RNA exit'), which link up to SL4 (U137-A164; beige). The peripheral structural elements, the RNA entry and exit sequences and the Sm site are each recognized by specific parts of one or several of the Sm proteins in a highly modular manner (contact regions 1–6 in Figure 3A).
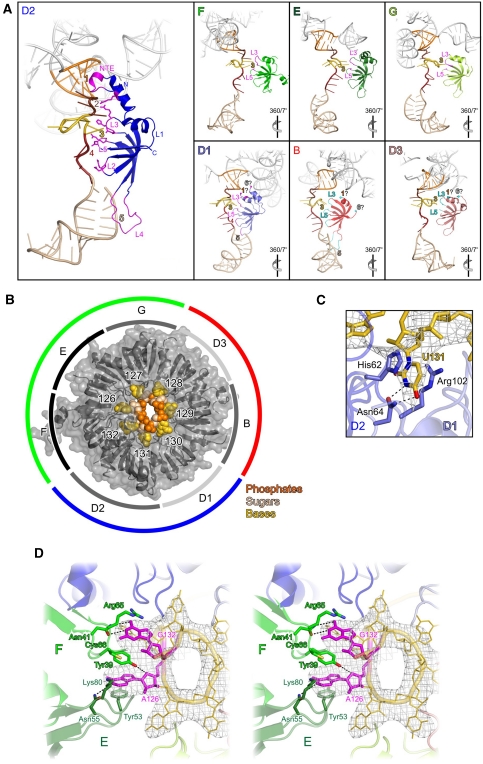
Figure 3.
Binding of U1 snRNA by the Sm proteins. (A) Ribbon plots of Sm proteins (colours as in Figure 1) showing RNA-binding elements (magenta except proteins D3 and B, cyan). RNA segments downstream of the four-way junction are colour coded. Helix H, orange; RNA entry, brown; Sm site, gold; RNA exit, red-brown; SL4, beige. The remainder of U1 snRNA is shown in light grey. Loops in Sm protein D2 are labelled L1–L5. Loops L3 and L5 build up the Sm pocket and are labelled in all panels. NTE, N-terminal extension. Protein–RNA contact regions are indicated by numbers coloured as the respective RNA element. (1) Contacts of N-terminal extensions of Sm proteins to elements of the four-way junction; (2) contacts to the RNA entry region; (3) Sm pockets; (4) contacts to the RNA exit region; (5) contacts to SL4; and (6) contacts of C-terminal extensions of Sm proteins to elements of the four-way junction. Contact regions that are inferred from the position of the Sm protein termini and their physicochemical properties, but not directly seen in the crystal structure, are labelled with a question mark. The view in the D2 panel is 130° about the y axis compared with Figure 1A. Relative rotations of the other panels by one-seventh of a turn about the y axis are indicated by icons. (B) Binding of the Sm site RNA (space-filling model) in the Sm ring (grey semitransparent surface with ribbons). Bases, gold; sugar units, beige; phosphates, orange. The Sm site nucleotides and Sm proteins are labelled. The inner circle indicates sectors corresponding to the Sm proteins. Light grey sections, Sm proteins with non-canonical Sm pockets; grey sections, Sm proteins with canonical Sm pockets; black sections, Sm proteins whose special Sm pockets form the buckle at the Sm site termini. The outer circle indicates sectors corresponding to the building blocks of Sm proteins during Sm core RNP assembly. Blue, D1-D2; green, F-E-G; red, D3-B. Viewed from the top of Figure 1A. (C) Canonical Sm pocket of protein D2 showing the Watson–Crick-like recognition by Asn64, π–π stacking by His62 and cation–π stacking by Arg102. Dashed lines indicate hydrogen bonds. Selected RNA and protein residues are shown as sticks and are colour coded by atom type. C, as the respective molecule; N, dark blue; O, red. Grey mesh, Fo−Fc ‘omit' electron density (with the RNA entry, Sm site and RNA exit omitted) contoured at the 2.5 σ level. Rotated to the right by 130° about the y axis and 10° about the x axis compared with Figure 1A. (D) Stereo plot of the Sm pockets of proteins E (dark green) and F (green) sealing the Sm site termini (magenta). Dashed lines indicate hydrogen bonds. Grey mesh—Fo−Fc ‘omit' electron density as in C. Viewed from the top of Figure 1A. Selected RNA and protein residues are shown as sticks and protein residues are colour coded by atom type as in C. S, yellow.
All Sm proteins contain an N-terminal α-helix followed by a strongly bent five-stranded β-sheet (Figure 1A). β-strands 4 and 5 form the right and left edges of the sheet, respectively. Viewed from the four-way junction, strand β5 of each Sm protein is paired with strand β4 of the clockwise neighbour, while its β4 strand is paired with strand β5 of the counterclockwise neighbour. Thus, the interaction mode originally seen in the crystal structures of Sm protein dimers (Kambach et al, 1999) is continued around the entire Sm ring in a Sm core RNP. With the exception of loop L1 (connecting the N-terminal α-helix to strand β1) at the periphery of the Sm ring, the loops, which connect secondary structure elements of the Sm proteins, as well as the terminal extensions, which precede and follow the canonical Sm folds in some Sm proteins, are facing the snRNA and engage in direct contacts (Figure 3A).
