Abstract
Motivation: Panels of cell lines such as the NCI-60 have long been used to test drug candidates for their ability to inhibit proliferation. Predictive models of in vitro drug sensitivity have previously been constructed using gene expression signatures generated from gene expression microarrays. These statistical models allow the prediction of drug response for cell lines not in the original NCI-60. We improve on existing techniques by developing a novel multistep algorithm that builds regression models of drug response using Random Forest, an ensemble approach based on classification and regression trees (CART).
Results: This method proved successful in predicting drug response for both a panel of 19 Breast Cancer and 7 Glioma cell lines, outperformed other methods based on differential gene expression, and has general utility for any application that seeks to relate gene expression data to a continuous output variable.
Implementation: Software was written in the R language and will be available together with associated gene expression and drug response data as the package ivDrug at http://r-forge.r-project.org.
Contact: riddickgp@mail.nih.gov
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Advances in high-throughput drug screening technologies in the past 20 years have enabled the testing of hundreds of thousands of drug candidates on panels of cancer cell lines. An important goal is applying this information to predict drug response in different cell lines specific to certain cancer types and ultimately clinical tumor samples. The enormous diversity of tumor biology, even within cancers from the same tissue, makes this aim especially challenging. Several attempts to create predictive models of drug response have made use of the NCI-60, a panel of human cell lines originally derived from human cancers spanning nine different tissues of origin (Covell et al., 2007; Shoemaker, 2006). The Developmental Therapeutics Program (DTP) at the National Cancer Institute has screened nearly 400 000 compounds against these cell lines since the origin of the NCI-60 program in the late 1980s. Cell lines from a particular tissue of origin were eventually shown not to be highly predictive of human tumors of the same tissue type. Nevertheless, the overall pattern of inhibitory responses from a particular drug across all 60 cell lines proved highly useful as a drug response ‘fingerprint’ that could help pinpoint drug mechanism and efficacy (Paull et al., 1989; Weinstein, 2006).
The NCI-60 has been extensively molecularly characterized using a variety of high-throughput assays, including five independent gene expression microarray experiments utilizing a number of different platforms. Staunton et al. (2001) first applied NCI-60 basal (resting) gene expression data, produced without application of a drug, to create predictive models of drug activity (Staunton et al., 2001). Gene signatures of 232 drugs from 6817 genes were created using a weighted voting algorithm, which predicted a binary response (sensitive or resistant) based on an association between gene expression and drug response. The contribution of each gene expression level in the training set to the model was weighted by the statistical correlation between the expression level for that gene across all cell lines and the experimentally determined drug sensitivity across all cell lines. Using this approach, one-third of the original 232 drugs could predict drug sensitivity at significant levels (P < 0.05) when tested on cell lines from the NCI-60 that had been held out from the original training set.
Lee et al. (2007) used differential gene expression between sensitive and resistant cell lines to define a drug signature (Lee et al., 2007). They showed that signatures produced from the top 12 and bottom 12 responding cell lines for each drug could be used to create a categorical model for cisplatin and paclitaxel in a panel of 40 bladder cancer cell lines. Then by screening 45 545 compounds in the DTP database against the same cell line panel, the authors identified 139 compounds predicted to be effective at inhibiting growth in >35% of cell lines. The top compound hit proved to be a potent inhibitor of growth in bladder cancer cell lines, although the rest of the predictions were not systematically tested.
Recently, Mori et al. (2009) developed a phenotype-based screen based on the NCI-60 (Mori et al., 2009). Signatures developed from Ras activation, PI3K activation, as well as tumor samples from basal-defined breast cancer were first compared with expression states in cell lines from the NCI-60. Cell lines were ranked by statistically defined similarity to signatures. The Pearson's correlation between signature similarity and drug response in the database was then used to identify drugs with similar response patterns. Although the results were not systematically experimentally tested, one drug (simvastatin) showed the ability to inhibit tumor growth of basal-type breast tumors in a mouse model.
