Abstract
Polynuclear platinum agents are a structurally unique class of anti-cancer drugs, distinct from the cisplatin family. To describe the chemistry and biology of this class, it was necessary to challenge the accepted paradigms for the structure–activity relationships; design new chemotypes and delineate the structures and consequences of their DNA binding modes. This article summarizes the structural changes induced in DNA by both covalent (bond-forming) and non-covalent (ligand recognition) adducts. Solution (Nuclear Magnetic Resonance), solid state (crystallography) and gas-phase (Electrospray Ionization Mass Spectrometry) techniques have all been used to describe the new DNA structures along with molecular biological techniques. The combined approaches allow molecular description of hitherto unobserved adducts such as long-range major-groove interstrand crosslinks; directional isomers on DNA and a third class of ligand–DNA binding, the phosphate clamp. The phosphate recognition is distinct from “classic” minor-groove recognition or intercalation.
1. Introduction
DNA has been a target for anti-cancer chemotherapy since the development of the alkylating agents in the 1940s. The target is, however, not specific for tumour cells, and currently anti-cancer drug development focuses to a great extent on identification and validation of new targets to produce more selective drugs capable of complementing those in current clinical use. Nevertheless, DNA reactivity is a fertile source of research for new analogs of existing drugs; interference with unusual DNA structures such as Holliday junctions and quadruplexes or DNA–protein complexes, notably telomerase and topoisomerase, and for novel target-activated and/or photo-activated approaches.1–3
DNA binding, and the consequences for structure and function, is the mechanistic paradigm by which cytotoxic platinum complexes are believed to exert their antitumour activity.4–6 As with most DNA-targeting agents used in cancer chemotherapy today, the mechanism of action of cisplatin was only determined after the observation of anticancer utility. The fact that the trans-isomer was not therapeutically active dictated the early fundamental structure–activity relationship which stressed the need for a neutral, cis-oriented geometry capable of bifunctional DNA binding. The modes of DNA binding, the structural consequences and protein recognition as well as effects of DNA binding on cellular signalling pathways have been extensively documented for the cisplatin family.4–6
The identification and acceptance of cellular DNA as target does raise the question of whether platinum chemistry can be extended beyond the cis-chemotype to produce further discrete alteration of DNA structure to affect the biological outcome of anticancer drugs. This remains the challenge for platinum chemists. In the broad picture, modification of DNA structure with subsequent therapeutic application is not restricted or unique to platinum compounds. Structural changes are manifested in many ways, both through covalent modification (the alkylating agents) and “non-covalent” intercalators and minor-groove binders as well as strand-breakage. Indeed, Pt–DNA adducts share similarities in DNA repair mechanisms with many other drugs, as might be expected.7 These considerations raise interesting questions—firstly, can chemists design new compounds capable of producing new adduct structures? This is now certainly the case. Secondly, given the role of DNA repair in clinical resistance to cisplatin,8,9 would the consequences of structurally discrete Pt–DNA adducts be eventually reflected in a different profile of biological and anticancer activity, especially in cisplatin-resistant tumors? It was in this vein that the work was started on polynuclear compounds, with the stated hypothesis that “Complexes capable of molecular interactions not accessible to monomeric complexes or acting by different mechanisms might also display a broader spectrum of clinical activity”.10 To achieve this goal it was necessary to challenge the accepted paradigms for the structure–activity relationships and design new chemotypes and delineate the structures and consequences of their DNA binding modes.
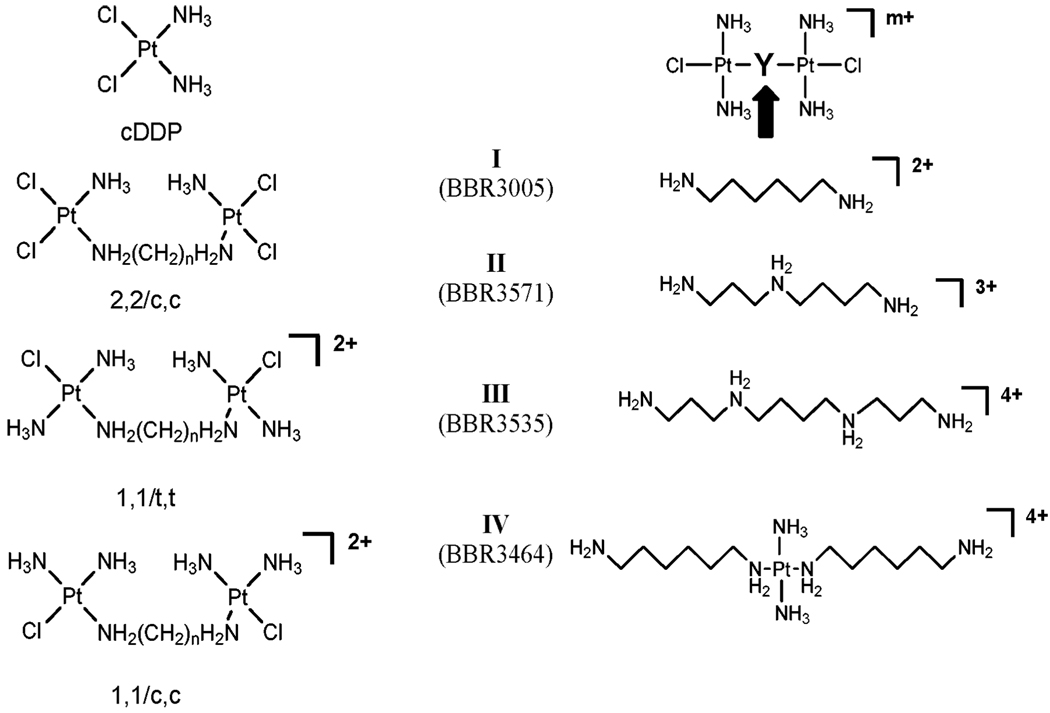
The first dinuclear compound reported by my group contained two cis-Pt units linked together, Fig. 1, and it was of considerable interest when these compounds showed activity in cisplatin-resistant cells—in structural terms, linking two mononuclear cis-[PtCl2(amine)2] units overcame resistance to the parent mononuclear entity.10–12 Quickly we established that a principal component of DNA binding of dinuclear compounds was the formation of {Pt,Pt} inter-strand cross-links (IXL), expected to be structurally distinct from those formed by cisplatin because each Pt unit will bind to one strand of the DNA duplex.13 It will be appreciated that to form a {Pt,Pt} IXL in a dinuclear compound, one needs only a monofunctional coordination sphere around each platinum—linking two cis-[PtCl2(amine)2] units produces tetrafunctional binding agents with a myriad of binding possibilities. When a model compound was not only a good cross-linking agent as expected but actually had biological activity with the ability to overcome cisplatin resistance and displayed in vivo antitumor activity in its’ own right, then a distinct new chemotype based on the structure in Fig. 1 was recognized.14,15 The structure is distinguished by the presence of two monofunctional coordination spheres giving a bifunctional DNA-binding agent with a minimal 2+ charge. Dinuclear compounds with straight-chain diamines as well as linear bridging polyamines based on spermidine (minimal charge 3+) and spermine (minimal charge 4+), as well as cis and trans geometric isomers are encompassed in the structure, Fig. 1. The incorporation of a third platinum tetram(m)ine group gives trinuclear compounds epitomised by the clinical agent BBR3464, the only platinum compound not based on the cisplatin chemotype to have entered human clinical trials.16 The poly ((di/tri)nuclear) motif is very diverse. Time and availability of personnel dictated that we focus on the bifunctional DNA–DNA crosslinking agents, especially BBR3464, and placed their chemistry and biochemistry in relation to their biological activity and that of the mononuclear agents.
Fig. 1.
Structures of cis-[PtCl2(NH3)2] (cisplatin, cis-DDP) and di and trinuclear platinum compounds studied. IV represents BBR3464.
This contribution therefore summarizes the structures and properties of new DNA structures produced by polynuclear platinum compounds (PPCs). We focus on three main points: the nature of (Pt,Pt) long-range interstrand crosslinks and their biological consequences; the effect of pre-association of charged complexes on the polymer backbone and the subsequent discovery of a new mode of ligand binding to DNA, the phosphate clamp, and thirdly, the preference for single-stranded over double-stranded DNA.
2. Global DNA binding
The properties of DNA globally modified by polynuclear complexes are highlighted by rapid binding, the formation of a significant percentage of long-range IXL and irreversible sequence-dependent conformational changes (B → Z; B → A).16 The nature and extent of cross-linking depends on a number of structural features of the compounds:
Geometry: the cis-geometry is a more effective cross-linking agent than the trans isomer. A notable distinction from mononuclear chemistry is that both geometries display useful anti-tumour activity.