Organization and recognition of the Sm site
The seven Sm site residues are tightly curled into a full, right-handed 360° turn. Phosphates, riboses and bases of the Sm site nucleotides are thereby arranged in three concentric circles, with the phosphates forming the inner surface of a pore through the Sm core RNP and the bases radiating outwards and coming to lie in pockets provided by the Sm proteins (Figure 3B). Consistent with this unusual arrangement, the backbone phosphates of the Sm site are accessible to modification by N-ethyl-N-nitrosourea within U1 snRNP, while the riboses are protected from hydroxyl radical attack (Hartmuth et al, 1999). The tight packing of phosphate moieties in the centre of the Sm core RNP will lead to charge repulsion, which may be overcome by metal ion binding. However, we could not locate metal ions in the electron density at the present resolution. The 2′-hydroxyl groups of Sm site nucleotides appear to be involved in intrastrand interactions, which stabilize the tight turn of the RNA, and in contacts to the Sm proteins. The multiplicity of these interactions explains why individual, but not all, 2′-hydroxyl groups of the Sm site sugars can be deleted (Raker et al, 1999). On the other hand, 2′-O-methyl groups are not tolerated at any position in the Sm site (Raker et al, 1999). This sugar modification reinforces the C3′-endo conformation, while the Sm site nucleotides may be required to adopt a C2′-endo sugar pucker (Törö et al, 2001; Kolev and Steitz, 2006). In addition, the bulky 2′-O-methyl groups may sterically interfere with the tight winding of the Sm site.
Each Sm protein binds a single nucleotide of the Sm site (A126—SmE; U127—SmG; U128—SmD3; U129—SmB; G130—SmD1; U131—SmD2; G132—SmF), consistent with previous crosslinking analyses (Urlaub et al, 2001). In each case, the nucleotide pocket on the Sm protein is formed by residues on loop L3 (β2-β3) and the very N terminus of strand β3, which are part of the Sm1 motif, and by loop L5 (β4-β5), which belongs to the Sm2 motif. While at the present resolution we cannot unequivocally unravel the interactions in atomic detail, the architecture of the Sm pockets generally matches the uridine-specific pockets seen in archaeal Sm proteins (Törö et al, 2001; Figure 3C). On the basis of this similarity, we presume that loop L3 (Sm1) provides one stacking partner for the bound nucleobase (typically an aromatic residue); a conserved asparagine at the N terminus of β3 (Sm1) is positioned by an aspartate in β2 (Sm1) to hydrogen bond to the Watson–Crick face of the base that comes to lie in the pocket; and loop L5 (Sm2) completes the pocket by providing a conserved arginine, which stacks on the other side of the base. A following glycine allows close approach of the base to the backbone of loop L5.
Only Sm proteins D2, G and B exhibit canonical Sm pockets, with an aromatic stacking partner and an asparagine in Sm1 and an Arg-Gly dipeptide in Sm2. In D1 and D3, the canonical aromatic stacking partners are replaced by a serine (Ser35) and an asparagine (Asn38), respectively, which are expected to engage in weaker interactions with bound nucleobases. In U1 snRNP G130 is bound at D1, and weaker electron density at this site suggests that the residue is bound in a more flexible manner than observed in a canonical Sm pocket. Thus, we suggest that the Sm pockets of Sm proteins D1 and D3 provide more relaxed binding sites that could accommodate non-uridine residues and thereby allow deviations from the Sm site consensus. Consistent with this notion, the position corresponding to G130 is the most variable within the Sm sites (Khusial et al, 2005).
Non-canonical nucleotide pockets on proteins F and E provide a buckle for the Sm site
The Sm pockets of proteins E and F bind the Sm site termini (Figure 3D) and bear non-canonical residues in their Sm2 motifs (Lys80 instead of an arginine in E; Cys66 instead of a glycine in F). The bulky Cys66 of protein F occupies part of one surface of G132. It slightly displaces Tyr39 of F, the canonical aromatic stacking partner of G132, towards A126. The smaller sized Lys80 of E leaves part of one A126 surface unoccupied and allows the approach of Tyr39 from F. Thus, the first and last nucleotides of the Sm site directly communicate with each other via Tyr39. In contrast, the bases of all other Sm site nucleotides appear to directly interact solely with residues from their cognate Sm pockets.
The special positioning of Tyr39 of F allows continuous stacking interactions across the nucleotide-binding pockets of proteins F and E. Efficient cross-pocket stacking apparently requires the large aromatic ring systems of purine bases. The inability of pyrimidines to effectively mediate a similar interaction explains the evolutionary conservation of purines as the bordering residues of the Sm sites and the loss of thermodynamic stability on replacement of A126 by a pyrimidine (Raker et al, 1999). An explanation for the absolute necessity for an adenine at the beginning of the Sm site (Khusial et al, 2005) has to await a structure at higher resolution.
