Summary
Wall teichoic acids (WTAs) are anionic polymers that play key roles in bacterial cell shape, cell division, envelope integrity, biofilm formation, and pathogenesis. B. subtilis W23 and S. aureus both make polyribitol-phosphate (RboP) WTAs and contain similar sets of biosynthetic genes. We use in vitro reconstitution combined with genetics to show the pathways for WTA biosynthesis in B. subtilis W23 and S. aureus are different. S. aureus requires a glycerol-phosphate primase called TarF in order make RboP-WTAs; B. subtilis W23 contains a TarF homolog, but this enzyme makes glycerol-phosphate polymers and is not involved RboP-WTA synthesis. Instead, B. subtilis TarK functions in place of TarF to prime the WTA intermediate for chain extension by TarL. This work highlights the enzymatic diversity of the poorly characterized family of phosphotransferases involved in WTA biosynthesis in Gram-positive organisms.
Introduction
Teichoic acids are highly abundant anionic polymers found in Gram-positive bacteria. There are two types of teichoic acids: the lipoteichoic acids (LTAs), which are embedded in the bacterial membrane and extend into the peptidoglycan layers; and the wall teichoic acids (WTAs), which are covalently attached to the peptidoglycan layers and extend beyond them (Figure 1A) (Neuhaus and Baddiley, 2003). Teichoic acids play important but as yet poorly understood roles in cell shape determination (D'Elia et al., 2006a; Pollack and Neuhaus, 1994; Soldo et al., 2002), cell division (Grundling and Schneewind, 2007; Oku et al., 2009; Schirner et al., 2009), biofilm formation (Fabretti et al., 2006; Fedtke et al., 2007; Vergara-Irigaray et al., 2008), cell adhesion (Gross et al., 2001; Weidenmaier et al., 2004), and other aspects of Gram-positive physiology (Swoboda et al., 2009a; Xia et al., 2009). Although neither type of TA is strictly essential for survival in vitro, organisms lacking either one display growth defects that range from mild to severe. Furthermore, it has been shown that pathogenicity is critically dependent on the expression of WTAs in S. aureus (Weidenmaier et al., 2004; Weidenmaier et al., 2005). A detailed understanding of WTA biosynthesis is important for exploring their roles in bacterial physiology and assessing their potential as antibacterial targets (May et al., 2005; Swoboda et al., 2009b).
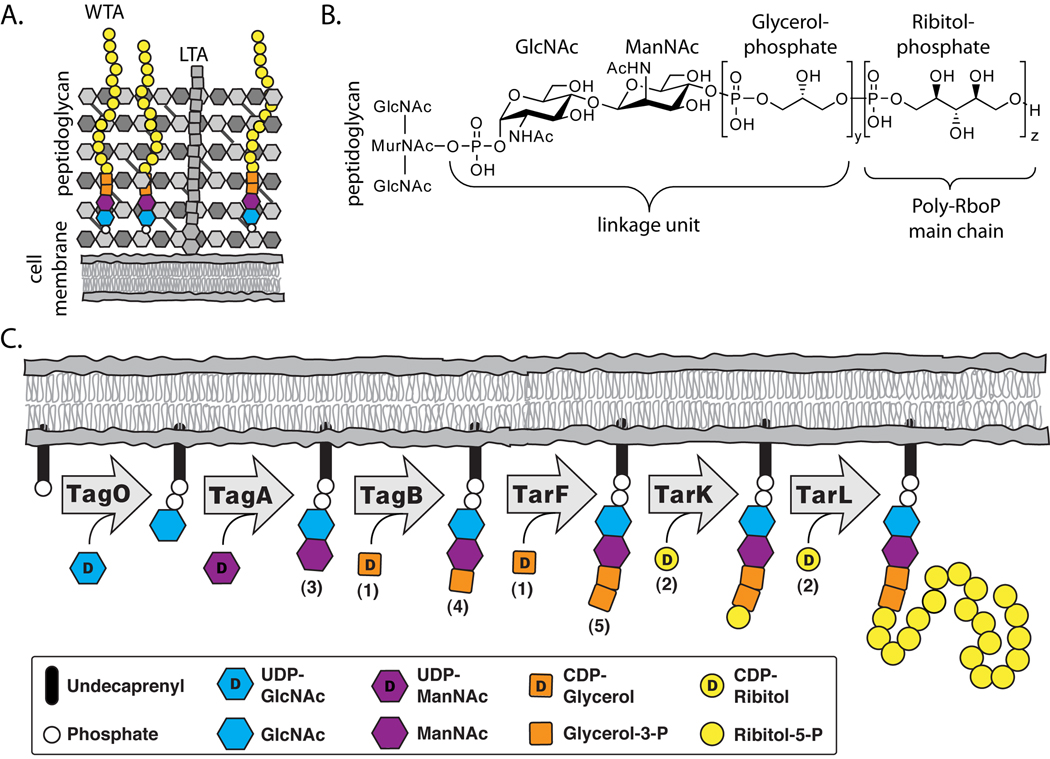
Figure 1. Teichoic acids are a major component of the gram-positive cell wall and the pathway for ribitol-phosphate wall teichoic acids has been proposed.
(A) A schematic of the Gram-positive cell wall depicting membrane anchored lipoteichoic acid and peptidoglycan anchored wall teichoic acid (WTA) polymers.
(B) The chemical structure of ribitol-phosphate wall teichoic acid as found in S. aureus and B. subtilis W23 (y = 1–2, z = 20–40). The tailoring modifications on the main chain hydroxyls are omitted for clarity.
(C) Lazarevic et al. (2002) proposed the biosynthetic pathway depicted above for polyribitol-phosphate WTAs. The numbers in parentheses correspond to the substrates utilized in our in vitro experiments (see Figure 2 for chemical structures).
Wall teichoic acids are attached via a phosphodiester linkage to the N-acetyl muramic acid sugars of peptidoglycan. WTAs typically consist of a disaccharide linkage unit followed by a polymeric main chain (Figure 1B). The B. subtilis W23 main chain is structurally identical to the main chain in S. aureus and contains ribitol-5-phosphate (RboP) repeats (Swoboda et al., 2009a). As shown in Figure 1C, a pathway for polyribitol phosphate WTA synthesis was proposed many years ago by Lazarevic et al. (Lazarevic et al., 2002). This model was based on comparing the genes for WTA biosynthesis in B. subtilis W23 to the genes in B. subtilis 168, which makes polyglycerol-phosphate WTAs (Neuhaus and Baddiley, 2003; Ward, 1981). Previous studies have confirmed the proposed functions of the first three steps in the RboP-WTA biosynthetic pathway. The first enzyme in this pathway, TagO, is an integral membrane protein that transfers phospho-GlcNAc from UDP-GlcNAc to an undecaprenyl carrier lipid embedded in the cytoplasmic surface of the bacterial membrane (D'Elia et al., 2006b; Weidenmaier et al., 2004). The lipid-linked monosaccharide is then converted to disaccharide 4 by the UDP-ManNAc transferase TagA (Brown et al., 2008; D'Elia et al., 2009; Zhang et al., 2006). A primase, TagB, then attaches a single GroP unit to the non-reducing end of the disaccharide (Brown et al., 2008). Following assembly of the disaccharide linkage unit, the pathway for polyRboP-WTAs was proposed to require three enzymes, TarF, TarK, and TarL, to complete the polymeric main chain (Lazarevic et al., 2002). The proposed functions of these three enzymes are shown in Figure 1C. Once WTA synthesis is complete, the RboP polymers, still attached to the undecaprenyl carrier lipid, are flipped to the external surface of the membrane where they are attached to peptidoglycan (Swoboda et al., 2009a).