In an attempt to build on these promising approaches, we asked whether the existing methods could be improved in three specific ways: (i) creation of drug gene expression signatures based on a multivariate model rather than a univariate test of differential gene expression. (ii) An automated means to remove outlying cell lines from the statistical model rather than manual curation. (iii) A multivariate regression model for predicting continuous drug response. We implemented this approach using Random Forest, an ensemble machine-learning approach that has been successfully applied to many different problems in computational biology (Breiman, 2001; Liaw and Wiener, 2002).
2 METHODS
2.1 Overview
Predictive models for each drug are individually created by combining two data sources: (i) drug sensitivity (IC50) for that drug across all cell lines in the NCI-60 and (ii) basal gene expression of each cell line in the NCI-60, which represents the ‘resting’ physiological state of the cell, before application of any drug.
Creating a model of drug responses using Random Forest consists of three steps (Fig. 1). First, IC50 response data for a particular drug are normalized to a [0, 1] interval. A Random Forest model is then trained on the basal gene expression data for the NCI-60 (16 644 probesets) with IC50 as the response variable (Fig. 1a). Variable importance generated by the model is then used to select a smaller subset of probesets that are highly predictive of drug response (typically 100–500 probesets). In the second step, another model is then fitted between the gene expression signature and the IC50 response. From this model, the case proximity matrix is then used to identify core cell lines associated with the drug (Fig. 1b, Equation 1). Once the IC50 values for outlying cell lines have been removed, a third model is fitted to the gene expression signature (Fig. 1c).
Fig. 1.
Overview of the model building algorithm. (A) A RANDOM FOREST model is fit between all the probesets in the training set (16 644) and the IC50 values for each drug. (B) PROBESETS that have a variable importance OF 2 SDs > mean of variable importance for all probesets are kept as a gene expression signature; a second Random Forest model is fit between this gene expression signature and the IC50 values for each drug. (C) CASE proximity values for each drug are generated from the second model using Equation (1), outlying cell lines are removed, and a third Random Forest model is fit with the remaining cell lines and the gene expression signature.
2.2 Datasets
2.2.1 Gene expression microarray
Gene expression microarray data for the NCI-60 cell lines were downloaded from the NCI DTP site (http://dtp.nci.nih.gov). Both Genelogic and Chiron NCI-60 datasets were originally generated using Affymetrix u133A/B microarrays and processed using MAS5. Probesets with a row-wise coefficient of variation (SD/mean) >0.06 were kept for further analysis. Probesets were further retained if they showed >0.2 Pearson's correlation coefficient across the 58 cell lines shared between Genelogic/Chiron datasets. Data for one glioma cell line (U251) that appeared both in the NCI-60 and 7 Glioma cell lines was dropped from the NCI-60 dataset before the generation of statistical models. Lastly, the arithmetic mean of matching probesets from Genelogic/Chiron datasets was taken, and these composite probesets were then z-normalized in a column-wise fashion for each cell line.
Affymetrix u133 2.0+ Gene expression microarray data for 19 breast cancer cell lines (GSE3156) was downloaded from the NCBI Geo Gene-Expression Database (http://www.ncbi.nlm.nih.gov/geo/). Probeset values were column-wise z normalized. Six cell lines from this dataset that also appeared in the NCI-60 were excluded from further analysis.
Gene expression microarray data for seven Glioma cell lines (A172, LN229, T98G, U87, U118, U251, U373) were measured on the Affymetrix u133 2.0+ platform. RNA extraction and analysis was performed as described previously (Li et al., 2008). Probeset values were processed using MAS5 and then z normalized in a column-wise fashion.
2.2.2 Drug sensitivity data
IC50 is defined as the concentration of a compound required to produce 50% growth inhibition after 48 h in a cell line relative to the control. NCI-60 IC50 data for a list of 40 federal drug administration (FDA)-approved oncology drugs were downloaded from the DTP web site and used as a training set. Values (previously −log 10 transformed) were normalized over the [0, 1] interval. If more than one experiment existed for each drug, the entry with the largest number of replicates was used.
