SUMMARY
Brassinosteroids (BRs) regulate a wide range of developmental and physiological processes in plants through a receptor-kinase signaling pathway that controls the BZR transcription factors. Here we use transcript profiling and chromatin-immunoprecipitation microarray (ChIP-chip) experiments to identify 953 BR-regulated BZR1 target (BRBT) genes. Functional studies of selected BRBTs further demonstrate roles in BR-promotion of cell elongation. The BRBT genes reveal numerous molecular links between the BR signaling pathway and downstream components involved in developmental and physiological processes. Furthermore, the results reveal extensive crosstalk between BR and other hormonal and light signaling pathways at multiple levels. For example, BZR1 not only controls the expression of many signaling components of other hormonal and light pathways, but also co-regulates common target genes with light-signaling transcription factors. Our results provide a genomic map of steroid hormone actions in plants, which reveals a regulatory network that integrates hormonal and light signaling pathways for plant growth regulation.
INTRODUCTION
Brassinosteroids (BRs) are essential hormones for a wide range of developmental and physiological processes throughout the life cycle of plants (Clouse and Sasse, 1998). BR deficient or insensitive mutants display a variety of growth defects, including reduced seed germination, dwarfism, dark green and curled leaves, delayed flowering, male sterility, and photomorphogenesis with expression of light-induced genes in the dark (Li et al., 1996; Szekeres et al., 1996). Physiological studies also showed that BRs promote cell elongation, enhance tolerance to environmental stresses and resistance to pathogens infections and thus increase crop yield (Divi and Krishna, 2009). BRs act through the BRI1 receptor-like kinase and its well-defined signal transduction pathway to activate members of the BZR family transcription factors (Kim et al., 2009). However, our understanding of BR functions remains incomplete, largely because the direct target genes regulated by BR signaling remain largely unknown (Gendron and Wang, 2007; Kim and Wang, 2010; Tang et al., 2010).
Genetic studies have revealed a central role for BZR1 and BZR2 (also named BES1) in BR regulation of plant growth (Wang et al., 2002; Yin et al., 2002). Activation of either BZR1 or BZR2/BES1 by dominant mutations (bzr1-1D, bes1-D, and bzr1-S173A) suppresses nearly all phenotypes of the BR-insensitive bri1 and bin2 mutants (Wang et al., 2002; Yin et al., 2002). BZR1 and BZR2 share 88% sequence identity at the protein level, and they bind to DNA through the conserved N-terminal DNA-binding domain (He et al., 2005; Yin et al., 2005). While BZR1 and BZR2/BES1 were shown to function as transcriptional repressor and activator, respectively (He et al., 2005; Yin et al., 2005), it is unclear whether opposite transcriptional activities were observed because of different target promoters analyzed or intrinsic differences between the BZR1 and BZR2 proteins (Gendron and Wang, 2007). Although microarray studies have identified a large number of BR-responsive genes (Goda et al., 2004; Nemhauser et al., 2006; Nemhauser et al., 2004; Vert et al., 2005), only a few of them have been shown to be direct target genes of BZR1 or BZR2/BES1. These include a pair of antagonizing helix-loop-helix transcription factors as targets of BZR1 (Zhang et al., 2009), and a MYB transcription factor (MYB30) as a target of BZR2/BES1 (Li et al., 2009), suggesting that BZR1 and BZR2/BES1 can regulate additional transcription factors to control secondary BR-responsive genes. BR can also affect other hormones (Nemhauser et al., 2006), and many BR-responsive genes also respond to auxin or light (Goda et al., 2004; Song et al., 2009), but the underlying mechanism remains unknown. A physical map of BZR factor-DNA interactions is required to understand the mechanisms for BR regulation of cellular and developmental processes and BR crosstalk with other pathways.
In this study, we performed genome-wide analysis of the direct targets of BZR1. Microarray analysis of bzr1-1D and bri1 mutants indicates that BZR1 regulates directly or indirectly the expression of about 80% of the genes downstream of BRI1. Using chromatin-immunoprecipitation followed by microarray (ChIP-chip), we identified 2260 high-confidence BZR1 binding sites, and about two third of them are associated with 953 BR-regulated genes, which are considered BR-regulated BZR1-target (BRBT) genes. The BRBT genes reveal numerous molecular links from BR signaling to known and new cellular and developmental processes as well as to other hormonal and light signaling pathways. This study provides a detailed global map of BR actions that reveals the molecular network through which BR regulates plant growth and development.
RESULTS
BZR1 is a major transcription factor responsible for BR-regulated gene expression and plant growth
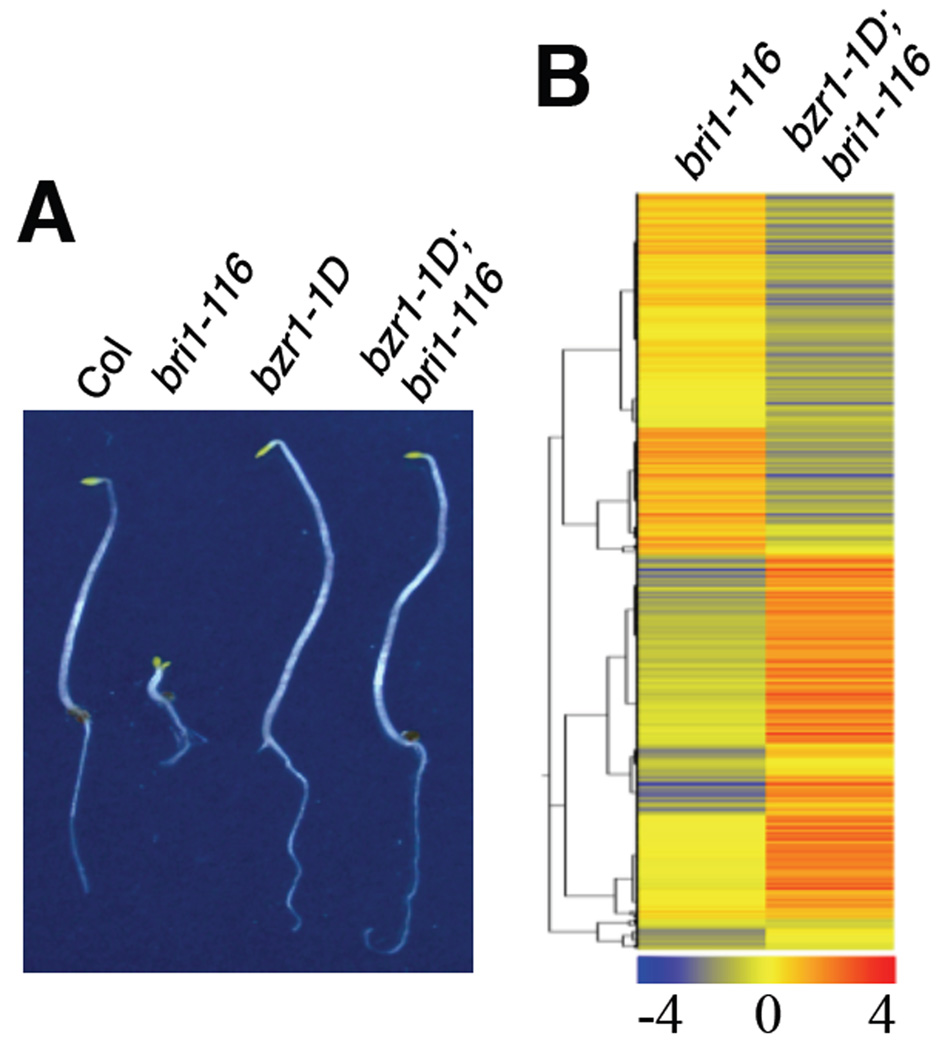
Molecular genetic analyses support a central role of BZR1 in BR regulation of plant development. The dominant bzr1-1D mutation, which stabilizes the BZR1 protein, completely suppresses the de-etiolated phenotype of the null bri1-116 mutant grown in the dark (Wang et al., 2002; Yin et al., 2002) (Figure 1A). Using 4.5-day-old etiolated seedlings, microarray analysis identified 3531 genes differentially expressed in bri1-116 compared to wild type and 6742 genes differentially expressed in the bzr1-1D;bri1-116 double mutant compared to the bri1-116 single mutant (1.5 fold cut off and P<0.01, Table S1a). Consistent with the phenotypic suppression of bri1-116 by bzr1-1D, 2796 (80%) of the genes affected in bri1-116 were affected in opposite ways by bzr1-1D (Figure 1B, Table S1b), indicating that BZR1 directly or indirectly mediates regulation of expression of about 80% of the genes controlled by BR through BRI1.
