Abstract
Summary: Characterizing genetic diversity through genotyping short amplicons is central to evolutionary biology. Next-generation sequencing (NGS) technologies changed the scale at which these type of data are acquired. SESAME is a web application package that assists genotyping of multiplexed individuals for several markers based on NGS amplicon sequencing. It automatically assigns reads to loci and individuals, corrects reads if standard samples are available and provides an intuitive graphical user interface (GUI) for allele validation based on the sequences and associated decision-making tools. The aim of SESAME is to help allele identification among a large number of sequences.
Availability: SESAME and its documentation are freely available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported Licence for Windows and Linux from http://www1.montpellier.inra.fr/CBGP/NGS/ or http://tinyurl.com/ngs-sesame.
Contact: emese.meglecz@univ-provence.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Since the breakthrough of PCR in the 1980s, researchers have been using various DNA markers and approaches to characterize the genetic diversity such as microsatellites, single strand conformation polymorphism (SSCP) and amplified fragment length polymorphism (AFLP). However, direct sequencing of DNA loci from a large number of individuals was costly and labor intensive. This was even further an issue for nuclear markers where cloning must precede Sanger sequencing to disentangle alleles. Technical advances involved in next-generation sequencing (NGS) technologies, including clonal amplification (454®, Roche) or single molecule sequencing (SMRT®, Pacific Bioscience), open new horizons where gigabases of DNA sequences can be obtained easily. One can take advantage from these innovations to directly and massively sequence amplicons. However, this approach necessitates tagging individual amplicons and sorting out real alleles from reads with sequencing errors.
We have created SESAME as a user-friendly web application package for analyzing amplicon sequences obtained through NGS, where the amplicon is completely covered by a read. It includes automatic sequence assignation to multiple markers and individuals via oligonucleotide tags. It provides an intuitive point-and-click interface to validate sequences as allele from individual-based variant alignments. The results are exported as genotypes or sequences of alleles. To our knowledge, no similar software has been published that accomplishes all these tasks.
2 IMPLEMENTATION
SESAME is a database oriented web application with an easy to use graphical user interface (GUI). The database can be accessed simultaneously by several users sharing their data within a project. An assistant guides the user for input data upload and through the sequence analysis steps (Supplementary Materials). All data are stored in a relational database. The user interacts with the database through an intuitive interface during the allele validation procedure.
2.1 Input data and automatic sequence analysis
Amplicons are individually identified by oligonucleotide tags linked as described in Galan et al. (2010). Although only one tag is strictly needed for assignation, using both forward and reverse tags increases the number of possible combinations. Furthermore, SESAME allows simultaneous analysis of several loci multiplexed in a single run. Sample (individual-locus combination) information is read from a comma-separated values (CSVs) file providing sample name, marker name, expected ploidy level, primer and tag sequences, status (sample or standard), population name and species (step 1). DNA sequences are read from one single FASTA file (step 2). Furthermore, users are asked to provide the list of DNA markers with corresponding reference sequences that will be used for assignation of the reads to loci and for trimming them from primers and tags. More precisely, DNA sequences are BLASTed (with user-defined E-value) against the reference sequences to assign each read to a locus and determine read orientation (step 3) then BLASTed against the list of tag sequences concatenated with primer sequences for sample assignation (step 4). Only reads with perfect match of both tags are assigned to sample, and thus all assigned reads cover the whole amplicon. All of the assigned reads are kept for further analyses. For each sample, all reads are aligned by MUSCLE (Edgar, 2004) or MAFFT (Katoh et al., 2005) according to the administrator's choice. Tags and primers are trimmed off (step 5). An optional position-specific correction of reads (step 6) can be performed if standard samples are provided by the user (typically a single cloned allele amplified and sequenced along with the samples; for details see Supplementary Materials).
2.2 Assisted allele validation
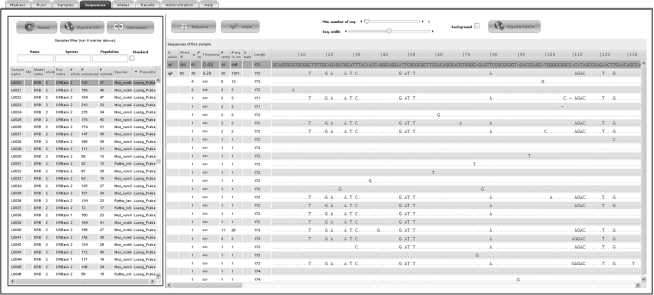
Users visualize the alignments of reads for each individual amplicon with an intuitive GUI. For a selected locus, all samples appear together with descriptive statistics, such as the number of sequences and variants assigned to the sample or the number of user-validated alleles. The number of sequences is displayed in red if too low to insure high-quality genotyping given the expected ploidy level (see User guide for details). Identical reads of a sample are pooled into variants and their numbers and frequencies are provided. Selecting a sample displays the alignment of all corresponding variants. Each variant is characterized by its length, its total occurrence in the whole run and the number of samples in which it is found (Fig. 1). Filters can be applied to help genotyping (e.g. the minimum number of reads for a variant; see User guide for details). Users can validate a variant as an allele by a simple click. A unique allele identifier is attributed allowing comparisons of genetic diversity among multiple runs within a project. After allele validation, the genotype table or the FASTA file of alleles can be exported. Filtering options are available to display or export only selected subsets (e.g. run, populations, species).
Fig. 1.
Screenshot of the allele validation step. For illustration, the selected sample L0020 (left frame) is a diploid species Mus cookii from the population Luang Prabang and has 130 sequences representing 27 variants of DRB gene with two validated alleles. These two validated alleles are in high frequency in the sample (right frame), respectively, 0.48 (63 sequences) and 0.29 (38 sequences), compared with other variants (frequency < 0.03). Analysis of the sequence alignment shows that these variants in low frequency display errors (indels, substitutions) or are chimeric recombination of the two validated alleles.
Information on a test dataset is available in the Supplementary Materials.
3 DISCUSSION
Obtaining high number of sequences for amplicons from nuclear markers brings new challenges with regard to deciphering alleles from each other and distinguishing weakly amplified alleles from sequencing errors. With this respect, bioinformatics tools did not cope yet with the analysis of amplicons sequenced in this high-throughput environment. Moreover, user-friendly tools are needed for sorting and analyzing this type of data, as end users are interested in the biological information and do not necessarily have programming or sharp bioinformatics skills. SESAME fills this gap and makes routine sequencing as DNA barcoding of a large number of amplicons at hand. SESAME is resolutely oriented toward biologists with regard to user-friendly interfacing and easy installation. Furthermore, most functions to support analyses are unique and provide a strong and productive framework for high-throughput amplicon sequencing.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan M, et al. A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics. 2010;11:269. doi: 10.1186/1471-2164-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, et al. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.