For the seven glioma cell lines in the test set, we measured percent growth inhibition relative to a control for the 40 drugs at five concentration points in triplicate: 50 μM, 5 μM, 500 nM, 50 nM and 5 nM. Cell lines U87, U373 and T98G were grown in modifified eagle's medium (MEM) 10% fetal bovine serum (FBS), LN229 was grown in Dulbecco's modified Eagle's medium 10% FBS, U251 was grown in RPMI 1640 5% FBS and both A172 and U118 were grown in DMEM 10% FBS. Cells were seeded at 10 000 cells/well in a 96-well plate in 150 μM media/well. Viability assays were performed after 48 h of initial seeding as described previously (Vichai and Kirtikara, 2006). IC50 calculations were performed by curve fitting of the data using the IC50 package for the R statistical computing environment.
For the 19 Breast cancer cell lines in the test set, IC50 data for simvastatin and pepleomycin were downloaded from Supplementary Material associated with Mori et al. (2009).
2.3 Signature generation
2.3.1 Using Random Forest variable importance to create a gene expression signature for each drug
Existing algorithms for producing gene expression signatures from drug response data compute univariate measures of differential gene expression between cell lines labeled sensitive or resistant. Two drawbacks exist with this approach: (i) definition of resistant and sensitive cell lines can be drug dependent, is arbitrarily defined and methods based on SD are only appropriate when IC50 values in the NCI-60 are normally distributed—which often not the case. (ii) Univariate differential gene expression cannot capture higher order gene–gene interactions that may be important for predicting drug response.
To improve on this approach, we used gene expression signature generated by fitting a regression model between drug IC50 and basal gene expression using Random Forest. This machine-learning algorithm combines many single regression or classification trees into a large ‘ensemble’ to improve performance and robustness. Two methods are used to inject randomness into the ensemble—(i) Bootstrap aggregation: each tree is grown from a randomly selected subset of the training cases (defaults to 70% for regression). (ii) Random Subspace Method: the splitting variable for each node in an individual tree is selected from a random subset of the input variables (defaults to 30% for regression). Supplementary Figure S1 provides a more detailed explanation of the Random Forest algorithm.
Each tree in the forest is trained on a random subset of the cases and then tested on the remaining cases, called the out-of-bag cases (OOB). To compute variable importance, individual values of variables in the OOB are randomly switched with another variable in the OOB. Decrease in performance of each regression tree, measured by R2 for each variable after its value has been permuted, provides a measure of its importance in the regression model.
To use the variable importance measure to create a signature, a Random Forest regression model using 25 000 trees was trained on the normalized IC50 values for each drug, using all the gene expression probeset values from the composite Genelogic/Chiron dataset. Probesets for the signature were selected if they showed variable importance values >2 SD above the mean of all variable importance values for each particular drug. To improve performance for signature generation, we used the Simple Network of Workstations (SNOW) package for R to process many drugs simultaneously on a computational cluster.
2.3.2 Using Random Forest case proximity to identify core cell lines associated with each drug
How to select an appropriate training set from a heterogeneous panel of cell lines derived from nine different tissues of origin remains a central challenge in using the NCI-60 to predict drug response. To accomplish this, we developed a novel computational approach using the case proximity metric originally developed in Random Forest. Case proximity in the model space is defined in the following way: both OOB and non-OOB cases (cell lines in our model) are put down in each regression tree after all the trees in the Random Forest have been grown. For each pair of j, k cases, count the number of times both cases are assigned to the same terminal node. Dividing by the total number of trees in the forest normalizes proximity counts.