The nature of the linker: the presence of charge affects the overall amount of inter-strand crosslinking. The IXL produced by any given compound is not chain length dependent but is charge dependent. Interestingly, the amount of inter-strand crosslinking appears inversely proportional to charge: IXL formation ability at equal (Pt/DNA) rb values is: IV < III < II < I, Fig. 1.16,17
The charge also enhances cellular accumulation and, thereby, cytotoxicity.18,19 Pre-association on Human Serum Albumin has also been observed.18
2.1 Structures and consequences of site-specific {Pt,Pt} interstrand crosslinks
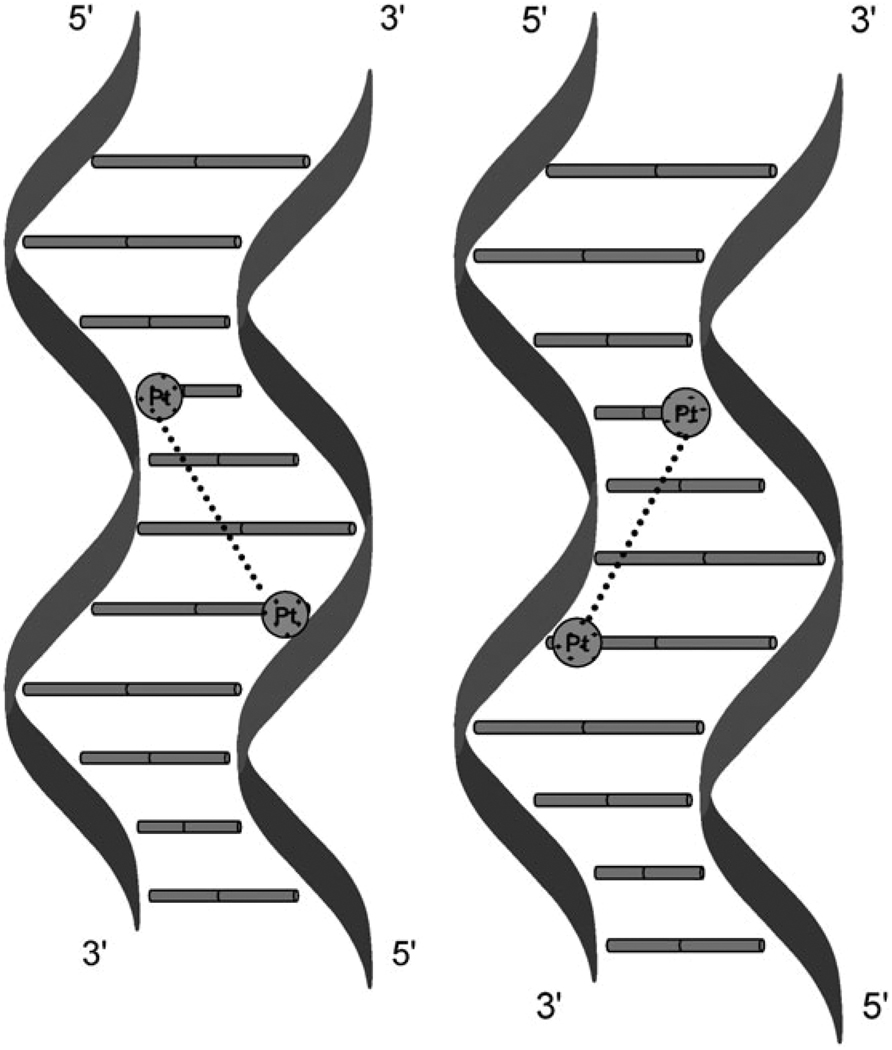
Site-specific crosslinks (or indeed adducts in general) are usually formed by preparing an adduct on one strand and then annealing the complementary strand in a process of “hybridization directed IXL formation”.20,21 Maxam–Gilbert sequencing and footprinting can then identify and confirm the exact mode of binding. Table 1 lists crosslinks identified and studied to date. For BBR3464, crosslinks occur not only in the “normal” 5′ → 5′ direction, since the DNA helix is normally read from the 5′-side, but also in the “opposite” antiparallel 3′ → 3′ direction, Fig. 2.22 The directionality is dependent on the nature of the cross-link. Consider the sequences where G are the platination sites:
| Duplex [1,2]: | 5′TCTCCTATTCGCTTATCTCTCAGAGGATAAGCGAATAGAGAG5′ |
| Duplex [1,4]: | 5′TCTCCTTCTTGTTCTTCCTCCGAGGAAGAACAAGAAGGAGG5′ |
| Duplex [1,6]: | 5′CTCTCTCTATTGTTATCTCTTCTAGAGAGATAACAATAGAGAAGA5′ |
Table 1.
Properties of site-specific crosslinks of polynuclear Pt compounds (see Fig. 1 for structures)
| Site-specific adductsa | ||||
|---|---|---|---|---|
| Compound | IXL, direction | Unwinding/° | Bending/° | HMG protein |
| Cisplatin | 1,2 | 6 | 40–45 | Yes |
| I (BBR3005) | 1,4 5′ → 5′ | 9 | 10 | No |
| II (BBR3571) | 1,4 5′ → 5′ | |||
| 1,4 3′ → 3′ | ||||
| IV (BBR3464) | 1,2 3′ → 3′ | — | — | — |
| 1,4 3′ → 3′ | 9 | 15 | No | |
| 1,4 5′ → 5′ | 10 | 21 | No | |
| 1,6 5′ → 5′ | — | — | — | |
Fig. 2.
Schematic of directional isomers formed in 1,4-interstrand crosslinks by polynuclear platinum complexes showing directionality.
The 1,2-IXL forms in only the 3′ → 3′ direction; the 1,4-IXL forms in both directions in approximately equal proportions while the 1,6-IXL forms in only the 5′ → 5′ direction. Further, the presence of a charged linker enhances directional isomer formation in the 1,4-IXL whereas the simple dinuclear compound with the hexanediamine linker (1,1/t,t, I Fig. 1) forms in only the 5′ → 5′ direction.23,24 To our knowledge, this is the only example of anti-cancer drugs behaving in this manner.25,26
Effect of DNA template on IXL formation and directional isomers
The nature of the DNA template actually affects the efficiency of IXL formation. When intact duplex is used as a template—“postsynthetic modification”20—there is, surprisingly, significantly less crosslinking observed but there was again a slight preference for the 5′–5′ 1,4-IXL over the 3′ → 3′.24 The experimental conditions of the two experiments may help to explain this anomaly. To keep the duplex double-stranded, rather high concentrations of NaClO4 (0.1 M) and lowered temperature (15 °C) are used—in this case the rate of binding and subsequent closure of monofunctional adducts to the bifunctional IXLs is apparently decreased.24
2.2 Structural properties of IXL: delocalization of adduct structure
Bending and unwinding
Both isomers produce similar unwinding but the 3′ → 3′ bends slightly more than its 5′ → 5′ analog in the direction of the major groove, 21° and 15° respectively. These results are significantly different from those of the cisplatin 1,2-interstrand crosslink. They also contrast with the adducts of the simple dinuclear compound (1,1/t,t, I) which, as stated, forms only 5′ → 5′ crosslinks.23
Chemical probes
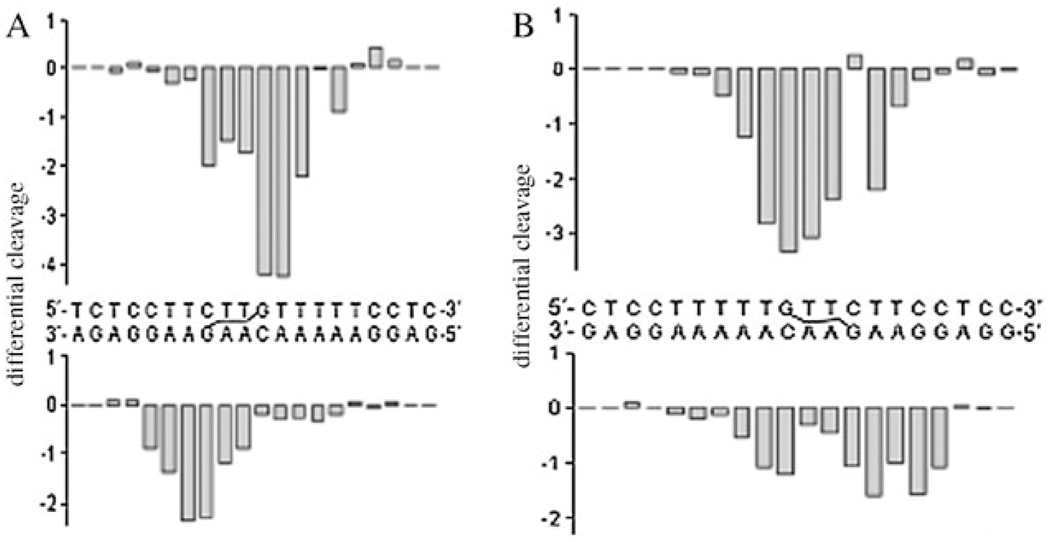
Chemical probes such as KMnO4, DEPC and KBr/KHSO5 which are base-specific (T, A and C respectively) give useful information on local conformational perturbation upon DNA adduct formation.27–29 The major conformational changes of the 5′ → 5′ IXLs occurred 5′ to the binding site, with somewhat weaker distortions on T residues within the binding site. In contrast, the 3′ → 3′ IXL showed most reactivity with the two central AA–TT base pairs separating the platinated bases, see Fig. 3.22,24
Fig. 3.
Properties of directional isomers of 1,4-IXLs. ● Strongly, ○ moderately affected. See ref. 22 and 24.
DNAse footprinting
The endonuclease DNAse I is a monomeric glycoprotein of molecular mass 30.4 kD that specifically cleaves the O3′–P bond of the phosphodiester backbone of the double-helical DNA substrate. Comparison of cleavage patterns in modified and non-modified DNA gives information on the structural perturbation caused by any adduct.30–32 The Pt-induced structural perturbations in the major groove translated into the minor groove, allowing their detection by DNase I with very high sensitivity, Fig. 4.33 The 5′–5′ IXL induces the larger conformational alteration in DNA extending over a wider range of nucleotides. DNase I inhibition by flanking sequences is also seen, mainly to the 3′ or 5′ sides of the 3′–3′ or 5′–5′ IXL respectively. Thus, the delocalized nature of these adducts is confirmed in the presence of a biologically relevant protein.
Fig. 4.
Differential histogram of A: the 3′ → 3′ 1,4-IXL and B: the 5′ → 5′ IXL of BBR3464. Vertical scales are in units of ln[(fa)/(fc)], where fa is the fractional cleavage at any bond in the presence of the drug and fc is the fractional cleavage in the control, nonmodified duplex. Negative values correspond to the adduct-protected and positive values to enhanced cleavage. Adapted from ref. 33.