Recent studies have shown that polyRboP-WTA polymer synthesis in S. aureus differs from the proposed pathway in Figure 1C in that only two enzymes are required to complete the polyRboP main chain (Brown et al., 2008; Meredith et al., 2008; Pereira et al., 2008a). One enzyme is TarF (TarFSa), which transfers a single glycerol-phosphate (GroP) to the linkage unit. The other enzyme is TarL, which combines the proposed functions of TarK and TarL shown in Figure 1C. That is, S. aureus TarL (TarLSa) is a ribitol-phosphate polymerase that can act directly on the TarF product without requiring a RboP-primed substrate. It functions to prime and extend the elaborated linkage unit (5) (Brown et al., 2008). Although we have shown that S. aureus contains two copies of tarL (one of which was the originally proposed TarK), only one of these copies is essential in cells (Meredith et al., 2008; Swoboda et al., 2009a). These discovered differences between the proposed and demonstrated S. aureus WTA pathway prompted us to investigate the WTA biosynthetic pathway for B. subtilis W23, the organism on which the pathway shown in Figure 1C for polyRboP-WTA biosynthesis is based. We show here that the B. subtilis pathway differs in several ways from the S. aureus pathway even though both organisms make a polyribitol-phosphate WTA polymer. Furthermore, it differs from the previously proposed B. subtilis pathway.
Results
Description of the approach
As described in the introduction, WTA precursors are synthesized in the cytoplasm on an undecaprenyl phosphate carrier lipid embedded in the membrane (Swoboda et al., 2009a). The first enzyme in the WTA biosynthetic pathway, TagO, is an integral membrane protein, but many of the other enzymes involved in assembly of the WTA polymer do not have predicted membrane-spanning regions, although it is presumed that they are membrane-associated either directly or indirectly (Bhavsar et al., 2007; Formstone et al., 2008). Membrane-associated enzymes in other lipid carrier-mediated biosynthetic pathways (e.g., peptidoglycan biosynthesis) will accept water-soluble substrates containing truncated lipid chains in the absence of biological membranes or membrane mimics (Chen et al., 2002; Chen et al., 2007; Faridmoayer et al., 2008; May et al., 2009; Men et al., 1998; Ye et al., 2001). Therefore, we synthesized WTA pathway intermediates containing short lipid chains and have previously shown that these intermediates are functional substrates for several WTA enzymes in vitro (Brown et al., 2008; Ginsberg et al., 2006; Zhang et al., 2006). Here, we use the short chain substrate analogs shown in Figure 2 to characterize the late steps of polyribitol-phosphate WTA biosynthesis in B. subtilis W23. As described below, analysis of the products formed in vitro by B. subtilis TarF (TarFBs), combined with the in vitro substrate preferences for B. subtilis TarK (TarKBs), suggested a revised pathway for polyRboP-WTA biosynthesis in B. subtilis W23. Using genetic approaches, we have confirmed the revised B. subtilis W23 WTA biosynthetic pathway.
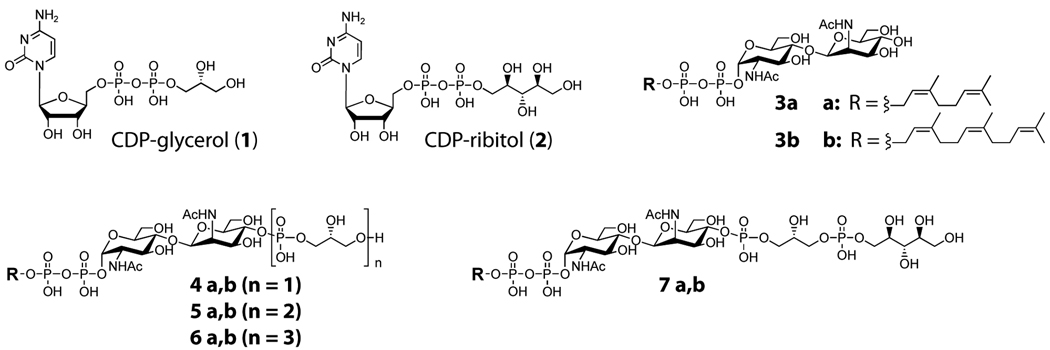
Figure 2. Structures of WTA substrates and intermediates.
The structures of the CDP-glycerol (1) and CDP-ribitol (2) donor and lipid acceptor substrates 3, 4, 5, 6, and 7. The natural acceptor substrates contain an undecaprenyl chain. Substrates having the “a” suffix contain a neryl chain and were used for LC/MS analysis; substrates having the “b” suffix contain a farnesyl chain and were used for PAGE analysis. Synthesis of substrates and incorporation of [14C] radiolabels for PAGE analysis were carried out as described previously (Brown et al., 2008).
Biochemical reconstitution of B. subtilis W23 TarF and comparison to S. aureus TarF
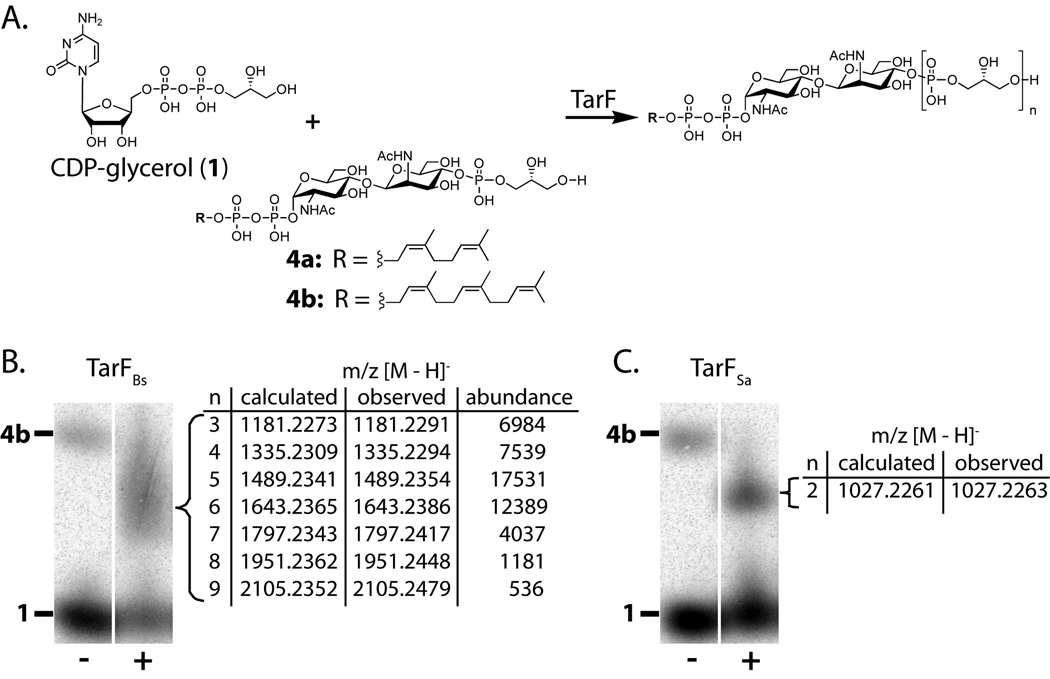
The first three steps in the pathways for WTA synthesis in B. subtilis W23 and S. aureus are identical and lead to the production of lipid-pp-GlcNAc-ManNAc-GroP (compound 4) (Figures 1C and 2). We have shown that S. aureus TarF is a primase that adds a single glycerol phosphate unit to 4 to produce lipid-pp-GlcNAc-ManNAc-GroP2 (compound 5) (Brown et al., 2008). The function of TarF in B. subtilis W23 has not previously been studied, but it was proposed, like its S. aureus ortholog, to act as a glycerol-phosphate primase (Lazarevic et al., 2002), which catalyzes the reaction of 4 with CDP-glycerol (1) to form WTA intermediate 5. To test this prediction, we cloned and expressed B. subtilis W23 TarF (TarFBs) as a C-terminal 6-His fusion from genomic DNA and purified it via Ni2+ chromatography. We incubated purified TarFBs with radiolabeled 4b in the presence of [14C]-CDP-glycerol (1) and analyzed the results by polyacrylamide gel electrophoresis. PAGE analysis showed that CDP-glycerol (1) reacted in the presence of TarFBs and lipid-pp-GlcNAc-ManNAc-GroP (4b) to generate a new radioactive spot (Figure S1A); see below for product identification. TarFBs under the same conditions did not react with 1 and 3b, or with CDP-ribitol (2) and 4b. These results suggested that TarFBs, like its S. aureus counterpart, catalyzes the addition of glycerol-phosphate, but not ribitol-phosphate, to primed substrate 4, but not to the unprimed precursor 3.