To identify cell lines that were outliers in the regression model, we examined the relationship between proximity and differences in IC50 values (Equation 1) for each drug. We reasoned that cell lines j showing a consistent gene expression/IC50 relationship should show a statistically significant correlation between these two sets of values. For all cell lines i and each particular cell line j, a vector of the
| (1) |
absolute value of the differences in IC50 values between that cell line and all other cell lines IC50j was first computed. The Pearson's correlation coefficient between the vector IC50j and proximity values Pj between this cell line and all other cell lines then defined this relationship. For each drug, cell lines j were kept in the training set that showed correlation coefficients P < 0.05. The Bonferonni method was applied to control for multiple hypothesis testing for each drug. To allow users to substitute alternative and less conservative measures of multiple hypothesis correction, we included an option in our software to disable Bonferonni correction. Figure 2 provides a visual representation of the algorithm for two drugs. The proximity matrices for pepleomycin (Fig. 2a) and simvastatin (Fig. 2b) are shown as heat maps. Cell lines showing similar behavior in the model space cluster together in blocks of high proximity values around the diagonal. After removing outlying cell lines, the proximity matrices (Fig. 2a and b) show higher degrees of internal consistency, and IC50 values for each cell line (upper bar) show a higher degree of correspondence with each cluster of cell lines along the diagonal.
Fig. 2.
Pairwise proximity matrixes for pepleomycin and simvastatin. Proximity matrices from Random Forest are defined as the number of instances in which two cases (cell lines) are assigned to the same terminal node of a tree, normalized over the [0, 1] interval. Proximity between a case and itself is not a meaningful value so these instances on the diagonal are set to zero (A1) proximity matrix for pepleomycin before reduction of cell-lines by Equation (1). (A2) Proximity matrix for pepleomycin after removal of outlying cell lines. (B1) Proximity matrix for simvastatin. (B2) Proximity matrix for simvastatin after removal of outlying cell lines.
After selecting core cell lines for each drug, the regression model was then built between the gene expression signature for these cell lines and the corresponding IC50 values for each drug using Random Forest with 10 000 trees. The model for each drug was then applied to the drug gene expression signature in the test set.
3 RESULTS
We tested the method on two external datasets: (i) 19 Breast Cancer Cell Lines tested with the two drugs simvastatin and pepleomycin (ii) Seven Glioma Cell lines tested with 40 FDA-approved oncology drugs.
3.1 Breast cancer cell lines
To test the predictive accuracy of the algorithm, cell lines were first assigned to sensitive or resistant groups based on experimentally determined IC50 results for simvastatin and pepleomyicin. Drug response predictions for sensitive and resistant subsets were generated using two methods: (i) full implementation of the algorithm and (ii) algorithm without the use of case proximity.
As shown in Figure 3, the full implementation of the algorithm outperforms the one-step approach. The difference between the predicted means of resistant and sensitive groups is larger for the two-step signature generation method.
Fig. 3.
Experimental confirmation of predictions for simvastatin and pepleomycin in 19 breast cancer cell lines. CELL lines were identified as resistant if showing −log(IC50) <4 and as sensitive if showing > 5.4−log(IC50) for simvastatin (A) and −log(IC50) <4 and −log(IC50)> 5.4 for pepleomycin (B). Y-axis shows the predicted IC50. Sensitive and resistant groups for the TWO-step method showed statistically significant differences in means using a two-tailed t-test (P < 0.05). The two-step method produced a greater separation of means (0.17, 0.40) versus (0.20, 0.30) for simvastatin and (0.28, 0.51) versus (0.31, 0.43) for pepleomycin.
3.2 Glioma cell lines
A statistical model of drug response based on the large database of drugs that have been tested against the NCI-60 may be particularly useful for in silico drug screening of external panels of cell lines specific to certain cancer types. To test the usefulness of our algorithm in such an application, we experimentally determined the response of seven glioma cell lines to 40 FDA-approved oncology drugs.
Out of the 280 drug/cell-type experiments, only 40 reached 50% growth inhibition required to calculate IC50. In order to use the information from all experiments, we substituted % growth at 48 h (relative to a control) for the 50 and 500 nM concentration points.