2.3 Kinetics of formation and structure of site-specific interstrand crosslinks
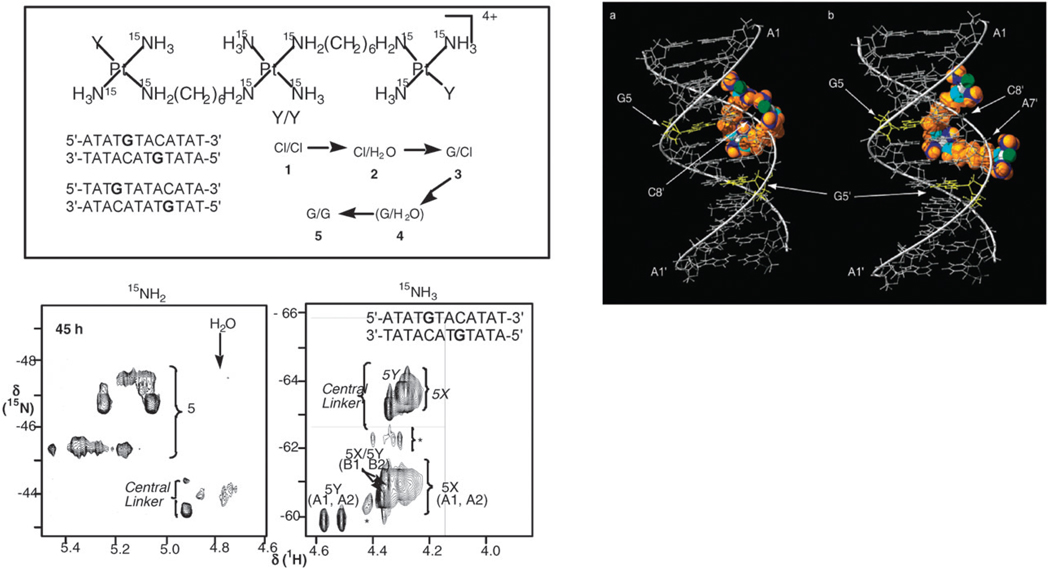
The nature of the directional preferences as well as the structure and kinetics of formation of the 1,4- and 1,6-crosslinks using fully-15N labelled IV (BBR3464) and the self-complementary 12-mer duplexes 5′-d(ATATGTACATAT)2 (1,4-IXL between G and the G of complementary C) and 5′-d(TATGTATACATA)2 (1,6-IXL) was examined by {1H,15N} HSQC NMR spectroscopy.34 This technique is especially useful for following the reactions of PPCs because 15N-labeling of both 15NH2 and 15NH3 groups is possible.35 1H (and 15N) shifts are also sensitive to H-bonding interactions with DNA and thus the local environment surrounding both ends of the molecule, as well as the central linker, can be probed throughout the process, including pre-association with the DNA. Initial electrostatic interactions (pre-association) with the DNA are observed for the Cl–Cl and Cl–H2O species and changes in the chemical shifts of certain DNA 1H resonances are consistent with binding of the central charged {PtN4} linker unit in the minor groove, Fig. 5.34 Whereas there is only one predominant conformer (one GH(8) signal) of the 1,6-crosslink, both the 1H and {1H,15N} NMR spectra show formation of two distinct and non-interconvertible conformers of the 1,4 cross-link. For the platination of the 1,4-IXL, molecular models indicate two distinct pathways for the terminal {PtN3Cl} groups to approach and bind the GN7 in the major groove with the central linker remaining anchored in the minor groove, Fig. 5. For guanine platination in the 1,6-IXL there is initial pre-association but the complex must diffuse off the DNA for covalent binding to occur. Thus, the IXL are conformationally flexible. Further, the implication is that, whilst remaining in the minor groove, directional isomers can be formed (as for 1,4-IXL) but once the compound diffuses off, the “normal” parallel cross-link is favored, as observed for the 1,6-IXL.22
Fig. 5.
{1H,15N} HSQC NMR provided unique insight into the mechanism of formation of 1,4- and 1,6-interstrand cross-links by BBR3464. Species 1–5 represent the stepwise formation of bifunctional IXL. The spectra (e.g. left) showed evidence for formation of two conformers of the 1,4-crosslink (5X,5Y), which stem from a pre-association of the central {PtN4} linker in the minor groove (illustrated in the molecular model above). From ref. 34.
The structure of site-specific interstrand crosslinks
The 5′ → 5′ 1,4-IXL of BBR3464 with the duplex 5′-d(ATGTACAT)2 was characterized by 1D and 2D NMR spectroscopy.36 Strong H8/H1′ intraresidue crosspeaks show a syn conformation of the platinated guanine nucleoside. More interestingly, a strong H8/H1′ intraresidue crosspeak for the A7 base is also consistent with the syn-conformation, which is unusual for adenine residues and bases “outside” and not directly involved in the cross-links. Within the sequence covered by the crosslink, the bases appear to be a mixture of syn and anti and Watson–Crick hydrogen bonding is maintained. This observation is unique and suggests how delocalization of lesions beyond the binding site may occur. Contacts between the central Pt–NH3 and Pt–NH2CH2 protons and the duplex confirm minor groove interactions with this unit. Interestingly, the structure for the same sequence with I, Fig. 1, shows very similar perturbations.37
Comparison of the 5′ → 5′ and 3′ → 3′ interstrand crosslinks
To examine the mechanistic details of the formation of the 3′–3′ 1,4-GG IXL, platination reactions of the self-complementary 12-bp duplex 5′-d(TATACATGTATA)2 (3′ → 3′ 1,4-IXL) with 15N-BBR3464 were also followed using 1D 1H and 2D [1H,15N] HSQC NMR spectroscopy. Note that this sequence is in direct analogy to that of the 5′ → 5′ 1,4-IXL above. The NMR experiments showed evidence for pre-association, aquation and monofunctional binding, suggesting that initially the complexes interact with the DNA similarly as for the formation of the 5′–5′ 1,4-IXL (as per Fig. 5). However, due to structural reasons a single 3′–3′ 1,4-GG IXL does not form under the conditions of these solution experiments and bifunctional DNA binding gives many products. Molecular modelling showed platination of the two guanine residues, separated by two residues (–AT–), would result in severe distortion of the duplex with opening of the major groove and severe fraying of the central base pairs. These results may reflect the chemical probe and DNAse I results where strong alterations of the central base pairs are observed for the 3′–3′ adduct (Fig. 4). Note also that altered conformational changes in the central AT base pairs is a significant feature of the 5′–5′ 1,4-GG IXL formed in the 8-mer and that distortions occur beyond the binding site. In contrast the 5′–5′ 1,4-GG IXL shows minimal distortion to the duplex, as expected.34,38 The overall results suggest that a 3′–3′ 1,4-GG IXL is likely to be a highly distorted adduct. Some evidence exists in the literature for conformational flexibility in closely related systems. Using an N4C–ethyl–N4C crosslink to mimic the IXL of alkylating agents, Noll et al. constructed plasmid DNAs where modified cytosines are incorporated into single CpG or GpC steps in small duplexes and then incorporated into plasmid DNA.39 Both sequences were substrates for DNA repair enzymes but there is a four-fold difference in NER efficiency between the two, the CpG step being more robust. NMR structures of oligonucleotide decamers containing the CpG (5′–5′) IXL showed a well-defined structure whereas disruptions caused by the CpG (3′–3′) produced considerable conformational flexibility which precluded structural analysis by NMR.
The 1,2-interstrand crosslink
In contrast to the difficulty in forming the 1,4-IXL the mol. wt. (ESIMS) of the BBR3464 adduct of 5′-d(ACGTATACGT)2 corresponded to two BBR3464 moieties per duplex. The sequence actually allows competition between a 1,2-IXL and a 1,6-IXL to be studied. NMR analysis suggested a structure composed of two simultaneous 1,2-interstrand crosslinks between the adjacent guanines, the first examples therefore of a structurally characterized 3′ → 3′ adduct.40 Relatively large chemical shifts of the H6 and H1′ protons of the corresponding C2 and C8 cytosines as well as the thymidine T4 and T10 bases indicated that guanine platination interrupted base pairing and stacking. The structure is quite distinct from the analogous cisplatin 1,2-interstrand (GC)2 adduct where the cytosines of the G–C base pairs are “flipped out” of the helix upon G-platination.41 It is also distinct from the 5′ → 5′ 1,2-IXL formed by [{trans-(PtCl(NH3)2}2μ-H2(CH2)4NH2]2+ (I, n = 4, Fig. 1).42 The conformational changes as elucidated by NMR studies on site-specific crosslinks are summarized in Table 2.
Table 2.
Summary of structural distortions of DNA interstrand cross-links of platinum complexes including nuceloside and sugar conformations
| Complexa | DNA | IXL |
syn orientation base |
N-type sugar |
G–C pair | Base de-stacking |
Weakened base stacking |
Reference |
|---|---|---|---|---|---|---|---|---|
| cis-DDP | CATAGCTATG | 5′ → 5′ 1,2- | G5 | G5 | No | G5CT | N/A | 41 |
| I, n = 4 | CATGCATG | 5′ → 5′ 1,2- | G4 | G4 | No | TG4C | C5A | 42 |
| I, n =6 | ATGTACAT | 5′ → 5′ 1,4- | A1,G3,A5,A7 | G3,A7 | Yes | TG3 | G3T | 37 |
| BBR3464 | ATGTACAT | 5′ → 5′ 1,4- | A1,G3,A7 | G3,A7 | Yes | ATG3 | G3T | 36 |
| BBR3464 | ACGTATACGTb | 3′ → 3′ 1,2- | G9 | G3,G9 | Yes, No | CG3 | A1C, CG9T | 40 |
See Fig. 1 for structures.
Simultaneous 1,2-IXL between adjacent CG base pairs.