We next analyzed the products formed by the reaction of radiolabeled 1 and 4b in the presence of TarFBs. For comparison, the same substrates were also incubated with S. aureus TarF (TarFSa). Products were analyzed via PAGE (Figures 3B and 3C). TarFSa produces a single well-resolved product, identified by exact mass analysis of a non-radioactive reaction as lipid-pp-GlcNAc-ManNAc-GroP2 (compound 5); this product corresponds to the transfer of a single glycerol-phosphate unit to 4. Under forcing conditions (high enzyme and substrate concentrations for prolonged reaction times), an additional product containing three glycerol-phosphate units (compound 6) can also be observed (Figure S2A). In marked contrast, under all conditions examined (low and high enzyme concentrations; short or long reaction times), the TarFBs reaction produces a smear of radioactivity without a distinct banding pattern. These results suggested the formation of multiple products. Reactions with TarFBs and substrates 1 and 4a were carried out using non-radiolabeled substrates and subjected to Q-TOF MS analysis. Discrete products with increasing numbers of glycerol-phosphates up to nine units were detected. Thus, while TarFSa is a primase that preferentially adds a single GroP unit to 4, TarFBs behaves as a polymerase, adding glycerol-phosphate units to 4 in rapid succession up to a detectable length of at least nine units.
Figure 3. TarF is a polymerase in B. subtilis W23 and a primase in S. aureus.
(A) Reaction carried out by TarF (n = 3 −9 for TarFBs; n = 2 for TarFSa).
(B, C) PAGE analysis of TarFBs (B) and TarFSa (C) reactions using 1 and substrate 4b in the presence of heat-treated (−) or active enzyme (+). Tabulated masses of purified TarF reaction products using substrate 4a are shown.
Reconstitution of B. subtilis W23 TarK and TarL
Lazarevic et al. proposed that TarK (TarKBs) and TarL (TarLBs) from B. subtilis W23 form a primase-polymerase pair, with the former transferring a single ribitol-phosphate unit to the TarFBs product and the latter adding multiple ribitol-phosphate units to build the polymer chain (Figure 1C) (Lazarevic et al., 2002). Using a heterologous complementation approach, we have previously provided evidence that B. subtilis W23 TarK and TarL function as a primase-polymerase pair since both are required to complement the deletion of S. aureus TarL, which combines both functions (Meredith et al., 2008; Pereira et al., 2008a). The genetic complementation experiments did not, however, provide detailed information on the enzymatic reactions carried out by TarKBs and TarLBs. Here, we have reconstituted the in vitro activities of these enzymes to characterize their substrate preferences and verify their proposed products.
TarKBs and TarLBs were cloned from genomic B. subtilis W23 DNA, expressed as C-terminal 6-His fusion proteins and purified by nickel chromatography. WTAs extracted from B. subtilis W23 cells are reported to contain one to two glycerol-phosphate units, and since TarKBs is a proposed ribitol-phosphate phosphotransferase, we first tested the activity of TarKBs with CDP-ribitol (2) and WTA intermediate 4a, which contains one GroP unit. The reaction was analyzed by HPLC because products could not be resolved by polyacrylamide gel electrophoresis (Figure S3A). A decrease in the CDP-ribitol peak, accompanied by the appearance of CMP, showed that TarKBs reacts with CDP-ribitol in the presence of 4a. We also tested the activity of TarKBs with 4a and CDP-glycerol (1). An unchanged peak area for CDP-glycerol with no appearance of CMP showed that TarKBs does not utilize 1 as a donor.
Exact mass analysis of a TarKBs reaction of 2 with 4a showed that the predominant product, compound 7a, contained one ribitol-phosphate unit (m/z [M-H]− expected: 1087.2472, actual: 1087.2509); a small amount of product (mass abundance less than 0.05% compared with compound 7a) containing two ribitol-phosphate units was also detected (m/z [M-H]− expected: 1301.2715, actual: 1301.2591). The MS data and HPLC traces confirm that TarK from B. subtilis W23 is a ribitol-phosphate primase that catalyzes the addition of a ribitol-phosphate unit from CDP-ribitol to WTA intermediate 4 to form lipid-pp-GlcNAc-ManNAc-GroP-RboP (7).
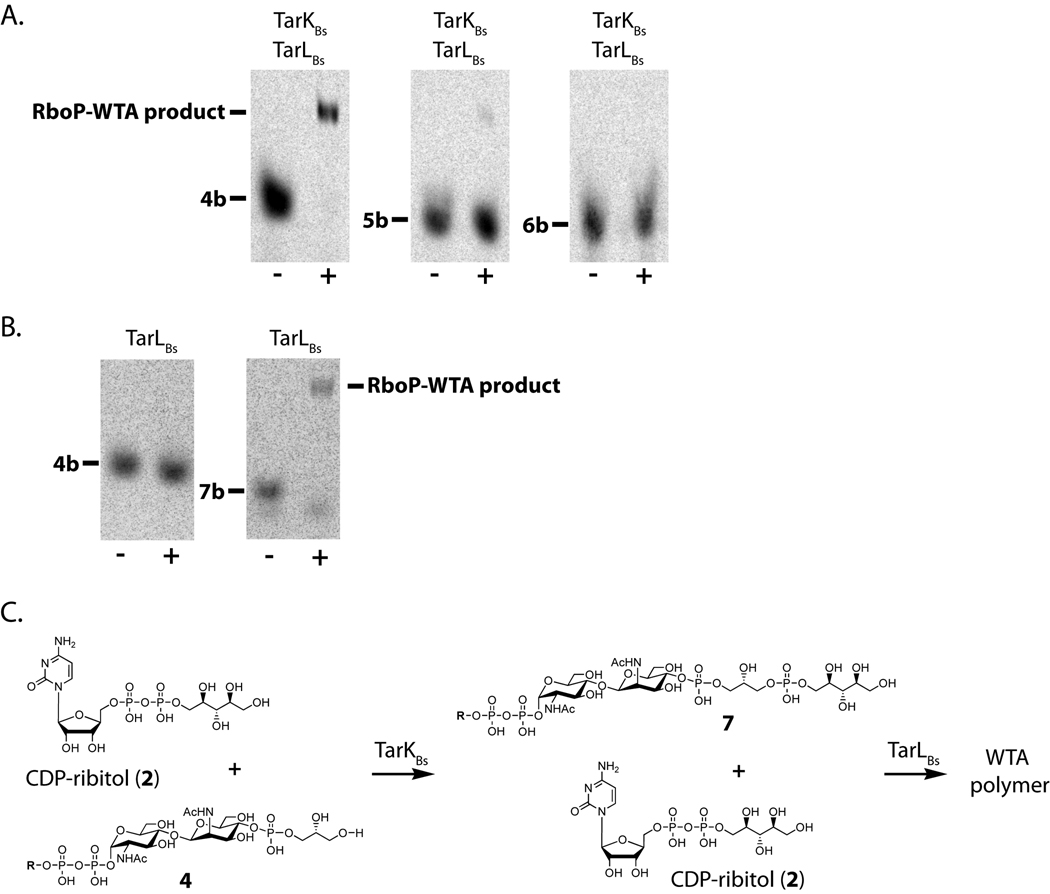
To evaluate the substrate specificity of TarKBs in more detail, we carried out tandem reactions of B. subtilis TarK and TarL. Tandem reactions were used to enable product analysis by PAGE, since the TarK products cannot be resolved from starting material. TarKBs and TarLBs were incubated with CDP-ribitol (2) and radiolabeled lipid intermediates 4b, 5b, and 6b. These intermediates differ in the number of glycerol phosphates appended to the ManNAc sugar. Reactions were analyzed for the appearance of polymeric product (Figure 4A). The TarKBs/TarLBs tandem reaction converted substrate 4b to polymeric product. Tandem reactions using substrates containing more than one GroP unit (e.g., 5b or 6b) resulted in little to no WTA polymer.
Figure 4. TarK and TarL react sequentially on the TarB product (4).