Figure 1. BZR1 is a major transcription factor of the BRI1 signaling pathway.
(A) Phenotypes of wild type (Col), bri1-116, bzr1-1D, and bzr1-1D;bri1-116 double mutant seedlings grown in the dark for 5 days. (B) Hierarchical cluster analysis of the genes differentially expressed in bri1-116 vs Col and bzr1-1D;bri1-116 vs bri1-116. The numerical values for the blue-to-red gradient bar represent log2-fold change relative to the control sample. Genes are listed in Table S1.
Genome-wide analysis of in vivo BZR1 binding sites
To identify genes directly regulated by BR signaling, we performed a ChIP-chip analysis using a polyclonal anti-YFP antibody and transgenic Arabidopsis plants expressing the BZR1-CFP fusion protein from the BZR1 promoter (He et al., 2005; Wang et al., 2002), with wild type plants as the negative control. The transgenic plants accumulate the BZR1-CFP protein at a level similar to the endogenous BZR1 of wild type plants, but the endogenous BZR1 protein at a much reduced level (Figure S1), presumably due to posttranslational control of total BZR1 protein level (He et al., 2002). Previous studies have indicated that the CFP fusion forms of BZR1 have similar activity as the non-fusion BZR1 (Gampala et al., 2007; He et al., 2005; Wang et al., 2002). We performed multiple chromatin-immunoprecipitation (ChIP) experiments and then combined the DNA to hybridize with the Affymetrix Arabidopsis tiling arrays.
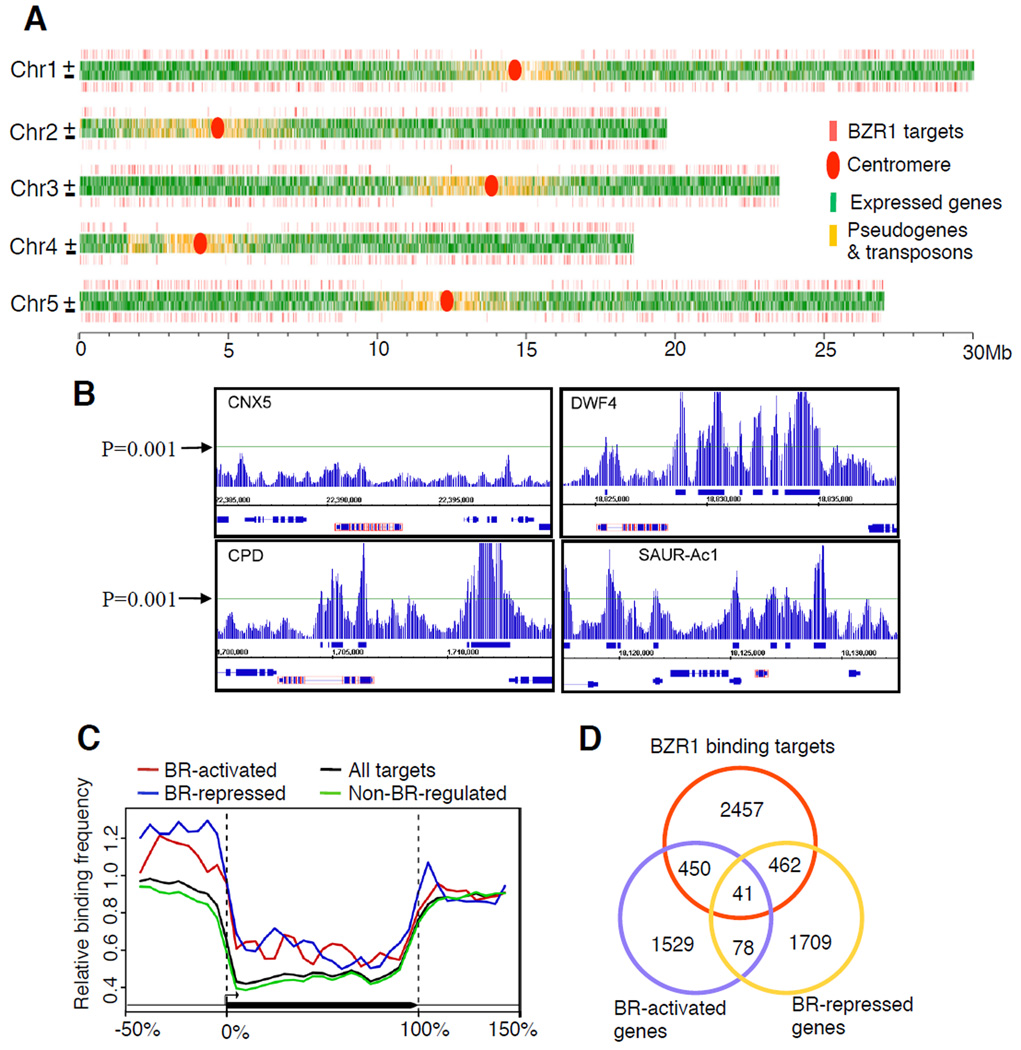
Analyses of the data using two statistic methods (see Experimental Procedures) identified 2260 high-confidence loci enriched in the BZR1-CFP samples over the wild type control. These in vivo BZR1 binding sites identified by both statistic methods were linked to 3410 nearest neighbor genes, which are considered high-confidence BZR1 binding target genes (Figure 2A, also see Table S2a). Additional 4800 genes identified by only one of the two statistic methods were considered low-confidence potential BZR1 binding target genes (Table S2b). The known BZR1 targets CPD and DWF4 are among the high-confidence BZR1 binding target genes, and two known non-targets of BZR1, CNX5 and UBC30 (He et al., 2005), were not enriched by BZR1-CFP in the ChIP-chip data sets (Figure 2B). Only the high-confidence binding target genes are included in subsequent studies, unless indicated otherwise.