The special binding of A126 correlates with an unusual susceptibility of its N7 position towards methylation by dimethylsulfate (DMS; Hartmuth et al, 1999). The N7 position of A126 is accessible to the bulk solvent, allowing access of the modifying reagent. In addition, we presume that the special binding situation for A126 modulates its electronic configuration to make it amenable to attack by DMS. This interpretation is in agreement with the notion that the unusual N7 reactivity of A126 is correlated with a commitment to Sm core formation and thus presumably a native-like positioning of the residue within the Sm ring (Hartmuth et al, 1999).
Guiding of the snRNA into and out of the Sm ring
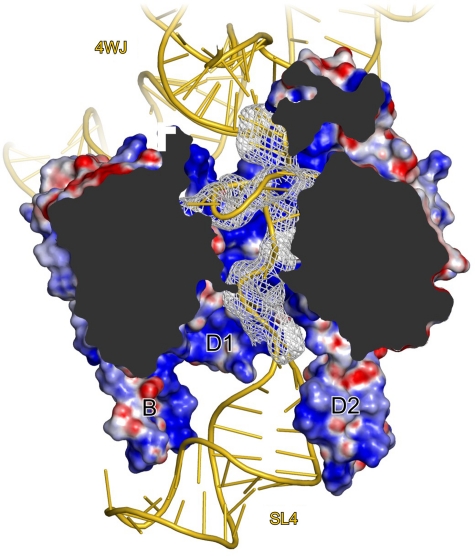
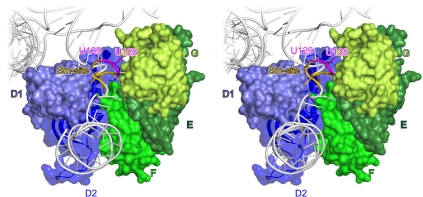
The RNA regions entering and exiting the Sm protein ring are positioned off-centre along its inner pore (Figure 4). The bulk of the electron density, corresponding to the RNA entry and exit regions, clearly abuts the inner surface formed by proteins D1, D2, F and E (Figure 4). As the proteins present strongly positively charged surface patches towards the inner pore (Figure 4), we placed the negatively charged backbone of the RNA entry and exit elements alongside proteins D1, D2, F and E. Conversely, the RNA entry and exit elements are more remote from the other side of the Sm ring formed by proteins G, D3 and B (Figure 4), and we cannot discern direct protein–RNA interactions between these components. However, it is conceivable that extended amino-acid side chains of Sm proteins G, D3 and/or B reach across the inner pore of the Sm ring to interact with the RNA regions neighbouring the Sm site, but are not seen at the present resolution.
Figure 4.
Threading of the snRNA through the Sm ring. Cut-away view into the Sm ring showing the displacement of the single-stranded RNA region towards the D1-D2 sector. Sm proteins are shown in surface representation with the electrostatic surface potential in the inner pore of the Sm ring. Blue, positive charge; red, negative charge. Selected elements are labelled and coloured as in Figure 1A. 4WJ, four-way junction. Grey mesh, Fo−Fc ‘omit' electron density (with the RNA entry, Sm site and RNA exit omitted) contoured at the 2.5 σ level. Rotated 150° about the y axis compared with Figure 1A.
In the structure of recombinant U1 snRNP, the D2 protein was seen to interact with helix H of the four-way junction (Pomeranz Krummel et al, 2009). We also observe that the N-terminal extension of D2 forms an additional α-helix that places a preceding loop across the minor groove of helix H (Figure 3A). Lys6 and Lys8 in this extension, Arg19 in the helix preceding the canonical Sm fold and Arg61 in loop L3 (connecting β2 to β3) of D2 seem to contact the backbone of A123, U124 and A125 directly preceding the Sm site. As there are no other obvious Sm protein contacts to the RNA entry, protein D2 is primarily responsible for guiding the snRNA into the pore of the Sm ring (Figure 3A). At the exit side, the backbone of the single-stranded nucleotides following the Sm site (around residue A135) is sandwiched between the L2 loops (β1-β2) of the D2 and D1 proteins, which thereby serve to guide the RNA exit region out of the Sm ring (Figure 3A).
Basic patches proximal to the Sm folds bind RNA secondary structure elements upstream of the Sm site
Similar to the N-terminal extension of protein D2, the very N termini of proteins D1 and B contain positively charged residues that approach the backbone of helix H and the RNA entry region. We suggest that these latter regions aid in positioning the four-way junction via electrostatic interactions. Proteins D1, D3 and B additionally exhibit positively charged residues in regions immediately C-terminal to their Sm folds. We suspected that these elements may also be used to mediate interactions with portions of U1 snRNA. Indeed, the C terminus of D1 folds back towards the four-way junction and approaches the ascending branch of SL1 in the region of residues G18 and A19 (Figure 3A). Conversely, electron density for the C-terminal extensions of proteins D3 and B ends more remote from snRNA elements (Figure 3A). Together, these contacts of the Sm protein termini to the four-way junction explain hydroxyl radical protection pattern at the ascending branch of SL3 and at the descending branch of helix H (Hartmuth et al, 1999).