(A) PAGE analysis of a tandem TarKBs/TarLBs reaction using either heat-treated (−) or active (+) enzymes incubated with CDP-ribitol and 4b, 5b, or 6b. The polymeric material in the middle panel is due to reaction with contaminating 4b. Polymer product only forms when 4b is used as the starting material (see text).
(B) PAGE analysis of TarLBs heat-treated (−) or active (+) enzyme incubated with 4b or 7b and CDP-ribitol (2). TarLBs reacts only with 7b. See also Figure S4.
(C) Reactions carried out by TarKBs and TarLBs with CDP-ribitol (2) and substrates 4 and 7 respectively.
To confirm that the resulting polymeric products observed in the TarKBs/TarLBs reactions were due to a tandem reaction of TarKBs acting as a primase and then TarLBs acting as a polymerase we tested the ability of B. subtilis W23 TarL to catalyze the addition of ribitol-phosphate units to different WTA intermediates. TarLBs was incubated with 4b, 5b, 6b, or 7b in the presence of CDP-ribitol (2). The reaction products were analyzed by PAGE (Figure 4B and S4A). No radioactive products were observed for reactions containing lipid substrates 4b, 5b, or 6b, showing that TarLBs is unable to use any of the glycerol-phosphate primed intermediates to make polymer. In contrast, the enzymatic reaction with compound 7b showed the disappearance of starting material and the appearance of a smear of radioactivity corresponding to higher molecular weight products. The gel analysis shows that TarLBs reacts with the ribitol-phosphate-primed substrate and is consistent with the addition of multiple RboP subunits. These data confirm that TarLBs is a ribitol-phosphate polymerase and that it requires for reaction a ribitol-phosphate-primed substrate produced by TarKBs.
The TarKBs/TarLBs tandem reactions showed that TarKBs has a strong preference for substrate 4, but it was not clear whether the small amount of polymeric product observed in the middle panel of Figure 4A resulted from reaction of TarKBs with 5b or with contaminating 4b since the starting material was estimated to be only 90% pure and the impurity could not easily be removed. Therefore, we incubated TarKBs with CDP-ribitol (2) and a mixture of 4a and 5a and analyzed the products by Q-TOF MS. The major peak proved to be unreacted lipid-pp-GlcNAc-ManNAc-GroP2 (compound 5a) (m/z [M-H]− expected: 1027.2261, actual: 1027.2264). Another peak corresponding to compound 7a (m/z [M-H]− expected: 1087.2472, actual: 1087.2468) was detected, resulting from TarKBs utilizing 4a as a substrate. We were unable to detect any product peak corresponding to the addition of RboP to 5a. These results imply that TarKBs is only able to accept lipid-pp-GlcNAc-ManNAc-GroP (4), the TagB product, as a substrate and imply that the small of polymeric product observed in the middle panel of Figure 4A is due to reaction with the small amount of substrate 4 in the starting material.
The results described in the previous paragraph confirm that TarKBs and TarLBs comprise a RboP primase-polymerase pair and carry out the reactions shown in Figure 4C. These reactions are not consistent with the model shown in Figure 1C (Lazarevic et al., 2002). In that model, TarFBs acts as a primase to synthesize a lipid-GlcNAc-ManNAc-GroP2 substrate for TarKBs, but our results show that TarKBs acts only on the TagB product, which contains a single glycerol-phosphate unit. Since TarKBs is unable to accept WTA intermediates containing more than a single GroP unit, and since TarFBs is a polymerase that adds multiple GroP units to 4, we surmised that TarFBs could not be on the pathway to polyribitol WTA formation in B. subtilis W23. If this supposition is correct, then one would predict that B. subtilis tarF can be deleted and polyribitol-phosphate WTAs will still be produced in strains containing the B. subtilis TarK-TarL primase-polymerase pair.
TarF is dispensable for polyribitol-phosphate WTA synthesis in pathways containing a TarK primase
To test the predicted dispensability of B. subtilis TarF in strains expressing B. subtilis TarK and TarL, we deleted tarF from two different strains expressing these B. subtilis enzymes. The first strain was a S. aureus hybrid strain (JT215) containing B. subtilis TarK-TarL in place of the native bifunctional S. aureus enzymes (TarL or TarL’) (Meredith et al., 2008). This hybrid strain contains the native S. aureus tarF, which is essential in the wild-type S. aureus background (D'Elia et al., 2006b). Despite its essentiality in the wild-type background, we were able to delete tarF from the hybrid S. aureus strain. In fact, the ΔtarF deletion allele was almost exclusively observed upon resolving the integrated tarF deletion vector, and the doubling time of the ΔtarF hybrid strain was faster than that of the hybrid parent strain, suggesting that removal of TarF actually increases strain fitness. We confirmed that WTAs extracted from the ΔtarF hybrid strain are identical in length and banding pattern to those extracted from the parent strain (Figure 5C). Thus, replacing S. aureus TarL with TarKBs-TarLBs renders S. aureus TarF dispensable for polyRboP-WTA synthesis in an S. aureus background.
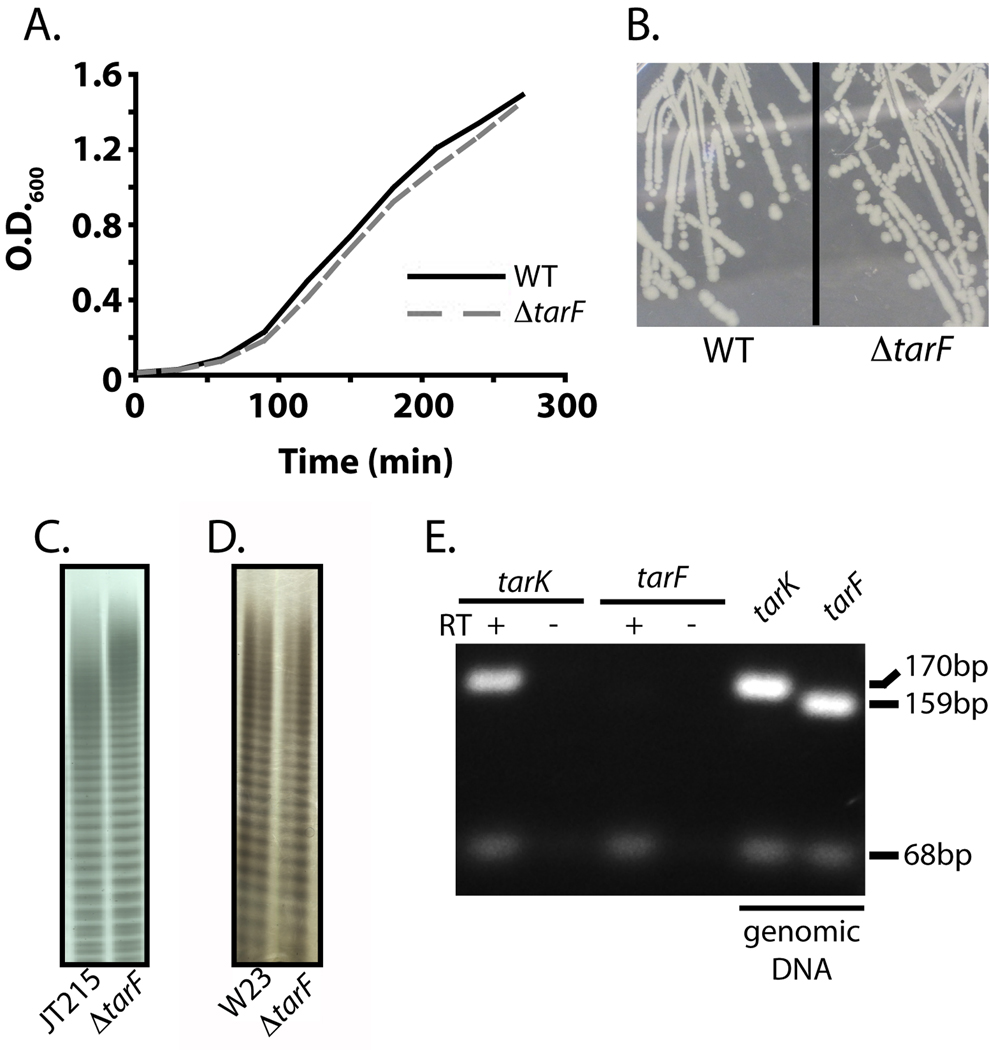
Figure 5. TarKBs and TarLBs make polyRboP-WTAs in vivo in the absence of TarFBs.
