SUMMARY
c-Myc (Myc) is an important transcriptional regulator in embryonic stem (ES) cells, somatic cell reprogramming, and cancer. Here, we identify a Myc-centered regulatory network in ES cells by combining protein-protein and protein-DNA interaction studies, and show that Myc interacts with the NuA4 complex, a regulator of ES cell identity. In combination with regulatory network information, we define three ES cell modules (Core, Polycomb, and Myc), and show that the modules are functionally separable, illustrating that the overall ES cell transcription program is comprised of distinct units. With these modules as an analytical tool, we have reassessed the hypothesis linking an ES cell signature with cancer or cancer stem cells. We find that the Myc module, independent of the Core module, is active in various cancers and predicts cancer outcome. The apparent similarity of cancer and ES cell signatures reflects in large part the pervasive nature of Myc regulatory network.
INTRODUCTION
The pluripotent state of embryonic stem (ES) cells is maintained through the combinatorial actions of core transcription factors, including Oct 4, Sox 2, and Nanog (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008; Loh et al., 2006), in addition to other regulatory mechanisms encompassing epigenetic regulation (Boyer et al., 2006; Lee et al., 2006), microRNAs (Marson et al., 2008; Melton et al., 2010), and signaling pathways (Niwa et al., 1998; Sato et al., 2004). The discovery that cocktails of core pluripotency factors and selected widely expressed factors, such as Myc and Lin28, reprogram differentiated cells to an ES-like state (Park et al., 2008; Takahashi and Yamanaka, 2006; Yu et al., 2007) underscores the central role of transcription factors in cell fate decisions (Graf and Enver, 2009). Comprehensive protein interaction and target gene assessment of core pluripotency factors has provided a framework for conceptualizing the regulatory network that supports the ES cell state. Striking among the features of this network is the extent to which the core factors physically associate within protein complexes, co-occupy target genes, and cross-regulate each other (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008; Loh et al., 2006; Wang et al., 2006).
Although its expression dramatically enhances induced pluripotent (iPS) cell formation, Myc is not an integral member of the core pluripotency network (Chen et al., 2008; Hu et al., 2009; Kim et al., 2008). Myc occupies considerably more genomic target genes than the core factors and Myc targets are involved predominantly in cellular metabolism, cell cycle, and protein synthesis pathways, whereas the targets of core factors relate more towards developmental and transcription associated processes (Kim et al., 2008). Interestingly, promoters occupied by Myc show a strong correlation with a histone H3 lysine 4 trimethylation (H3K4me3) signature, and a reverse correlation with histone H3 lysine 27 trimethylation (H3K27me3), suggesting a connection between Myc and epigenetic regulation (Kim et al., 2008). It is notable that the H3K4me3 signature has a positive correlation with active genes, and an open chromosomal structure, a distinctive feature of ES cells (Meshorer et al., 2006). Studies in non-ES cells have also revealed that Myc interacts with histone acetyltransferases (HATs) (Doyon and Cote, 2004; Frank et al., 2003). Improved iPS cell generation by addition of histone deacetylase inhibitors implies that global changes in epigenetic signatures are critical to efficient somatic cell reprogramming (Huangfu et al., 2008).
While remaining pluripotent, ES cells are capable of indefinite self-renewal. Both blocked differentiation and the capacity for self-renewal, hallmarks of ES cells and adult stem cells, are shared in part by cancer cells (Clarke and Fuller, 2006; Reya et al., 2001). Although contested in the literature, expression of pluripotency factors, such as Oct 4, and Nanog, has been described in some cancers (Kang et al., 2009; Schoenhals et al., 2009). The involvement of Myc in many cancers (Cole and Henriksson, 2006), taken together with its effects in iPS cell generation, raises important issues regarding the relationship between cancer and embryonic stem cell states. Moreover, renewed focus on tumor subpopulations that initiate tumor formation on transfer to a suitable host (cancer stem cells) has contributed to the comparison of cancers and stem cells, and to the potential resemblance of metastatic cancer cells to stem cells.
These relationships have been reinforced by reports of "stem cell" or "embryonic stem cell" (ESC-like) signatures in human and mouse cancers (Ben-Porath et al., 2008; Wong et al., 2008a; Wong et al., 2008b). The properties of such "ESC-like signatures" have thus far not been clearly defined, leaving open the possibility that they are comprised of multiple gene expression signatures that are the outcomes of functionally independent transcriptional regulatory networks Cancer cells may share only one or few of these subdivided signatures observed in ES cells, and thus have relatively less in common with the "embryonic state" than recently suggested.
In the present study, we sought to define how the regulatory network controlled by Myc relates to the previously defined core pluripotency network (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008; Loh et al., 2006). We first identified a Myc-centered regulatory network in ES cells, and revealed that this Myc-centered network is largely independent of the core ES cell pluripotency network. Based on these findings, we propose that the overall ES cell specific gene expression signature is comprised of smaller sets of sub-signatures which are represented as ‘modules’ - modules for the core pluripotency factors (Core module); the Polycomb complex factors (PrC module); and the Myc related factors (Myc module). We provide evidence that these modules are functionally independent in ES cells, as well as during somatic cell reprogramming. With these modules as analytical tools, we observe that ES cells and cancer cells share Myc module activity, but generally do not share Core module activity. These findings argue against the hypothesis that cancer cells often reactivate an embryonic stem cell gene signature, even as they progress to a more highly invasive or metastatic state. Instead the common features of ES cells and cancer cells reflect in large part the pervasive nature of the Myc regulatory network.