Other snRNAs bear different secondary structure elements neighbouring their Sm sites. These RNA elements may be supported by differently oriented Sm protein termini. The latter notion is corroborated by the observation that the accumulation of positive charges at the termini and in terminal extensions of Sm proteins D1, D2, D3 and B is phylogenetically conserved, while the exact sequence patterns are variable (for example, RG-repeats in higher eukaryotic Sm proteins D1, D3 and B are replaced by K/R-rich elements in yeast; Supplementary Figure S3).
Tentacle-like L4 loops of Sm proteins D2 and B suspend the 3′-terminal SL4
The majority of U snRNAs contain RNA secondary structure elements on both sides of their Sm sites. In the case of U1 snRNP, the presence of a 3′-terminal stem-loop, SL4, additionally stabilizes the Sm core RNP and kinetically aids in its assembly (Raker et al, 1999) by a so far unknown mechanism. The L4 loops in the Sm2 motif of proteins D2 and B are unusually long and are partially disordered in isolation (Kambach et al, 1999), bearing a number of conserved positively charged side chains at their tips (Supplementary Figure S3). Although the electron density is fragmented in this region, our present structure unequivocally shows that the elongated L4 loops of D2 and B reach towards SL4 and contact the element remote from the core of the Sm ring (Figures 1A, B and 3A; Supplementary Figure S2). The L4 loops of proteins D2 and B are reminiscent of a clamp that secures the Sm ring against the 3′-terminal SL (Figure 1B). The existence of such clamp-like features was previously postulated based on nucleotide analogue interference mapping, which implied important Sm protein contacts to SL4 in U1 snRNP (McConnell et al, 2003).
The importance of this additional remote recognition is corroborated by the observation that the long L4 loops are not cleaved during in situ limited proteolysis (Supplementary Figure S1A), suggesting that they engage in stable interactions with SL4 in the framework of native U1 snRNP. In the complexes of the recombinant U1 snRNP structure (Pomeranz Krummel et al, 2009), SL4 is positioned variably with respect to the Sm ring (Figure 2B). As L4 contacts to SL4 are not contained in that structure, this observation suggests that the L4 interactions we observe in the present structure reinforce a stable positioning of SL4.
Discussion
We have determined the crystal structure of a trimmed U1 snRNP obtained natively from HeLa nuclear extract. Apart from the U1-C protein, this structure presumably encompasses the entire stably folded core of U1 snRNP and provides a picture of how U1 proteins are assembled on a full-length native U1 snRNA. In particular, our structure elucidates how the Sm proteins bind the Sm site, neighbouring single-stranded regions of the snRNA and peripheral secondary structure elements in a manner that is most likely paradigmatic for all Sm core RNPs. Although we cannot unequivocally discern details of atomic contacts at the present resolution, the quality of the electron density suffices to deduce general architectural principles. Most importantly, our work elucidates an intricate interplay between RNA and protein elements within and around the Sm core RNP on different organizational levels.
Stable Sm core formation includes contacts beyond the Sm site
Nine nucleotides containing the Sm site are sufficient for initial Sm core assembly (Raker et al, 1999). However, the thermodynamic stability and the kinetics of assembly of the Sm core RNP are strongly enhanced by inclusion of flanking single-stranded regions and the neighbouring secondary structure elements of the snRNA (Raker et al, 1999). Our structure revealed that these neighbouring RNA elements are recognized by special motifs of some Sm proteins. The single-stranded RNA entry and exit sequences directly neighbouring the Sm site are contacted by an N-terminal extension on D2 and by the L2 loops of the D1 and D2 proteins, respectively. N- and C termini of Sm proteins D1, D2 and B bearing conserved basic patches support the four-way junction upstream of the Sm site. In addition, the long L4 loops of D2 and B interact with SL4. Our structure lacks the very C-terminal RG-rich tails of proteins D1, D3 and B. Arginines in these tails are di-methylated during regulated Sm core RNP formation in vivo (Friesen et al, 2001; Meister et al, 2001). It is possible that di-methylated RG-rich tails solely improve initial Sm protein–snRNA contacts. Additionally, assembled Sm core RNPs may employ the modified extensions to establish flexible, sequence-independent interactions with each other and with the substrate pre-mRNA on spliceosome assembly.