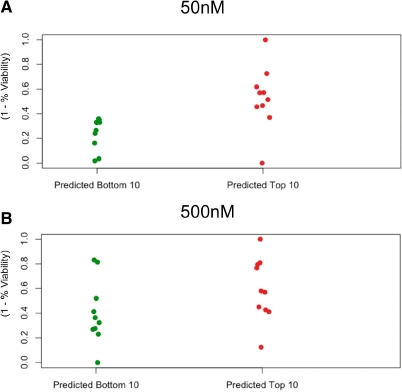
We applied the algorithm to generate gene expression signatures drug response models from the NCI-60 for each of the 40 drugs. The model proved successful in predicting the top 10 (Table 1) and the bottom 10 responding drugs at the 50 and 500 nM concentration points (Fig. 4).
Table 1.
Top 10 predicted most potent drugs in seven glioma cell lines
| NSC | Drug names |
|---|---|
| 613 327 | Gemcitabine |
| 266 046 | Oxaliplatin |
| 606 869 | Clofarabine |
| 49 842 | Vinblastine sulfate |
| 821 51 | Daunorubicin HCL |
| 312 887 | Fludarabine |
| 127 716 | Decitabine |
| 67 574 | Vincristine sulfate |
| 119 875 | Cisplatin |
| 740 | Methotrexate |
Fig. 4.
Experimental confirmation of predictions for 40 FDA-approved cancer drugs in seven glioma cell lines. FOR each drug, the mean of predicted IC50 response over the seven cell lines was computed. The percent viability of cell lines relative to a control was measured at 50 and 500 nm of drug concentration after growth of 48 h normalized over the [0, 1] interval. A two-tailed significance test (correlation test in R) of the Pearson product moment correlation between predicted and measured IC50 values across all 37 cell lines showed significance for both concentration points at P < 0.001.
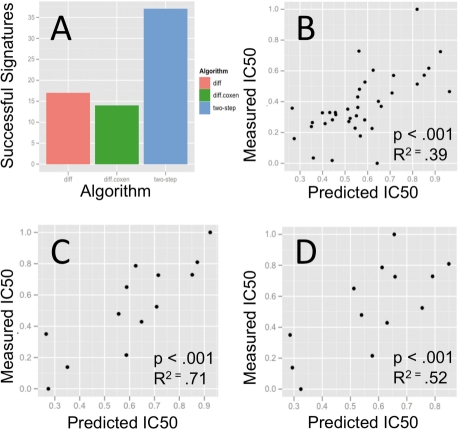
To compare the performance of previous methods, we tested the predictive capability of signatures generated from differential gene expression between the top 12 and bottom 12 responding cell lines for each drug using the Significance Analysis of Microarrays (SAM, FDR ≤ 0.1; Fig. 5). In addition, we also applied the co-expression extrapolation technique (Supplementary Fig. S2) developed by Lee et al. (2007) to identify and retain genes that display consistent expression between training and test sets. Signatures from the differentially expressed genes were used to build statistical models using Random Forest regression. Results are plotted as [0, 1] normalized x−y scatter plots of predicted versus actual drug sensitivity at the 50 nM concentration point (Fig. 5), and show that the two-step method outperforms differential gene expression both in the total number of signatures created as well as the accuracy of those signatures at predicting measured IC50 response.
Fig. 5.
Performance evaluation of the two-step algorithm. (A) THE two-step method successfully created 37 signatures from the 40 FDA-approved drugs while the signature generation based on differential gene expression produced 17 and differential gene expression + co-expression extrapolation produced 14. (B) SCATTER plot of two-step algorithm predictions for 37 drugs versus measured IC50 values. (C) Scatter plot of two-step algorithm predictions for 14 drugs. (D) Scatter plot of predicted versus actual IC50 values for the same 14 drugs predicted using the co-expression extrapolation method.
4 DISCUSSION
We have created a novel algorithm for predicting in vitro drug response from a signature of basal gene expression. Unlike previous methods, this approach incorporates multivariate interaction of input variables (gene expression levels), automatically indentifies core cell lines associated with each drug, and models drug response as a continuous variable. As demonstrated, this approach outperforms a comparable method based on univariate differential gene expression.