RESULTS
Construction of a Myc-centered protein-protein interaction network in ES cells
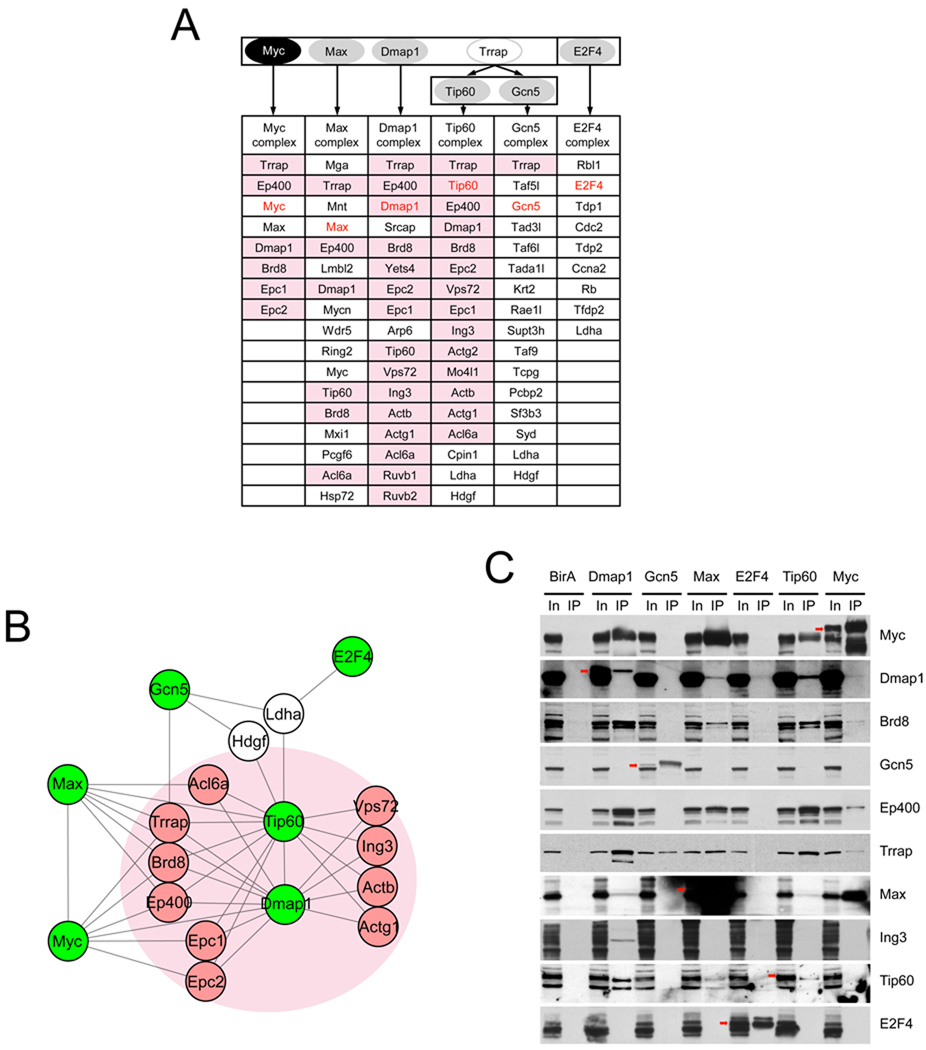
Previous protein-DNA interaction studies in ES cells indicated that targets occupied by the core pluripotency factors differ from genes bound by Myc (Chen et al., 2008; Kim et al., 2008). A recent RNA interference based functional screen additionally suggested the existence of a second network linked functionally with Myc (Hu et al., 2009). Since coregulators that function with Myc have not been characterized previously in ES cells, we first sought to identify protein complexes that contain Myc with Myc-associated factors in ES cells. Using the in vivo metabolic biotin tagging method (de Boer et al., 2003; Wang et al., 2006), protein complexes containing tagged Myc in ES cells were affinity purified and analyzed by mass-spectrometry. We identified several proteins known to interact with Myc in other cell types, including Max, Ep400, Dmap1 and Trrap (Figure 1A) (Cai et al., 2003; Fuchs et al., 2001; McMahon et al., 1998). To expand and validate the protein-protein interaction network encompassing Myc, we subsequently generated ES cell lines expressing tagged Max and tagged Dmap1. ES cells expressing tagged Tip60 and tagged Gcn5 were also generated because they are histone acetyltransfeases (HATs) and known interacting partners of Trrap (Ikura et al., 2000; McMahon et al., 2000). We also generated tagged E2F4 ES cells, since another E2F family member E2F1 shares many common targets with Myc (Chen et al., 2008). E2F1 and E2F4 have many common targets and interchangeable roles in normal and tumor cells (Xu et al., 2007). Among E2F family proteins, E2F4 shows strongest expression in ES cells. In summary, we established ES cell lines expressing tagged Myc, Max, Dmap1, Tip60, Gcn5, and E2F4 (Figure 1A and Figure S1A), and identified their interacting partner proteins (summarized in Table S1). Figure 1A shows lists of high confidence interacting partner proteins of each factor tested. Interactions were independently validated by co-immunoprecipitation (Figure 1C and Figure S1B).
Figure 1. Myc-centered protein-protein interaction network in ES cells.
(A) Schematic representation of the strategy for mapping a Myc-centered protein–protein interaction network in ES cells. High-confidence components of multiprotein complexes were identified and listed in the table. Pink cells represent NuA4 complex proteins.
(B) Depiction of the features of the Myc-centered protein-protein interaction network. Proteins with green labels are biotin tagged proteins and pink circles indicate NuA4 complex proteins Proteins identified by multiple biotin tagged factors are shown. Entire protein interaction network is shown in Figure S1C. See also Table S1.
(C) Validation of the interaction network by co-immunoprecipitation.
Myc interacts with the NuA4 HAT complex in ES cells
We did not observe overlap of proteins existing between the core protein interaction network (Wang et al., 2006) and the Myc-centered protein interaction network (Figure S1C). Although this may be due to the stringency of our conditions for recovery of protein complexes, within each network we observed a high degree of interactions, strongly suggesting that these two networks, and their protein complexes, are physically separate. Interestingly, we observed that Myc interacts with many proteins in a recognized conserved protein complex known as NuA4 HAT (or the Tip60-Ep400 complex) (Doyon and Cote, 2004) as shown in Figure 1A (pink cells) and Figure 1B (proteins in a pink circle). Myc, Max, Dmap1, Tip60, Trrap, and Ep400 are tightly interconnected within the network; however, Gcn5 and E2F4 show a lower degree of association, suggesting their weak or indirect interaction with Myc/NuA4. It has been suggested that transcription factors, such as Myc, p53, and E2Fs, require the NuA4 complex to activate downstream targets in non-ES cell contexts (Ard et al., 2002; McMahon et al., 1998). Our data (Figure 1 and Table S1) strongly support the view that Myc interacts with an intact NuA4 HAT complex in ES cells, also implying that histone 3 and 4 acetylation (AcH3, and AcH4, respectively) signatures may also be generated in part by the Myc/NuA4 complex via Tip60 in ES cells. Previous RNAi based phenotypic analyses in ES cells revealed that factors in the NuA4 HAT complex, including Ep400, Dmap1, Tip60, Trrap, Ruvb1 and Ruvb2, are critical to ES cell identity (Fazzio et al., 2008) (also our observation, Figure S1D and S1E). These findings imply a crucial role for the Myc/NuA4 complex in ES cells.