The single-stranded flanking regions of different snRNAs exhibit different lengths. For example, while U1 snRNA contains three nucleotides before and four nucleotides behind the Sm site, U5 snRNA exhibits a longer entry and a shorter exit region. The exit funnel of the Sm ring is rather wide, suggesting that it could accommodate parts of a secondary structure element that closely follows 3′-end of the Sm site. Conversely, it is easily conceivable that elongated entry (or exit) sequences may loop out at the top or bottom of the Sm ring, explaining the mild phenotypes elicited by insertions in the single-stranded region of U5 snRNA (Jones and Guthrie, 1990).
The U7 snRNP involved in 3′-end maturation of replication-dependent histone pre-mRNAs exhibits a special Sm core RNP, in which the D1-D2 dimer is replaced by the LSm10-LSm11 dimer (Pillai et al, 2003). The LSm11 protein that replaces D2 carries a unique domain insertion that is functionally involved in 3′-end cleavage (Pillai et al, 2003). We suggest that LSm11-specific elements may functionally substitute for D2 elements that contact RNA secondary structures neighbouring the Sm site. For example, they could recognize 5′-portions of the U7 snRNA when base-paired to the histone downstream element in the pre-mRNA, thereby aiding correct recognition and positioning of the substrate.
Extended termini and elongated L4 loops are not present in prokaryotic Sm/LSm-type proteins, showing that formation of homo-oligomeric Sm core RNPs rely primarily on the most fundamental recognition element, the Sm site. Thus, higher-order aspects of the protein–RNA network in eukaryotic Sm core RNPs are not represented in the prokaryotic counterparts.
Modularity lends robustness and specificity to Sm core RNP formation
Each Sm protein presents a modular RNA-binding surface that can interact with multiple RNA elements. The situation is best illustrated with Sm protein D2 that truly resembles a Swiss army knife for RNA recognition (Figure 3A). The protein contains an N-terminal extension that binds across the minor groove of helix H at the base of the four-way junction, guides the RNA into the Sm ring via this extension and its loop L3 (contact regions 1 and 2 in Figure 3A), uses portions of its Sm1 and Sm2 motifs to provide a pocket for the Sm site nucleotide U131 (contact region 3), conducts the RNA exit region out of the Sm ring through its L2 loop (contact region 4) and binds the 3′-terminal SL via its unusually long L4 loop (contact region 5). In contrast to protein D2, which uses all of its RNA-binding elements to interact with different segments of the snRNA, not all RNA-binding elements of the other Sm proteins are engaged in direct RNA contacts in the framework of U1 snRNP. For example, besides D2 only D1 also uses its L2 loop to guide the RNA out of the Sm ring, and protein B is the only other Sm protein exhibiting an elongated L4 loop that contacts SL4.
We suggest that the above-mentioned recognition modules have an additive effect on Sm core stability and, thus, stable RNA binding does not require perfect fitting of all modules. For example, some Sm pockets may be occupied by non-canonical nucleotides, such as G130 in the variant Sm pocket of protein D1 in our present structure, if sufficient Sm pockets bind canonical uridines and/or if in addition other protein–RNA contacts (for example, between the termini and the four-way junction or between long L4 loops and SL4) exist. This idea is supported by yeast U4 snRNA, which does not exhibit a 3′-terminal SL and is highly sensitive towards point mutations in the Sm site (Hu et al, 1995). Thus, the peripheral interactions at the 3′-SL can apparently make up for some deficits in other RNA–protein contacts, for example, in the Sm site, effectively buffering the stability of the system.
The above-mentioned concept of a modular protein–RNA recognition in the Sm core RNP can explain the unexpected robustness of the yeast U5 snRNP against presumed destabilizing mutations in the Sm site (Jones and Guthrie, 1990). Apparently, destabilizing mutations can be tolerated if sufficient compensating interactions are present. By the same reasoning, it becomes understandable that Sm sites cannot be transplanted at will between snRNPs (Jarmolowski and Mattaj, 1993). The U1 Sm site bears a non-canonical G130, which does not fit perfectly into its Sm pocket but is apparently allowed because of the multitude of RNA contacts beyond the Sm site (Figure 3A). Such additional RNA contacts may be less pronounced in U5 snRNP so that stable Sm core assembly in U5 snRNP may not be possible with a sub-optimal U1-like Sm site. Conversely, transplantation of the canonical U5 Sm site to U1 snRNA is expected to give rise to a functional snRNP as observed (Jarmolowski and Mattaj, 1993).
Some of the Sm protein–RNA contacts can apparently not be compromised. For example, the occupation of a minimum number of the U-specific pockets on the Sm proteins by uridines and the presence of terminal purines that serve as a buckle for the Sm site appear to be indispensable for stable Sm core formation (Jones and Guthrie, 1990; Raker et al, 1999). Most likely, the single-stranded connectors neighbouring the Sm site also require a minimum length to link up to the peripheral secondary structure elements when present.