Figure 2. Identification of BZR1 direct target genes using ChIP-chip.
(A) Distribution of BZR1 binding sites along the five chromosomes of Arabidopsis. (B) ChIP-chip data displayed by Integrated Genome Browser software at selected chromosomal regions around known BZR1 target and non-target genes (red-box). The horizontal line indicates the cut off P-value (0.001). (C) Frequency of BZR1 binding sites along the virtually normalized gene models (promoter is −50-0%, coding region 0–100%, 3’ region 100–150%) of each class of BZR1 target genes. (D) Venn diagram shows the overlaps of BZR1 target genes (Table S2a) with BR-activated and BR-repressed genes (Table S5c).
Thirty-three selected binding sites that represent the range of statistic confidence of the ChIP-chip data set were confirmed by ChIP followed by quantitative PCR (ChIP-qPCR) (Table S3). BZR1 binding to 21 loci was analyzed in three independent ChIP-qPCR experiments: (1) ChIP experiments performed using wild type plants and anti-MBP-BZR1 antibody with anti-MBP antibody as control, (2) ChIP experiments using anti-YFP antibody and five-day-old light-grown BZR1-CFP plants with 35S-GFP transgenic plants as control, and (3) ChIP using five-day-old dark-grown BZR1-CFP with 35S-GFP as control (Table S3). Additional 12 loci were analyzed in experiment (1). The results show that all 33 loci were enriched compared to the control loci, although the folds of enrichment were variable in different experiments. The ChIP-qPCR data from different tissues also suggest that BZR1 binds to mostly the same genes at different developmental stages or under different light conditions.
As expected for a transcription factor with specific functions, the in vivo binding sites of BZR1-CFP are distributed throughout the genome but are rare in the centromere regions (Figure 2A), which is similar to the distribution of expressed genes. The binding sites were substantially enriched in the 5’, and to a less degree 3’, intergenic regions compared to the transcribed regions of genes (Figure 2C).
Identification of BR-regulated BZR1 target (BRBT) genes
Because BZR1’s DNA binding activity and nuclear localization are tightly controlled by BR signaling (Gendron and Wang, 2007; Kim and Wang, 2010), the genes regulated by BZR1 should be responsive to BR treatment or affected in BR mutants. To determine the BR-regulated BZR1 targets (BRBTs), we compared our ChIP-chip data to expression profiling microarray data of genes affected by BR treatment or BR mutants.
Many genes that respond to BR treatment have been identified by several microarray studies (Goda et al., 2008; Goda et al., 2004; Goda et al., 2002; Mouchel et al., 2006; Mussig et al., 2002; Nemhauser et al., 2006; Nemhauser et al., 2004; Vert et al., 2005) (Data summarized in Table S4a). However, the datasets from these studies overlap only partially (Table S4a), presumably due to different conditions used and tissue specific effects of BR. Such partial overlap among available datasets suggests that additional BR-responsive genes are yet to be found, particularly in specific tissues. We therefore performed a microarray experiment of the inflorescence tissues of the BR-deficient mutant det2-1 after a 3-hour treatment with brassinolide (the most active form of BRs), and identified 283 BR responsive genes, with 225 not identified previously (Table S4b).
All of the microarray datasets of BR-responsive genes showed significant overlaps with the high-confidence BZR1 targets dataset (Table S5). For example, over 80.8% of the early BR-repressed genes, 51.4% of early BR-induced genes, 37.7% of late BR-repressed genes, and 27.6% of late BR-induced genes identified by Goda et al. (Goda et al., 2004) are BZR1 target genes, whereas a random gene would have about 11% probability of being a BZR1 target and only 3% of the 100 constitutively expressed non-BR-responsive genes (Czechowski et al., 2005) flank BZR1 binding sites.
Combining all available data of BR microarray experiments, 1957 genes respond to BR treatment (Table S4a) and 2796 genes are affected oppositely by bri1-116 and bzr1-1D mutations (Table S1b). Together these experiments identified 4265 BR-regulated genes, including 1977 BR-activated genes, 2169 BR-repressed genes and 119 genes that behaved oppositely in different microarray experiments potentially due to different conditions or time points of the experiments (Table S4c). These genes include 953 high-confidence BZR1 target genes, which are considered BRBT genes (Figure 2D, Tables S2a, BR-regulated BZR1 targets (BRBTs)). There are 450 BRBT genes activated, 462 repressed, and 41 affected in complex ways by BR, indicating BZR1 can be activator and repressor for different genes. There are also 872 low-confidence BZR1 binding target genes that are regulated by BR (Table S2b). Since the ATH1 expression array contains only about two third of the genes in the Arabidopsis genome, only 1601 of the 2260 high-confidence BZR1 binding sites have all of their flanking genes present on the ATH1 expression array, of these 994 (62%) are associated with at least one BRBT. The 33 binding peaks confirmed by ChIP-qPCR also include about 2/3 BRBTs (Table S3). Therefore, about two third of the high-confidence BZR1 binding sites identified in our ChIP-chip experiments flank BR-responsive genes and thus appear to be functional.
BZR1 and BZR2/BES1 have similar function as both transcriptional activator and repressor
Previous studies of individual target genes suggested a transcription repressor function of BZR1 and an activator activity of BZR2/BES1. BZR1 binds to the BR-response element (BRRE, CGTGT/CG) of BR-repressed genes (He et al., 2005), whereas BZR2/BES1 interacts with the bHLH transcription factor BIM1 and together bind to the E-box element (CANNTG) of a BR-induced promoter (Yin et al., 2005). However, BZR1 and BZR2 share 88% sequence identity at the protein level. The BRBTs include approximately equal numbers of BR-induced and BR-repressed genes (Figure 2D), suggesting that BZR1 functions as a transcription activator and repressor on different promoters. ChIP-qPCR experiments show that both BZR1 and BZR2/BES1 bind to the BR-repressed gene DWF4 and the BR-induced gene SAUR-AC1 (Figure 3A), and BZR2/BES1 binds to 18 of the 19 BZR1 binding sites analyzed (Table S3). Competition DNA binding assays showed that BZR1 and BZR2/BES1 have similar binding sequence specificity in vitro, both having high affinity for BRRE but lower affinity for the E-box of the SAUR-AC1 promoter (Figure 3B). Together, our results indicate that BZR1 and BES1 have very similar biochemical functions, but may play overlapping yet distinct biological roles by targeting overlapping sets of genes, perhaps with overlapping tissue specificities or different co-factors.