Cooperative effects in long-range 1,4-crosslinks
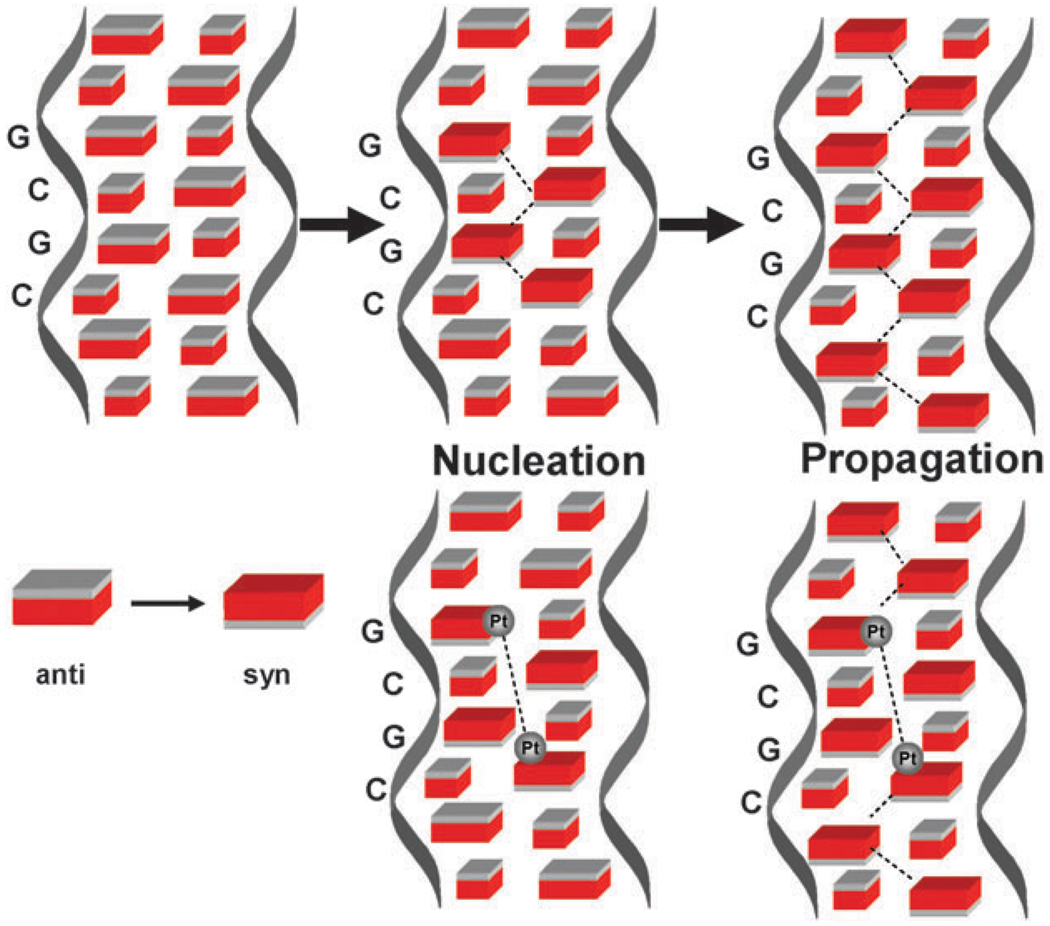
The combined effects of the molecular biology probes as well as the NMR structures indicate a delocalized, conformationally flexible lesion, with structural changes transmitted beyond the binding site. This is in contrast to the major 1,2-intrastrand adduct of cisplatin which, although bending the helix significantly, maintains the B-form in solution.43,44 So, how might these conformational changes be transmitted beyond the physically identifiable platination sites? The conformational changes suggest “Z-like” properties because of the appearance of the syn purine sugar conformation, an absolute requirement for the left-handed helix.45 The B → Z transition is a highly cooperative one and prominent amongst the explanations proposed for this transition is a nucleation step involving formation of a short Z-DNA sequence at a B → Z junction followed by propagation through flipping of the intervening base pairs from anti to syn.46,47 It is provocative therefore that the 4-base-pair sequence covered by a 1,4-IXL also displays all nucleoside conformations deviating from the anti conformation of B-DNA. An analogy may be made as shown in Fig. 6. Once the purine nucleoside conformations are altered it is possible to suggest, given the right sequence, that the 1,4-IXL represents a nucleation site primed to transmit further distortion along the helix eventually resulting in the observed delocalised lesions.
Fig. 6.
Schematic for propagation of conformational changes in DNA. B-DNA has anti conformation of nucleosides. Top: the ‘classical’ nucleation of a B → Z sequence followed by propagation. Bottom: how a 4-base pair sequence involving a {Pt,Pt} IXL can propagate similar conformational changes.
2.4 Biological consequences of long-range interstrand crosslinks: protein recognition
The DNA-binding event is essentially a signal for invocation of the cellular injury response. It is the “downstream” cellular processing of Pt–DNA distortions which is ultimately responsible for cytotoxicity and anti-tumor activity. It is axiomatic that inter-strand crosslinks, especially delocalized ones as depicted above, will present significant challenges to cellular repair processes involving nucleotide excision repair.48,49 This is especially true for PPCs where a family of crosslinks will form on DNA; the specifics of DNA repair may vary with the specific structure of the adduct.50 Therefore the consequences of the long-range IXLs have been studied in this context and their effects compared with those of cisplatin.
High mobility group proteins do not recognize the 1,4-interstrand crosslink
Gel retardation assays showed only very weak recognition of the 3′ → 3′ and 5′ → 5′ 1,4-IXLs by high mobility group HMG1 proteins, whose action is implicated in the cytotoxicity of cisplatin. Logically, this lack of recognition indicates that this event is not critical to the cytotoxicity of BBR3464. HMGB1a or HMGB1b binding to the cisplatin 1,2-GG intrastrand crosslink induces further bending of the DNA.51 Plausible explanations for the differential recognition may be that the prebending due to the 1,4-IXL is too small to be recognized by HMG-domain proteins or that the trinuclear complex restricts the additional DNA bending required for protein binding. Phenylalanine intercalation into the DNA minor groove is the recognition motif for HMG binding to cisplatin-adducted DNA.52 The presence of the central Pt(tetraam(m)ine) unit in the minor groove may represent a steric block to protein recognition and a biological consequence of the structure is observed.
Importance of p53 protein in activation by BBR3464
BBR3464 displays high activity in human tumor cell lines and in vivo murine xenografts characterized by both wild type and mutant p53 genes.53 The role of p53 as tumor suppressor protein is intimately related to its involvement in DNA repair and induction of apoptosis by DNA-damaging agents. Cellular recognition of DNA damage upregulates protein expression, whose action is then crucially related to its DNA binding activity.54,55 Modification of DNA fragments containing a consensus DNA response element (CDRE) of p53, and also smaller oligodeoxyribonucleotide duplexes (21bp), by BBR3464 and cisplatin in a cell free medium resulted in reduced binding affinity of both active and latent p53 protein, with BBR3464 being considerably more effective than cisplatin.56
Long-range interstrand crosslinks are poor substrates for nucleotide excision repair
Nucleotide excision repair (NER) is used by human cells for the removal of damaged nucleotides and drug-induced helix-distorting adducts from DNA.48,49 Excision repair assays where the site specific 3′ → 3′ or 5′ → 5′ 1,4-IXLs were incubated with repair-proficient human or rodent cell-free extracts (CFE) showed no excision products, implying that the IXLs are resistant to repair.22 These results validate nicely the finding that overexpression of the human NER complex (ERCC1) was not detrimental to the cellular sensitivity of two ovarian cancer cell lines to BBR3464.57 Cellular pharmacology studies in L1210 and osteosarcoma cells using alkaline elution show the persistence of inter-strand crosslinks with time, consistent with a slower rate of repair.58
In summary, the biological consequences of the 1,4-IXLs differ significantly from those produced by cisplatin and suggest that indeed a “molecular approach” to control downstream effects of protein recognition and apoptosis pathways may consist in design of structurally unique DNA adducts as cell signals.
3. The phosphate clamp: a new mode of DNA binding
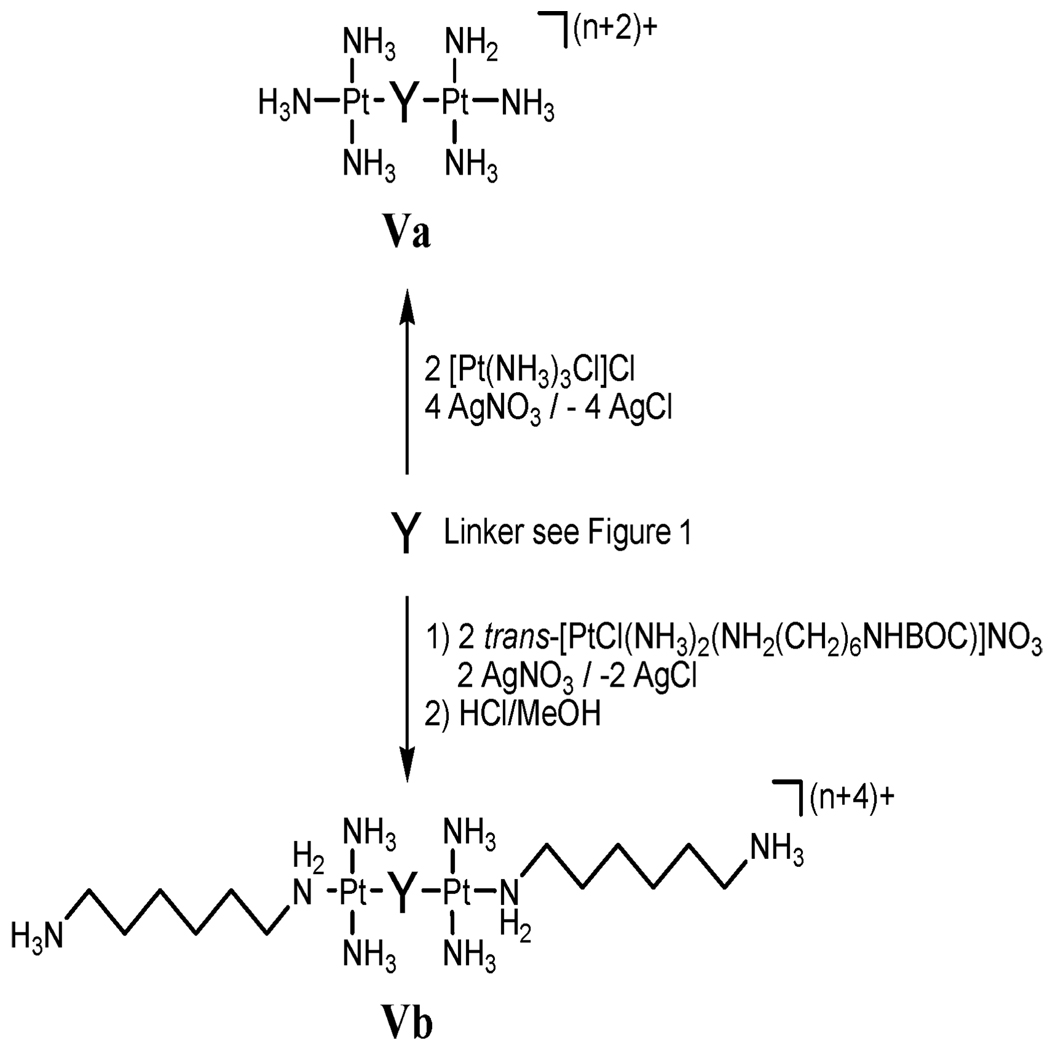
Synthesis and DNA binding of high-affinity non-covalent DNA binding agents: as stated, pre-association (or at least the presence of charge in the central linking moiety) may dictate many chemical and biological properties of PPCs. To examine pre-association only, a series of high-affinity “non-covalent” compounds were prepared by substitution of Cl− with either NH3 (Va) or a “dangling amine” (Vb) where one end of the diamine is linked to Pt with the other end free and protonated, Fig. 7. The intent was to allow examination of pre-association phenomena in the absence of Pt–DNA bond formation. Replacement of Cl− by inert ligands increases total charge to 6+ or 8+.