The distribution of the canonical nucleotide-binding pockets in the Sm ring does not appear to be random (Figure 3B). First, each of the Sm core building blocks (D1-D2, F-E-G and D3-B) contains one Sm protein that exhibits a canonical U-specific nucleotide-binding pocket (in proteins D2, G and B, respectively). These canonical interactions are presumably important to ensure stable integration of each building block into the Sm core RNP. In particular, the canonical contact involving protein B may aid in the addition of the D3-B dimer, which is the last step in Sm core formation. Second, in the assembled Sm core RNP, two of the canonical interactions (protein G-U127 and protein D2-U131) are directly bordering the non-canonical pockets on proteins F and E, possibly reinforcing the sealing of the Sm site termini.
The highly complex and modular protein–RNA interactions at the Sm site can explain the exceptional thermodynamic and kinetic stability of Sm core RNPs (Raker et al, 1996, 1999). However, high stability per se does not explain specificity in Sm core formation. We suggest that imperfection or lack of some recognition modules and compensation by other modules is an important proofreading mechanism during assembly that ensures specificity in the protein–RNA interactions. If initial, sub-optimal contacts have to be consolidated by peripheral contacts to generate a stable Sm core RNP or give rise to a core RNP in a given timeframe, cognate interactions can be efficiently distinguished from non-cognate binding events. In this manner, it is understandable, how specific Sm sites can, in the framework of their respective snRNAs, associate with specific sets of Sm/LSm proteins.
Implications for the biogenesis of the Sm core domain
In vitro assembly studies have revealed that the Sm proteins D1-D2 spontaneously associate with the F-E-G trimer and with U1 snRNA to form a stable sub-core that is committed to complete Sm core assembly by addition of the D3-B dimer (Raker et al, 1999). The unusual N7 DMS reactivity of A126 is observed in this sub-core, showing that the RNA–protein interactions within the sub-core closely resemble the interactions in the fully assembled Sm core RNP (Hartmuth et al, 1999). The idea that the sub-core is a bona fide intermediate during Sm core assembly is further corroborated by the observation that in vivo assembly of Sm core RNPs via the PRMT5 and SMN complexes passes through the same intermediate (Kolb et al, 2007; Chari et al, 2009). Based on our structure, the sub-core Sm proteins fix the single-stranded region of the snRNA along its entire path through the Sm core RNP (primarily via protein D2) and fasten the tight helical turn of the Sm site RNA (via proteins E and F; Figure 5), while the D3-B sector is dispensable for this scaffolding. As a consequence, Sm site nucleotides U128 and U129 of U1 snRNA in the sub-core are presented for D3-B binding (Figure 5). Addition of the D3-B dimer will be aided by the L4 loop of B, completing the clamp-like binding of SL4 (Figure 1B). The conservation of the Sm site and the presence of flanking secondary structures in most other snRNAs suggest that the assembly of other Sm core RNPs will follow the same scenario.
Figure 5.
Sub-core RNP. Stereo plot showing a model of the sub-core RNP after removal of the D3-B sector. Other Sm proteins are labelled and coloured as in Figure 1A. D2 is shown as a ribbon plot, with semitransparent surface highlighting the distal contacts to the snRNA. U1 snRNA is in light grey with the Sm site in gold and residues U128 and U129, presented to D3-B, in magenta. Rotated 120° about the y axis and 45° about the x axis compared with Figure 1A.
A multi-layered RNA–protein interaction network underlying full snRNP assembly
The structure of U1 snRNP reveals an intricate interplay between RNA and proteins on multiple levels. On the most fundamental level, the Sm proteins organize the Sm site, as well as the entry and exit of neighbouring RNA regions into and out of the Sm core RNP. This crosstalk generates novel binding sites for specific proteins, as illustrated by the U1-70K protein. The extended N terminus of 70K sneaks along the stem of SL1 to the Sm core RNP, reaches around the side formed by D2 to the underside of the Sm ring and interacts with the U1 snRNA exiting the core (Figure 1B and C). These higher-order U1-70K contacts will reinforce the Sm core assembly. In addition, the U1-70K protein is apparently recruited to recognize and thus consolidate a U1-specific Sm site exit. In other snRNPs, the Sm proteins may interact differently with the respective snRNAs and provide other binding sites for specific proteins. Direct evidence for such higher level interplay comes from EM studies of U5 snRNP. While 10S U5 snRNP (comprising the U5 snRNA and the Sm proteins) clearly shows the doughnut shape of the Sm core structure (Kastner et al, 1990), the core cannot be discerned in the 20S U5 snRNP (comprising in addition the U5-specific proteins; Kastner et al, 1990; Sander et al, 2006), presumably because it is tightly ‘packaged' by interactions with specific proteins.