Figure 3. BZR1 functions as both transcriptional activator and repressor.
(A) ChIP-qPCR analyses of BZR1-CFP and BES1-GFP binding to DWF4 and SAUR-AC1 promoters relative to the control gene (CNX5). Error bars indicate standard error for three biological repeats. (B) Quantitative data of competition electrophoretic mobility shift assays of BZR1 and BES1 binding to the BRRE elements in the CPD and SAUR-AC1 (BRRE) promoters and the E-box of the SAUR-AC1 promoter. (C) Motif analysis of BR-activated and repressed BZR1 target genes. All data are significantly different from genomic control (binomic P-value <0.01)
Whether BZR1 activates or represses a target gene is presumably determined by the structure of the promoter. It has been shown that the transcriptional activation or repression is mediated by the transcription factor’s binding site or flanking sequence. (Blauwkamp et al., 2008; MacDonald et al., 2009; Valentine et al., 1998). We performed motif analyses to identify cis-elements enriched in the BZR1 binding regions associated with BR-induced or BR-repressed BZR1 target genes. Consistent with previous study (He et al., 2005), BRRE is the most enriched motif in BR-repressed BZR1 targets, and is also enriched in the BR-induced BZR1 targets albeit to a lower degree (Figure 3C). Additional enriched cis-elements include the G-box (CACGTG, which contains two inverted repeats of the BRRE core sequence, CGTG, and is also a type of E-box), CATGTG (a type of E-box), and a GGTCC motif. Although no cis-element was identified exclusively in BR-repressed or BR-induced genes, the BRRE and G-box elements are more enriched in the BR-repressed targets, while the CATGTG motif is more enriched in the BR-induced genes (Figure 3C). These results raise a possibility that the homodimer of BZR1 represses gene expression through BRRE and heterodimer of BZR1 with other TFs mediates activation through other motifs.
BRBTs play important roles in BR promotion of cell elongation
Functional studies were performed for selected BRBTs. DREPP was recently identified by a proteomic study as a BR-induced plasma membrane-associated protein that can partly suppress the det2 mutant when overexpressed (Tang et al., 2008). Our ChIP-chip data indicated that DREPP is a BR-induced BZR1 target gene (Figure 4A). BZR1 binding to the promoter of DREPP was confirmed by ChIP-qPCR (Figure 4B), and the microarray study showed that expression of DREPP was reduced over 4-fold in bri1-116 but increased in bzr1-1D;bri1-116 (Table S1). The expression of DREPP genes was also induced by brassinolide treatment with peak level detected at 6 hours after treatment (Figure 4C). The DREPP-GFP protein expressed in transgenic Arabidopsis confirmed its localization on the plasma membrane (Figure 4D). Overexpression of DREPP dramatically increased the cell length in the petioles of det2-1 (Figure 4E). These results indicate that BZR1 directly activates the expression of the membrane-associated DREPP protein to promote cell elongation.
Figure 4. BZR1 directly activates the expression of DREPP to promote cell elongation.
(A) ChIP-chip data displayed by Integrated Genome Browser software shows BZR1 binding at the DREPP promoter. (B) ChIP-qPCR analysis of BZR1 binding to DREPP relative to the control gene CNX5. Error bars indicate standard deviation in 3 biological repeats. (C) Reverse transcription PCR (RT-PCR) analysis of DREPP RNA levels in 7-day-old seedlings after various time of treatment with 100 nM BL or mock solutions. (D) Confocal microscopy image of DREPP-GFP in epidermal cells of transgenic Arabidopsis. (E) Overexpression of DREPP increases cell elongation. Three-week-old seedlings of det2 (upper images) and transgenic det2 overexpressing DREPP (lower images) and scanning electron microscopy images of epidermal cells of the petioles.
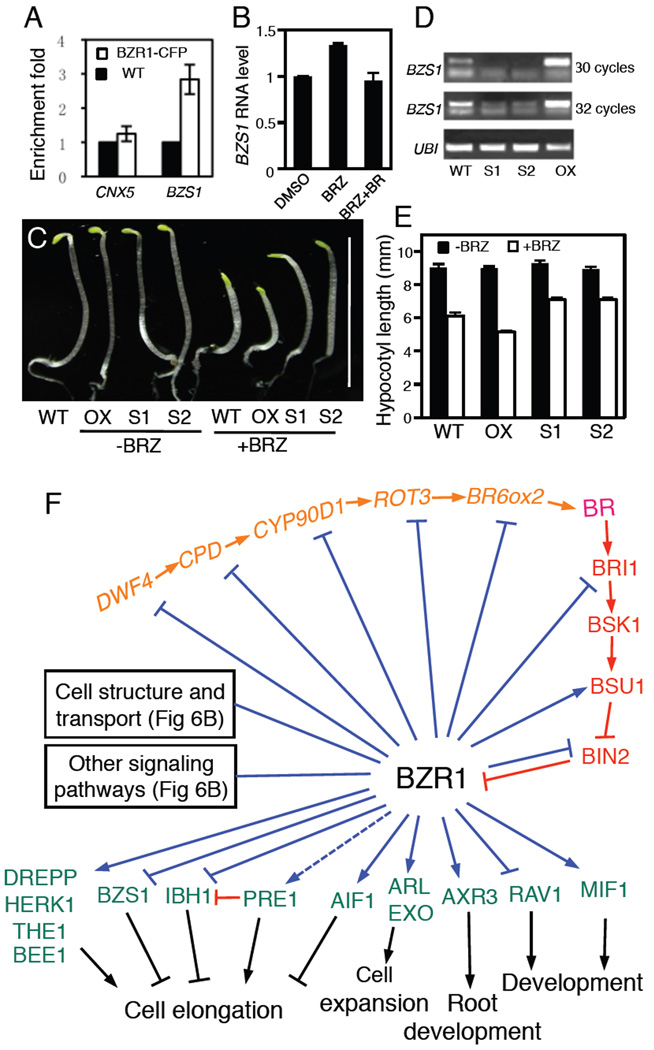
A BRBT gene (BZS1, At4g39070) encodes a putative zinc finger transcription factor (Figure 5). We confirmed the ChIP-chip data by qPCR analysis (Figure 5A), and showed that expression of BZS1 was increased under BR-deficient condition and repressed by BR (Figure 5B). Transgenic Arabidopsis plants overexpressing BZS1 showed a hypersensitivity to the BR biosynthetic inhibitor brassinazole (BRZ) (Figure 5C-5E). In contrast, transgenic plants expressing reduced level of BZS1 due to co-suppression had longer hypocotyls than wild type when grown on BRZ (Figure 5C-5E). These results demonstrate that the elevated expression of BZS1 contributes to hypocotyl inhibition under BR-deficient conditions, and BZR1 represses BZS1 expression to promote BR-induced hypocotyl elongation. These results indicate that BR signaling promotes cell elongation through multiple mechanisms, including cell membrane activities and downstream transcription factors.