Fig. 7.
Synthesis of non-covalent DNA binding agents. For Y, see Fig. 1. Where Y is a PtN4 unit, Va and Vb are analogues of BBR346 where Vb is TriplatinNC.
3.1 Crystal and molecular structure of a duplex DNA with a non-covalent trinuclear platinum agent
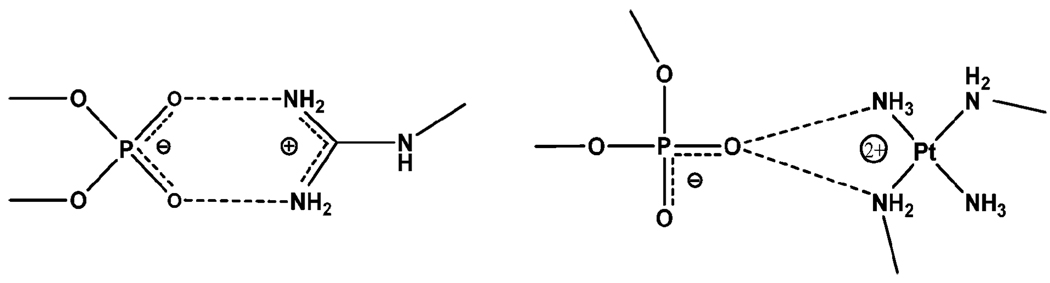
The crystal and molecular structure of a double-stranded B-DNA dodecamer 5′-d(CGCGAATTCGCG)2 (the DDD Dickerson Dodecamer), associated with complex Vb in Fig. 7 (abbreviated as TriplatinNC) has been solved at 1.2 Å resolution.59 Hydrogen bonding with phosphate oxygens results in either backbone tracking or groove spanning. The square-planar tetra-am(m)ine Pt(II) coordination units all form bidentate N–O–N complexes with OP atoms, phosphate clamps. The geometry is remarkably conserved and the interaction is selective for O2P atoms (the frequency of interaction is O2P > O1P, base and sugar oxygens > N) in all phosphate clamps throughout the structure. The TriplatinNC–DNA interactions are similar in some ways to those of the guanidino group of arginine which also shows an analogous, but attenuated, OP clamping ability in which two OP atoms form a clamp-like structure with a single guanidino group, see Scheme 1.60 A second structure using Va has also been resolved and shows similar features.61
Scheme 1.
Note that polyamines themselves (spermine, spermidine) would not be strong modifiers of phosphate interactions in the same manner due in part to the distances between amino groups and the lack of the rigidity provided by either the planar arginine or the square-planar Pt unit.
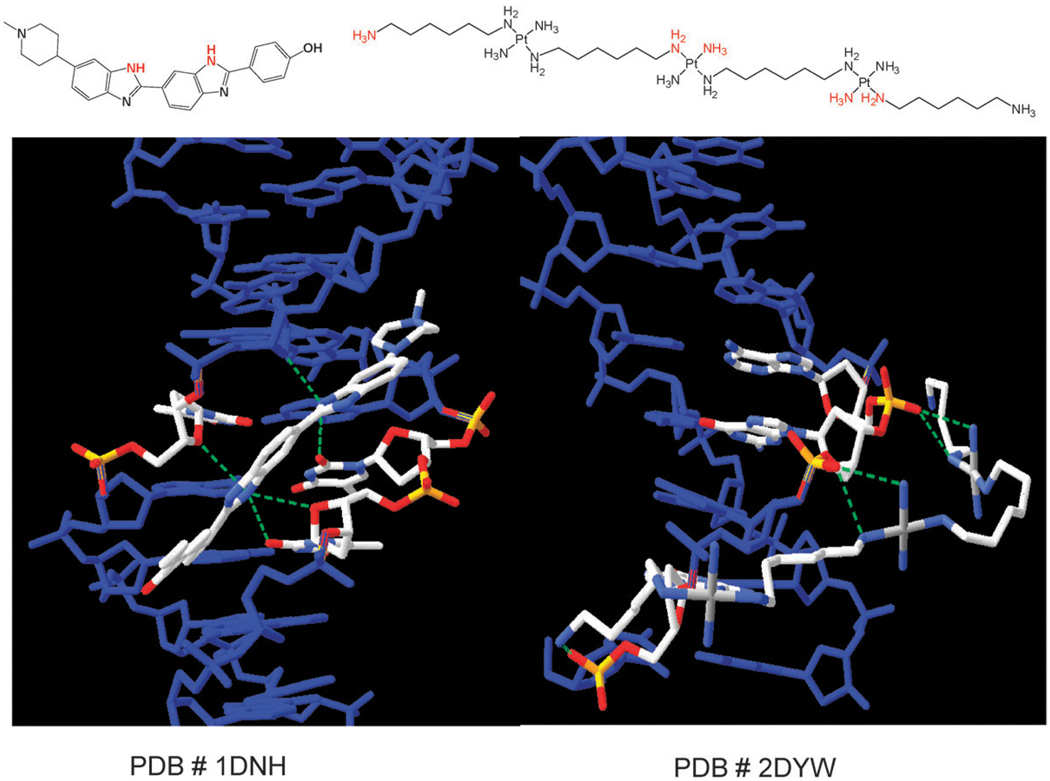
The DNA conformation in the DDD–TriplatinNC complex differs significantly from that of the native DDD structure (NDB entry bdl084). The axial bend is greater, helical parameters are perturbed and the minor groove width profile is modestly impacted.59 Electrostatic forces induce modest DNA bending into the major groove. The phosphate recognition is distinct from “classic” minor-groove recognition or intercalation and is a third, discrete mode of ligand binding to DNA. A comparison of the molecular recognition motifs between the platinum complex and a minor-groove binder Hoechst Dye (PDB#1NH62a) on the same DDD duplex is shown in Fig. 8. The complex does not bind by either of the conventional intercalation and/or groove binding modes. Whereas the “classic” minor groove binders like HD interact through H-bonds and exocyclic purine and pyrimidine atoms, especially thymine oxygens, the hydrogen-bonding of the phosphate clamp is a discrete new mode of ligand–DNA binding.
Fig. 8.
Comparison of molecular recognition motifs of a minor groove binder (PDB#1NH) showing H-bonds from heterocycle Ns to exocyclic O atoms on T and sugar backbone (left).62a In contrast TriplatinNC binding involves H-bonds to phosphate oxygens of the backbone.59,61 Figure adapted from PDB files.
The structure is the first described for a non-covalent platinum complex. The NDB database shows only a very limited number of crystal structures of duplex DNA bound with cisplatin (intrastrand GG and the cisplatin–DNA duplex associated with the HMG protein43,51; interstrand (GC)2 62b; oxaliplatin (intrastrand GG)63 and cis-[Pt(NH3)-(cyclohexylamine)Cl2] (intrastrand GG)64 and a Z-DNA-forming sequence d(CGTNH2ACG)2 stabilized by [Pt(NH3)3]+.65 With the exception of the latter, all of the DNA–platinum structures belong to the cis-[PtX2(amine)2] family.
Biological relevance
The “non-covalent” complexes may have important biological activity in their own right, as a new class of DNA-binding cytotoxic agents.1–3 Cellular accumulation of Triplatin NC is significantly enhanced even over the “parent” BBR3464. Although the formal charge is +8, TriplatinNC displays equivalent cytotoxicity to cisplatin against A2780 and the p53-null SKOV human ovarian cancer cell lines.66–68 Non-covalent compounds are also likely to engage in hydrogen-bonding and electrostatic interactions with proteins such as Human Serum Albumin but no decomposition of the di/trinuclear structure in the presence of sulfur nucleophiles is anticipated. It is noteworthy that compounds such as TriplatinNC induce apoptosis in tumor cells—suggesting that covalent Pt–DNA bond formation is not a prerequisite for antitumor activity—for Pt compounds with high DNA affinity.