Implications for spliceosome assembly
U1 snRNP is the first snRNP to land on the pre-mRNA and is a major organizer of spliceosome assembly. In cooperation with U2 snRNP, it has to achieve the functional pairing of the intron ends already during early steps of spliceosome assembly (Dönmez et al, 2007). Therefore, it requires free docking sites for components of the U2 snRNP (and vice versa). The Sm core RNPs may have key roles in providing such docking sites by the same principles, through which they generate binding sites for the specific proteins during snRNP assembly. That is, the combinations of Sm proteins and specific snRNAs may give rise to binding platforms for elements of other snRNPs and/or non-snRNP splicing factors. It has been shown that hydroxyl radical generators specifically attached to the 5′-region of U2 snRNA induce cleavages in the U1 SL3 (Dönmez et al, 2007). Thus, one of the U2 snRNP docking sites on U1 could be the protein-free SL3. The Sm proteins restrain, directly or indirectly, other parts of the U1 snRNA structure, exposing this loop at the top of the fully assembled U1 snRNP in proximity to the 5′-SS-binding region (Figure 1A and B).
Several of the common and all of the specific proteins of U1 snRNP bear long extensions, whose distal regions are not visible in our structure. These regions include long C-terminal extensions of Sm proteins D1, D3 and B (residues 86–119, 95–126 and 91–240, respectively) that are rich in Arg-Gly repeats and, in the case of B, also contain Pro-rich regions; a long C-terminal stretch that contains Arg-Ser repeats in the U1-70K protein (residues 184–437), resembling SR-type splicing factors (Graveley and Maniatis, 1998); a second RRM at the C terminus of the U1-A protein (residues 208–282), which is connected to the N-terminal RRM by a flexible linker (residues 115–207); and a C-terminal extension of the U1-C protein (residues 62–159, not contained in the crystal structure of recombinant human U1 snRNP). Combining the present structure with the previous recombinant U1 snRNP structure (Pomeranz Krummel et al, 2009), we constructed a full model of the U1 snRNP encompassing intrinsically unstructured or flexibly attached regulatory elements to illustrate the remarkable size of these elusive parts (Supplementary Figure S5). Some of these extensions may adopt a more compact conformation under certain conditions and may fold back onto the structured part of U1 snRNP, which could explain additional masses seen in the cryo-EM structure of native human U1 snRNP (Stark et al, 2001), compared with the crystal structures.
On the basis of the observation that Sm proteins D1, D3 and B can be crosslinked to the pre-mRNA during early spliceosome assembly steps in yeast (Zhang and Rosbash, 1999), and that removal of their positively charged C-terminal tails affects cell viability and commitment complex formation (Zhang et al, 2001), it was suggested that the tails consolidate short intermolecular base-pairing interactions critical for spliceosome assembly. The long extensions are also reminiscent of the C-terminal domain of the largest subunit of eukaryotic RNA polymerase II, which serves as a landing pad for RNA processing factors (Meinhart et al, 2005). Thus, in an analogous manner, the extensions of the U1 snRNP may also mediate close approximation of snRNPs during spliceosome assembly and/or may serve to establish contacts to other splicing factors. For example, the alternative splicing factor ASF/SF2 binds to the RS-like domain of U1-70K (Xiao and Manley, 1997).
Comparison of the full U1 snRNP model (Supplementary Figure S5) with its structured core (Figure 1A and B) demonstrates the potential of our in situ trimming strategy for crystallizing RNPs purified from native sources. U1 snRNP may afford a test case for even more complex assemblies that bear long, unstructured regions at their peripheries, such as entire spliceosomes. Owing to their complexity, spliceosomes resist reconstitution from recombinant parts. However, stable intermediates can be stalled and purified (Bessonov et al, 2008; Fabrizio et al, 2009; Warkocki et al, 2009). Limited proteolysis may in the future allow crystallization of these particles.
Materials and methods
Detailed procedures are provided in the Supplementary data. Briefly, U1 snRNP was isolated from HeLa nuclear extract by immunoaffinity purification, glycerol gradient centrifugation and ion exchange chromatography and concentrated by ultracentrifugation (Kastner and Lührmann, 1999). The particle was co-crystallized with a DNA nonamer (5′-AGGTAAGTA-3′) mimicking a 5′-SS in the presence of chymotrypsin. A model of a heptameric ring of Sm proteins and the structure of the N-terminal RRM of U1-A in complex with the tip of SL2 were positioned sequentially in molecular replacement searches. Molecular replacement phases enabled the localization of heavy atom sites in derivative crystals by difference Fourier analyses. An atomic model was built into experimental electron density maps, aided by the backbone model of a recombinant truncated U1 snRNP (Pomeranz Krummel et al, 2009). Final rounds of refinement made use of deformable elastic network restraints (Schröder et al, 2010).
Accession codes
Coordinates and structure factors have been deposited with the RCSB Protein Data Bank (http://www.rcsb.org/pdb/) under accession code 3PGW and will be released on publication.