(A) Growth curves of B. subtilis W23 wildtype (WT) or ΔtarF strains.
(B) Photograph of bacterial colonies of B. subtilis W23 wildtype (WT) or ΔtarF strains. (C,D) WTAs of the parent wildtype strain or the ΔtarF strain extracted from the B.subtilis/S. aureus hybrid strain JT215 (C) or B. subtilis W23 (D) analyzed by PAGE analysis and alcian blue/silver staining.
(E) Agarose gel analysis of PCR amplified fragments from B. subtilis W23 cDNA or genomic DNA (positive control). (+) and (−) indicate the presence or absence of reverse transcriptase during cDNA synthesis. The sizes of the amplified fragments are 170 bp (tarK), 159 bp (tarF), and 68 bp (16S rRNA) (internal control).
We next constructed a marked tarF deletion in a wildtype B. subtilis W23 strain containing only the native B. subtilis enzymes. The ΔtarF strain (DM1) had a similar growth rate as the parent strain and the colony morphology was unchanged (Figures 5A and 5B). There were no apparent differences in the amounts of extracted WTAs in the ΔtarF and parent strains. Furthermore, WTAs extracted from the two strains were indistinguishable by PAGE analysis (Figure 5D). Degradation of extracted WTAs from each strain followed by Q-TOF MS analysis confirmed that in both strains the subunits of the main chain polymer are composed of ribitol-phosphates covalently modified with glucose (m/z [M-H]− expected: 393.0804; actual wildtype: 393.0813, ΔtarF: 393.0811; Figure S5). These deletion experiments are fully consistent with the in vitro experiments showing that TarKBs reacts preferentially with the TagB product, and not with intermediates containing additional GroP units. Thus, genetic studies in two different strain backgrounds confirm the biochemical work showing that TarFBs is not on the pathway to polyRboP WTAs in B. subtilis W23.
B. subtilis tarF is not expressed during exponential growth
We have shown that tarFBs, which is located within the WTA biosynthetic gene cluster, encodes a functional enzyme that makes short GroP-WTA polymers, but it is not involved in polyRboP WTA synthesis. Since it has been reported that strains in the W23 group (Nakamura et al., 1999), S. xylosus, S. saprophyticus (Endl et al., 1983; Endl et al., 1984), and an S. aureus biofilm producing strain (Vinogradov et al., 2006) produce both ribitol-phosphate and glycerol-phosphate WTAs, we considered the possibility that B. subtilis W23 might contain a small proportion of polyGroP-WTAs. We were unable to confirm the presence of GroP-WTAs by MS analysis of WTAs extracted from B. subtilis W23 (the detection limit is 5 pmol), so we attempted to generate deletions of tarK in B. subtilis W23 to determine whether TarFBs was capable of making polyGroP-WTAs in a ΔtarK background. All efforts to delete tarK were unsuccessful, suggesting that this gene, unlike tarF, is essential in B. subtilis W23. We therefore examined the relative levels of tarF and tarK gene expression using RT-PCR. Under normal laboratory conditions the tarF transcript is barely detectable in cells harvested during late exponential growth, while the tarK transcript is abundant (Figure 5E). Increased amounts of template cDNA in the PCR reaction did not change the levels of detectable transcript. Although TarFBs is a functional enzyme, it is not expressed under normal laboratory growth conditions. Its cellular function, if any, remains a mystery, but it is not involved in polyRboP-WTA biosynthesis.
Discussion
Wall teichoic acids comprise a large fraction of the Gram-positive cell wall. They play important roles in cell envelope integrity and there is increasing evidence that they are involved in fundamental aspects of cell growth and division (Xia et al., 2009). Understanding how these polymers are made is a necessary step in dissecting their cellular functions and in exploring their potential as antimicrobial targets in pathogens such as S. aureus. Although pathways for polyglycerol-phosphate and polyribitol-phosphate WTA biosynthesis in B. subtilis were proposed many years ago (Lazarevic et al., 2002; Ward, 1981), verification and detailed characterization of the enzymatic steps was not possible until recently. The development of chemoenzymatic approaches to obtain functional WTA intermediates has made the in vitro reconstitution of both individual enzymes and entire pathways for WTA biosynthesis feasible (Brown et al., 2008; Ginsberg et al., 2006; Sewell et al., 2009; Zhang et al., 2006). In this paper, we have carried out an extensive biochemical and genetic analysis of polyRboP-WTA synthesis in B. subtilis W23. Although some steps of the proposed model for polyRboP-WTA biosynthesis were confirmed, we have revised others.
Characterization of the enzymes proposed to be involved in the late steps of the B. subtilis polyribitol-phosphate WTA pathway revealed two unexpected results. First, unlike S. aureus TarF, B. subtilis TarF is not a primase. Rather than adding a single glycerol phosphate unit, it adds several units in rapid succession. Therefore, it is a polymerase that makes short glycerol-phosphate WTA polymers. Second, B. subtilis TarK, although it is indeed a phosphoribitol primase, acts only on the TagB product. It cannot use substrates made by B. subtilis TarF, which adds multiple GroP units. This finding implies that B. subtilis TarF is not on the pathway to polyRboP-WTAs.
We confirmed this surprising conclusion by first deleting S. aureus tarF from an S. aureus hybrid strain (Meredith et al., 2008) expressing the primase-polymerase pair from B. subtilis, TarKBs/TarLBs. Although S. aureus TarF is essential in a background containing S. aureus TarL (D'Elia et al., 2006b), we showed that it is dispensable in a strain containing the complementing B. subtilis enzymes. In fact, its deletion in the hybrid strain appears to confer a modest fitness advantage. A plausible explanation for the growth advantage of the ΔtarF S. aureus strain is that TarF activity impedes the maturation of WTAs since the product it makes is not a preferred substrate for the complementing B. subtilis enzymes. We were also able to delete tarFBs from the B. subtilis W23 wildtype strain without any apparent effects on growth, colony morphology, or the production of WTAs. In contrast, all similar attempts to delete tarKBs from B. subtilis W23 failed, suggesting that this gene cannot be deleted without affecting viability. This result is consistent with other studies showing that enzymes required to complete the WTA main chain are conditionally essential (D'Elia et al., 2006a; D'Elia et al., 2006b; Swoboda et al., 2009a). The ability to delete tarFBs, but not tarKBs, supports the hypothesis that the latter is required for RboP-WTA synthesis but the former is not. RT-PCR analysis further supports this hypothesis since it shows that tarK is expressed at high levels during exponential growth while tarF transcription is almost undetectable.