4. Binding to single-stranded DNA
DNA as a template affects kinetics of substitution reactions occurring within its domain. We have seen that the nature of the DNA template affects inter-strand cross-linking efficiency and the relative ratios of directional isomers in 1,4-inter-strand crosslinks. The nature of template DNA (single-stranded vs. double-stranded) may also uniquely affect simple coordination chemistry processes such as aquation kinetics leading to substrate specificity. The third feature of unique DNA binding modes for PPCs is the kinetic preference for single-strand over double-stranded DNA. Mass spectrometric studies using 18-mers showed a kinetic preference for binding to (ss) DNA over (ds) DNA.69 For both dinuclear (I) and trinuclear (BBR3464, IV) the aquation rate constant is of the same order of magnitude to that of cisplatin, but the chloride anation rate constant is much higher so that the equilibrium favours the dichloro form.70,71 There is a 3-fold slowing of the aquation of I and IV in the presence of double-stranded (ds) DNA but not for single-stranded (ss) DNA.72 This is in contrast to cis-DDP.73,74 This feature may account for the kinetic binding preference and may also be relevant in stabilization of G-quartet quadruplex structures using I.75 The results emphasize how the alteration of chemical properties of small molecules in the presence of large host interactions is dependent on the conformation and nature of that host.
Non-covalent compounds also bind avidly to ssDNA. Indeed noncovalent interactions on DNA are important not only as a potential source of new drugs.1–3 Interactions of protonated amino acid residues with the negative charge of DNA phosphates represent an important motif for DNA–protein interactions. The protonated natural polyamines such as spermidine and spermine are present in cells and may interact with DNA. Further, cellular transport of genes or oligonucleotides may be facilitated by packaging in the presence of transfection agents containing polybasic compounds—the eventual success of gene and related therapies is critically dependent on successful delivery of the biological weapon. Many of these interactions are facilitated through arginine–phosphate recognition. Indeed the arginine moiety mediates many biological noncovalent events, including protein/metal, protein/protein, protein/peptide, and protein/oligonucleotide interactions.76–78 Phosphorylation and dephosphorylation of amino acid residues in serine, threonine and tyrosine govern numerous biochemical and physiological processes.79 The interaction of ssDNA and simple polybasic compounds has thus been studied from these aspects. In further pursuit of the arginine fork–phosphate clamp analogy we have also examined the interaction of the non-covalent compounds in Fig. 5 with ssDNA.
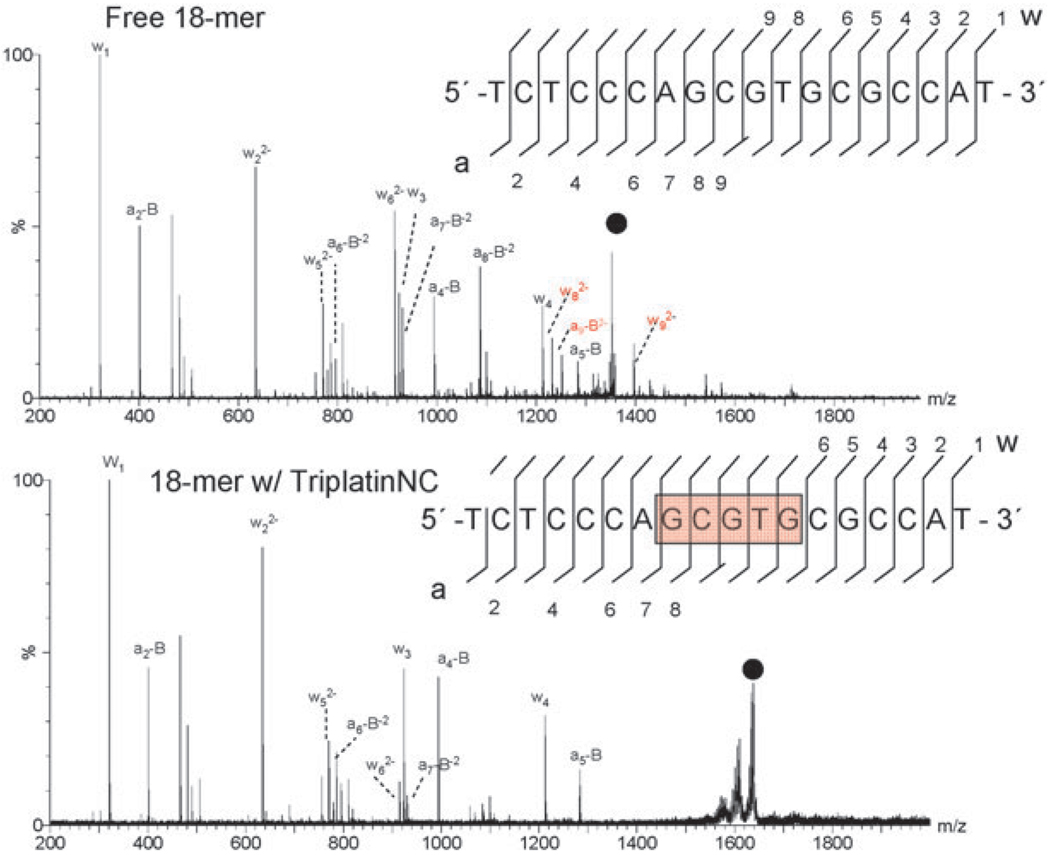
Mass spectrometry has proved to be especially useful in analysis of these interactions and tandem MS dissociation studies may even pinpoint the binding sites of small molecules, Fig. 9. The usefulness of tandem mass spectrometry (MS/MS) as a tool for the characterization of biopolymers such as DNA is now well established and derives from both the accuracy of measuring parent/product ions and the ability to obtain structural information through unimolecular dissociation chemistry.80,81 Tandem MS–MS of a single-stranded 18-mer oligonucleotide (5′-TCTCCCAGCGTGCGCCAT-3′) complexed with TriplatinNC (Vb) illustrates this principle. Given the high charge and size of the TriplatinNC (Vb), the most abundant stoichiometry observed with the oligonucleotide was 1 : 1. Fragment ions of free ss oligonucleotide, Fig. 9 (top), may be assigned using the standard McLuckey nomenclature.82,83 For the free oligonucleotide, complete sequence identification is achieved following cleavage of the phosphate backbone by collision induced dissociation. The addition of 1 equivalent of TriplatinNC (Vb) on the oligonucleotide results in the number and intensity of product ions observed to be noticeably less for the same applied translational energy, Fig. 9 (bottom). Examination of the sequence coverage in presence of the 8+ polynuclear platinum complex reveals clear areas of increased stabilization including an intact five-nucleobase stretch in the internal sequence of the base which is protected from cleavage. Positive ESI-MS/MS of the 18-mer oligonucleotide with TriplatinNC under the same conditions did not show evidence of any platinated oligonucleotides or free Pt agent.
Fig. 9.
ESI-MS/MS of free (top) and TriplatinNC adducted (bottom) 5′-d(TCTCCCAGCGTGCGCCAT) at 100 and 120 V of collisional energy, respectively. The region of enhanced stability is in red. Fragmentation of the glycosidic bonds is prevalent throughout the free oligonucleotide, while the associated fragment ions, (w82−, w92−, and a9-B shown in red) are absent in the adduct. Additionally, the intensity of a7-B and a8-B is dramatically reduced.
Fragmentation pathway for oligonucleotide-polynuclear platinum complexes
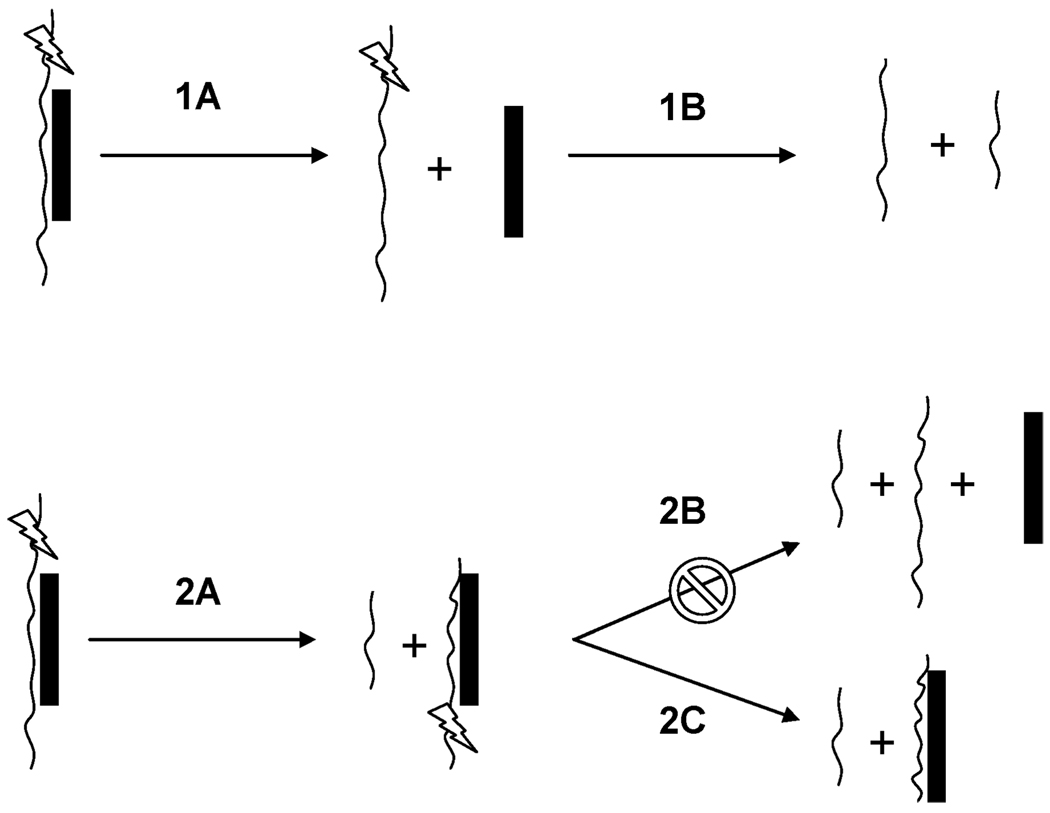
Analysis of cleavage patterns of polybasic peptides and polyamines complexed with ssDNA allowed Terrier et al. to suggest limiting fragmentation pathways based on the size and type of polybasic peptide.76,77 Based on the observed fragmentation patterns and binding site location of the non-covalent complexes of platinum, two possible fragmentation pathways may occur in the gas phase. Pathway 1A involves the noncovalent dissociation of the platinum moiety from the intact oligonucleotide which is then succeeded by pathway 1B where secondary fragmentations result in the production of the a-B and w type ions. This pathway is more likely for polybasic compounds that have the ability to carry a negative charge upon dissociation, as the increase in negative repulsion between the oligonucleotide and compound would make dissociation favorable (Fig. 10).76 Pathway 2A represents a more favorable dissociation scheme for the highly charged and stable platinum compounds—noncovalent dissociation is unfavored and cleavage of the phosphodiester backbone is the primary pathway. As seen in Fig. 9, small singly charged oligonucleotide a-B and w-type fragments dominate the spectra, indicative of cleavage occurring on either side of the stabilized region of platinum binding. The absence of observable free Pt compound suggests pathway 2C as the most likely mode of dissociation. The most plausible explanation is that the phosphate interaction is sufficiently strong that there is no discernible dissociation. Indeed there is literature precedent for strong arginine(guanidinium)–phosphate interactions in the gas phase.84 The results presented here complement the structural studies as it is reasonable to assume that similar interactions will be responsible for the arginine–phosphate interaction. Again, the modular nature of the interaction will increase the overall strength of the interaction. It is of interest that fragmentation pathway 2C for other cationic species is favored only under certain conditions. The results suggest that the noncovalent polynuclear platinum compounds may have advantages over polybasic peptides and polyamines in stabilization and packaging of ssDNA. Interestingly, Martin et al. showed that the covalent adduction of a platinum moiety helped stabilize the oligonucleotide resulting in a significant decrease in fragment ion abundance when studied by ESI-MS/MS.85,86 Additionally, cis-DDP alters the observed fragmentation pattern of oligonucleotides, compared to the free species.86
Fig. 10.