Supplementary Material
Acknowledgments
We are grateful for the help of many people at the beginning of the project, in particular Alain O Miller (CIL BIOTECH s.a., Mons, Belgium) for his support in HeLa cell cultivation. We thank Thomas Conrad, Peter Kempkes and Hossein Kohansal (MPI for Biophysical Chemistry, Göttingen, Germany) for maintenance of the HeLa cell culture and help with snRNP preparation; Christian Stegmann (MPI for Biophysical Chemistry), Gleb Bourenkov (EMBL Outstation, Hamburg, Germany) and Alexandre Urzhumtsev (University of Strasbourg, France), for advice in crystallographic computing; Henning Urlaub and Monika Raabe (MPI for Biophysical Chemistry), for mass spectrometric analyses; the team of Clemens Schulze-Briese for support during diffraction data collection at beamlines PXI and PXII of the Swiss Light Source (Villigen, Switzerland); and Klaus Hartmuth (MPI for Biophysical Chemistry) for fruitful discussions. This work was supported by the Max-Planck-Gesellschaft, the Freie Universität Berlin, the Deutsche Forschungsgemeinschaft (to BK and RL), the Fonds der Chemischen Industrie (to RL) and the Ernst-Jung-Stiftung (to RL).
Footnotes
The authors declare that they have no conflict of interest.
References
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′- end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18: 5789–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D (1996) Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol 257: 398–411 [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Branlant C, Krol A, Ebel JP, Lazar E, Haendler B, Jacob M (1982) U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J 1: 1259–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Paknia E, Fischer U (2009) The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 21: 387–393 [DOI] [PubMed] [Google Scholar]
- Dönmez G, Hartmuth K, Kastner B, Will CL, Lührmann R (2007) The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell 25: 399–411 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AI, Lilley DM (1995) The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell 83: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R (2009) The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fury MG, Zhang W, Christodoulopoulos I, Zieve GW (1997) Multiple protein: protein interactions between the snRNP common core proteins. Exp Cell Res 237: 63–69 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T (1998) Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell 1: 765–771 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Raker VA, Huber J, Branlant C, Lührmann R (1999) An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J Mol Biol 285: 133–147 [DOI] [PubMed] [Google Scholar]
- Hu J, Xu D, Schappert K, Xu Y, Friesen JD (1995) Mutational analysis of Saccharomyces cerevisiae U4 small nuclear RNA identifies functionally important domains. Mol Cell Biol 15: 1274–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Mattaj IW (1993) The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J 12: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Guthrie C (1990) Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J 9: 2555–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Lührmann R, Li J, Nagai K (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387 [DOI] [PubMed] [Google Scholar]
- Kastner B, Bach M, Lührmann R (1990) Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc Natl Acad Sci USA 87: 1710–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Lührmann R (1999) Purification of U small nuclear ribonucleoprotein particles. Methods Mol Biol 118: 289–298 [DOI] [PubMed] [Google Scholar]
- Keel AY, Rambo RP, Batey RT, Kieft JS (2007) A general strategy to solve the phase problem in RNA crystallography. Structure 15: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusial P, Plaag R, Zieve GW (2005) LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci 30: 522–528 [DOI] [PubMed] [Google Scholar]
- Kolb SJ, Battle DJ, Dreyfuss G (2007) Molecular functions of the SMN complex. J Child Neurol 22: 990–994 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA (2006) In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat Struct Mol Biol 13: 347–353 [DOI] [PubMed] [Google Scholar]
- Krol A, Westhof E, Bach M, Lührmann R, Ebel JP, Carbon P (1990) Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res 18: 3803–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liautard JP, Sri-Widada J, Brunel C, Jeanteur P (1982) Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J Mol Biol 162: 623–643 [DOI] [PubMed] [Google Scholar]
- McConnell TS, Lokken RP, Steitz JA (2003) Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. RNA 9: 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P (2005) A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U (2001) Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11: 1990–1994 [DOI] [PubMed] [Google Scholar]
- Nelissen RL, Will CL, van Venrooij WJ, Lührmann R (1994) The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J 13: 4113–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K (1994) Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schumperli D (2003) Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev 17: 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Lührmann R (1999) Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol 19: 6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Lührmann R (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J 15: 2256–2269 [PMC free article] [PubMed] [Google Scholar]
- Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Lührmann R, Stark H (2006) Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell 24: 267–278 [DOI] [PubMed] [Google Scholar]
- Schröder GF, Levitt M, Brunger AT (2010) Super-resolution biomolecular crystallography with low-resolution data. Nature 464: 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield DG, Lynch M (2008) Evolutionary diversification of the Sm family of RNA-associated proteins. Mol Biol Evol 25: 2255–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J 14: 2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Lührmann R, Kastner B (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409: 539–542 [DOI] [PubMed] [Google Scholar]
- Törö I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J 20: 2293–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Kostka S, Grelle G, Lührmann R (2000) A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. J Biol Chem 275: 41458–41468 [DOI] [PubMed] [Google Scholar]
- Urlaub H, Raker VA, Kostka S, Lührmann R (2001) Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J 20: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R (2009) Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev 11: 334–344 [DOI] [PubMed] [Google Scholar]
- Zhang D, Abovich N, Rosbash M (2001) A biochemical function for the Sm complex. Mol Cell 7: 319–329 [DOI] [PubMed] [Google Scholar]
- Zhang D, Rosbash M (1999) Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev 13: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.