Construction of a Myc-centered protein-DNA interaction network in ES cells
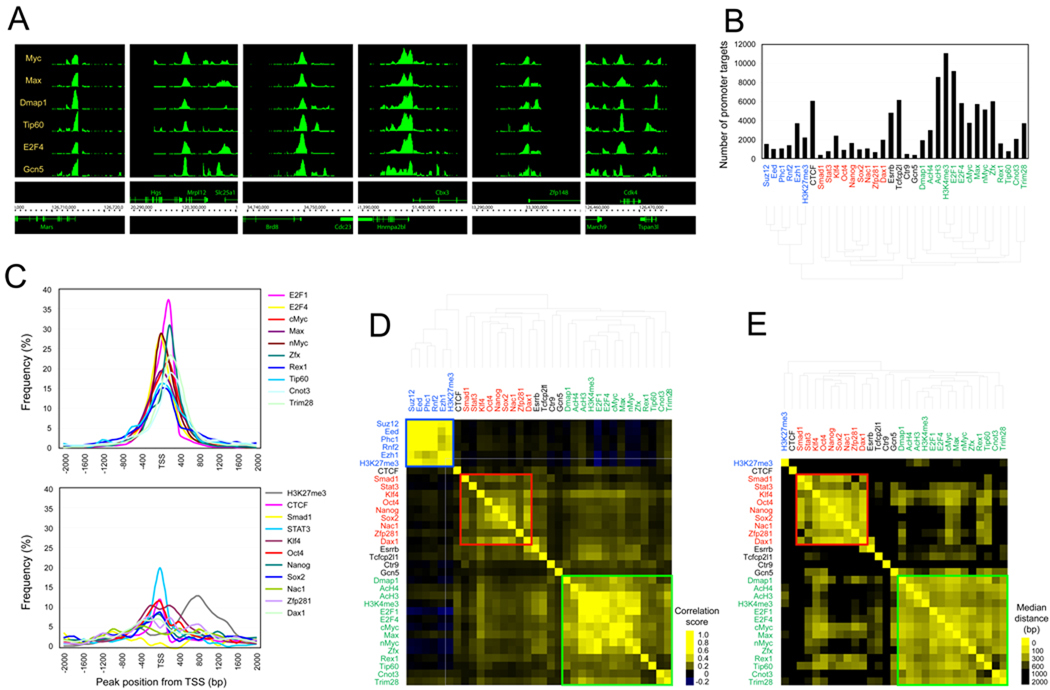
To identify genomic targets of Myc and its associated factors tested in Figure 1, we performed bioChIP-chip (Kim et al., 2008). Since Tip60 and Gcn5 generate AcH3 and AcH4 histone modification signatures, we also performed ChIP reactions using native antibodies against AcH3 and AcH4. We found that the six factors we tested (Myc, Max, Dmap1, Tip60, E2F4, and Gcn5) co-occupy many target promoters in close proximity (Figure 2A).
Figure 2. Myc-centered protein-DNA interaction network in ES cells.
(A) Representative view of Myc, Max, Dmap1, Tip60, E2F4, and Gcn5 occupancy at the target loci.
(B) Number of target promoters bound by each factor or associated with each histone modification signature. Blue represents factors or histone signatures involved in PrC complexes. Red represents factors involved in ES cell core factors, and Green represents Myc and Myc-related factors or histone signatures (D–E). See also Figure S2 and Table S2.
(C) Relative position of chromosomal target loci of each factor in the Myc cluster (upper panel) and Core cluster (bottom panel) shown in (B) and (C) to the TSS.
(D) Target correlation map: The degree of target co-occupancy of each pair of factors (either transcription factor or histone modification signature) is shown. Yellow indicates more frequent colocalization of each pair of factors.
(E) Median distance map: Median distances between the loci co-occupied by two tested factors (except PrC complex proteins) shown in (D). Yellow indicates closer colocalization.
To obtain a global view of individual and multiple transcription factor occupancy, we combined this new data set with previously published ChIP-chip or ChIP-sequencing data sets (Boyer et al., 2006; Chen et al., 2008; Ding et al., 2009; Hu et al., 2009; Kim et al., 2008; Shen et al., 2008), and tested the factor occupancy or histone modification signatures (see Supplemental Data). The numbers of genes that are occupied by a tested factor or marked by a tested histone modification signature are summarized in Figure 2B and Table S2 with a hierarchical clustering image based on target co-occupancy. We then calculated the degree of target co-occupancy of each pair of factors. As shown in the target correlation map in Figure 2D, we observed three major clusters. Factors in Polycomb complexes are associated with the H3K27me3 signature to form a distinct cluster (PrC cluster, blue-colored box in Figure 2D and blue letters in Figure 2B and 2D). Core pluripotency factors, including Nanog, Sox2 and Oct4 and others, form an independent cluster (Core cluster, red-colored box and red letters). Myc forms a cluster with other factors and AcH3, AcH4, and H3K4me3 signatures (Myc cluster, Green-colored box and green letters).
We calculated the median distances between binding peaks of each pair of factors using the same cluster information shown in Figure 2D (except for the PrC cluster due to availability of the processed data). The target distance map demonstrates that the factors within the Core or Myc clusters regulate their common targets in close proximity, whereas the factors belonging to a different cluster regulate their common targets in a relatively remote manner (Figure 2E).
Previously, we observed that Myc occupies more target genes than the ES cell core factors (Kim et al., 2008). Similarly, we observed that the factors in the Myc cluster, such as Max, nMyc, E2F4, and Dmap1, tend to occupy more targets than factors in the Core or PrC clusters (Figure 2B), suggesting more global roles in their target gene regulation. The majority of binding peaks generated by the factors in the Myc cluster are more centered at the transcription start site (TSS) compared to the target binding peaks of the factors in the Core cluster (Figure 2C). The factors in the Myc cluster may interact with basal transcription machinery, whereas core factors have both promoter and upstream enhancer targets as described (Chen et al., 2008; Loh et al., 2006). In summary, our data suggest that the factors belonging to each of the distinct clusters (Core, PrC, and Myc) regulate their own rather similar downstream targets in close proximity and may be functionally separated in regulating aspects of ES cell identity.