Figure 5. BRBT genes play important roles in BR-regulated plant growth.
(A) ChIP-qPCR analysis of BZR1 binding to the BZS1 promoter relative to the control gene CNX5. Average of 3 biological repeats with standard deviation. (B) Expression of BZS1 is induced by BRZ and repressed by BR. Error bars indicate standard error of the mean. (C) Phenotypes of 5-day-old BZS1 over-expression line (OX) and two co-suppressed lines (S1 and S2) grown in the dark on medium with or without 1 µM BRZ. Scale bar indicates 10 mm. (D) RT-PCR analysis of BZS1 RNA level of the transgenic lines in panel (C). (E) Measurement of hypocotyl lengths of seedlings as shown in panel C, represented as mean +/− standard error from over 30 seedlings. (F) A diagram of the BR pathway. The BR biosynthetic enzymes are shown in orange color, the BR signaling components in red, and downstream components with demonstrated BR-related functions in green. Red arrows and bar ends show activation and inhibition at the protein level. Blue arrows and bar ends show BZR1 binding to BR-activated and repressed genes, respectively, and dashed line indicates finding from previous studies.
BZR1 directly regulates genes that function in BR biosynthesis, BR signaling, and BR-regulated growth
We searched the BRBTs for genes with known functions in BR responses. BZR1 has been shown to directly feedback regulates BR biosynthetic genes (He et al., 2005). The ChIP-chip and expression microarray data show that BZR1 inhibits at least five BR biosynthetic genes, as well as BRI1, providing negative feedback. However, BZR1 appears to positively regulate downstream BR signaling by inhibiting transcription of BIN2 and activating BSU1 and BZR1 itself (Figure 5F, also see Table S2a).
Genetic and transgenic studies have demonstrated functions for many genes in BR-regulated cell elongation and plant development. Many of these genes turn out to be direct targets of BZR1 based on our ChIP-chip data (Table S2a), which reveal BZR1-binding as the likely mechanism for BR regulation of these functional genes, thus establishing connections between BR signaling and downstream genes that mediate growth responses. Together the functional studies and the BZR1 binding data allow us to assemble an expanded BR regulatory pathway (Figure 5F, also see Table S2a, BRBTs with demonstrated function).
BZR1 regulates a wide range of cellular activities and biological processes
Functional classification of the BRBTs and BR-regulated non-BZR1 target genes (BR-regulated genes excluding both high- and low-confidence BZR1 binding targets) based on Gene Ontology categories showed that BR directly and indirectly regulates a range of biological processes and cellular activities (Figure 6A, Figure S2). For example, genes involved in transcription, development, and cell growth are highly enriched in BRBT genes (Figure 6A). Genes involved in cell wall organization are equally enriched in BRBTs and BR-regulated non-BZR1 target genes. In contrast, BR-regulated genes involved in response to heat are only significantly enriched in non-BZR1 target genes but not in BRBTs (Figure 6A).
Figure 6. Functional classification analysis of the BR-regulated BZR1 target and non-target genes.
(A) Enrichment of selected GO categories in BRBT and BR-regulated non-BZR1 target genes. Numbers on the right are P-values corresponding to the categories on the left (P values >0.01 are in red). (B) Representative BRBTs with known functions (Table S2a, BRBTs with demonstrated function) in various cellular processes and response/regulatory pathways. Genes repressed and activated by BR or in the bzr1-1D mutant are in blue and red color, respectively; genes responded in opposite ways in different microarray experiments are in orange. Genes only differently expressed in the bzr1-1D mutant but not affected by bri1 or BR treatment are marked with asterisk.
Based on the genome annotation at The Arabidopsis Information Resource (TAIR), about 380 (~40%) of the BRBT genes have been studied experimentally and their functions elucidated (Table S2a, BRBTs with demonstrated function). These BRBTs with known functions provide direct evidence and molecular mechanisms for BR regulation of a wide range of cellular, developmental, and regulatory processes (Figure 6B). BR activates a large number of BZR1 target genes encoding cell wall-related enzymes, such as cellulose synthase, pectinesterases, xyloglucosyl transferases, and expanins (Figure 6B), which are likely to mediate BR promotion of cell elongation and differentiation. BR also directly activates a large number of genes involved in cellular transport and cytoskeleton organization, which are likely to contribute to BR promotion of solute uptake and directional cell elongation (Figure 6B).
BRBT genes reveal nodes of crosstalk between BR and other regulatory pathways
Functional interactions between BR and other hormones have been observed at the physiological and gene expression levels (Gendron et al., 2008; Goda et al., 2004; Hardtke et al., 2007; Nemhauser et al., 2004). However, the underlying molecular mechanisms of hormone crosstalk remain largely unknown. We found that there is a significant enrichment among the BRBTs genes related to hormone levels, hormone mediated signaling, responses to auxin, light, wounding, water deprivation, ABA, BR, cold, and ethylene (Figure 6A and 6B, Table S2a). Some of the BRBT genes encode key components of other hormone pathways. These include the ABA receptor PYR1 (Park et al., 2009), several ethylene signaling components (CTR1, EIL1, EBF2, and ERFs) (Stepanova and Alonso, 2009); a large number of components of the auxin pathway (Chapman and Estelle, 2009); the GA biosynthetic gene GA5 and four of the five GA-signaling DELLA proteins (RGA, GAI, RGL1, RGL3) (Schwechheimer, 2008); and the early jasmonate signaling protein JAZ10 (Chung and Howe, 2009) (Figure 6B, Table S2a: BRBTs with demonstrated function). A close relationship between BR and auxin has been recognized (Hardtke et al., 2007). Our ChIP-chip analysis discovered that BZR1 not only directly regulates auxin responsive genes but also targets multiple auxin signaling components and genes involved in auxin metabolism (Figure 6B, Figure S4a).
Comparison of the BR-regulated genes (Table S4a) with expression microarray data for other hormones (Nemhauser et al., 2006) showed a significant number of co-regulated genes (Figure S3b). The transcriptional overlap is especially high with GA, auxin, and ethylene, three hormones that also regulate cell elongation. Interestingly, the co-regulated genes are enriched with BRBTs, suggesting that the other hormone pathways also regulate BR signaling or transcriptionaly co-regulate common target genes with the BR pathway. Consistent with the later notion, the binding site for auxin response factors (AuxRE) is enriched, together with an E-box, in the promoters of BRBTs that are induced by both BR and auxin (Figure S3c). These results indicate that BR cross talks with auxin and other plant hormones at multiple levels (e.g. Figure S3a).
Crosstalk between BR and light signaling
The de-etiolation phenotypes of BR mutants demonstrated an essential role of BR in photomorphogenesis (Li et al., 1996; Szekeres et al., 1996). How BR regulates light responses has been a long-standing question in plant biology. Our ChIP-chip and RNA profiling data indicate that BR negatively regulates the transcription of several key components of the light response pathways, including photoreceptors phytochrome B (PHYB), phototropin1, and the phytochrome-interacting proteins PIF3 and FHL (Figure 6B). The bri1-116 mutant accumulate a higher level of PHYB protein than wild type (Figure 7A), which likely contributes to the enhanced photomorphogenesis.