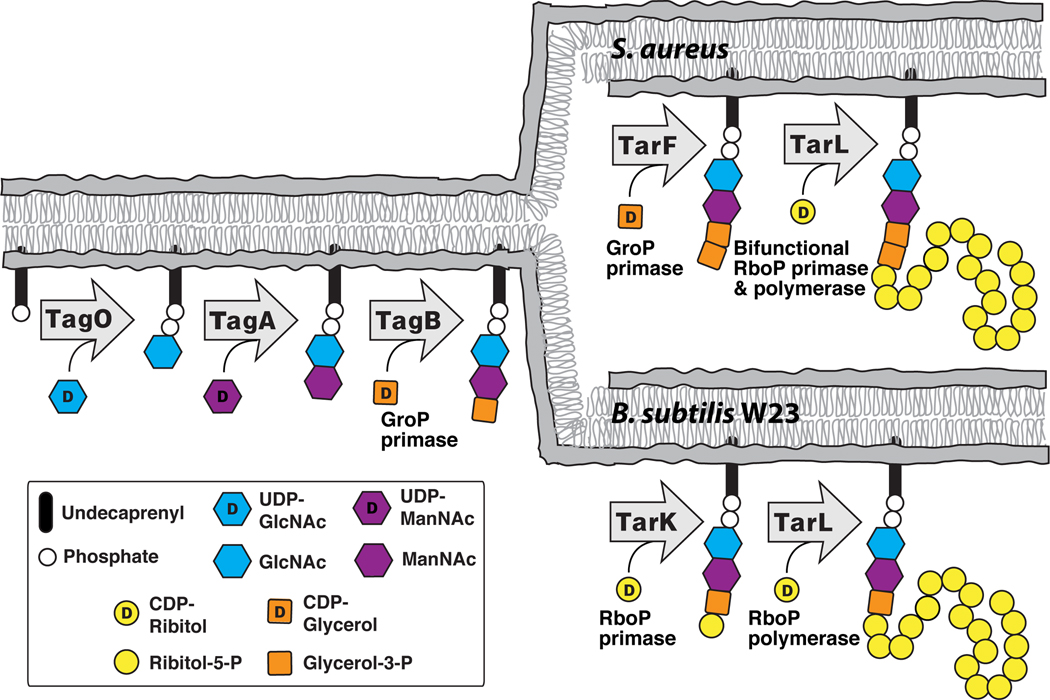
The pathways for polyribitol-phosphate WTA biosynthesis in B. subtilis W23 and S. aureus are now established, as shown in Figure 6. These pathways proceed through the same three initial steps and result in the formation of a lipid-diphospho-GlcNAc-ManNAc-phosphoglycerol unit (4) (Bhavsar et al., 2005; Ginsberg et al., 2006; Soldo et al., 2002; Zhang et al., 2006). Following the third step, catalyzed by TagB, the B. subtilis W23 and S. aureus pathways diverge. B. subtilis W23 uses a RboP primase (TarK) followed by an RboP polymerase (TarLBs) to complete the WTA main chain; S. aureus uses a GroP primase (TarF) followed by a bifunctional RboP primase/polymerase (TarLSa) to complete the chain. The revised pathways, shown in Figure 6, are in line with previously reported data by Yokoyama and coworkers on the isolation of WTA intermediates from B. subtilis W23 and S. aureus (Yokoyama et al., 1986).
Figure 6. There are two pathways for polyribitol-phosphate wall teichoic acid formation in bacteria.
A schematic of the verified late stage intracellular steps in wall teichoic acid biosynthesis in S. aureus and B. subtilis W23.
In addition to establishing the pathways for polyribitol-phosphate WTA polymer synthesis, we note that this work highlights the enzymatic diversity of the large family of CDP-phosphotransferases involved in WTA biosynthesis. These enzymes include both GroP and RboP primases and polymerases, and it does not yet appear possible to predict their functions or substrate preferences accurately based on protein sequences (see Figure S6). For example, S. aureus TarF is a GroP primase that adds one GroP subunit to the linkage unit whereas B. subtilis 168 TagF is a GroP polymerase that adds 45–60 GroP units (Pereira et al., 2008b). B. subtilis W23 TarF makes short GroP polymers in vitro, adding about eight GroPs to the linkage unit. The RboP transferases are similarly diverse, with the primase TarKBs having stringent specificity for singly GroP-primed substrates and the sequence-related polymerase TarLBs having similarly stringent specificity for RboP-primed substrates, while S. aureus TarL functions both as an RboP primase and polymerase, and is selective for doubly GroP-primed substrates. The first structure of a member of this diverse family of phosphotransferases was recently reported (S. epidermidis TagF, (Lovering et al., 2010)) and it may, when combined with accurate information about enzymatic function, facilitate the identification of the features that determine which substrates are selected or whether a particular enzyme functions as a primase or a polymerase. In closing, we note that there is a strong argument to be made for a new systematic nomenclature to designate the CDP-phosphotransferases involved in WTA biosynthesis now that the diversity of enzymatic function has been recognized and the first ascribed gene names do not accurately describe these functions.
Significance
Wall teichoic acids are important cell surface polymers in many Gram-positive organisms, both pathogens and non-pathogens alike. They play central roles in cell growth, division, morphology, adhesion and envelope integrity. Understanding how they are made is important for conducting detailed studies of their cellular functions (Swoboda et al., 2009a). We have developed chemoenzymatic approaches to obtain WTA intermediates to enable the detailed enzymatic characterization of WTA biosynthesis. Here, we have used an extensive set of purified WTA intermediates to compare the pathways for polyribitol-phosphate WTA synthesis in B. subtilis W23 and S. aureus. We show that these organisms use different sets of enzymes to make similar WTA polymers. Hence, bacteria employ two distinct pathways to make polyribitol-phosphate WTAs as outlined in Figure 6. This work should facilitate continued efforts to predict teichoic acid gene functions and determine pathway order in sequenced genomes, will enable well-grounded biological studies to establish the roles of WTA enzymes in the growth and division of S. aureus and B. subtilis, and may facilitate antibiotic discovery.
Experimental Procedures
Materials and Methods
B. subtilis W23 is from ATCC (ATCC 23059). Restriction enzymes were from New England Biolabs. The plasmid used for cloning, pET-24b(+), and His-Bind Resin are available from EMD Chemicals. PCR was performed using KOD HotStart (EMD Chemicals). The pKM074a vector was generous gift from David Rudner (Harvard Medical School). Radiolabeled L-[14C]-glycerol-3-phosphate was purchased from GE Healthcare. All other reagents and chemicals used were from Sigma. The wall teichoic acid extraction and analysis by polyacrylamide gel electrophoresis and alcian blue/silver staining has been described previously (Meredith et al., 2008).
Cloning of B. subtilis W23 genes
The S. aureus enzymes used in this study were cloned, overexpressed, and purified as described previously (Brown et al., 2008). Supplemental Information contains a table listing all primers and strains used. The tarF, tarK, and tarL genes were PCR amplified using B. subtilis W23 genomic DNA as a template and the primers tarFp24bF and tarFp24bR, tarKp24bF and tarKp24bR, tarLp24bF and tarLp24bR. The PCR products were digested with the appropriate restriction enzymes and ligated into a similarly digested pET-24b(+) vector. The constructed plasmids were verified by restriction digest and DNA sequencing (Dana-Farber/Harvard Cancer Center DNA Resource Core).
Protein overexpression and purification
S. aureus proteins used in this study were overexpressed and purified as described previously (Brown et al., 2008). TagB from B. subtilis 168 was used to construct various WTA intermediates and was overexpressed and purified as described previously (Ginsberg et al., 2006). All constructed Tar protein-pET-24b plasmids were transformed into Rosetta2(DE3)pLysS cells for overexpression with IPTG. The cell pellets were lysed with rLysozyme and benzonase in 100mM TrisHCl pH 7.5, 500mM NaCl, 0.6% CHAPS and 5% glycerol. The clarified lysate was purified over charged nickel His-Bind resin. More detailed procedures can be found in Supplemental Information. The proteins were stored as 50% glycerol stocks at either −20°C or −80°C. The yield is approximately 5mg/L for TarFBs, 10mg/L for TarKBs and 4mg/L for TarLBs.
Construction of WTA intermediates
Radiolabeled [14C]-CDP-glycerol (1) was prepared as described previously using TarD (Badurina et al., 2003). CDP-ribitol (2) was prepared as described previously (Brown et al., 2008; Pereira and Brown, 2004). Due to issues with degradation upon long-term storage CDP-ribitol was purified by HPLC after chemoenzymatic synthesis using a strong anion exchange column from Phenomenex (Phenosphere SAX 80A, 250 × 4.6mm 5 micron). For purification details see Supplemental Information. WTA intermediates 3, 4, and 5 were prepared as described previously (Brown et al., 2008). To prepared radiolabeled compound 6b, an enzymatic reaction (30µL) containing 2µM TagB, 2µM TarFSa, 20µM of purified 3b, and 80µM [14C]-CDP-glycerol were incubated in 20mM Tris pH 7.5, 100mM NaCl and 10mM MgCl2 for 30 minutes at room temperature. This allows formation of intermediate 5b. TarFBs (2µM) was then added to utilize the remaining [14C]-CDP-glycerol to form compound 6b. 6b was purified using a Phenomenex Strata X 33 µm polymeric reversed phase (30mg/1mL) column. To prepare radiolabeled compound 7b a 30µL reaction containing 500nM TagB, 500nM TarFSa, 5µM TarKBs, 50µM of purified 3b, 100µM of [14C]-CDP-glycerol, and 100µM of CDP-glycerol were incubated in 20mM Tris pH 7.5, 100mM NaCl and 30mM MgCl2 for 2 hours at room temperature. The reaction was purified as described for compound 6b. Cold reactions performed simultaneously and analyzed by LC/MS confirmed the formation of compounds 6 and 7.