Target co-occupancy of factors within the Myc cluster has a positive correlation with histone H3 and H4 acetylation signatures
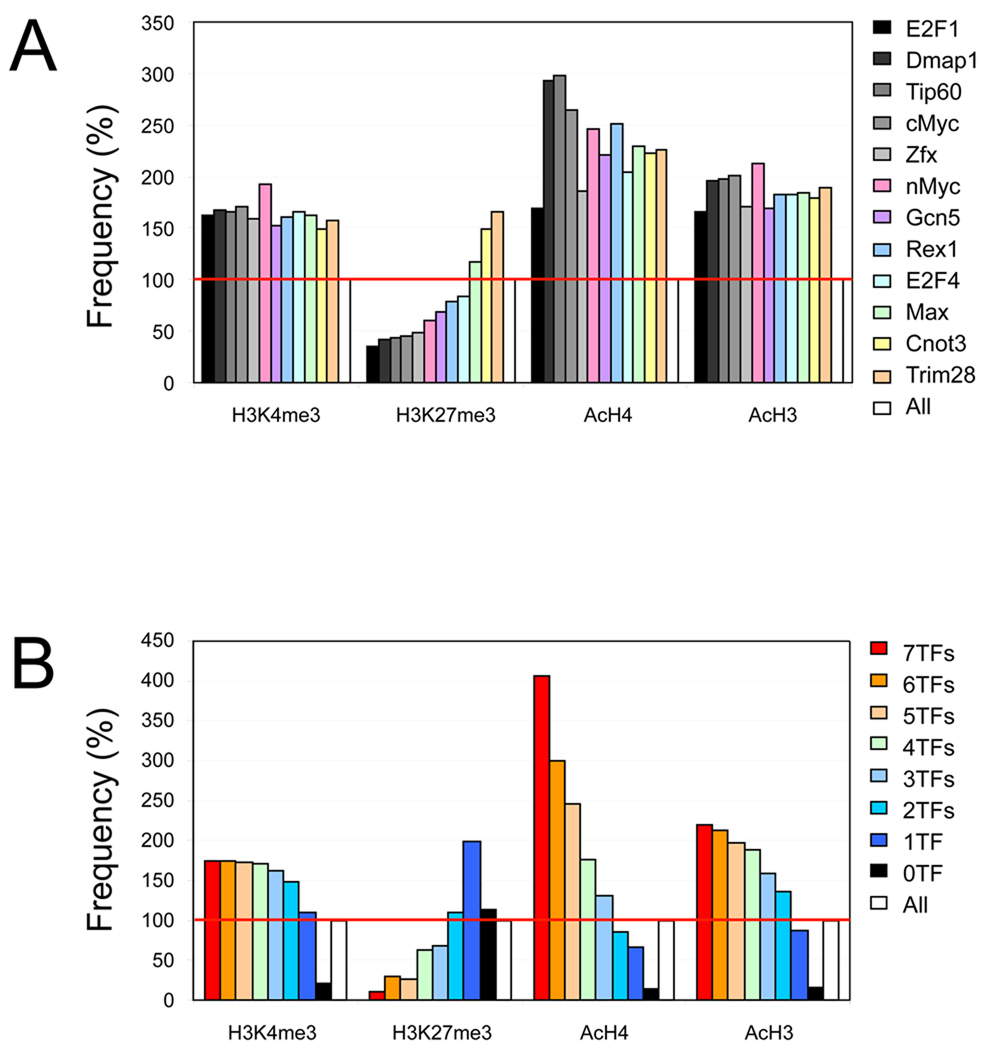
Our prior work revealed that Myc target promoters correlate positively with an active H3K4me3 signature, and negatively with a repressive H3K27me3 signature (Kim et al., 2008). Since Myc is associated with histone acetylation (Frank et al., 2001), we tested the correlation between target occupancy of each factor in the Myc cluster and the histone modification status of their target promoters. As shown in Figure 3A, the majority of the factors in the Myc cluster harbor significantly higher levels of H3K4me3, AcH3, and AcH4 signatures on their target promoters over background (at least >150%). On the contrary, the H3K27me3 signature is significantly underrepresented on the target promoters of approximately half of the factors in the Myc cluster. Interestingly, Cnot3 and Trim28 target promoters show bivalent modifications (both H3K4me3 and H3K27me3 positive), suggesting that, although these factors share many common targets with Myc, they may have different functions compared to the other factors in the cluster.
Figure 3. Histone modification signatures on the target promoters of Myc cluster proteins.
(A) Histone marks on the target promoters of each factor in the Myc cluster. “All” represents all promoters.
(B) Histone marks and target co-occupancy of 7 factors in Myc cluster (Myc, Max, nMyc, Dmap1, E2F1, E2F4, and Zfx).
Additionally, we tested the relationship between the factor co-occupancy (7 factors in the Myc cluster shown in Figure 2D and Figure 2E, including Myc, Max, nMyc, Dmap1, E2F1, E2F4, and Zfx) and histone modification signatures. As shown in Figure 3B, target promoters co-occupied by multiple factors in the Myc cluster show a higher level of histone acetylation than the common targets of fewer factors. Targets occupied by 7 factors show approximately 400%, and 220% of AcH4, and AcH3 signatures, respectively, over the background level. Upon the decrease of co-occupancy, the level of these signatures decreased on their common targets. We failed to observe correlation between co-occupancy and the H3K4me3 signature, presumably due to the abundance of H3K4me3 marks across many promoters (>60 % of all promoters)(Kim et al., 2008). The repressive signature H3K27me3 displays a reverse correlation with the Myc cluster factor co-occupancy (Figure 3B).
Modules defined by transcriptional regulatory subnetworks in ES cells
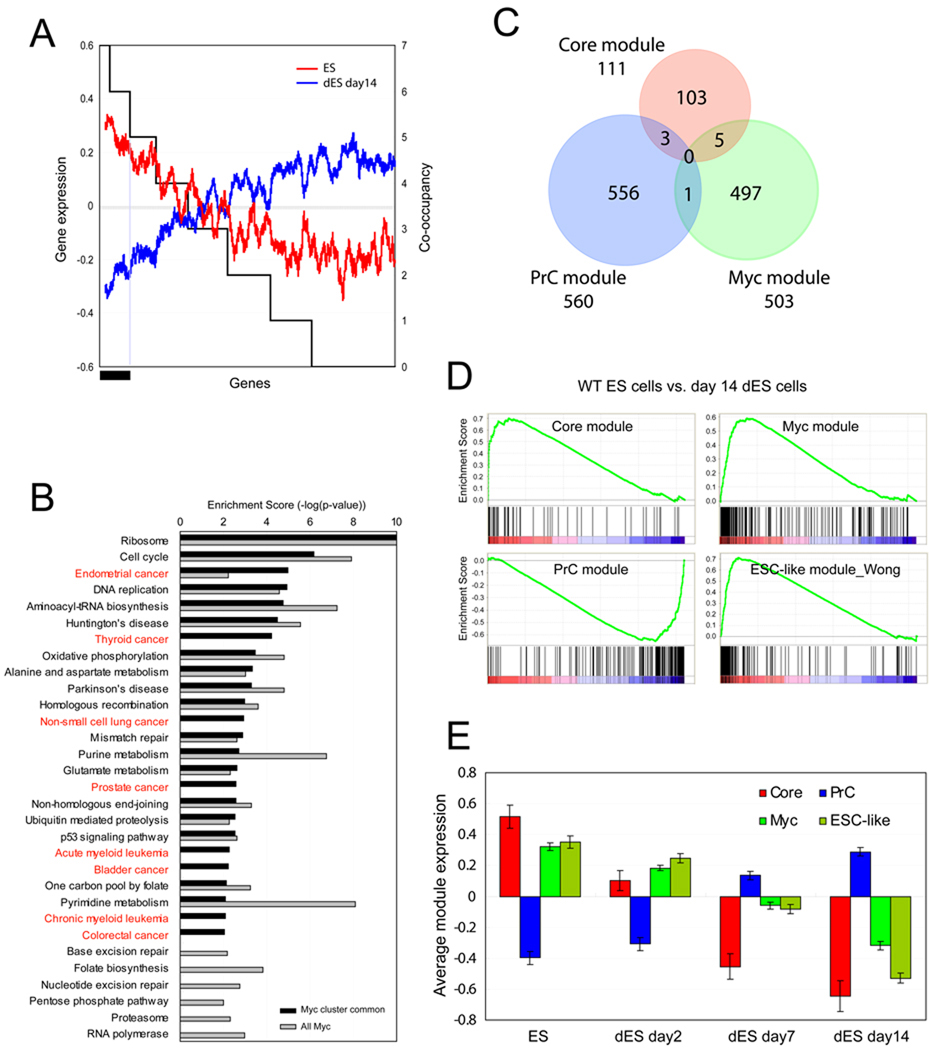
Since we observed a strong positive correlation between target co-occupancy of the factors in the Myc cluster and histone acetylation signatures, we examined the correlation between target co-occupancy and gene expression. As shown in Figure 4A, targets co-occupied by 7 or 6 factors in the Myc cluster are more active than the common targets of five or fewer factors in ES cells (red line), and repressed upon differentiation (blue line). To test if the information generated from the factor co-occupancy in the Myc cluster is functionally relevant, we compared KEGG pathways (Dennis et al., 2003; Ogata et al., 1999) enriched in the genes that are common targets of at least 6 factors among 7 factors in the Myc cluster (Myc, Max, nMyc, Dmap1, E2F1, E2F4, and Zfx; black bar in Figure 4A), and the global target genes of Myc. Many cancer-related pathways (red letters in Figure 4B and Table S3) are enriched in the genes co-occupied by the factors in the Myc cluster. In contrast, these cancer-related pathways are not enriched within the global set of genes occupied by Myc. This observation strongly suggests that factor co-occupancy in the Myc cluster does not represent a random subset of Myc targets, and may provide additional information in understanding the combinatorial function of factors in the Myc cluster in ES cells and in cancer cells (Figure 4B).