Figure 7. Crosstalk between light and BR pathways in regulating gene expression.
(A) Western analysis of PHYB protein level in 5-day-old etiolated BR mutants. (B) Venn diagram shows the overlap of differentially expressed genes in BR mutants (as in Table S1a) and light. (C) Hierarchical cluster analyses of selected genes differentially expressed in either wild type seedlings grown under red light (RL) or dark-grown BR mutants (bri1-116 and bzr1-1D;bri1-116, genes listed in Table S1a), compared to dark-grown wild type control seedlings. Genes are selected as differentially expressed in both red light and the BR mutants. The numerical values for the blue-to-red gradient bar represent log2-fold change relative to the control sample. (D) Overlap of BZR1 targets with HY5 targets, the overlapped genes are listed in Table S6b column A. (E) Regulation of BZR1-HY5 co-targeted genes by BR and RL. (F) Overlap of BZR1 target genes with PIL5 target genes (genes listed in Table S6c). (G) Regulation of co-target genes of BZR1 and PIL5 by BR and PIL5. In (B, D, F), “+” indicates activation and “−” indicates repression by BR or RL. (H) A model for BR-light interaction in seed germination and seedling growth, with arrows showing positive regulation and bar ends showing negative regulation.
A recent microarray study identified 2542 genes differentially expressed between de-etiolated seedlings grown under red light and etiolated seedlings grown in the dark (Hu et al., 2009). A comparison of these red light-regulated genes with genes affected in both bri1-116 and bzr1-1D grown in the dark (Table S1b) demonstrates a significant overlap of gene expression changes caused by BR deficiency and red light (Figure 7B). An overlap of 812 genes was observed between the two data sets, and 93% of these genes are affected similarly by red light and bri1-116 (oppositely by bzr1-1D) (Figure 7C, Table S6a). These results demonstrate that phytochrome and BR pathways antagonistically regulate seedling photomorphogenesis largely by regulating common downstream genes.
Direct target genes have been identified for two key transcription factors of the phytochrome signaling pathways. The basic leucine zipper (bZIP) transcription factor HY5 acts as a positive regulator downstream of multiple photoreceptors including phytochromes. A recent ChIP-chip study identified 3984 HY5 binding target genes (Lee et al., 2007), and 1170 of them are also BZR1 targets (Figure 7D, Table S6b). This is 3-fold higher than by random chance. Of the co-binding target genes 76 showed similar changes and 12 genes showed opposite changes caused by the bri1 mutation and red light, indicating that the co-target genes are mostly regulated in opposite ways by BR/BZR1 and light/HY5 (Figure 7E). These data support a hypothesis that BZR1 and HY5 antagonistically regulate common target genes downstream of the BR and light signaling pathways.
The phytochrome interacting factor3-like5 (PIL5) is a bHLH transcription factor that interacts with phytochrome and negatively regulates light promotion of seed germination (Oh et al., 2004). Light releases PIL5 inhibition of germination through phytochrome-mediated PIL5 degradation (Oh et al., 2009). A ChIP-chip analysis identified 750 PIL5 binding target genes (Oh et al., 2009). Surprisingly, over 52% of these PIL5 target genes are also BZR1 target genes (75.5% if low stringency BZR1 targets are included) (Figure 7F, Figure S5, Table S6c). Comparison of microarray expression data of the pil5 mutant (Oh et al., 2009) and bzr1-1D;bri1-116 (Table S1b) showed that 38 co-targeted genes were regulated by both BR and red light/PIL5 (13 expected by random chance), with 28 of them oppositely and only 10 similarly regulated by PIL5 and BR/BZR1 (Figure 7G, Table S6c). In particular, most of the co-regulated genes were repressed by BZR1 and activated by PIL5 (Figure 7G, Table S6d), which include RGA, Phytochrome B, FHL, and the auxin anabolic gene NIT1. Such antagonistic actions of a positive regulator of the BR pathway (BZR1) and a negative regulator of the light pathway (PIL5) are consistent with the similar effects of BR and light on promoting seed germination. Together these results demonstrate a convergence between the BR and light signaling pathways at the levels of transcription regulation of common target genes in addition to BR regulation of light signaling components (Figure 7H).
DISCUSSION
RNA expression microarray studies have identified over four thousand BR-responsive genes; however, many of them are affected due to BR’s effects on other signaling pathways and thus do not reflect a function of BR signaling per se. Our ChIP-chip study indicates that about a quarter of the BR-responsive genes can be directly regulated by BZR1. The number of BZR1 target genes identified in this study appears surprisingly large but is consistent with the wide range of developmental and physiological functions of BR. Many pieces of evidence indicate that the ChIP-chip data identified mostly real BZR1 in vivo binding sites. First, the BZR1-CFP protein appears to behave the same as the native BZR1, because CFP fusions to BZR1 containing either bzr1-1D or bzr1-S173A mutations (Gampala et al., 2007) cause same BR-activation phenotypes as the non-fusion mutant proteins. Second, the ChIP-chip data obtained from three pools of nine repeats appear to be of high quality and the statistic analysis indicated a very low false discovery rate (FDR). In addition to low FDR based on genes enriched in the control sample, the percent overlap with BR-regulated genes was also used as a parameter to optimize the statistic cutoffs. When more stringent statistic criteria (TAS P<0.0001 and tilemap PP>0.9) were used, the number of BZR1 target genes was reduced from 3410 to 1609. But the 1801 genes removed by this higher stringency cutoff still contain 26% BR-regulated genes, which is much higher than expected for a random set of genes or none-BR-responsive genes (6% or 1.6% for 1801 genes). Similarly when we increased the run length from 100 to 150, 411 high stringency BZR1 targets including 104 (25%) BRBTs were removed, whereas decreasing the gap size from 100 to 70 only decreased the number of binding intervals by less than 2%. The high percentage of BRBTs among genes removed by these more stringent statistic cutoffs indicate that the statistic cutoffs used in our analysis were appropriate, and a more stringent cutoff would cause a large number of type 2 errors (false negatives). Third, ChIP-qPCR experiments confirmed BZR1 binding to all 33 selected binding sites identified by ChIP-chip. Finally, the identified binding sites of BZR1-CFP show significant co-distribution with BR-regulated genes. The binding sites are preferentially in the intergenic regions of expressed genes, and about 62% of them are flanked by at least one BR-regulated gene, which is a higher percent of functional targets than reported in previous ChIP-chip or ChIP-seq studies (Kaufmann et al., 2010; Lee et al., 2007; Oh et al., 2009; Thibaud-Nissen et al., 2006). These analyses indicate that our ChIP-chip data are of high confidence.