Schematic of possible dissociation pathways for the fragmentation of noncovalent platinum complexed oligonucleotides.
5. Conclusions and the caveat
Target binding is not the only determinant of useful anti-cancer activity. Ultimately, the therapeutic index is the final arbiter of any chemical’s suitability as a medicine. In the case of platinum complexes cellular accumulation and the extent of metabolizing interactions, especially with sulfur nucleophiles, determine the overall biological profile. In some respects, the study of target interactions tells us how a drug works but it is the combination of pharmacokinetics and pharmacodynamics that may tell us what makes a particular molecular entity a useful drug. This is a distinction which is very useful to remember for those involved in “rational” drug development. I benefitted early on (although forgetting to take full advantage at regular intervals) by being told by Jim Williamson of MRC, Mill Hill, to read Adrian Albert’s classic “Selective Toxicity” if I were truly interested in drug development.87 Nevertheless, space dictates that this article is restricted to DNA structure studies.
The exceptional DNA binding properties of BBR3464 allied to its high potency made it an attractive drug for clinical development. The overall profile certainly allows us to state that we have achieved the goal of modifying biological activity through design of new DNA binding modes—and from a “molecular” basis. Drug development is fraught with difficulties which we have described as the 4Ps’—politics, personalities, patents and pharmacokinetics.88 BBR3464 has not advanced as of yet to full human use—the pharmacokinetics and plasma decomposition were deemed non-optimal despite the drug showing some promise in Phase I and II clinical trials. Newer analogs,89,90 including the unique Vb, may resolve these issues. Nevertheless, the journey through DNA binding has provided wonderful scenery, significantly expanding the description of platinum drug–polynucleotide interactions and showing the marvellous diversity of interactions of the molecule of life.
Acknowledgements
NF sincerely acknowledges the long-term contributions of Dr Yun Qu to the development of this project. The work presented here would not be possible without the contributions of excellent and enthusiastic collaborators Sue Berners-Price, Viktor Brabec and Loren Williams. It is a pleasure to acknowledge graduate students and postdoctoral fellows who have contributed, as well as past collaborators (in alphabetical order) Miles Hacker, John Roberts, Kirsten Skov and Ben VanHouten. This work has been supported by The American Cancer Society, National Science Foundation and The National Institutes of Health (RO1CA78754).
Biography

John B. Mangrum received his BS in Chemistry and Biology from Longwood University. He obtained his PhD from Virginia Commonwealth University in 2010 working under the auspices of Professor Nicholas P. Farrell. His research utilized ESI-MS to characterize the gas phase interactions of polynuclear platinum complexes with biological targets. While working on his PhD, he also served as interim director for the Mass Spectrometry Resource Center.
Professor Farrell received his BSc from University College Dublin and PhD in Chemistry from Sussex University. He has written and/or co-edited three books in the area of medicinal inorganic chemistry. His work on polynuclear platinum agents led to human clinical trials of a new class of anticancer agents. He was Distinguished Research Scholar of Virginia Commonwealth University in 2003–2004.
Notes and references
- 1.Hurley LH. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 2.Boer DR, Canals A, Coll M. J. Chem. Soc., Dalton Trans. 2009:399–414. doi: 10.1039/b809873p. [DOI] [PubMed] [Google Scholar]
- 3.Palchaudhuri R, Hergenrother PJ. Curr. Opin. Biotechnol. 2007;18:497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Kelland L. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 5.Kelland LR, Farrell N. In: Platinum-Based Drugs in Cancer Therapy, in Cancer Drug Discovery and Development. Teicher BA, editor. Humana Press; 2000. ISBN 0896035999. [Google Scholar]
- 6.Wang D, Lippard SJ. Nat. Rev. Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A, Reardon JT. Adv. Protein Chem. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- 8.Reed E. Clin. Cancer Res. 2005;11:6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 9.Stewart DJ. Crit. Rev. Oncol. Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Farrell N, Qu Y, Hacker MP. J. Med. Chem. 1990;33:2179. doi: 10.1021/jm00170a021. [DOI] [PubMed] [Google Scholar]
- 11.Farrell NP, de Almeida SG, Skov KA. Bisplatinum Complexes Containing Two cis-Pt(amine)2 Units. Synthesis and Initial DNA-Binding Studies. J. Am. Chem. Soc. 1988;110:5018. [Google Scholar]
- 12.Kraker AJ, Hoeschele JD, Elliott WL, Showalter HDH, Sercel AD, Farrell N. J. Med. Chem. 1992;35:4526–4532. doi: 10.1021/jm00102a003. [DOI] [PubMed] [Google Scholar]
- 13.Roberts JD, van Houten B, Qu Y, Farrell NP. Nucleic Acids Res. 1989;17:9719. doi: 10.1093/nar/17.23.9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeschele JD, Kraker AJ, Qu Y, Van Houten B, Farrell N. Bis(platinum) Complexes: Chemistry, Antitumor Activity and DNA-Binding. In: Pullman B, Jortner J, editors. Molecular Basis of Specificity in Nucleic Acid–Drug Interactions. Dordrecht: Kluwer Academic Publishers; 1990. pp. 301–321. [Google Scholar]
- 15.Farrell N, Appleton TG, Qu Y, Roberts JD, Soares Fontes AP, Skov KA, Wu P, Zou Y. Biochemistry. 1995;34:15480. doi: 10.1021/bi00047a013. [DOI] [PubMed] [Google Scholar]
- 16.Farrell N. Met. Ions Biol. Syst. 2004;41:252–296. [PubMed] [Google Scholar]
- 17.McGregor TD, Kasparkova J, Neplechova K, Novakova O, Penazova H, Vrana O, Brabec V, Farrell N. J. Biol. Inorg. Chem. 2002;7:397–404. doi: 10.1007/s00775-001-0312-4. [DOI] [PubMed] [Google Scholar]
- 18.Montero EI, Benedetti BT, Mangrum JB, Oehlsen MJ, Qu Y, Farrell NP. J. Chem. Soc., Dalton Trans. 2007:4938–4942. doi: 10.1039/b708433c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts JD, Peroutka J, Farrell N. J. Inorg. Biochem. 1999;77:51–57. doi: 10.1016/s0162-0134(99)00147-6. [DOI] [PubMed] [Google Scholar]
- 20.Noll DM, Noronha AM, Wilds CJ, Miller PS. Front. Biosci. 2004;9:421–437. doi: 10.2741/1246. [DOI] [PubMed] [Google Scholar]
- 21.Brabec V. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- 22.Kasparkova J, Zehnulova J, Farrell N, Brabec V. J. Biol. Chem. 2002;277:48076–48086. doi: 10.1074/jbc.M208016200. [DOI] [PubMed] [Google Scholar]
- 23.Kasparkova J, Farrell N, Brabec V. J. Biol. Chem. 2000;275:15789–15798. doi: 10.1074/jbc.M000777200. [DOI] [PubMed] [Google Scholar]
- 24.Ruhayel RA, Moniodis JJ, Yang X, Kasparkova J, Brabec V, Berners-Price SJ, Farrell NP. Chem.-Eur. J. 2009;15:9365–9374. doi: 10.1002/chem.200900958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajski SR, Williams RM. Chem. Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 26.Noll DM, Mason TM, Miller PS. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailly C, Gentle D, Hamy F, Purcell M, Waring MJ. Biochem. J. 1994;300(Pt 1):165–173. doi: 10.1042/bj3000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailly C, Waring MJ. Diethylpyrocarbonate and Osmium Tetroxide as Probes for Drug-Induced Changes in DNA Conformation In Vitro. Totowa, NJ: Humana Press Inc.; 1997. [DOI] [PubMed] [Google Scholar]
- 29.Ross SA, Burrows CJ. Nucleic Acids Res. 1996;24:5062–5063. doi: 10.1093/nar/24.24.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox KR, Waring MJ. Methods Enzymol. 2001;340:412–430. doi: 10.1016/s0076-6879(01)40434-4. [DOI] [PubMed] [Google Scholar]
- 31.Lahm A, Weston SA, Suck D. Nucleic Acids Mol. Biol. 1991;5:171–186. [Google Scholar]