Figure 4. ES cell modules.
(A) Gene expression profiles (log2, left y-axis) upon J1 ES cell differentiation (wild type ES cells: ES, differentiated ES cells for 14 days: dES day 14) are shown as moving window averaged lines (ES; red line, dES day 14; blue line, bin size 100 and step size 1). Randomized genes are sorted (x-axis) by the target co-occupancy of 7 factors in the Myc cluster (right y-axis). Black bar represents target genes co-occupied by at least 6 factors among the 7 factors in the Myc cluster.
(B) Enrichment of KEGG pathways. All Myc target genes (gray bars, total 3733 genes) and genes co-occupied by at least 6 factors among the 7 factors tested marked by black bar in (A) (black bars, total 1756 genes). See also Figure S3A, S3B and Table S3.
(C) ES cell modules: Three ES cell modules are defined based on the target co-occupancy within each cluster shown in Figure 2D. See also Table S3.
(D) GSEA analyses show the gene activity of the three ES cell modules (Core, PrC and Myc modules) as well as the previously defined ESC-like module (Wong et al. 2008a) upon ES cell differentiation (wild type ES cells; ES vs. 14 days differentiated ES cells; dES).
(E) Average gene expression values (log2) of each module (C) are tested upon ES cell differentiation (ES day0, dES day2, dES day7, and dES day14, respectively). Data are represented as mean ± SEM. See also Figure S2B and Figure S3C.
We previously demonstrated that common targets of multiple factors in the core pluripotency network are significantly active in ES cells. However, when these factors occupy targets alone or with few factors they are not associated with activation of target genes (Kim et al., 2008). Since the targets co-occupied by 7 factors in the Myc cluster show the strongest gene activity (Figure 3B and Figure 4A), we classified common target gene ‘modules’ in ES cells based on the target co-occupancy within the clusters shown in Figure 2; the PrC module, the Core module, and the Myc module (Figure 4C and listed in Table S3). Definition of each module is as follows; the Core module is comprised of genes co-occupied by at least 7 factors among 9 factors shown in the Core cluster (Smad1, Stat3, Klf4, Oct4, Nanog, Sox2, Nac1, Zfp281, and Dax1), depicted in the red box in Figure 2D. The PrC module genes are the common targets of PrC cluster proteins, Suz12, Eed, Phc1, and Rnf2 (blue box in Figure 2D). The Myc module is comprised of genes that are common targets of 7 factors (Myc, Max, nMyc, Dmap1, E2F1, E2F4, and Zfx) in the Myc cluster (green box in Figure 2D). For construction of the Myc module, we excluded Tip60, Gcn5, and Rex1 due to their relatively small number of targets (Figure 2B), and Trim28, and Cnot3 due to the bivalent signature on their target promoters (Figure 3A), as well as the discrepancy of their target similarity within the Myc cluster (Figure S2A). Additional gene sets co-occupied by different combinations of factors in Myc cluster were also tested but showed no significant difference, because the majority of target genes among the tested sets are shared (see below and Figure S2B). Lists of gene sets tested are summarized in Table S3. Indeed, the Core module includes previously known factors in core regulatory circuitry, such as Nanog, Oct4, Rest, Sox2, Tcf3, and Rex1. The PrC module includes genes generally repressed in ES cells, including Hox cluster genes. As shown in Figure 4C, the overlap between each module is minimal and they are involved in different pathways (Figure S3A and S3B).
We then tested activity of each module (hereafter ‘module activity’, averaged expression of all genes in each module in a given expression data set) in ES cells as compared to module activity in differentiated cells. Gene activity of a previously identified ‘Core ESC-like gene module’(hereafter ‘ESC-like module’) (Wong et al., 2008a) was also tested. Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) revealed that the Core, Myc, and previously identified ESC-like modules are highly active in ES cells. As anticipated, the PrC module is repressed in ES cells (Figure 4D). We additionally tested the activity of each module during a time-course of ES cell differentiation. As shown in Figure 4E and Figure S3C, in ES cells the Core module is most active, yet the Myc and ESC-like modules show some activity; these modules become repressed with time during differentiation, whereas the PrC module shows an opposite pattern.
Functional separation of Core and Myc modules in partial iPS cells
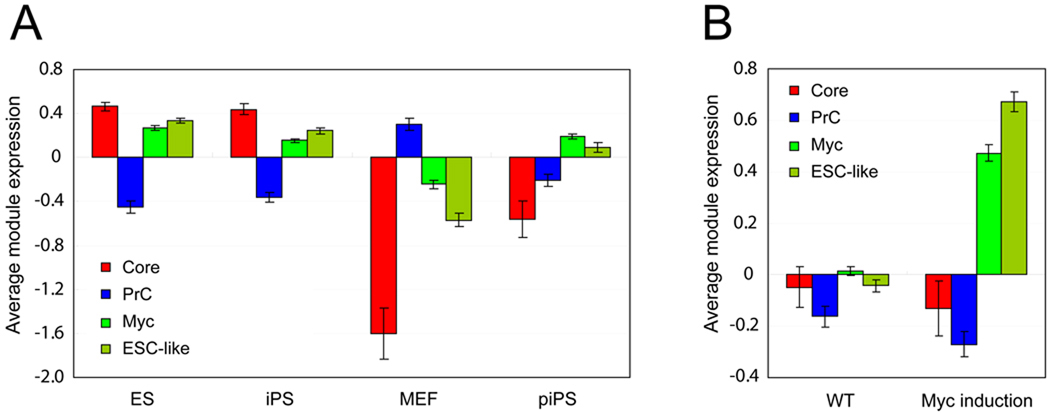
Although we observed that both the Core and Myc modules are active in ES cells, the genes that comprise the Core module are distinct from those of the Myc module (Figure 4C). To test if the modules can be functionally separable, we tested the module activity of our three ES cell modules, along with the ESC-like module (Wong et al., 2008a) in other cell types, including iPS cells, partial iPS (piPS) cells, and mouse embryonic fibroblasts (MEFs). Global gene expression profiles of ES and iPS cells are highly similar (Takahashi and Yamanaka, 2006). Relying on a publicly available data set (Sridharan et al., 2009), we tested if the module activity is similar between ES and iPS cells. Similar to the data shown in Figure 4E, the Core and Myc modules are highly active in both ES and iPS cells (Figure 5A and Figure S3D). The PrC module is inactive in both cell types, as expected. In MEFs, the module activity pattern is similar to the module activity shown in differentiated ES cells shown in Figure 4E, suggesting that strongly active Core and Myc modules, as well as an inactive PrC module, may characterize the pluripotent state of cells, such as ES and iPS cells.