HPLC Analysis of TarK reactions
For HPLC analysis a Phenomenex Phenosphere SAX 80 A, 250 × 4.6mm 5 micron strong anion exchange column was used on an Agilent 1100 HPLC at flow rate of 1 mL/min with a linear gradient of 0% to 20%B over 15min (buffer A: 5mM ammonium acetate, pH 3.8; buffer B: 750mM ammonium dihydrogen phosphate pH 3.7). The UV signal was monitored at 271nM. TarKBs (30µL) reactions contained 200µM 4a, 200µM 1 or 2 and 1µM enzyme in 20mM Tris pH 7.5, 100mM NaCl and 10mM MgCl2. After 30 minutes at room temperature the reactions were quenched with 30µL methanol and analyzed by HPLC. Reactions containing heat-treated enzymes served as negative controls.
PAGE analysis of enzymatic reactions
To assay TarFBs substrate specificity, reactions (3µL) contained 500nM or 5µM enzyme, 1µM 4b and 6µM of either 2 or [14C]-1 in a buffer of 100mM NaCl, 20mM Tris pH 7.5, and 10mM MgCl2. A reaction was also set up with 2µM 3b and 2µM [14C]-1. Heated treated enzyme served as a negative control. Reactions were quenched after 3 hours with 3µL DMF. 2uL of 4× loading dye (80% glycerol, 0.02% bromophenol blue) was added and 4uL of the mixture was loaded on a 7 × 8 × 0.1 cm, 0.25M TBE / 20% acrylamide gel (preparation described previously (Brown et al., 2008)). The gel was electrophoresed at 100V for 110 min, dried, exposed to a phosphor screen and analyzed by radiometry.
TarF reactions (3µL) were performed using 500nM enzyme (S. aureus or B. subtilis W23) with 3µM 4b, 18µM [14C]-1 and 18µM 1 in 100mM NaCl, 20mM Tris pH 7.5, and 10mM MgCl2. Reactions were quenched with 3µL DMF after 30 minutes or 12 hours at room temperature. Reactions containing the same composition, but with 2µM enzyme were allowed to proceed for 12 hours at room temperature. Identical reactions using heat-treated enzyme served as negative controls. 2uL of 4× loading dye was added and 8uL of the mixture was loaded on a 16 × 16 × 0.1 cm 0.25M TBE / 20% acrylamide gel. After running for 260 minutes at 225V the gel was dried, exposed to phosphor screen and analyzed by radiometry.
TarLBs reactions (3µL) were performed using 5µM enzyme with 2µM 4b, 5b, 6b, or 7b and 200µM 2. TarKBs reacting in tandem with TarLBs (3µL reactions) contained 1µM of each enzyme with 2µM 4b, 5b, or 6b and 200µM 2. Reaction buffer was 100mM NaCl, 20mM Tris pH 7.5, and 30mM MgCl2. Each substrate combination was also incubated with a heat-treated enzyme to serve as a negative control. After 2 hours at room temperature the reactions were quenched with 3µL DMF and analyzed as described for TarFBs substrate specificity reactions.
Q-TOF LC/MS Analysis
All analysis was performed on an Agilent 6520 Q-TOF LC/MS using a Phenomenex Gemini 5 micron C18 100 A column (50 mm × 4.6 mm) at a flow rate of 1 mL/min (solvent A: water; solvent B: methanol both with 0.1% ammonium hydroxide as a solvent modifier) with a linear gradient from 0 to 100% B over 10 minutes. Detailed reaction conditions and purification procedures are located in Supplemental Information.
Construction of S. aureus tarF deletion strain
The tarF deletion cassette was obtained by assembly PCR of two amplicons encoding approximately 1 kB DNA regions flanking the tarF open reading frame (P1tarF-P2tarF and P3tarF-P4tarF), digested with ApaI, and ligated into pKOR1 (digested with ApaI/EcoRV) to generate pKOR1–223. The deletion vector was introduced into strain JT215 by electroporation (Schenk and Laddaga, 1992), and the tarF deletion strain JT230 was subsequently isolated by double crossover as described (Meredith et al., 2008).
Construction of B. subtilis W23 tarF deletion strain
The marked tarF gene deletion in B. subtilis W23 was made using a protocol developed for B. subtilis 168 gene deletion (Jarmer et al., 2002). Assemby of the linear fragment, Z, containing the CAT cassette flanked by 1kB DNA flanking of B. subtilis W23 tarF is described in Supplemental Information. The B. subtilis W23 strain to be transformed was streaked on LB/agar and grown overnight at 37°C. A single colony was used to inoculate a 5mL culture of B. subtilis W23 and grown overnight at 37°C in modified Spizizen minimal salt medium with glutamate (Jarmer et al., 2002). At OD450 ~ 1, 50µL of cells were diluted into 5mL of fresh minimal media and grown at 37°C. At OD450 ~ 0.6, 200µL cells were incubated with at least 2µg of linear DNA Z. After 2 hours shaking at 37°C the cells were plated on LB/agar containing 5µg/mL chloramphenicol and incubated overnight at 37°C. A marked tarF gene deletion was confirmed by PCR analysis using primers that anneal outside the deletion region.
WTA extraction and degradation
Wall teichoic acids were extracted and broken into monomeric units following a combination of previously published protocols (Bernal et al., 2009; Sadovskaya et al., 2004; Tomita et al., 2009; Wickham et al., 2009). A detailed protocol can be found in Supplemental Information. Briefly, the cell pellet was disrupted by sonication. Noncovalently bound components were removed from the cell wall by incubation with SDS and covalently bound protein and nucleic acids were removed by treatment with trypsin, DNase, and RNase. The WTA was extracted by 10% trichloroacetic acid. Following centrifugation to remove peptidoglycan, the WTAs were precipitated in 95% ethanol. To prepare monomeric units, the WTAs were treated with 1N NaOH at 100°C for 3 hours. The yield was between 5 to 9mg from 500mL cultures. The monomeric units were analyzed by Q-TOF MS using direct inject with Solvent A (described above) at a flow rate of 0.2 mL/min.
RT-PCR
3 mL of B. subtilis W23 culture at an OD600 of ~0.8 was harvested by centrifugation, resuspended in RNALater reagent (Qiagen) and stored at −20°C until further use. RNA was extracted from the cell pellets using the Qiagen RNeasy Mini Kit with an initial treatment of 0.3µg/mL lysostaphin and 100µg/mL Proteinase K. RQ1 DNase (Promega) was used to degrade DNA. 400ng of RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) using random hexamer primers. RNA without reverse transcriptase treatment was used to detect potential DNA contamination. 0.3 – 5 µL of this reaction was used as template for PCR using KOD Hot Start (Novagen). Amplification of a fragment of the 16s rRNA transcript was used as an internal reference to ensure equal amounts of cDNA template in each PCR reaction. RT-PCR was performed in both biological and analytical duplicate. PCR of B. subtilis W23 genomic DNA was performed to confirm the primers are able to amplify the desired fragment of cDNA. PCR products were analyzed by electrophoresis on an ethidium bromide stained 2% agarose gel. The primers used are listed in Supplemental Information.