Because transcription factor can act from various distances at both 5’ and 3’ side of a gene, we included both genes flanking a BZR1 binding site as BZR1 binding targets although only one of them is likely regulated by BR/BZR1. On the other hand, some BR-regulated genes are not identified by the microarray experiments performed so far. ATH1 microarray contains only about two third of Arabidopsis genes, and thus about one third of BR-regulated genes are yet to be identified. In addition, the BR microarray datasets from various laboratories overlap only partly, suggesting that each experiment only identified a subset of the BR-responsive genes under a specific condition. It is conceivable that some BR-regulated genes are expressed in specific cell types and require more selective experiments to detect. In fact, about 80% of the BR-responsive genes identified in our microarray analysis of the det2 inflorescence tissues were not identified by previous microarray experiments. A recent study identified 188 genes affected by both treatments with BR and bikinin, an inhibitor of BIN2 (De Rybel et al., 2009), but 64 (34%) of them were not identified in previous microarray experiments, of which 23 (36%) are BZR1 target genes. Therefore, more BRBTs will likely be identified by additional BR RNA profiling experiments, particularly those performed with specific tissue types.
BZR1 is a major regulator of the BR pathway
Microarray analyses of gene expression in the bri1-116 and bzr1-1D;bri1-116 mutants indicates that BZR1 regulates about 80% of the genes downstream of the BRI1-mediated BR signaling pathway. Thus, the BRBT genes should represent a significant portion of the genomic targets of the BR signaling pathway. Although for most BRBT genes the amount of physiological contributions to BR responses are yet to be evaluated by genetic and physiological analysis, a number of them have been shown to play important roles in the BR pathway. In this study, we provide genetic evidence for important functions of two BRBTs, DREPP and the BZS1 zinc finger protein, in BR-regulation of cell elongation. The homolog of DREPP was recently shown to be a microtubule-associated protein that functions in directional cell growth by destabilizing cortical microtubules (Wang et al., 2007). Therefore, DREPP is likely to mediate BR regulation of cytoskeleton organization or interaction between cytoskeleton and plasma membrane, as DREPP is localized at the plasma membrane (Tang et al., 2008). BZS1 appears to be a negative regulator of cell elongation, and BZR1 inhibits BZS1 expression to promote cell elongation.
BRBTs include about a dozen genes previously shown to play a role in BR-regulated plant growth. These include AXR3 involved in BR-regulated root development (Kim et al., 2006); MIF1 (Hu and Ma, 2006) and RAV1 ((Hu et al., 2004) transcription factors that affect both leaf and root development; the transcription factor ARGOS-like (ARL) (Hu et al., 2006) and extracellular protein EXORDIUM (EXO) that promotes cell expansion (Schroder et al., 2009); the HERK1 receptor kinase (Guo et al., 2009), and bHLH transcription factor BEE1 (Friedrichsen et al., 2002), which promote cell elongation; and IBH1 (Zhang et al., 2009) and AIF1 (Wang et al., 2009), which inhibit cell elongation. The BES1-target gene MYB30 (Li et al., 2009) and THESEUS1 (THE1) receptor kinase (Guo et al., 2009) are identified as low-confidence BZR1 targets (Table S2b). This study identified additional BR-biosynthetic genes as BZR1 targets, confirming previous finding that BZR1 mediates feedback inhibition of BR biosynthesis. Interestingly, BZR1 also directly feedback regulates genes encoding upstream components of the BR signaling pathway. Although the activities of the BR signaling proteins are tightly controlled at the protein level, extensive feedback regulation by BZR1 at the transcriptional level is likely to provide homeostasis and robustness to the system. The ChIP-chip data reveal the overall organization of the BR pathway.
BRBTs reveal crosstalks between BR and other signaling pathways at multiple levels
The BRBTs also identify molecular links for BR interaction with other hormonal signals. First, our data shows that BZR1 directly regulates a number of genes involved in biosynthesis of other hormones, such as auxin, GA, ethylene, and JA. Second, BZR1 direct regulates the PIN genes encoding auxin efflux carriers (Li et al., 2005). Third, BR directly modulates the expression levels of key components of other signaling pathways, thereby affecting the sensitivity of plants to other signals and indirectly alters the expression of genes downstream of these pathways. The fourth level of crosstalk is co-regulation of common transcriptional targets.
Our results demonstrate that BR regulates light signaling pathways at multiple levels. The essential role of BR in light regulation of plant development was initially revealed by the BR-deficient mutants (Li et al., 1996; Szekeres et al., 1996), which not only display light-grown morphology but also express light-induced genes in the dark (Chory et al., 1991; Song et al., 2009; Szekeres et al., 1996). The molecular mechanisms underlying this antagonizing relationship have remained unknown. The initial hypothesis that light modulates BR synthesis or signal transduction to inhibit cell elongation has not been supported by experimental evidence. Our study shows that BZR1 directly regulates the expression of key components of the light signaling pathways, including photoreceptors and downstream effecters. In addition, BZR1 and the transcription factors of the light-signaling pathway, HY5 and PIL5, co-regulate a significant number of common target genes in ways that are consistent with the antagonistic functions of BR and light in seedling morphogenesis and their similar actions on seed germination. Together our results demonstrate that BR signaling, through the transcriptional activity of BZR1, directly regulates the signal transduction components as well as common target genes of the phototransduction pathways, whereby BR modulates the sensitivity of plants to light and helps maintain a transcriptional profile essential for skotomorphogenesis in the dark.
Among the 953 BRBT genes identified, about 380 genes have been functionally studied previously. These genomic targets with known functions potentially explain how BR specifically regulates various cellular, developmental, and physiological processes, or crosstalks with other pathways. Future functional studies of the BRBTs with unknown function will further advance our understanding of the BR pathway.
EXPERIMENTAL PROCEDURES
Plant material and growth condition
Arabidopsis, unless indicated otherwise, is wild type Arabidopsis thaliana Columbia ecotype. The det2-1, bzr1-1D, bri1-116 and bzr1-1D;bri1-116 plants are in Arabidopsis thaliana Columbia ecotype background. The pBZR1::BZR1-CFP transgenic plants were described previously (He et al., 2005). For dark-grown seedlings, seeds were treated in cold (4°C) for 2 days and at 22°C under light for 8 hours to promote germination before growing in the dark.
Microarray gene expression analysis
For the microarray analysis of gene expression in dark-grown BR mutants, bzr1-1D;bri1-116, segregating bri1-116, and wild type Col were grown on solid ½ MS medium with 1% sucrose in the dark for 108 hours. The seedlings were frozen in liquid nitrogen in complete darkness and then the homozygote seedlings of bri1-116 were picked individually from the heterozygous population. For the expression array using det2 inflorescence tissues, det2 plants were grown on soil for 5 weeks. About half of the collected tissues contained flower buds younger than stage 8 and the other half were stage 8 to open flowers. Tissues were treated with 100 nM brassinolide or mock solution for 3 hours. Total RNA was isolated, biotin-labeled, and hybridized with Arabidopsis ATH1 genome array, according to the manufacturer’s instruction (Affymetrix). Arrays were scanned using Agilent GeneArray Scanner. All microarray experiments included three biological repeats. Microarray data were analyzed using Genespring software (version 7 and version 10, Silicon Genetics).