Figure 5. Module activity in various cells.
(A) Average gene expression values (log2) (Sridharan et al., 2009) of ES cell modules (Core, Prc and Myc) and previously defined ESC-like module are tested in ES cells (ES), iPS cells (iPS), MEFs (MEF), and partial iPS cells (piPS). See also Figure S3D.
(B) Average gene expression values (log2) (Bild et al., 2006) of each module tested in (A) upon induction of Myc in human epithelial cells (Myc induction) and in control cells (WT). Human orthologues of genes in three ES cell modules are tested (listed in Table S3) and data are represented as mean ± SEM (A–B).
Previous work has shown that piPS cells exist at an intermediate stage in the reprogramming process (Maherali et al., 2007). The endogeneous ES cell core regulators Oct4 and Nanog are not re-activated in piPS cells, whereas they are reactivated in fully reprogrammed iPS cells. To test if the ES cell modules we have defined are functionally separable in piPS cells, we analyzed ES module activity using gene expression data from piPS cells (Figure 5A and Figure S3D) (Sridharan et al., 2009). We found that the activity of the Myc module in piPS cells is comparable to that in ES cells and iPS cells, but the Core module is not re-activated in piPS cells. These data demonstrate that the regulatory modules defined in ES cells may be considered functionally separable units, not arbitrary subdivisions of the overall ES cell signature. Of particular note, the ESC-like module (Wong et al., 2008a) shows similar module activity to our Myc module, but not to the Core module in piPS cells.
ES cell module activity in cancer
Others have described ESC-like gene modules (Wong et al., 2008a) or ES-cell like gene expression signatures (Ben-Porath et al., 2008) that have been widely used in assessment of cancer gene signatures. With the three ES cell modules we defined as new analytical tools, we readdress the relatedness of ES cell and cancer gene signatures as a series of case studies. For analyses of human data, human orthologues of mouse genes were used (Table S3).
Myc induction does not activate the Core module in human epithelial cells
We tested ESC-like modules (both Core ESC-like gene and mouse ESC-like gene modules) (Wong et al., 2008a) and found that they behave similarly to our Myc module in various settings (Figure 4E, Figure 5A and data not shown). Since we observed that our defined Core and Myc modules can be functionally separated in piPS cells (Figure 5A), we examined whether the induction of Myc may activate the Core module in a different cellular context. It has been reported previously that the induction of Myc activates the ESC-like module in adult human epithelial cells (Wong et al., 2008a). As shown in Figure 5B, upon reanalysis of this dataset (Bild et al., 2006), we find that the Core module is not activated following Myc induction, whereas the Myc module is strongly represented. In addition, core factors in ES cells, such as Nanog and Oct4, are also not activated by Myc induction (Figure S4A). These observations confirm that the Myc and Core modules are functionally separable, and also support the view that the overall ES cell expression signature can be subdivided into functionally distinct units. Our refined analysis argues against the prior conclusion that Myc induction leads to activation of an ESC-like gene module in human epithelial cells (Wong et al., 2008a).
ES cell modules in mouse MLL myeloid leukemia models
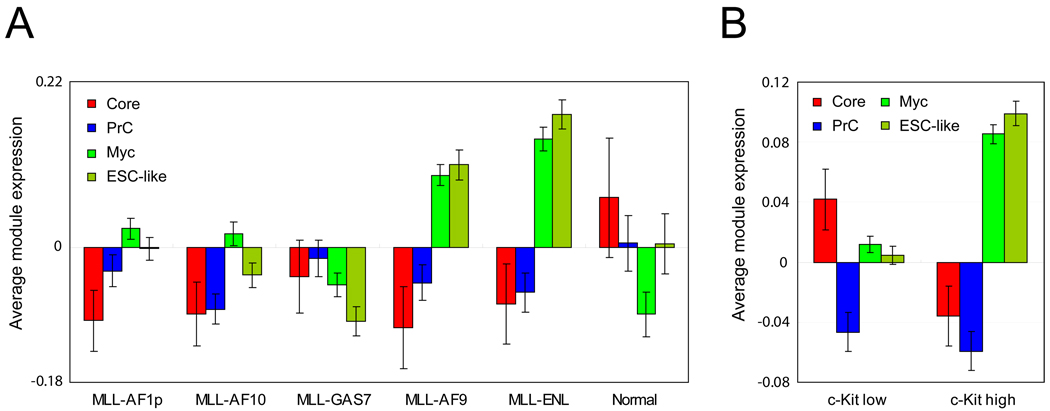
We have assessed the relevance of our ES cell modules within a mouse model of acute myeloid leukemia (AML). Expression of MLL alleles leading to expression of fusion products, such as MLL-AF9, MLL-ENL, MLL-AF10, MLL-AF1p, and MLL-GAS7, initiates leukemia. MLL associated leukemia models in mice have served as platforms for purifying and examining the gene expression profiles of leukemia stem cells (LSCs; also called LIC, leukemia initiating cells) (Krivtsov et al., 2006). It has also been suggested that LSCs are present at a higher frequency in leukemic mice in which AML was initiated by MLL-ENL or MLL-AF9 as compared with MLL-AF10, MLL-AF1p, and MLL-GAS7 (Somervaille et al., 2009). We tested the activity of our defined modules in these leukemias. We first observed that the Core module is not active in any of the AMLs as compared to the Core module activity of a control group (Figure 6A). Moreover, we failed to detect an active Core module in AMLs demonstrated to have high LSC frequency (MLL-ENL and MLL-AF9) (Figure 6A). In contrast, we observed active Myc module expression in high frequency LSC AMLs (MLL-ENL and MLL-AF9), but not in low frequency LSC AMLs (MLL-AF10, MLL-AF1p, and MLL-GAS7) or control.
Figure 6. ES cell modules in mouse MLL myeloid leukemia models.
(A) Average gene expression values (log2) of ES cell modules and the previously defined ESC-like module are tested in various mouse models of acute myeloid leukemia (AML) initiated by MLL-AF9, MLL-ENL, MLL-AF10, MLL-AF1p, and MLL-GAS7 (Somervaille et al., 2009). (B) Average gene expression values (log2) of each module are tested in a c-kit high MLL-AF10 leukemia cell population (MLL-AF10 c-kit high) and a c-kit low MLL-AF10 leukemia cell population (MLL-AF10 c-kit low) (Somervaille et al., 2009). Data are represented as mean ± SEM (A–B). See also Figure S4.
It has been reported that the previously defined ESC-like gene module (Wong et al., 2008a) is prominent in a MLL-AF10 leukemia cell population enriched for LSCs (c-kit high) as compared to c-kit low cells (Somervaille et al., 2009). As shown in Figure 6B, we observed a stronger Myc module activity in the LSC-enriched population. However, this cell population shows relatively inactive Core module activity. In both of the tests shown in Figure 6A and Figure 6B, we observed that the activity of the previously defined ESC-like gene module (Wong et al., 2008a) is similar to the activity of the Myc module rather than the Core module. If the gene expression findings are functionally relevant to self-renewal of LSCs, our findings undermine the notion that reactivation of an ESC-like pattern is critical for LSCs in this setting. In contrast, Myc module activity alone appears to correlate with LSC frequency in mouse AML models. Core module activity does not appear to be a major determinant of LSC frequency in AML.