- 32.Suck D, Oefner C. Nature. 1986;321:620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 33.Chvalova K, Kasparkova J, Farrell N, Brabec V. FEBS J. 2006;273:3467–3478. doi: 10.1111/j.1742-4658.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- 34.Hegmans A, Berners-Price SJ, Davies MS, Thomas D, Humphreys A, Farrell NP. J. Am. Chem. Soc. 2004;126:2166–2180. doi: 10.1021/ja036105u. [DOI] [PubMed] [Google Scholar]
- 35.Berners-Price SJ, Ronconi L, Sadler PJ. Prog. Nucl. Magn. Reson. Spectrosc. 2006;49:65–98. [Google Scholar]
- 36.Qu Y, Scarsdale NJ, Tran M-C, Farrell N. J. Biol. Inorg. Chem. 2003;8:19–28. doi: 10.1007/s00775-002-0383-x. [DOI] [PubMed] [Google Scholar]
- 37.Qu Y, Scarsdale NJ, Tran M-C, Farrell N. J. Inorg. Biochem. 2004;98:1585–1590. doi: 10.1016/j.jinorgbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Cox JW, Berners-Price SJ, Davies MS, Qu Y, Farrell N. J. Am. Chem. Soc. 2001;123:1316–1326. doi: 10.1021/ja0012772. [DOI] [PubMed] [Google Scholar]
- 39.Noll DM, Webba daSilva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Biochemistry. 2005;44:6764–6775. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- 40.Qu Y, Tran M-C, Farrell NP. J. Biol. Inorg. Chem. 2009;14:969–977. doi: 10.1007/s00775-009-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Science. 1995;270:1842–1845. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- 42.Yang D, van Boom S, Reedijk J, van Boom J, Farrell N, Wang AH-J. Nat. Struct. Biol. 1995;2:577. doi: 10.1038/nsb0795-577. [DOI] [PubMed] [Google Scholar]
- 43.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 44.Gelasco A, Lippard SJ. Biochemistry. 1998;37:9230–9239. doi: 10.1021/bi973176v. [DOI] [PubMed] [Google Scholar]
- 45.Rich A. Nat. Struct. Biol. 2003;10:247. doi: 10.1038/nsb0403-247. [DOI] [PubMed] [Google Scholar]
- 46.Ha SC, Lowenhaupt K, Rich A, Kim Y-G, Kim KK. Nature. 2005;437:1183. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 47.Ho PS. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9549. doi: 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reardon JT, Sancar A. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 49.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Chem. Rev. 2006;106:233. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 50.Brabec V, Kašpárková J, Vrána O, Nováková O, Cox J, Qu Y, Farrell N. Biochemistry. 1999;38:6781–6790. doi: 10.1021/bi990124s. [DOI] [PubMed] [Google Scholar]
- 51.Ohndorf U-M, Rould MA, He Q, Pabo CO, Lippard SJ. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 52.Ohndorf UM, Whitehead JP, Raju NL, Lippard SJ. Biochemistry. 1997;36:14807–14815. doi: 10.1021/bi9717643. [DOI] [PubMed] [Google Scholar]
- 53.Pratesi G, Perego P, Polizzi D, Righetti SC, Supino R, Caserini C, Manzotti C, Giuliani FC, Pezzoni G, Tognella S, Spinelli S, Farrell N, Zunino F. Br. J. Cancer. 1999;80:1912–1919. doi: 10.1038/sj.bjc.6690620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 55.Ahn J, Prives C. Nat. Struct. Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- 56.Kasparkova J, Fojta M, Farrell N, Brabec V. Nucleic Acids Res. 2004;32:5546–5552. doi: 10.1093/nar/gkh896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Servidei T, Ferlini C, Riccardi A, Meco D, Scambia G, Segni G, Manzotti C, Riccardi R. Eur. J. Cancer. 2001;37:930–938. doi: 10.1016/s0959-8049(01)00061-2. [DOI] [PubMed] [Google Scholar]
- 58.Roberts JD, Peroutka J, Beggiolin G, Manzotti C, Piazzoni L, Farrell N. J. Inorg. Biochem. 1999;77:47–50. doi: 10.1016/s0162-0134(99)00137-3. [DOI] [PubMed] [Google Scholar]
- 59.Komeda S, Moulaei T, Woods KK, Chikuma M, Farrell NP, Williams LD. J. Am. Chem. Soc. 2006;128:16092–16103. doi: 10.1021/ja062851y. [DOI] [PubMed] [Google Scholar]
- 60.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 61.Komeda SJ, Moulaei T, Chikuma M, Odani A, Kipping R, Farrell NP, Williams LD. Nucleic Acids Res. doi: 10.1093/nar/gkq723. DOI: 10.1093/nar/gkq723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Wood AA, Nunn CM, Czarny A, Boykin DW, Neidle SJ. Nucleic Acids Res. 1995;23:3678–3684. doi: 10.1093/nar/23.18.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spingler B, Whittington DA, Lippard SJ. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 64.Silverman AP, Bu W, Cohen SM, Lippard SJ. J. Biol. Chem. 2002;277:49743–49749. doi: 10.1074/jbc.M206979200. [DOI] [PubMed] [Google Scholar]
- 65.Parkinson GN, Arvanitis GM, Lessinger L, Ginell SL, Jones R, Gaffney B, Berman HM. Biochemistry. 1995;34:15487–15495. doi: 10.1021/bi00047a014. [DOI] [PubMed] [Google Scholar]
- 66.Harris AL, Yang X, Hegmans A, Povirk L, Ryan JJ, Kelland L, Farrell NP. Inorg. Chem. 2005;44:9598–9600. doi: 10.1021/ic051390z. [DOI] [PubMed] [Google Scholar]
- 67.Harris AL, Qu Y, Farrell NP. Inorg. Chem. 2005;44:1196–1198. doi: 10.1021/ic048356p. [DOI] [PubMed] [Google Scholar]
- 68.Harris AL, Ryan JJ, Farrell NP. Mol. Pharmacol. 2006;69:666–672. doi: 10.1124/mol.105.018762. [DOI] [PubMed] [Google Scholar]
- 69.Kloster MBG, Hannis JC, Muddiman DC, Farrell N. Biochemistry. 1999;38:14731–14737. doi: 10.1021/bi991202e. [DOI] [PubMed] [Google Scholar]
- 70.Davies MS, Cox JW, Berners-Price SJ, Barklage W, Qu Y, Farrell N. Inorg. Chem. 2000;39:1710–1715. doi: 10.1021/ic991104h. [DOI] [PubMed] [Google Scholar]
- 71.Davies MS, Thomas DS, Hegmans A, Berners-Price SJ, Farrell N. Inorg. Chem. 2002;41:1101–1109. doi: 10.1021/ic010851n. [DOI] [PubMed] [Google Scholar]
- 72.Davies MS, Berners-Price SJ, Cox JW, Farrell N. Chem. Commun. 2003:122–123. doi: 10.1039/b209661g. [DOI] [PubMed] [Google Scholar]
- 73.Sadler PJ, Barnham KJ, Berners-Price SJ, Frey U. Chem.-Eur. J. 1996;2:1283. [Google Scholar]
- 74.Reeder F, Guo Z, Murdoch PdS, Corazza A, Hambley TW, Berners-Price SJ, Chottard JC, Sadler PJ. Eur. J. Biochem. 1997;249:370. doi: 10.1111/j.1432-1033.1997.00370.x. [DOI] [PubMed] [Google Scholar]
- 75.Ourliac-Garnier I, Elizondo-Riojas M, Redon S, Farrell NP, Bombard S. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]
- 76.Terrier P, Tortajada J, Buchmann W. J. Am. Soc. Mass Spectrom. 2007;18:346–358. doi: 10.1016/j.jasms.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 77.Terrier P, Tortajada J, Zin G, Buchmann W. J. Am. Soc. Mass Spectrom. 2007;18:1977–1989. doi: 10.1016/j.jasms.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 78.Ohara K, Smietana M, Vasseur J-J. Characterization of Specific Noncovalent Complexes between Guanidinium Derivatives and Single-Stranded DNA by MALDI. J. Am. Soc. Mass Spectrom. 2006;17(3):283–291. doi: 10.1016/j.jasms.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 79.Woods AS, Ferre S. J. Proteome Res. 2005;4:1397–1402. doi: 10.1021/pr050077s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hofstadler SA, Griffey RH. Chem. Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 81.Loo JA. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 82.McLuckey SA, Van Berker GJ, Glish GL. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 83.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 84.Jackson SN, Wang H-YJ, Woods AS. J. Proteome Res. 2005;4:2360–2363. doi: 10.1021/pr050261d. [DOI] [PubMed] [Google Scholar]
- 85.Martin LB, Schreiner AF, vanBreemen RB. Anal. Biochem. 1991;193:6–15. doi: 10.1016/0003-2697(91)90035-r. [DOI] [PubMed] [Google Scholar]
- 86.Nyakas A, Eymann M, Schurch S. J. Am. Soc. Mass Spectrom. 2009;20:792–804. doi: 10.1016/j.jasms.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 87.See http://www.asap.unimelb.edu.au/bsparcs/aasmemoirs/albert.htm.
- 88.Farrell N. ACS Symp. Ser. 2005;vol. 903:62–79. [Google Scholar]
- 89.Zerzankova L, Suchankova T, Vrana O, Farrell NP, Brabec V, Kasparkova J. Biochem. Pharmacol. 2010;79:112–121. doi: 10.1016/j.bcp.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Billecke C, Finniss S, Tahash L, Miller C, Mikkelsen T, Farrell NP, Bögler O. Neuro-Oncology. 2006;8:215–226. doi: 10.1215/15228517-2006-004. [DOI] [PMC free article] [PubMed] [Google Scholar]