Although statistical tests of differential gene expression have been an important tool for the analysis of microarray data, interactions between the biological pathways that drive gene expression levels provide another layer of information that can be mined using multivariate approaches such as Random Forest. Since regression trees incorporate variable interactions as a natural consequence of data partitioning, they provide an ideal algorithmic approach for incorporating variable interactions in the creation of a gene expression signature. Techniques for explicitly encoding gene–gene interactions, such a multifactor dimensionality reduction (MDR), may also be worthwhile to investigate in future work. Although single trees do not generally provide the statistical power of other multivariate techniques, ensemble methods such as Random Forest that randomly sample from both cases and input variables have shown to be competitive with class-leading techniques such as support vector machines and stochastic gradient boosting (Diaz-Uriarte and Alvarez de Andres, 2006). In addition, Random Forest requires little or no parameter tuning and is therefore suitable for machine-learning tasks such an in silico screen that require the creation of a large number of statistical models.
The heterogeneity of cell line panels such as the NCI-60 presents a challenge to the creation of drug gene expression signatures. Previous workers have created models using only cell lines from the NCI-60 showing extreme values of IC50 response to any particular drug. However, defining resistant and sensitive cell lines becomes problematic when many drugs show IC50 distributions across the NCI-60 that are not normally or uniformly distributed. Using a ranked-based definition of drug sensitivity may also produce non-optimal training sets for drugs in which the IC50 distribution is skewed. To overcome these obstacles, we created a novel approach to identify core cell lines for each drug using the case proximity metric in Random Forest. We note that another group has recently published a method for associating drugs with sets of core cell lines (Kutalik et al., 2008). However, this approach was based on a fully linear method, does not incorporate variable interactions and was not used to develop predictive models of drug response.
Functional screens that combine basal gene expression and drug response from panels of cell lines such as the NCI-60 may prove to be an important tool for the discovery of compound leads—especially for complex and heterogeneous diseases such as cancer. By experimentally testing the inhibitory profiles of 40 FDA-approved cancer drugs in seven glioma cell lines, we have provided one of the most complete validation tests to date of this approach. The predictive algorithm that we have developed can be generalized to other problems in machine learning that require the generation of predictive signatures from large numbers of input variables that may exhibit a high degree of noise and self-correlation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Yuri Kotliarov, Aiguo Li and Serdar Bozdag for their helpful discussions. We are also grateful to the DTP at the NCI for supplying FDA-approved oncology drugs used for testing. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD, USA (http://biowulf.nih.gov).
Funding: Intramural Research Program of the National Institutes of Health; National Cancer Institute.
Conflict of Interest: none declared.
REFERENCES
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Covell DG, et al. Anticancer medicines in development: assessment of bioactivity profiles within the National Cancer Institute anticancer screening data. Mol. Cancer Therap. 2007;6:2261–2270. doi: 10.1158/1535-7163.MCT-06-0787. [DOI] [PubMed] [Google Scholar]
- Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutalik Z, et al. A modular approach for integrative analysis of large-scale gene-expression and drug-response data. Nat. Biotechnol. 2008;26:531–539. doi: 10.1038/nbt1397. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomforest. R News. 2002;2:18–22. [Google Scholar]
- Li A, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol. Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- Lee JK, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc. Natl Acad. Sci. USA. 2007:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, et al. Utilization of genomic signatures to identify phenotype-specific drugs. PLoS ONE. 2009;4:e6772. doi: 10.1371/journal.pone.0006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull KD, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Potti A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat. Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Staunton JE, et al. Chemosensitivity prediction by transcriptional profiling. Proc. Natl Acad. Sci. USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nature Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Weinstein JN. Spotlight on molecular profiling: ‘Integromic’ analysis of the NCI-60 cancer cell lines. Mol. Cancer Therap. 2006;5:2601–2605. doi: 10.1158/1535-7163.MCT-06-0640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.