ES cell modules in human cancers
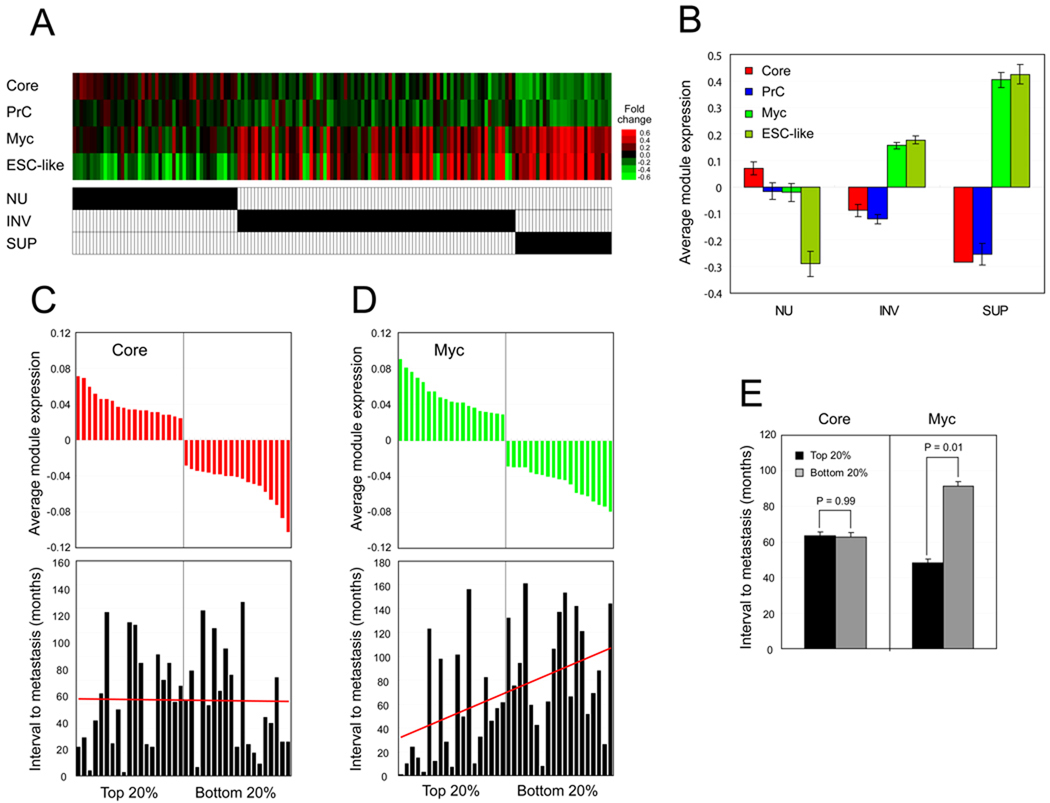
To test the activity of ES cell modules more generally, we tested module activity in gene expression profiles acquired from human bladder carcinoma samples including superficial and invasive carcinomas, as well as a control group of normal urinary tract cells (Sanchez-Carbayo et al., 2006). Figure 7A represents each module activity from total 157 patient samples (each column). Figure 7B represents combined module activity from different groups of patient samples. In both superficial and invasive carcinomas, the Myc module is more active compared to its level of activity in control samples. However, the Core module activity is repressed in both grades of cancers. Of note, we observed a more active Myc module in superficial carcinoma samples compared to more advanced stage of invasive carcinoma samples. Heterogeneity of invasive carcinoma samples may underlie this observation or the active Myc module may be critical in initiating invasive behavior, and not necessarily active afterwards. Importantly, the previously defined ESC-like gene module activity is again similar to the activity of the Myc module. However, the Core module seems to be even more repressed in carcinoma samples compared to control group (Figure 7A and Figure 7B).
Figure 7. ES cell modules in human cancers.
(A–B) Average gene expression values (log2) of ES cell modules and previously defined ESC-like module are tested in human bladder carcinoma samples including superficial (SUP), and invasive carcinomas (INV), as well as normal urothelium (NU) as a control group (marked by black bars) (Sanchez-Carbayo et al., 2006). Each column represents one patient sample (total 157 samples) (A). Averaged module activities within the sample group (NU, INV, and SUP) (B). Data represented as mean ± SEM.
(C–E) Average gene expression values (log2) of ES cell Core (C) and Myc (D) module are tested from 97 human breast cancer patient samples (van 't Veer et al., 2002). (C) Core module activities were calculated, and top and bottom 20% of samples (19 samples each) were further analyzed. Bar graph represents the corresponding interval to metastases (months, bottom panel). (D) Samples showing top and bottom 20% of Myc module activity were further analyzed. Bar graph represents the corresponding interval to metastases (months) for each patient (bottom panel). (E) For each tested group (C–D), interval to distant metastases is calculated as mean ± SEM, and p-values are from Student’s T-tests. See also Figure S5.
We next tested module activity within a human primary breast cancer expression data set (van 't Veer et al., 2002) containing fifty eight samples from patients who developed distant metastases within 5 years (poor prognosis group), and 39 samples from patients who continued to be disease free for at least 5 years (good prognosis group). First, we calculated Core module activity of all samples and further analyzed samples showing the strongest Core module activity (top 20% of samples, n=19), and the weakest Core module activity (bottom 20%, n=19). As shown in Figure 7C and Figure 7E, no correlation was observed between Core module activity and patient outcome (interval to metastasis). On the other hand, Myc module activity correlates positively with a poor prognosis (Figure 7D). On average, metastasis occurs within 47 months in breast cancer patients with strong Myc module activity (top 20%, n=19). In contrast, it took on average 89 months for the patients with weak Myc module activity (bottom 20%) to progress to metastasis (Figure 7E), suggesting that Myc module activity predicts patient outcome. We observed that Myc module activity in human breast cancer patient samples is very similar to the previously defined ESC-like modules (Wong et al., 2008a) (Figure S5A). Additional analyses using independent breast cancer data sets also revealed that tumor samples with a more active Myc module tend to be highly proliferative basal-like tumors (Figure S5B, S5C, and S5E; middle panel) or ER negative tumors (Figure S5D and S5E; left panel). These results are consistent with findings of others demonstrating a correlation of Myc activity with poor outcome in breast cancer (Wolfer et al., 2010). Interestingly, we observed that highly proliferative cells show stronger Myc module activity (Figure 6, Figure 7, Figure S4, and Figure S5), suggesting a link between the Myc module activity and cell proliferation.
DISCUSSION
By integrating protein-protein interaction and protein-DNA interaction studies, we constructed a Myc-centered transcriptional regulatory network in an effort to complement the previously identified core regulatory, and Polycomb networks in ES cells. Our approach, analyzed together with data of others, delineates three major transcriptional regulatory subnetworks in ES cells. Based on the target co-occupancy of factors in each network, we defined three functionally separable regulatory modules (Figure 4 and Figure 5), and showed that the overall ES cell gene transcription program can be subdivided largely into functionally independent regulatory units.