Highlights
-
--
B. subtilis and S. aureus make polyribitol wall teichoic acids by different pathways
-
--
B. subtilis W23 TarF is not involved in polyribitol wall teichoic acid biosynthesis
-
--
B. subtilis W23 TarF has a different enzymatic function from S. aureus TarF
Supplementary Material
Acknowledgements
We would like to thank Yuriy Rebets and David Rudner for helpful discussions on gene deletions; Emma Doud for assistance with mass spectrometry analysis; Jennifer Campbell for allowing us to modify her WTA schematic; and Deborah Perlstein for a critical reading of this manuscript. This research was funded by the NIH (1P01AI083214 and 5R01M078477 to S.W., and F3178727 to J.G.S.).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badurina DS, Zolli-Juran M, Brown ED. CTP:glycerol 3-phosphate cytidylyltransferase (TarD) from Staphylococcus aureus catalyzes the cytidylyl transfer via an ordered Bi-Bi reaction mechanism with micromolar K(m) values. Biochim Biophys Acta. 2003;1646:196–206. doi: 10.1016/s1570-9639(03)00019-0. [DOI] [PubMed] [Google Scholar]
- Bernal P, Zloh M, Taylor PW. Disruption of D-alanyl esterification of Staphylococcus aureus cell wall teichoic acid by the {beta}-lactam resistance modifier (−)-epicatechin gallate. J Antimicrob Chemother. 2009;63:1156–1162. doi: 10.1093/jac/dkp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, D'Elia MA, Sahakian TD, Brown ED. The Amino terminus of Bacillus subtilis TagB possesses separable localization and functional properties. J Bacteriol. 2007;189:6816–6823. doi: 10.1128/JB.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Truant R, Brown ED. The TagB protein in Bacillus subtilis 168 is an intracellular peripheral membrane protein that can incorporate glycerol phosphate onto a membrane-bound acceptor in vitro. J Biol Chem. 2005;280:36691–36700. doi: 10.1074/jbc.M507154200. [DOI] [PubMed] [Google Scholar]
- Brown S, Zhang YH, Walker S. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem Biol. 2008;15:12–21. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Men H, Ha S, Ye XY, Brunner L, Hu Y, Walker S. Intrinsic lipid preferences and kinetic mechanism of Escherichia coli MurG. Biochemistry. 2002;41:6824–6833. doi: 10.1021/bi0256678. [DOI] [PubMed] [Google Scholar]
- Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2007;46:14342–14348. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia MA, Henderson JA, Beveridge TJ, Heinrichs DE, Brown ED. The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:4030–4034. doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia MA, Millar KE, Beveridge TJ, Brown ED. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol. 2006a;188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol. 2006b;188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endl J, Seidl HP, Fiedler F, Schleifer KH. Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch Microbiol. 1983;135:215–223. doi: 10.1007/BF00414483. [DOI] [PubMed] [Google Scholar]
- Endl J, Seidl PH, Fiedler F, Schleifer KH. Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch Microbiol. 1984;137:272–280. doi: 10.1007/BF00414557. [DOI] [PubMed] [Google Scholar]
- Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect Immun. 2006;74:4164–4171. doi: 10.1128/IAI.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem. 2008;283:34596–34604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Gotz F, Zahringer U, Peschel A. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol. 2007;65:1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone A, Carballido-Lopez R, Noirot P, Errington J, Scheffers DJ. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J Bacteriol. 2008;190:1812–1821. doi: 10.1128/JB.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg C, Zhang YH, Yuan Y, Walker S. In vitro reconstitution of two essential steps in wall teichoic acid biosynthesis. ACS Chem Biol. 2006;1:25–28. doi: 10.1021/cb0500041. [DOI] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189:2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmer H, Berka R, Knudsen S, Saxild HH. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett. 2002;206:197–200. doi: 10.1111/j.1574-6968.2002.tb11009.x. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Abellan FX, Moller SB, Karamata D, Mauel C. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology. 2002;148:815–824. doi: 10.1099/00221287-148-3-815. [DOI] [PubMed] [Google Scholar]
- Lovering AL, Lin LY, Sewell EW, Spreter T, Brown ED, Strynadka NC. Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat Struct Mol Biol. 2010;17:582–589. doi: 10.1038/nsmb.1819. [DOI] [PubMed] [Google Scholar]
- May JF, Splain RA, Brotschi C, Kiessling LL. A tethering mechanism for length control in a processive carbohydrate polymerization. Proc Natl Acad Sci U S A. 2009;106:11851–11856. doi: 10.1073/pnas.0901407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JJ, Finking R, Wiegeshoff F, Weber TT, Bandur N, Koert U, Marahiel MA. Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall. FEBS J. 2005;272:2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- Men HB, Park P, Ge M, Walker S. Substrate synthesis and activity assay for MurG. Journal of the American Chemical Society. 1998;120:2484–2485. [Google Scholar]
- Meredith TC, Swoboda JG, Walker S. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J Bacteriol. 2008;190:3046–3056. doi: 10.1128/JB.01880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura LK, Roberts MS, Cohan FM. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int J Syst Bacteriol. 1999;49(Pt 3):1211–1215. doi: 10.1099/00207713-49-3-1211. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol. 2009;191:141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MP, Brown ED. Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus. Biochemistry. 2004;43:11802–11812. doi: 10.1021/bi048866v. [DOI] [PubMed] [Google Scholar]
- Pereira MP, D'Elia MA, Troczynska J, Brown ED. Duplication of teichoic acid biosynthetic genes in Staphylococcus aureus leads to functionally redundant poly(ribitol phosphate) polymerases. J Bacteriol. 2008a;190:5642–5649. doi: 10.1128/JB.00526-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MP, Schertzer JW, D’Elia MA, Koteva KP, Hughes DW, Wright GD, Brown ED. The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. Chembiochem. 2008b;9:1385–1390. doi: 10.1002/cbic.200800026. [DOI] [PubMed] [Google Scholar]
- Pollack JH, Neuhaus FC. Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J Bacteriol. 1994;176:7252–7259. doi: 10.1128/jb.176.23.7252-7259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr Res. 2004;339:1467–1473. doi: 10.1016/j.carres.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28:830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell EW, Pereira MP, Brown ED. The wall teichoic acid polymerase TagF is non-processive in vitro and amenable to study using steady state kinetic analysis. J Biol Chem. 2009;284:21132–21138. doi: 10.1074/jbc.M109.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldo B, Lazarevic V, Karamata D. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology. 2002;148:2079–2087. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- Swoboda JG, Campbell J, Meredith TC, Walker S. Wall Teichoic Acid Function, Biosynthesis, and Inhibition. Chembiochem. 2009a doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem Biol. 2009b;4:875–883. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Furihata K, Nukada T, Satoh E, Uchimura T, Okada S. Structures of two monomeric units of teichoic acid prepared from the cell wall of Lactobacillus plantarum NRIC 1068. Biosci Biotechnol Biochem. 2009;73:530–535. doi: 10.1271/bbb.80582. [DOI] [PubMed] [Google Scholar]
- Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, Lasa I. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology. 2008;154:865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E, Sadovskaya I, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus aureus MN8m, a biofilm forming strain. Carbohydr Res. 2006;341:738–743. doi: 10.1016/j.carres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Ward JB. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol Rev. 1981;45:211–243. doi: 10.1128/mr.45.2.211-243.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR, Bayer AS. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis. 2005;191:1771–1777. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- Wickham JR, Halye JL, Kashtanov S, Khandogin J, Rice CV. Revisiting magnesium chelation by teichoic acid with phosphorus solid-state NMR and theoretical calculations. J Phys Chem B. 2009;113:2177–2183. doi: 10.1021/jp809313j. [DOI] [PubMed] [Google Scholar]
- Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2009 doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. Better substrates for bacterial transglycosylases. J Am Chem Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Miyashita T, Araki Y, Ito E. Structure and functions of linkage unit intermediates in the biosynthesis of ribitol teichoic acids in Staphylococcus aureus H and Bacillus subtilis W23. Eur J Biochem. 1986;161:479–489. doi: 10.1111/j.1432-1033.1986.tb10469.x. [DOI] [PubMed] [Google Scholar]
- Zhang YH, Ginsberg C, Yuan Y, Walker S. Acceptor substrate selectivity and kinetic mechanism of Bacillus subtilis TagA. Biochemistry. 2006;45:10895–10904. doi: 10.1021/bi060872z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.