Chromatin immunoprecipitation-microarray
For ChIP-chip experiments, the BZR1-CFP transgenic plants and the wild type control plants were grown together on soil under long-day conditions for 4 weeks and processed in parallel as described below. Rosette tissues were treated with 100 nM brassinolide for 2 hrs and then cross-linked by 1% formaldehyde. Chromatin immunoprecipitation using anti-YFP antibodies and qPCR was performed according to He et al. (He et al., 2005). For ChIP-chip, the precipitated DNA was amplified by Whole Genome Amplification Kit following the manufacturer’s instruction (Sigma). Enrichment of CPD and DWF4 promoter DNA in the BZR1-CFP sample over the wild type control (>5 fold) was confirmed by qPCR before and after amplification. Nine ChIP experiments were performed using three batches of plants, and the amplified DNAs were combined into three pools, then labeled and hybridized, according to Affymetrix Chromatin Immunoprecipitation Protocol, to Affymetrix GeneChip Arabidopsis Tiling 1.0R arrays, which contains a 25-nucleotide oligo for every 35 base pairs of the Arabidopsis genome.
ChIP-chip data analysis
To identify the statistically enriched regions, microarray hybridization data were analyzed by two independent statistic methods: Tiling Analysis Software (TAS version 1.1, Affymetrix) and TileMap (Ji and Wong, 2005). With TAS software, the analysis was performed using the “two sample comparison analysis” option. Probes were analyzed using signal from both PerfectMatch and MisMatch with a bandwidth 250 bps, and the enriched intervals were defined by p <0.001 and signal ratio >2, maximum gap 100 bps and minimum run 100 bps. The analysis using TileMap software was performed using HMM option with posterior probability 0.5 and maximum gap 100 bps.
The identified binding regions were linked to the nearest neighbor genes, including the two genes to either side of the binding site in addition to any gene that overlaps with the binding region. Analysis using TAS identified 2539 BZR1 binding regions with 4075 neighbor genes. Analysis using TileMap identified 5039 binding regions with 7545 neighbor genes. Genes identified by both TAS and TileMap were considered high-confidence BZR1 target genes (3410 genes, Table S2a). The genes identified by only one of the two softwares were considered low-confidence BZR1 binding targets (4800 genes, Table S2b).
To estimate the false positive rate, the raw data from control and sample were switched in a mock analysis. TileMap and TAS identified 76 and 6 regions, respectively, as intervals enriched in wild type control compared to the BZR1-CFP samples, which are about 1.5% and 0.24% of the number of intervals enriched in BZR1-CFP over wild type. No genomic region enriched in wild type was identified by both softwares. Therefore, the false positive rate of the high-confidence BZR1 binding data (Table S2a) is close to zero (1.5% × 0.24%=0.0036%) and that for the low-confidence data (Table S2b) is about 2.57% (1.5% × 7545+0.24% ×4075)/4800).
The microarray data are available at National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under Series GSE23774.
Distribution of BZR1 binding sites relative to genes
The enrichment of BZR1 binding sites was calculated by the averaged ratio of ChIP hybridization intensity of BZR1-CFP to that of wild type control along the gene structure. The gene length was first converted into a percentile scale. Zero and 1 indicate the 5’ transcription start site and 3’ end of a gene respectively; −1 and 2 indicate the boundary of the promoter region and 3’ intergenic region respectively. Then all target genes were aligned and a sliding window approach with initial size as 0.1 and step width as 0.05 was used to calculate the average ratio values along the gene structure. The algorithm and plot were implemented in an R ChIP-chip analysis package NTAP (http://ntap.cbi.pku.edu.cn).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Shauna Somerville and Bi-Huei Hou for technical assistance in microarray analyses, and Dr. Peter Quail for providing the anti-PHYB antibody. This study was supported by grants from NIH (R01GM066258) and NSFC (30470169). S.Z. was supported by the Chinese Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Blauwkamp TA, Chang MV, Cadigan KM. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J. 2008;27:1436–1446. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Krishna P. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. N Biotechnol. 2009;26:131–136. doi: 10.1016/j.nbt.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Haque A, Gendron N, Chang T, Asami T, Wang Z-Y. Chemical Genetic Dissection of Brassinosteroid-Ethylene interaction. Mol Plant. 2008;1:368–379. doi: 10.1093/mp/ssn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2009;106:7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Dorcey E, Osmont KS, Sibout R. Phytohormone collaboration: zooming in on auxin-brassinosteroid interactions. Trends Cell Biol. 2007;17:485–492. doi: 10.1016/j.tcb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- He J-X, M J, Gendron, Sun YLSS, Gampala, Gendron N, Sun CQ, Wang Z-Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Ma H. Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 2006;45:399–422. doi: 10.1111/j.1365-313X.2005.02626.x. [DOI] [PubMed] [Google Scholar]
- Hu W, Su Y-S, Lagarias J. A lignt-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcription networks. Molecular Plant. 2009;2:166–182. doi: 10.1093/mp/ssn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 2006;47:1–9. doi: 10.1111/j.1365-313X.2006.02750.x. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Ji H, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- Kim H, Park PJ, Hwang HJ, Lee SY, Oh MH, Kim SG. Brassinosteroid signals control expression of the AXR3/IAA17 gene in the cross-talk point with auxin in root development. Biosci Biotechnol Biochem. 2006;70:768–773. doi: 10.1271/bbb.70.768. [DOI] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J, Sun Y, Burlingame AL, Wang Z-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Bio. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17:2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- Mussig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder F, Lisso J, Lange P, Mussig C. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 2009;9:20. doi: 10.1186/1471-2229-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. Understanding gibberellic acid signaling--are we there yet? Curr Opin Plant Biol. 2008;11:9–15. doi: 10.1016/j.pbi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol Plant. 2009;2:755–772. doi: 10.1093/mp/ssp039. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM. Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol. 2009;12:548–555. doi: 10.1016/j.pbi.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tang W, Deng Z, Oses-Prieto JA, Suzuki N, Zhu SW, Zhang X, Burlingame AL, Wang Z-Y. Proteomic studies of brassinosteroid signal transduction using prefractionation and Two-dimensional DIGE. Mol Cell Proteomics. 2008;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Wang ZY. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr Opin Plant Biol. 2010;23:27–33. doi: 10.1016/j.pbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006;47:152–162. doi: 10.1111/j.1365-313X.2006.02770.x. [DOI] [PubMed] [Google Scholar]
- Valentine SA, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey AJ. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis Brassinosteroid Signaling by Atypical Basic Helix-Loop-Helix Proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M. Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell. 2007;19:877–889. doi: 10.1105/tpc.106.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. Antagonistic HLH/bHLH Transcription Factors Mediate Brassinosteroid Regulation of Cell Elongation and Plant Development in Rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.