It is interesting to note that a previous RNAi-based screen revealed that members of the NuA4 HAT complex (or Tip40-Ep400 complex) are critical in ES cell identity (Fazzio et al., 2008). Upon knockdown of some of NuA4 HAT complex proteins, as well as Myc, we also observed that ES cells display flattened morphology (Figure S1E). Of note, knockdown of Ep400 or Dmap1 did not change the expression level of Oct4 and Nanog proteins, nor did knockdown of Nanog change the protein level of Ep400 and Dmap1 ((Fazzio et al., 2008), also Figure S1D). These data support the conclusion that the Core and Myc-centered subnetworks in ES cells are separable units with unique roles in maintaining ES cell self-renewal.
Previous studies have suggested that Myc is critical at an early stage in somatic cell reprogramming (Sridharan et al., 2009). Our work suggests that, beyond Myc itself, reactivation of a larger module comprised of more than 500 genes is critical to achieve partially or fully reprogrammed stem cell-like cells. It is particularly interesting that the Core module, which is composed of more than 100 genes, remains inactive in piPS cells, again implying that the reactivation of an entire functional module by a limited set of factors is critical to achieving induced pluripotency. It will be of interest to determine whether specific small molecules or genes selectively modulate the activity of the ES cell modules in efforts to identify new chemicals or factors not only for replacing Myc or other factors in somatic cell reprogramming, but also for selection of putative therapeutic targets in cancer. Since Myc interacts with NuA4 complex proteins in ES cells, recruitment of the NuA4 HAT complex by Myc may be a critical step in somatic cell reprogramming.
The relationship between ES cell and cancer signatures has been a focus of attention given that self-renewal is a hallmark of both cell types. It has been proposed that the activation of an ESC-like gene expression program in adult cells may confer self-renewal to cancer cells or cancer stem cells (Ben-Porath et al., 2008; Wong et al., 2008a). It is noteworthy that we observed very similar patterns of module activity between our Myc module and the previously defined ESC-likes (Core ESC-like gene module and mouse ESC-like gene module) (Wong et al., 2008a), but not with our Core module, in situations we tested. In accordance with this observation, approximately 60% of genes in the previously defined Core ESC-like module (Wong et al., 2008a) are Myc targets that we identified (Kim et al., 2008). Notably, 57% of genes in the Core ESC-like module (Wong et al., 2008a) are common targets of at least 5 factors among 7 factors in the Myc cluster (Figure 4). In contrast, less than 2% of genes in the previously defined ESC-like module are shared with the Core module. These findings argue that the previously described ESC-like module (Wong et al., 2008a) conveys information largely contributed by the Myc module, and conversely that the ESC-like module is quite distinct from the Core module. The simple interpretation that the presence of ESC-like module activity in cancer reflects dedifferentiation or regression to an embryonic or ES-like state (Wong et al., 2008a) is inconsistent with our analysis.
In their recent work, Ben-Porath et al. (2008) compiled 13 partially overlapping gene sets belonging to four groups (ES-expressed, active NOS (Nanog, Oct4, and Sox2) targets, Polycomb targets, and Myc targets) which are similar to the modules utilized in our analysis. They showed that poorly differentiated tumors show preferential expression of ES cell specific genes, in addition to preferential repression of Polycomb target genes. Interestingly, their analysis revealed that ES-expressed and Polycomb-target sets show the most significant degree of enrichment in most tumors, while the other gene sets are not a major determinant of their ES cell-like gene expression signature. Of special note, we find that 38% and 52% genes in their ES-expressed gene sets (ES exp1 and ES exp2, respectively) contain the common targets of at least 5 factors among 7 factors in the Myc cluster, suggesting that a large portion of genes in their ES-expressed gene sets are in turn Myc-related genes. It is noteworthy that the PrC module defined in ES cells is also largely repressed in most cancers we tested, suggesting a role of Polycomb complex proteins and their targets in cancer initiation and/or progression.
Our analysis is conceptually different from prior approaches in that we have stringently defined regulatory modules based on common gene targets of multiple factors. By this strategy we have defined modules that serve as powerful analytical tools to interrogate different cellular states and the relatedness of gene expression signatures of ES cells and cancers. Reanalysis of prior datasets in this manner raises concern regarding the hypothesis that cancer cells, or cancer stem cells, recapitulate regulatory programs characteristic of embryonic stem cells. As a unifying view the hypothesis is attractive and has gained considerable attention in recent literature. Nonetheless, our findings should temper enthusiasm and stimulate further reassessment of these issues. Moreover, our findings reemphasize the critical nature of regulatory pathways controlled by Myc in cancer.
EXPERIMENTAL PROCEDURES
ES Cell Lines and culture
Mouse J1 ES cell lines were maintained in ES medium as documented in Supplemental Data.
Protein complex pulldown and mass-spectrometry
One-step affinity purification and protein complex identification using nuclear extracts from ES cell lines expressing BirA only (reference) or both BirA and biotin tagged proteins (sample) with streptavidin-agarose were performed as described previously (Kim et al., 2009; Wang et al., 2006). Further details are documented in Supplemental Data.
ChIP-chip
At least three biological replicates of ChIP and bioChIP reactions were performed for each factor as described previously (Kim et al., 2009; Kim et al., 2008). Detailed procedure and a list of antibodies used for native antibody ChIP reactions are available in Supplemental Data.
Microarray and data Processing
Amplification of ChIP samples, and microarray hybridizations were performed as described previously (Kim et al., 2008). The raw and processed ChIP-chip data set can be found on the public server GEO under the accession number of GSE20551. Further details are available in Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Trowbridge for critical reading of the manuscript, the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for mass-spectrometry and peptide identification, and the Microarray Core Facility at the Dana Farber Cancer Institute for ChIP sample processing. The project described is partially supported by Award Number K99GM088384 to J.K. from the NIH/NIGMS. S.H.O. is an investigator of the Howard Hughes Medical Institute.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include Extended Experimental Procedures, 5 Figures, 3 Tables, and Supplemental References and can be found with this article at http://www.
REFERENCES
- Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol. 2002;22:5650–5661. doi: 10.1128/MCB.22.16.5650-5661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Cole MD, Henriksson M. 25 years of the c-Myc oncogene. Semin Cancer Biol. 2006;16:241. doi: 10.1016/j.semcancer.2006.08.003. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Kang J, Shakya A, Tantin D. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 2009;34:491–499. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schoenhals M, Kassambara A, De Vos J, Hose D, Moreaux J, Klein B. Embryonic stem cell markers expression in cancers. Biochem Biophys Res Commun. 2009;383:157–162. doi: 10.1016/j.bbrc.2009.02.156. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledgebased approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, et al. MYC regulation of a "poor-prognosis" metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008a;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Segal E, Chang HY. Stemness, cancer and cancer stem cells. Cell Cycle. 2008b;7:3622–3624. doi: 10.4161/cc.7.23.7104. [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.